Abstract
The urokinase receptor (uPAR), expressed on the surface of many cell types, coordinates plasmin-mediated cell surface proteolysis for matrix remodeling and promotes cell adhesion by acting as a binding protein for vitronectin. There is great clinical interest in uPAR in the cancer field as numerous reports have demonstrated that up-regulation of the uPA system is correlated with malignancy of various carcinomas. Using both stable cell lines overexpressing uPAR and transient gene transfer, here we provide evidence for a non-reported role of uPAR in the phagocytosis of apoptotic cells, a process that has recently been termed efferocytosis. When uPAR was expressed in human embryonic kidney cells, hamster melanoma cells, or breast cancer cells (BCCs), there was a robust enhancement in the efferocytosis of apoptotic cells. uPAR-expressing cells failed to stimulate engulfment of viable cells, suggesting that uPAR enhances recognition of one or more determinant on the surface of the apoptotic cell. uPAR-mediated engulfment was not inhibited by expression of mutant β5 integrin, nor was αvβ5 integrin-mediated engulfment modulated by cleavage of uPAR by phosphatidylinositol-specific phospholipase C. Further, we found that the more aggressive BCCs had a higher phagocytic capacity that correlated with uPAR expression and cleavage of membrane-associated uPAR in MDA-MB231 BCCs significantly impaired phagocytic activity. Because efferocytosis is critical for the resolution of inflammation and production of anti-inflammatory cytokines, overexpression of uPAR in tumor cells may promote a tolerogenic microenvironment that favors tumor progression.
The urokinase plasminogen activator receptor (uPAR3 or CD87) is a multidomain glycosylphosphatidylinositol (GPI)-anchored protein (1) implicated in many cellular processes, ranging from wound repair, inflammation, motility, tumor invasion, to metastasis (2–6). Although the best understood functions of uPAR lie in its ability to act as a saturable receptor for urokinase (uPA) resulting in plasminogen activation and subsequent pericellular proteolysis and matrix remodeling (7, 8), its ability to regulate cell adhesion and migration acting as a vitronectin (VN) receptor is likely equally important (9, 10). The human uPAR cDNA encodes a 335-amino acid polypeptide that undergoes extensive post-translational glycosylation upon cell sorting through the secretory pathways, and the mature protein is covalently attached to the plasma membrane via a C-terminally modified GPI anchor (11, 12). Because uPAR lacks a trans-membrane domain, the signal transduction originating from this receptor, at least at the plasma membrane, requires lateral interactions with co-receptors. In this capacity, a wide range of surface proteins have been implicated as uPAR co-receptors, including integrins (13, 14), receptor tyrosine kinases (15), G-protein-coupled receptors (16), chemokine receptors (17, 18), and low density lipoprotein receptors (19). This not only allows for increased signal complexity, but also co-localizes uPAR with a number of diverse extracellular ligands within the extracellular environment (20, 21).
In addition to the physiological roles in tissue remodeling, numerous studies have reported up-regulation of both uPAR and uPA/tissue plasminogen activator in malignant carcinomas (22–26). In both knockout and transgenic mouse models, overexpression of either uPAR or uPA is associated with enhanced metastasis and poor survival, and efforts aimed at inhibiting the protease activation of uPA are considered promising therapeutic strategies (27). Paradoxically, however, expression of the primary inhibitors of endogenous uPA, the type-1 plasminogen activator inhibitors (28), also correlate with poor prognoses in cancer patients (29, 30), suggesting there may be non-proteolysis-mediated uPAR activation events (31). Indeed, uPAR is a saturable receptor for VN, which binds this ligand in an RGD-independent manner via the somatomedin-B domain (14). The structures of the extracellular domains of uPAR in complexes with peptide substrates of either uPA or VN reveal these proteins bind to overlapping but clearly distinct domains in uPAR (32, 33), although all three domains in uPAR (D1, D2, and D3) are required for binding. Binding of VN to uPAR induces alterations in the actin cytoskeleton, cell shape changes, and activation of signaling pathways (ERK, FAK, and Rac1) that favor cell migration (34). These data suggest that uPAR can modify integrin function that lead to enhanced matrix binding, adhesion and migration, and cell signaling.
In the present study, we have found that uPAR has a strong ability to promote efferocytosis of apoptotic cells. This was demonstrated using transient overexpression and creation of stable cell lines, as well as inhibitors of uPAR function. Expression of uPAR promoted engulfment of apoptotic T cells (CEM-1 and Jurkat cell lines), but not viable T cells, and this effect was abrogated using phosphatidylinositol-specific phospholipase C to cleave the GPI anchor of uPAR. Because uPAR and β5-integrin cooperate to promote migration and metastasis in breast cancer cells (BCCs), and both uPAR and β5-integrin are often elevated in metastatic cancers, we also investigated whether BCCs might have acquired greater phagocytic ability and investigated whether uPAR might regulate the process of β5-integrin-mediated phagocytosis. Interestingly, we found that BCCs that natively overexpress uPAR had significantly increased phagocytic capacity compared with less transformed or non-transformed epithelial cells. Surprisingly, although uPAR expression modified the expression and distribution of αvβ5 integrins, we discovered that uPAR apparently mediates phagocytosis independently of αvβ5 or αvβ3 integrins. The studies presented here reveal a novel function of uPAR in the uptake of apoptotic cells.
MATERIALS AND METHODS
Plasmids, Antibodies, and Reagents
The generation of bicistronic β5-integrin expression plasmids encoding either wild type (pCx-β5-EGFP), C-terminal deletion mutant of β5 (pCx-β5ΔC-EGFP), and point mutant (pCx-β5Y750A-EGFP) are previously described (35). pN1-uPAR construct was kindly provided by Dr. Francesco Blasi (Milan, Italy). For generation of uPAR domain truncation mutants, the cDNA variants encoding domain I (D1, amino acids 1–92), domain II (D2, amino acids 93–191), domain III (D3, amino acids 192–281), and domain II plus domain III (D2D3, amino acids 93–281) were constructed by PCR using full-length uPAR as a template. Each PCR product was subcloned into pcDNA3 at the HindIII and EcoRI sites. Monoclonal and polyclonal anti-uPAR antibodies Mab3932 and 399R were obtained from American Diagnostica (Stamford, CT), anti-αvβ5 antibody P1F6 was from Chemicon (Temecula, CA). Horseradish peroxidase, fluorescein isothiocyanate, and phycoerythrin-conjugated anti-mouse IgG and anti-rabbit IgG secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Glycophosphatidylinositol-specific phospholipase C (PIPLC) was purchased from Sigma.
Cell Culture and Transfection
HEK293-uPAR pooled stable cells were established by transfection of pN1-uPAR cDNA and selection by G418 (Invitrogen). HEK293 and HEK293-uPAR stable cells, the breast cancer cell lines, MCF7 and MDA-MB231, were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C in a humidified atmosphere containing 5% CO2. HEK293-uPAR cells were continuously maintained in selection medium by addition of 400 μg/ml G418 (Invitrogen), and medium was changed every 2–3 days. Breast epithelial cell line MCF10A was maintained according to protocol published by Debnath et al. (36) in 1:1 DMEM/F-12 medium supplemented with 5% horse serum, 20 ng/ml EGF, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxin, and 10 μg/ml insulin. After trypsinization, cells were harvested in 1:1 DMEM/F-12 medium supplemented with 20% horse serum. CS-1 hamster melanoma, CEM-1, and Jurkat T cells were similarly maintained in RPMI 1640 supplemented with 10% fetal bovine serum. CEM-1 cells were additionally supplemented with 1 mm sodium pyruvate. CS-1 cells were transfected using Lipofectamine 2000 (Invitrogen) at a DNA:liposome ratio of 1:5.
Phagocytosis Assay
CEM-1 or Jurkat T-cells were used as apoptotic targets. Cells were labeled red using PKH26 dye (Sigma) according to manufacturer's protocol prior to apoptosis induction. For apoptosis induction, cells were resuspended as 3 × 106 per ml of PBS in a tissue culture dish and irradiated with 25 mJ/cm2 ultraviolet B. Cells were then washed and resuspended as 5 × 106 per ml of complete medium and incubated further for 6–8 h. Apoptosis induction was verified by Annexin-fluorescein isothiocyanate versus propidium iodide staining according to the manufacturer's protocol (Clontech). 1 × 106 CS-1 cells (phagocytes) were color-coded green by (co-)transfection of an EGFP-encoding vector. Fresh medium was added after 6 h of transfection, and cells were harvested for phagocytosis assay after 24 h of culture using 10 mm EDTA-PBS solution. Cells were resuspended as 1 × 106 of fresh medium, and 100 μl (i.e. 100,000 cells) was co-cultured with 100 μl (i.e. 500,000 cells) apoptotic cells for 2 h. Cells were then harvested with ice-cold 10 mm EDTA, fixed in 2% paraformaldehyde, and analyzed by flow cytometry (BD Biosciences). The percentage of green phagocytes that have internalized red apoptotic material detected as double-positive cells was obtained. For PIPLC treatment of phagocytes, transfected CS-1 cells were supplemented with 2 units/ml PIPLC in the medium 1.5 h prior to co-culture with apoptotic cells, washed with PBS, and resuspended in complete medium. When adherent cells were used as phagocytes, membrane labeling green dye PKH67 (Sigma) was used. After labeling, 100,000 HEK293 and HEK293-uPAR cells were plated in 6-cm tissue culture dish and allowed to revive for 36 h in high serum (20% fetal bovine serum) medium. MCF10A, MCF7, and MDA-MB231 were similarly labeled and cultured in respective media for 24 h. Medium was then replaced by 1 ml (1 million cells) of red-labeled apoptotic Jurkat cell suspension and co-cultured for the time points indicated in the figure under “Results.” At the end of co-culture, apoptotic Jurkat cell suspension was aspirated out. Phagocytes were washed with 1× PBS, trypsinized for 5 min at 37 °C to remove bound but non-internalized cells, collected with 500 μl of medium, fixed in 2% paraformaldehyde, and analyzed by flow cytometry.
Amnis ImageStream Data Analysis
ImageStream data were acquired using the Amnis ImageStream Analyzer instrument equipped with the Amnis INSPIRE software. Analysis was performed with the Amnis IDEAS software. Briefly, apoptotic cell uptake was determined utilizing a three-step method. First, transfected cells were identified by plotting the area of the cells versus intensity of the EGFP expression. EGFP-positive cells were then analyzed for PKH26 uptake by plotting area versus intensity of the PKH26. PKH26-positive cells were then processed to determine how many of the cells had internalized the PKH26. Cells with an internalization score above 0 are shown to have internalized PKH26 material. Red apoptotic material that overlapped with two peripheral pixels of green phagocytes was considered as bound but not internalized particles. The rest of the apoptotic material was considered as internalized and was observed as yellow spots in the merged images.
Detection of Cell Surface Proteins by Flow Cytometry
Because uPAR is sensitive to trypsin treatment (37), cells were harvested from tissue culture dish using a cell scraper and washed with ice-cold 1× PBS. 1–5 × 106 cells were resuspended in 100 μl of ice-cold 3% bovine serum albumin-PBS solution. 1 μg of primary antibody was added, and cells were incubated at room temperature for 1–2 h with intermittent mixing. Cells were then washed thrice with ice-cold PBS and re-suspended in ice-cold 3% bovine serum albumin solution. Fluorescein isothiocyanate- or phycoerythrin-conjugated secondary antibodies were added in the ratio of 1:50 and 1:100, respectively, and incubated in dark at room temperature for 1 h. Cells were washed thrice with ice-cold PBS and fixed in 0.5% paraformaldehyde, and the percentage of fluorescently labeled cells was measured by flow cytometry.
Electron Microscopy
HEK cells were co-cultured with apoptotic T cells for 1 h, after which the cells were harvested by trypsinization and centrifuged at low speed for 5 min. The resulting cell pellets were fixed with cold 1% glutaraldehyde and after a brief rinse in the buffer, were postfixed for 1 h with cold 1% osmium tetroxide also buffered in phosphates at pH 7.3 prior to treatment with 0.5% cold uranyl acetate at pH 5 for 60–90 min. Both dehydration in ethanol and propylene oxide and embedding in Epon 812 were done by routine procedures.
Western Blotting
Protein expression was analyzed by Western blotting. Briefly, cells were washed with ice-cold 1× PBS, lysed in radioimmune precipitation buffer (25 mm Tris·HCl, pH 7.6, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, and 20 μg/ml aprotinin), and kept on ice for 10 min followed by cold centrifugation at 14,000 rpm. The supernatant containing solubilized protein was used for further analysis. Total protein was estimated by Bradford Lowry method. Proteins were resolved by SDS-PAGE gel electrophoresis, transferred to a polyvinylidene difluoride membrane, incubated with appropriate primary antibodies and horseradish peroxidase-conjugated secondary antibodies, and detected by chemiluminescence.
Reverse Transcriptase-PCR
Total RNA was prepared using TRIzol® reagent, followed by DNase I (1 μg/ml) treatment to clean the preparation of genomic DNA. Two-step reverse transcriptase-PCR reaction (Invitrogen Thermoscript RT-PCR System catalog no. 11146-024) was performed according to manufacturer's protocol. Forward primer (5′-CATGCAGTGTAAGACCAACG-3′) and the reverse primer (5′-CTCTCACAGCTCATGTCTGATGAGCCAC-3′) for human uPAR yielded a 311-bp product. The forward and reverse primers for β-actin, 5′-GGACTTCGAGCAAGAGATGG-3′ and 5′-AGCACTGTGTTGGCGTACAG-3′, respectively, yielded a 234-bp product. The forward and reverse primers for β5-integrin, 5′-AGGATGCACTGCATTTGCTG-3′ and 5′-TCCACCGTTGTTCCAGGTAT-3′, yielded a 274-bp product.
RESULTS
αvβ5 and αvβ3 integrins have important roles in apoptotic cell phagocytosis, acting as tethering receptors not only to link apoptotic cells and phagocytes but also as signaling receptors to activate Rac1 (38) via the phosphatidylserine opsonin factor MFG-E8 (39). A physical interaction between αvβ5 integrin and uPAR has been observed based on the co-localization of these proteins at focal adhesions and the stable co-immunoprecipitation in detergent lysates (40). In addition, functional studies showed that either anti-uPAR or anti-αvβ5 antibodies inhibit human breast carcinoma and pancreatic carcinoma cell migration toward vitronectin (41, 42). uPAR and αvβ5 integrin also interact with a shared ligand, VN (10), providing a molecular rationale for the cooperation between these molecules in mediating cell adhesion, actin reorganization, and migration. Considering the critical role that αvβ5 integrin plays in apoptotic cell phagocytosis through the activation of Rac1, we examined the possibility that uPAR might modulate efferocytosis of apoptotic cells.
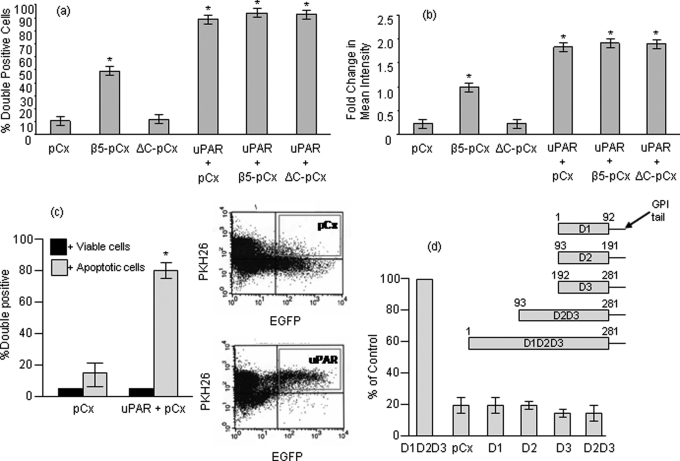
We first analyzed the effect of uPAR on efferocytosis of apoptotic T cells in hamster melanoma CS-1 cells in the presence and absence of β5 integrin expression. Previous studies showed that the β5/β3-deficient CS-1 cells express αv integrin and that reconstitution of β5 results in re-expression of a functional αvβ5 heterodimer on the CS-1 cell surface (43). Fig. 1 shows that when CS-1 cells, which do not express endogenous uPAR (data not shown), were transfected with wild-type β5 integrin (β5-pCx), αvβ5 integrin-expressing CS-1 cells increased their capacity to engulf apoptotic cells, whereas a mutant β5 (deleting the intracellular domain, ΔC-pCx) failed to stimulate engulfment. Interestingly, expression of uPAR produced a potent phagocytosis-promoting effect in the CS-1 cells, independent of β5 expression, whereby almost 90% of the uPAR-expressing cells contained engulfed apoptotic cells. Surprisingly, this effect of uPAR was not influenced by expression of the wild-type β5-pCx, nor did the dominant negative β5 integrin, ΔC-pCx, affect uPAR-mediated efferocytosis indicating that this mutant was unable to sequester uPAR into a non-functional complex. We observed similar effects when we examined the percentage of double positive cells as a geometric mean, indicating that uPAR-expressing cells had internalized more apoptotic material compared with controls of β5-expressing cells (Fig. 1b). In contrast, overexpression of uPAR in cells had no effect on engulfment of viable cells, suggesting it did not make these cells more entotic (Fig. 1c). Because uPAR has well characterized Lys/neurotoxin D1, D2, and D3 domains that possess unique activities, we next engineered cDNA variants encoding domain I (D1, amino acids (1–92), domain II (D2, amino acids 93–191), domain III (D3, amino acids 192–281), and domain II plus domain III (D2D3, amino acids 93–281)). Each domain was constructed by PCR using full-length uPAR as a template. Fragments were subcloned into pcDNA3 such that all variants were GPI-linked and associated with the plasma membrane. Single domain transfectants failed to show an increase in efferocytosis over vector transfected cells. Moreover, phagocytic capacity of the truncated mutant D2D3 was also similar to vector-transfected cells suggesting that all the three domains are required for engulfment (Fig. 1d).
FIGURE 1.
uPAR enhances phagocytosis independent of αvβ5 integrin. a, percentage of transfected green CS-1 cells that are positive for red-labeled apoptotic cells after 2 h of co-culture. CS-1 cells were transfected with pCx-EGFP or co-transfected with pCx-EGFP and pN1-uPAR or pN1-uPAR and β5-pCx or pN1-uPAR and the non-functional deletion mutant of β5 (ΔC-pCx). b, fold change in mean geometric intensity of the red channel (Y geometric mean) in the upper right quadrant that is representative of double positive population was measured. Values represent the amount of apoptotic material engulfed by phagocytes. Data are representative of at least three different experiments. c, uptake of viable or apoptotic T cells by EGFP and uPAR overexpressing cells after 2 h of co-culture. Co-culture of viable CEM-1 cells with transfected CS-1 cells resulted in ∼5% double-positive cells that correspond to basal cell death level in culture conditions in vitro. Panels on the right show graphical output of flow cytometry analysis showing engulfment represented in the upper right quadrant. d, the amino acid residues spanning each of the truncation mutants are depicted schematically. CS-1 cells were transfected with pCx-EGFP alone or together with the GPI-anchor containing pcDNA3-uPAR domain constructs as indicated, and a phagocytosis assay was performed. Values were normalized against D1D2D3, which was set at 100%. Asterisk, p < 0.05.
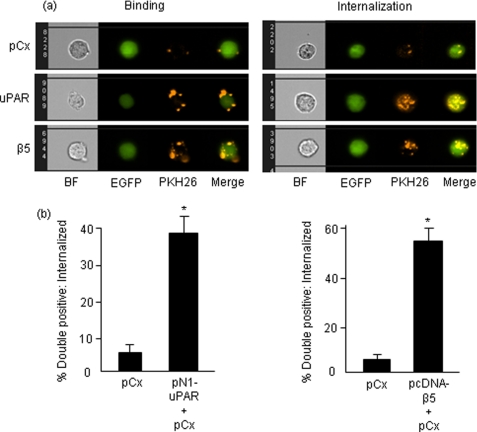
To investigate whether uPAR promoted phagocytosis by increasing cell binding or by inducing internalization and degradation, we used Amnis cellular imaging technology to decipher surface association versus internalization. This approach allowed us to witness interactions of individual cells with confocal accuracy but using fluorescence-activated cell sorting-based quantitation to sample enough cells for statistical analysis. Engulfment was specifically quantitated by subtracting two peripheral green pixels from double positive phagocyte images. Shown in Fig. 2a are representative panels of CS-1 cells expressing pEGFP (upper panel), and pEGFP co-transfected with β5 integrin (lower panel), or uPAR (middle panel) and co-cultured with red-labeled apoptotic T cells. Data are quantified in Fig. 2b. As shown in their respective panels, both αvβ5 integrin and uPAR increased both the surface association of apoptotic bodies (left panels), as well as the internalization (right panels). Moreover, it is noteworthy that uPAR transfected cells showed a pattern of internalizing both apoptotic blebs and bodies similar to αvβ5 integrin, a well studied phagocytic receptor, and uPAR expression promoted equally potent engulfment of apoptotic material.
FIGURE 2.
Analysis of phagocytosis by Amnis ImageStream System - Comparison of binding versus internalization. a, output images after Amnis acquisition. CS-1 cells were transiently transfected as shown in the figure and co-cultured with apoptotic cells for 2 h. Panels on the left depict peripheral binding of apoptotic components. Right panels were obtained after subtraction of two peripheral green pixels from phagocytes and depict internalized apoptotic material. BF, brightfield. b, quantification of internalized apoptotic material. Note that quantification does not include peripheral binding values and only represent phagocytes that have completely engulfed apoptotic bodies. Asterisk, p < 0.05.
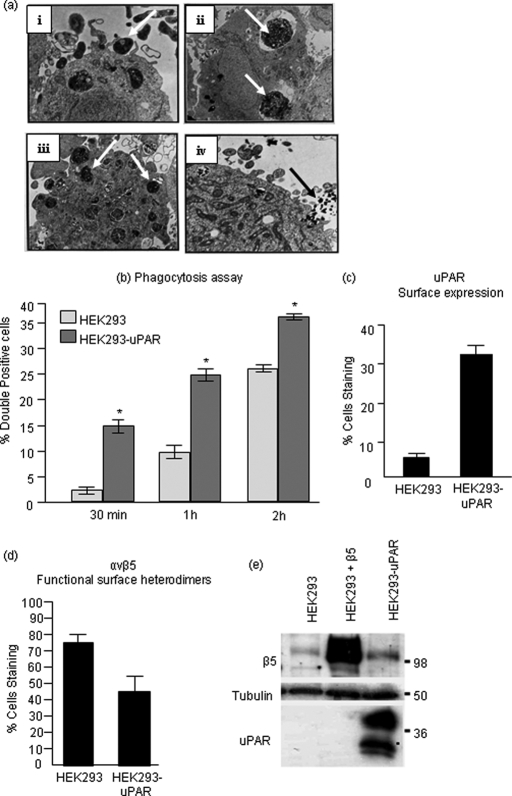
To explore how uPAR promotes efferocytosis in more detail and circumvent variability due to transient transfection, we generated stable uPAR-expressing HEK cells (Fig. 3). HEK293 cells are classified as non-professional phagocytes, because they are of a non-myeloid origin. They are nevertheless capable of engulfing apoptotic cells by a typical mechanism, which involves extension of filopodia and formation of phagocytic cups (Fig. 3a). HEK293 cells do express functional αvβ5 integrin but are negative for uPAR expression (58). Functional expression of uPAR on the cell surface of HEK293 cells stably expressing uPAR was confirmed by immunostaining of live cells and flow cytometric detection (Fig. 3c) and by adhesion assay on vitronectin (data not shown). Consistent with the results of transient expression experiments, stable uPAR-HEK cells showed increased phagocytic activity upon co-culture with apoptotic cells at each of the time points analyzed (Fig. 3b). However, although stable expression of uPAR also increased total β5 expression by immunoblotting (Fig. 3e, lanes 1 versus 3), it decreased expression of functional αvβ5 heterodimers on the cell surface, as evident from surface staining with anti-PIF6 mAb that recognizes a shared activation-specific epitope on the αvβ5 integrin (Fig. 3d). Moreover, when we transfected uPAR-expressing cells with pC3-EGFP-Rac1, we noted a co-localization of uPAR and Rac1 in specific membrane patches (data not shown), suggesting that uPAR may be poised to activate Rac1-dependent phagocytosis when primed with apoptotic cells.
FIGURE 3.
Stable overexpression of uPAR in HEK293 cells accelerates corpse uptake. a, electron micrograph of HEK293 cell, a non-professional phagocyte engulfing apoptotic bodies indicated by arrowheads in respective panels. i, filopodial extensions and formation of phagocytic cup around an apoptotic body; ii and iii, presence of multiple apoptotic bodies within HEK293 cell; iv, a phagocytic portal engulfing multiple apoptotic blebs. b, HEK293 and HEK293-uPAR cells were labeled with membrane labeling dye PKH67 and co-cultured with the phagocytes at a ratio of 1:10. As cells were harvested by trypsinization, the above data represents mostly internalization. c, surface uPAR expression in HEK293-uPAR stable cells was confirmed by rabbit polyclonal (399R) antibody staining of live cells. d, cell surface expression of functional αvβ5 heterodimers in HEK293 and HEK293-uPAR cells was analyzed by monoclonal P1F6 antibody that recognizes a shared epitope in the heterodimer. e, Western blot analysis following extraction in SDS-containing radioimmune precipitation buffer. Asterisk, p < 0.05.
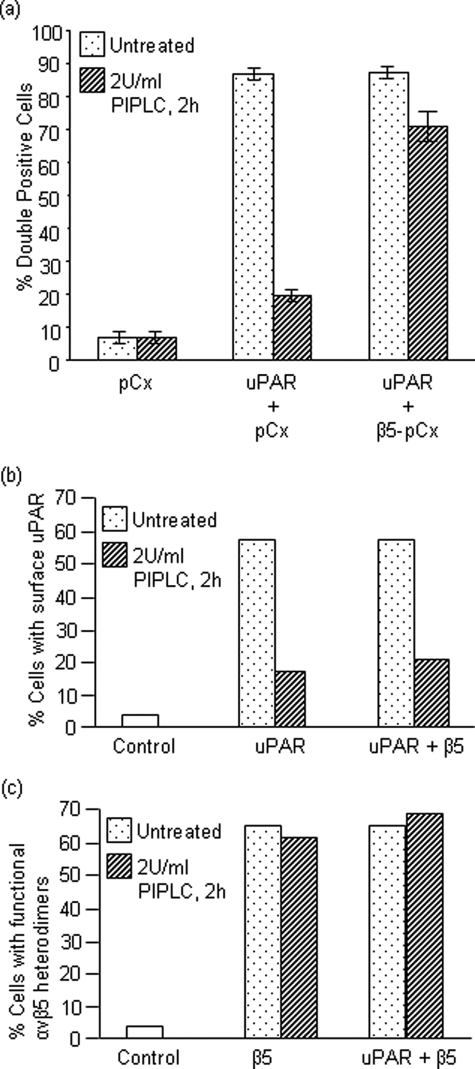
The data above suggested that uPAR might effect efferocytosis independently of αvβ5 integrin. To more directly investigate the relationship between uPAR and β5 integrin and phagocytosis, we treated uPAR-expressing cells with PIPLC under conditions known to cleave the GPI-anchor and release soluble uPAR. As shown in Fig. 4, pretreatment of CS-1 cells with phosphatidylinositol-specific phospholipase C either expressing uPAR alone or uPAR co-expressed with β5 integrin only abolished phagocytosis in the former, suggesting that uPAR may act as a “stand-alone” receptor upstream of Rac1, and relay on a non-αvβ5 integrin dependent pathway. Consistent with this view, PIPLC effectively reduced expression of uPAR from the surface, but had no effect on functional αvβ5 integrins (Fig. 4, B and C, respectively).
FIGURE 4.
Deletion of cell-surface uPAR abrogates the observed phagocytic effect due to uPAR expression. a, CS-1 cells were transfected as indicated. The cells were either left untreated or treated with 2 units/ml glycophosphatidylinositol (GPI)-specific phospholipase C (PIPLC) that cleaves uPAR from the cell surface. Cells were then assessed for their phagocytic efficiency. b and c, surface expression of overexpressed proteins in transiently transfected CS-1 cells was confirmed by staining of live cells with primary antibodies followed by fluorescent secondary antibodies and analyzed by flow cytometry. Surface uPAR before and after PIPLC treatment was detected using a rabbit polyclonal 399R antibody (b). Functional αvβ5 heterodimers on the surface were confirmed by mouse monoclonal P1F6 antibody and were found to be intact after PIPLC treatment (c).
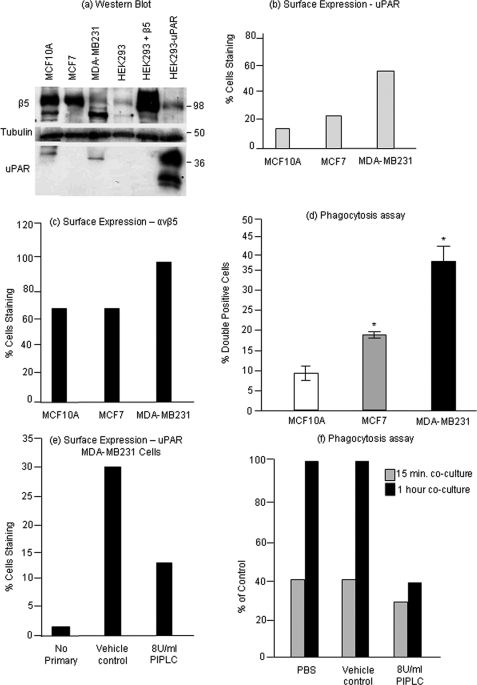
Having established that uPAR overexpression enhanced recognition and clearance of apoptotic T cells, we next wanted to evaluate the potential significance of this finding in a system relevant to uPAR overexpression. As alluded to above, the plasminogen activator system, including overexpression of uPAR and αvβ5 integrin, is often correlated with malignant breast carcinoma and associated with poorer prognoses for long term survival. To explore the relationship between uPAR expression on BCCs and their phagocytic capacity, we examined human BCC lines of varying degrees of invasiveness. MCF7 and MDA-MB231 cell lines were reported to be increasingly invasive in that order and also have increasing levels of β5-integrin and uPAR mRNA and protein. MCF10A, a non-transformed breast epithelial cell line, was used as normal cell control. Shown in Fig. 5 are expression profiles of MCF10A, MCF7, and MDA-MB231 cells interrogated for β5 integrin and uPAR protein by Western blotting (Fig. 5a), and surface distribution (Fig. 5, b and c). Although these different cells expressed variable amounts of mRNA (data not shown) and total protein, curiously the most transformed cells expressed higher levels of both αvβ5 integrin and uPAR on their surface, which corresponds to the most functionally active. To investigate whether these transformed cells had higher phagocytic capacity, each of the above cell lines was co-cultured with apoptotic Jurkat cells for 2 h. Interestingly, the more transformed BCC lines MCF7 and MDA-231 had higher phagocytic activities compared with less transformed cells (Fig. 5d). Treatment of MDA-MB231 cells with PIPLC, which led to a 50% reduction in cell surface uPAR (Fig. 5e), resulted in decreased phagocytic activity when measured at the 1-h time point (Fig. 5f) suggesting that uPAR, in addition to its well known function in motility and invasion, co-opts an additional function involving efferocytosis of tumor cells.
FIGURE 5.
Relationship between uPAR expression and phagocytosis in BCCs. Two breast cancer cell lines (BCCs) viz. MCF7 and MDA-MB231 (in order of increasing invasiveness) and a normal cell line MCF10A were characterized for uPAR and β5 expression. a, protein expression was assessed by Western blotting. Note that in MDA-MB231 cells the major β5-integrin band runs as a faster migrating form. b and c, expression at the cell surface was analyzed by antibody staining of live cells followed by flow cytometric analysis. d, phagocytosis assay was performed by co-culturing MCF10A, MCF7, and MDA-MB231 cells with apoptotic T-cells for 2 h and analyzed by flow cytometry. e, MDA-MB231 cells were treated with vehicle or 8 units/ml PIPLC, and the reduction in the level of cell surface uPAR was assessed by antibody staining of live cells followed by flow cytometry. f, phagocytosis assay was carried out by co-culturing MDA-MB231 cells pre-treated with 8 units/ml PIPLC with apoptotic Jurkat cells for 15 min and 1 h in a phagocyte:apoptotic cell ratio of 1:5. Phagocytosis was compared with both PBS-treated and vehicle-treated cells. Asterisk, p < 0.05.
DISCUSSION
In the present study, we report that uPAR has a dominant role in the efferocytosis of apoptotic cells, promoting engulfment of apoptotic corpses in transiently overexpressing model phagocytes as well as in breast cancer lines that acquire increased expression of uPAR by epigenetic changes. Because uPAR and the plasminogen activator system are often overexpressed in malignant epithelial carcinomas that include breast, prostate, colon, brain, and others, these data imply that uPAR might confer a phagocytic advantage of uPAR-expressing tumor cells in the tumor microenvironment. We posit that, as breast cancer tumors grow, the luminal cells that die by apoptosis would be preferentially engulfed by uPAR-expressing cells, increasing both the net amount of phagocytosis as well as cytokines associated with the process. Because efferocytosis is typically associated with the resolution of inflammation and the production of non-inflammatory cytokines, enhanced engulfment may promote a tolerogenic environment that favors tumor progression.
We initially undertook this study to identify relevant co-receptors that impinge on the αvβ5 integrin-mediated pathway of engulfment. In a previous study, we found that αvβ5 integrin utilized an unconventional NPXY-independent activation signal to promote phagocytosis, and from these studies we posited that αvβ5 integrins may alternate between mutually exclusive pools that includes a pool of integrins participating in focal adhesions, and a second pool of integrin in membrane rafts that activate Rac1. Because uPAR is a membrane raft-associated protein, we anticipated that its association with αvβ5 integrin might be a requisite for its engulfment-promoting effects. However, despite confident predictions that uPAR would require αvβ5 integrin to stimulate engulfment, several lines of evidence suggest the contrary. First, uPAR had a dominant effect on efferocytosis in cells expressing dominant negative integrins. Second, treatment of cells with phosphatidylinositol-specific phospholipase C blocked uPAR-mediated engulfment but had no effect on αvβ5 integrin-mediated engulfment. Third, increased expression of uPAR did not enhance αvβ5 integrin expression on the surface of transfected cells.
The mechanism by which uPAR stimulates efferocytosis is likely related to Rac activation. uPAR is up-regulated for example in cancer cells under hypoxic conditions and promotes cell invasion by activating Rac1 (44). Conversely, uPAR knockout or knockdown cells are deficient in Rac-1 activation (19, 44). Systematic functional analyses of signaling pathways operating downstream of uPAR have shown that uPAR activates Rac1 via the β3-integrin-p130Cas/CrkII/Dock180 pathway (45), which we have also previously identified as a downstream module for αvβ5-integrin dependent phagocytosis (38, 39). However, in our hamster melanoma model, uPAR promotes phagocytosis independently of αvβ3/β5-integrins. The apparent dissociation of uPAR from αv-integrin-dependent phagocytosis could be explained on the basis of pre-activated Rac1 requirement during phagocytosis. Nakaya et al. (48) found that apoptotic cells did not induce Rac-1 activation upon interaction with phagocytes; rather apoptotic cells entered through pre-activated Rac1-containing lamellipodial sites termed ports of entry. The authors argue that Rac1 would be activated via other means such as growth factor receptor signaling and that the presence of apoptotic cells might lead to receptor clustering and recruitment of activated Rac1 at the phagosome. It is therefore possible that uPAR promotes phagocytosis by creating ports of entry for apoptotic cell uptake via integrin-independent Rac1 activation. There is also increasing evidence that GPI-anchored uPAR might independently activate downstream signaling through yet unidentified mechanisms. uPAR-mediated integrin-independent adhesion to vitronectin and protein tyrosine phosphorylation has been reported (10, 49). In both cases, vitronectin ligation and GPI-anchor were found to be necessary and sufficient to induce morphological changes and downstream signaling. We have made a similar observation that shows that Rac1 and uPAR are co-localized at discrete membrane ruffles in the presence of vitronectin (not shown).
It is not clear whether uPAR recognizes epitope(s) directly or indirectly on the apoptotic cell membrane. In the former respect, it will be interesting to determine whether elements of the uPA/tissue plasminogen activator complex are recruited to the apoptotic cell membrane during apoptosis, and could serve as opsonins for apoptotic cell engulfment. This might implicate a protease-specific function of uPAR on the phagocyte possibly to unmask latent cytokines or generate cleavage products of proteins associated with the extracellular matrix and the apoptotic cells. A very interesting study suggests that the uPA inhibitor plasminogen activator inhibitor-1 serves as a potent modulator of neutrophil apoptosis, acting as a “don't eat me” receptor (50). These studies further showed that the increased phagocytosis associated with plasminogen activator inhibitor-1 deficiency or blockade was dependent on both LRP1 and calreticulin suggesting plasminogen activator inhibitor-1 may inhibit this endogenous pathway for clearance. This is quite relevant in lieu of previous reports that uPAR also interacts with LRP1/CED-1 (51), another bona fide engulfment receptor that interacts with calreticulin and C1q on apoptotic cells (52, 53). The involvement of these receptors could be identified using specific inhibitors of LRP1, including Rap1. Moreover, LRP1 has been implicated in phagocytosis by acting on the Rac1 pathway (52), and further studies are required to determine whether uPAR and LRP1 are functional co-receptors in the context of efferocytosis.
Equally important to the net increase in efferocytosis in uPAR-overexpressing cells will be to profile the cytokines and chemokines concomitant with engulfment of the dying breast cancer cells. It is generally well established that efferocytosis is associated with the production of non-inflammatory cytokines such as interleukin-10 and transforming growth factor-β that would promote tolerance and immune subversion (54, 55). However, other studies have demonstrated that tumor cell death may or may not elicit an immune response. Under certain conditions, referred to as immunogenic cell death, tumor cells express markers that permit them to stimulate an adaptive immune response that involves dendritic cell activation and cross-presentation of tumor antigens (56). During phagocytosis, phagocytes can actively secrete cytokines and chemokines, many of which have the capacity to alter the microenvironment to influence progression and metastasis (57). Further studies, aimed at identifying key cytokines or proteokines as a result of uPAR engagement during phagocytosis should help address this question.
In summary, we have identified a novel role for uPAR as a phagocytic receptor. In this emerging picture, this study raises the intriguing question as to whether altered interactions between phagocytes and apoptotic cells and engulfment may contribute to the tumor-promoting capacities of uPAR. A detailed molecular analysis of the uPAR-mediated phagocytosis in tumor cells may lead to the development of novel therapeutic strategies that specifically target downstream signaling and cytokine production thereby modulating tumor invasion.
Footnotes
- uPAR
- urokinase plasminogen activator receptor
- GPI
- glycosylphosphatidylinositol
- VN
- vitronectin
- ERK
- extracellular signal-regulated kinase
- BCC
- breast cancer cell
- PIPLC
- GPI-specific phospholipase C
- DMEM
- Dulbecco's modified Eagle' medium
- EGF
- epidermal growth factor
- PBS
- phosphate-buffered saline
- EGFP
- enhanced GFP.
REFERENCES
- 1.Roldan A. L., Cubellis M. V., Masucci M. T., Behrendt N., Lund L. R., Danø K., Appella E., Blasi F. ( 1990) EMBO J. 9, 467– 474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasi F. ( 1993) Bioessays 15, 105– 111 [DOI] [PubMed] [Google Scholar]
- 3.Rømer J., Bugge T. H., Pyke C., Lund L. R., Flick M. J., Degen J. L., Danø K. ( 1996) Nat. Med. 2, 725. [DOI] [PubMed] [Google Scholar]
- 4.Romer J., Bugge T. H., Pyke C., Lund L. R., Flick M. J., Degen J. L., Dano K. ( 1996) Nat. Med. 2, 287– 292 [DOI] [PubMed] [Google Scholar]
- 5.Saksela O., Rifkin D. B. ( 1988) Annu. Rev. Cell Biol. 4, 93– 126 [DOI] [PubMed] [Google Scholar]
- 6.Vassalli J. D., Sappino A. P., Belin D. ( 1991) J. Clin. Invest. 88, 1067– 1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pöllänen J., Stephens R., Salonen E. M., Vaheri A. ( 1988) Adv. Exp. Med. Biol. 233, 187– 199 [DOI] [PubMed] [Google Scholar]
- 8.Vaheri A., Stephens R. W., Salonen E. M., Pöllänen J., Tapiovaara H. ( 1990) Cell Differ. Dev 32, 255– 262 [DOI] [PubMed] [Google Scholar]
- 9.Kjøller L., Hall A. ( 2001) J. Cell Biol. 152, 1145– 1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madsen C. D., Ferraris G. M., Andolfo A., Cunningham O., Sidenius N. ( 2007) J. Cell Biol. 177, 927– 939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kjøller L. ( 2002) Biol. Chem. 383, 5– 19 [DOI] [PubMed] [Google Scholar]
- 12.Ploug M., Rønne E., Behrendt N., Jensen A. L., Blasi F., Danø K. ( 1991) J. Biol. Chem. 266, 1926– 1933 [PubMed] [Google Scholar]
- 13.Chapman H. A., Wei Y., Simon D. I., Waltz D. A. ( 1999) Thromb. Haemost. 82, 291– 297 [PubMed] [Google Scholar]
- 14.Wei Y., Waltz D. A., Rao N., Drummond R. J., Rosenberg S., Chapman H. A. ( 1994) J. Biol. Chem. 269, 32380– 32388 [PubMed] [Google Scholar]
- 15.Liu D., Aguirre Ghiso J., Estrada Y., Ossowski L. ( 2002) Cancer Cell 1, 445– 457 [DOI] [PubMed] [Google Scholar]
- 16.Resnati M., Pallavicini I., Wang J. M., Oppenheim J., Serhan C. N., Romano M., Blasi F. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 1359– 1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alfano M., Sidenius N., Blasi F., Poli G. ( 2003) J. Leukoc Biol. 74, 750– 756 [DOI] [PubMed] [Google Scholar]
- 18.Furlan F., Orlando S., Laudanna C., Resnati M., Basso V., Blasi F., Mondino A. ( 2004) J. Cell Sci. 117, 2909– 2916 [DOI] [PubMed] [Google Scholar]
- 19.Ma Z., Thomas K. S., Webb D. J., Moravec R., Salicioni A. M., Mars W. M., Gonias S. L. ( 2002) J. Cell Biol. 159, 1061– 1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyetko M. R., Sitrin R. G., Fuller J. A., Todd R. F., 3rd, Petty H., Standiford T. J. ( 1995) J. Leukoc Biol. 58, 533– 538 [DOI] [PubMed] [Google Scholar]
- 21.Wei Y., Yang X., Liu Q., Wilkins J. A., Chapman H. A. ( 1999) J. Cell Biol. 144, 1285– 1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hienert G., Kirchheimer J. C., Pflüger H., Binder B. R. ( 1988) J. Urol. 140, 1466– 1469 [DOI] [PubMed] [Google Scholar]
- 23.Kirchheimer J. C., Pflüger H., Ritschl P., Hienert G., Binder B. R. ( 1985) Invasion Metastasis 5, 344– 355 [PubMed] [Google Scholar]
- 24.Miyake H., Hara I., Yamanaka K., Gohji K., Arakawa S., Kamidono S. ( 1999) Prostate 39, 123– 129 [DOI] [PubMed] [Google Scholar]
- 25.Reuning U., Magdolen V., Wilhelm O., Fischer K., Lutz V., Graeff H., Schmitt M. ( 1998) Int. J. Oncol. 13, 893– 906 [DOI] [PubMed] [Google Scholar]
- 26.Schmitt M., Wilhelm O., Jänicke F., Magdolen V., Reuning U., Ohi H., Moniwa N., Kobayashi H., Weidle U., Graeff H. ( 1995) J. Obstet. Gynaecol. 21, 151– 165 [DOI] [PubMed] [Google Scholar]
- 27.Sidenius N., Blasi F. ( 2003) Cancer Metastasis Rev. 22, 205– 222 [DOI] [PubMed] [Google Scholar]
- 28.Wun T. C., Reich E. ( 1987) J. Biol. Chem. 262, 3646– 3653 [PubMed] [Google Scholar]
- 29.Croucher D. R., Saunders D. N., Lobov S., Ranson M. ( 2008) Nat. Rev. Cancer 8, 535– 545 [DOI] [PubMed] [Google Scholar]
- 30.Croucher D. R., Saunders D. N., Stillfried G. E., Ranson M. ( 2007) Biochem. J. 408, 203– 210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stefansson S., Lawrence D. A. ( 1996) Nature 383, 441– 443 [DOI] [PubMed] [Google Scholar]
- 32.Huai Q., Mazar A. P., Kuo A., Parry G. C., Shaw D. E., Callahan J., Li Y., Yuan C., Bian C., Chen L., Furie B., Furie B. C., Cines D. B., Huang M. ( 2006) Science 311, 656– 659 [DOI] [PubMed] [Google Scholar]
- 33.Huai Q., Zhou A., Lin L., Mazar A. P., Parry G. C., Callahan J., Shaw D. E., Furie B., Furie B. C., Huang M. ( 2008) Nat. Struct. Mol. Biol. 15, 422– 423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang H., Kerins D. M., Hao Q., Inagami T., Vaughan D. E. ( 1998) J. Biol. Chem. 273, 18268– 18272 [DOI] [PubMed] [Google Scholar]
- 35.Singh S., D'mello V., van Bergen en Henegouwen P., Birge R. B. ( 2007) Biochem. Biophys. Res. Commun. 364, 540– 548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Debnath J., Muthuswamy S. K., Brugge J. S. ( 2003) Methods 30, 256– 268 [DOI] [PubMed] [Google Scholar]
- 37.Høyer-Hansen G., Rønne E., Solberg H., Behrendt N., Ploug M., Lund L. R., Ellis V., Danø K. ( 1992) J. Biol. Chem. 267, 18224– 18229 [PubMed] [Google Scholar]
- 38.Albert M. L., Kim J. I., Birge R. B. ( 2000) Nat. Cell Biol. 2, 899– 905 [DOI] [PubMed] [Google Scholar]
- 39.Akakura S., Singh S., Spataro M., Akakura R., Kim J. I., Albert M. L., Birge R. B. ( 2004) Exp. Cell Res. 292, 403– 416 [DOI] [PubMed] [Google Scholar]
- 40.Franco P., Vocca I., Carriero M. V., Alfano D., Cito L., Longanesi-Cattani I., Grieco P., Ossowski L., Stoppelli M. P. ( 2006) J. Cell Sci. 119, 3424– 3434 [DOI] [PubMed] [Google Scholar]
- 41.Carriero M. V., Del Vecchio S., Capozzoli M., Franco P., Fontana L., Zannetti A., Botti G., D'Aiuto G., Salvatore M., Stoppelli M. P. ( 1999) Cancer Res. 59, 5307– 5314 [PubMed] [Google Scholar]
- 42.Yebra M., Parry G. C., Strömblad S., Mackman N., Rosenberg S., Mueller B. M., Cheresh D. A. ( 1996) J. Biol. Chem. 271, 29393– 29399 [DOI] [PubMed] [Google Scholar]
- 43.Wu Y., Singh S., Georgescu M. M., Birge R. B. ( 2005) J. Cell Sci. 118, 539– 553 [DOI] [PubMed] [Google Scholar]
- 44.Lester R. D., Jo M., Montel V., Takimoto S., Gonias S. L. ( 2007) J. Cell Biol. 178, 425– 436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith H. W., Marra P., Marshall C. J. ( 2008) J. Cell Biol. 182, 777– 790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deleted in proof
- 47.Deleted in proof
- 48.Nakaya M., Kitano M., Matsuda M., Nagata S. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 9198– 9203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petzinger J., Saltel F., Hersemeyer K., Daniel J. M., Preissner K. T., Wehrle-Haller B., Kanse S. M. ( 2007) Cell Commun. Adhes. 14, 137– 155 [DOI] [PubMed] [Google Scholar]
- 50.Park Y. J., Liu G., Lorne E. F., Zhao X., Wang J., Tsuruta Y., Zmijewski J., Abraham E. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 11784– 11789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Czekay R. P., Kuemmel T. A., Orlando R. A., Farquhar M. G. ( 2001) Mol. Biol. Cell 12, 1467– 1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardai S. J., McPhillips K. A., Frasch S. C., Janssen W. J., Starefeldt A., Murphy-Ullrich J. E., Bratton D. L., Oldenborg P. A., Michalak M., Henson P. M. ( 2005) Cell 123, 321– 334 [DOI] [PubMed] [Google Scholar]
- 53.Ogden C. A., deCathelineau A., Hoffmann P. R., Bratton D., Ghebrehiwet B., Fadok V. A., Henson P. M. ( 2001) J. Exp. Med. 194, 781– 795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fadok V. A., Bratton D. L., Konowal A., Freed P. W., Westcott J. Y., Henson P. M. ( 1998) J. Clin. Invest. 101, 890– 898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel V. A., Longacre-Antoni A., Cvetanovic M., Lee D. J., Feng L., Fan H., Rauch J., Ucker D. S., Levine J. S. ( 2007) Autoimmunity 40, 274– 280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Obeid M., Tesniere A., Ghiringhelli F., Fimia G. M., Apetoh L., Perfettini J. L., Castedo M., Mignot G., Panaretakis T., Casares N., Métivier D., Larochette N., van Endert P., Ciccosanti F., Piacentini M., Zitvogel L., Kroemer G. ( 2007) Nat. Med. 13, 54– 61 [DOI] [PubMed] [Google Scholar]
- 57.Golpon H. A., Fadok V. A., Taraseviciene-Stewart L., Scerbavicius R., Sauer C., Welte T., Henson P. M., Voelkel N. F. ( 2004) Faseb J. 18, 1716– 1718 [DOI] [PubMed] [Google Scholar]
- 58.Wei Y., Lukashev M., Simon D. I., Bodary S. C., Rosenberg S., Doyle M. V., Chapman H. A. ( 1996) Science 273, 1551– 1555 [DOI] [PubMed] [Google Scholar]