Abstract
NAD+ (nicotinamide adenine dinucleotide) is an essential cofactor involved in various biological processes including calorie restriction-mediated life span extension. Administration of nicotinamide riboside (NmR) has been shown to ameliorate deficiencies related to aberrant NAD+ metabolism in both yeast and mammalian cells. However, the biological role of endogenous NmR remains unclear. Here we demonstrate that salvaging endogenous NmR is an integral part of NAD+ metabolism. A balanced NmR salvage cycle is essential for calorie restriction-induced life span extension and stress resistance in yeast. Our results also suggest that partitioning of the pyridine nucleotide flux between the classical salvage cycle and the NmR salvage branch might be modulated by the NAD+-dependent Sir2 deacetylase. Furthermore, two novel deamidation steps leading to nicotinic acid mononucleotide and nicotinic acid riboside production are also uncovered that further underscore the complexity and flexibility of NAD+ metabolism. In addition, utilization of extracellular nicotinamide mononucleotide requires prior conversion to NmR mediated by a periplasmic phosphatase Pho5. Conversion to NmR may thus represent a strategy for the transport and assimilation of large nonpermeable NAD+ precursors. Together, our studies provide a molecular basis for how NAD+ homeostasis factors confer metabolic flexibility.
The pyridine nucleotide NAD+ and its reduced form NADH are primary redox carriers involved in metabolism. In addition to serving as a coenzyme in redox reactions, NAD+ also acts as a cosubstrate in protein modification reactions including deacetylation and ADP-ribosylation (1, 2). NAD+ also plays an important role in calorie restriction (CR)2-mediated life span extension via regulating NAD+-dependent longevity factors (3, 4). CR is the most effective regimen known to extend life span in various species (5, 6). CR also ameliorates many age-related diseases such as cancer and diabetes (5). The Sir2 family proteins are NAD+-dependent protein deacetylases, which have been shown to play important roles in several CR models in yeast (3, 7) and higher eukaryotes (8, 9). By coupling the cleavage of NAD+ and deacetylation of target proteins, the Sir2 family proteins serve as a molecular link relaying the cellular energy state to the machinery of life span regulation. Mammalian Sir2 family proteins (SIRT1–7) have also been implicated in stress response, cell survival, and insulin and fat metabolism (8–10), supporting a role for SIRT proteins in age-related metabolic diseases and perhaps human aging.
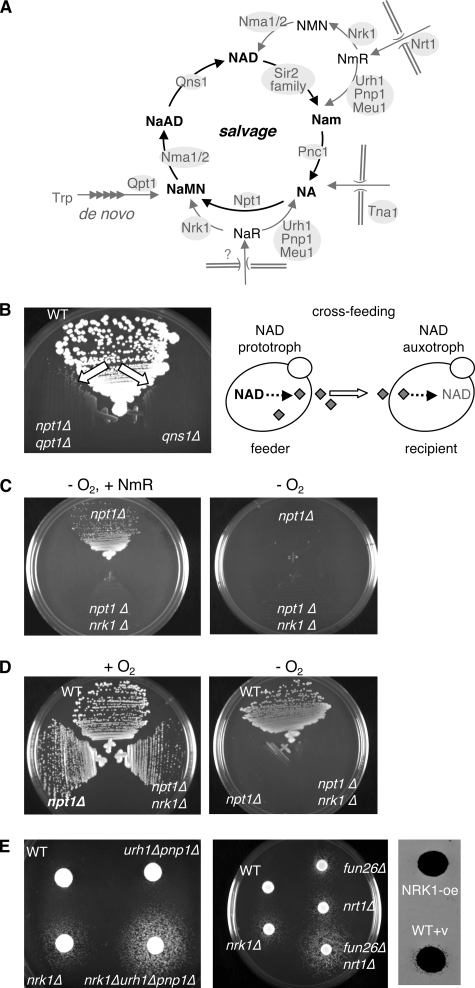
In eukaryotes, NAD+ is generated by de novo synthesis and by salvaging various intermediary precursors (see Fig. 1A). In yeast, the de novo pathway is mediated by Bna1–5 and Qpt1 (Bna6), which produces nicotinic acid mononucleotide (NaMN) from tryptophan (11). Because the de novo pathway requires molecular oxygen as a substrate, cells grown under anaerobic growth conditions would rely on exogenous NAD+ precursors for the nicotinamide (Nam) moiety (11). Yeast cells also salvage Nam from NAD+ consuming reactions or nicotinic acid (NA) from environment via Tna1, Pnc1, and Npt1, leading to NaMN production. NaMN is then converted to NAD+ via Nma1/2 and Qns1 (see Fig. 1A). Nma1/2 are adenylyltransferases with dual specificity toward NMN and NaMN (12, 13), and Qns1 is a glutamine-dependent NAD+ synthetase. Recent studies also showed that supplementing nicotinamide riboside (NmR) and nicotinic acid riboside (NaR) to growth medium rescued the lethality of NAD+ auxotrophic mutants (14–16). Assimilations of exogenous NmR and NaR are mainly mediated by a conserved NmR kinase (Nrk1) and three nucleosidases (Urh1, Pnp1, and Meu1). Nrk1 phosphorylates NmR and NaR to produce nicotinamide mononucleotide (NMN) and NaMN, respectively (14, 16). Urh1, Pnp1, and Meu1 catabolize NmR and NaR to generate Nam and NA (15, 16).
FIGURE 1.
Nicotinamide riboside (NmR) is an endogenous metabolite in yeast. A, the current model of the NAD+ biosynthesis pathways. Extracellular NmR enters the salvage cycle through Nrk1, Urh1, Pnp1, and Meu1. B, NAD+ prototrophic cells release metabolites into growth medium to cross-feed NAD+ auxotrophic cells (the npt1Δqpt1Δ and qns1Δ mutants). Micro-colonies of the NAD+ auxotrophic mutants become visible after 2-day incubation at 30 °C, which show “gradient” growth patterns descending from the side adjacent to WT. C, Nrk1 is required for NAD+ auxotrophic cells to utilize NmR. Anaerobic growth conditions (−O2) are utilized to block de novo NAD+ biosynthesis in the npt1Δ and npt1Δnrk1Δ mutants. D, Nrk1 is required to utilize cross-feeding metabolites. E, cross-feeding activity is modulated by factors in NmR metabolism. Cells defective in NmR utilization (left panel) or transport (middle panel) show increased cross-feeding in spot assays. Overexpressing Nrk1 decreases cross-feeding activity (right panel). The results show growth of the npt1Δqpt1Δ recipient (plated on YPD at a density of ∼9000 cells/cm2) supported by feeder cells (∼2 × 104 cells spotted directly onto the recipient lawn). oe, overexpression.
NmR supplementation has recently been shown to be a promising strategy for prevention and treatment of certain diseases (17). For example, NmR protected neurons from axonal degeneration via functioning as a NAD+ precursor (18, 19). Given that several NmR assimilating enzymes and NmR transporters have been characterized and many are conserved from fungi to mammals (14, 15, 20–22), NmR has been speculated to be an endogenous NAD+ precursor (17, 23). Here, we provided direct evidence for endogenous NmR as an integral part of NAD+ metabolism in yeast. We also determined the biological significance of salvaging endogenous NmR and studied its role in CR-induced life span extension. Moreover, we demonstrated that the NmR salvage machinery was also required for utilizing exogenous NMN, which has recently been shown to increase NAD+ levels in mammalian cells (24). Finally, we discussed the role of Sir2 in modulating the flux of pyridine nucleotides between alternate routes.
EXPERIMENTAL PROCEDURES
Strains and Media
Yeast strain BY4742 MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 was acquired from Open Biosystems (25). Rich media YPD and synthetic media SD were made as described (26). The growth medium used for replicative life span analysis was YPD (2% bacto peptone, 1% yeast extract, 1.5% agar) supplemented with filter-sterilized glucose at a final concentration of 2 or 0.5%. Growth medium used for chronological life span analysis was either YPD or SD (supplemented with 4× auxotrophic amino acids leucine, histidine, lysine, and uracil). All gene deletions were made by replacing wild type genes with a reusable loxP-Kanr-loxP marker as described (27) and verified by PCR using oligonucleotides flanking genes of interest. Multiple deletions were carried out by popping out the Kanr marker using a galactose inducible Cre-recombinase. The Nrk1 overexpression construct was made as follows: a pair of oligonucleotides adding a NotI site to the 5′ end and a NheI site to the 3′ end of the NRK1 gene was designed to amplify the coding region of NRK1 via PCR. After PCR amplification, DNA was digested with NotI and NheI and then cloned into ppp81 (7), resulting in pADH1-NRK1, which was verified by DNA sequencing. Anaerobic growth conditions were achieved by using the BBL GasPak anaerobic chamber system.
NAD+, NmR, and NaR Measurements
Total intracellular levels of NAD+ were determined using enzymatic cycling reaction as described (7). NmR and NaR were determined by LC-MS as described (28) at the metabolomics core laboratory at University of California, Davis. The cell extracts were prepared from 1010 cells grown to late log phase by beads beating in ice-cold 50 mm ammonium acetate solution (29). Culture supernatants (50 ml) were collected and lyophilized along with cell extracts at −80 °C. Lyophilized products were resuspended in 100 μl (cell lysate) or 2 ml (culture supernatant) of 13 mm ammonium acetate/acetonitrile (1:1, v/v). 10 μl was used for LC-MS analysis. Chemically synthesized NmR and NaR, which were kindly provided by Dr. A. Sauve (30), were used to establish standard curves. Detection and quantification of NmR and NaR were performed using the MS-multiple reaction mode methods (NmR, retention time 6 min; NaR, retention time 8 min).
Chronological Life Span
Four single colonies from each strain were analyzed in each experiment as described (31) with a few modifications. The cells were grown in YPD or SD supplemented with 2 or 0.5% glucose at starting A600 of 0.1. The cells were grown in 50-ml tubes on a roller drum set at maximum speed to ensure proper aeration. After 2 days, the cells were collected by centrifugation and washed three times with sterile water. The cells were then resuspended in 10 ml of sterile water to A600 of 1 and then were incubated at 30 °C. Cell viability was monitored every 1–2 days by plating a fraction of culture onto fresh growth medium to determine the colony-forming units. The cell survival rate was calculated by normalizing the colony-forming units to the cell number obtained at day 2 (maximum A600).
Heat Shock Resistance
The cells were grown in SD at a starting A600 of 0.1. After 2 days, the cells were spotted onto YPD plates in 5-fold serial dilutions (started at A600 of 1) and then were incubated at 55 or 25 °C for 45 or 60 min. After heat shock, the plates were transferred to 30 °C for another 2–3 days.
Replicative Life Span
All RLS analyses were carried out on YPD plates supplemented with glucose at a final concentration of 2 or 0.5% with 50 cells/strain/experiment (7) using a micromanipulator. Statistical analysis was carried out using the JMP statistics software (SAS), and the Wilcoxon rank sum test p values were calculated for each pair of life spans.
Deamidase Activity Assay
300 A600 unit cells grown overnight in YPD were harvested, and cell lysate was obtained by beads beating in breaking buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, Roche protease inhibitors). Cell lysate containing 80 μg of total cellular proteins was added to 300 μl of deamidase reaction mix (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm MgCl2) (32, 33) using 8 mm of NAD+, NMN, NmR, or Nam as substrates followed by incubation at 30 °C for 45 min. 100 μl of the deamidase reaction mix was then added to 1 ml of ammonia assay mix (3.4 mm α-ketoglutarate, 0.23 mm NADPH, 50 mm phosphate buffer, pH 7.4, 10 units of glutamate dehydrogenase) followed by a reaction at room temperature for 15 min (32, 33). The amount of ammonia was calculated by the decrease in absorbance at 340 nm using standard curve derived from the ammonia standard solutions (Sigma).
RESULTS
NmR Is a Major NAD+ Precursor Released by NAD+ Prototrophic Cells
Yeast cells lacking both the NPT1 and QPT1 genes or the QNS1 gene are inviable in regular growth media because a functional salvage or de novo NAD+ biosynthesis pathway is essential for growth (Fig. 1A) (3, 14). Our previous studies (3) led to a fortuitous discovery that the lethality of the npt1Δqpt1Δ and qns1Δ mutants could be rescued by growing these cells adjacent to wild type (WT) cells (Fig. 1B, left panel). These data suggested that NAD+ prototrophic yeast cells (feeders) constantly released certain NAD+ precursors, which rendered the growth of NAD+ auxotrophic cells (recipients) via cross-feeding (Fig. 1B, right panel). Because NmR supplementation could rescue the lethality of the qns1Δ mutant (14), it was possible that WT cells cross-fed the npt1Δqpt1Δ and the qns1Δ mutants with NmR. We first examined whether cross-feeding would be prevented by deleting NRK1 (NmR kinase) in recipient cells. Anaerobic growth conditions (−O2) were utilized to block de novo NAD+ biosynthesis (11) in the npt1Δ mutants. As shown in Fig. 1C, npt1Δ cells grown anaerobically became auxotrophic for NAD+, and as expected, utilization of exogenous NmR required functional Nrk1 (Fig. 1C, left panel). Nrk1 was also required for the npt1Δ mutant to utilize cross-feeding molecules released by WT cells (Fig. 1D). Furthermore, deleting NmR assimilation enzymes Nrk1, Urh1, and Pnp1 in WT cells dramatically enhanced cross-feeding activity (Fig. 1E, left panel). Conversely, overexpressing NRK1 reduced the cross-feeding ability of WT cells (Fig. 1E, right panel). Interestingly, preventing NmR import by deleting NmR transporters NRT1 and FUN26 also conferred strong cross-feeding (Fig. 1E, middle panel), indicating that yeast cells constantly released NmR. Together, these data showed that cross-feeding activity of the feeder cells appeared to be inversely correlated with NmR salvage and import activities, supporting NmR as the major released NAD+ precursor that rescued the lethality of recipients.
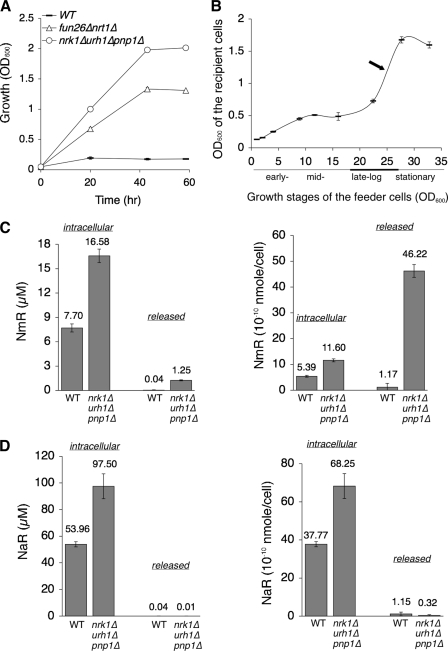
To understand the significance of the NmR pool, we first exploited our cross-feeding reporter system to determine the amount of NmR released by WT and NmR assimilation mutants. Fractions of cell-free culture supernatants of the feeders were collected to supplement the recipients. Fig. 2A showed that the magnitude of cross-feeding conferred by the supernatants of WT and nrk1Δurh1Δpnp1Δ and nrt1Δfun26Δ mutants in liquid culture-based assays was correlated with that determined by agar plate-based assays (Fig. 1E). Using a standard curve derived from defined concentrations of NmR and their corresponding abilities to support growth of the recipients (supplemental Fig. S1), the cumulative concentration of NmR released by the nrk1Δurh1Δpnp1Δ mutant was estimated to be ∼6.74 μm. The amount of NmR released by each nrk1Δurh1Δpnp1Δ mutant cell was about 250 × 10−10 nmol (∼0.3 mm). For comparison, the level of total NAD+ in a WT cell was ∼840 × 10−10 nmol (1.2 mm) (see Fig. 4B). These data highlighted the significance of the NmR pool in NAD+ metabolism.
FIGURE 2.
Quantification of NmR. A, liquid culture-based cross-feeding assays. The results show the ability of the culture supernatants of WT and the NmR salvage mutants to support growth of the recipient (the npt1Δqpt1Δ mutant) over time. 50 μl of cell-free culture supernatant of the feeder is added to the recipient culture (8 ml). B, the rate of NmR release of the nrk1Δurh1Δpnp1Δ mutant at different growth stages. NmR release peaks at the late log phase. One set of representative experiments conducted in triplicate is shown. The error bars denote standard deviations. C, quantification of intracellular and released NmR by LC-MS. The results show steady state concentrations (μm) (left panel) and amount (nmol/cell) (right panel) of intracellular NmR and released NmR. D, quantification of intracellular and released NaR by LC-MS. The results show steady state concentrations (μm) (left panel) and amount (nmol/cell) (right panel) of intracellular NaR and released NaR. The volume of yeast cell is set at 70 femtoliter for calculation.
FIGURE 4.
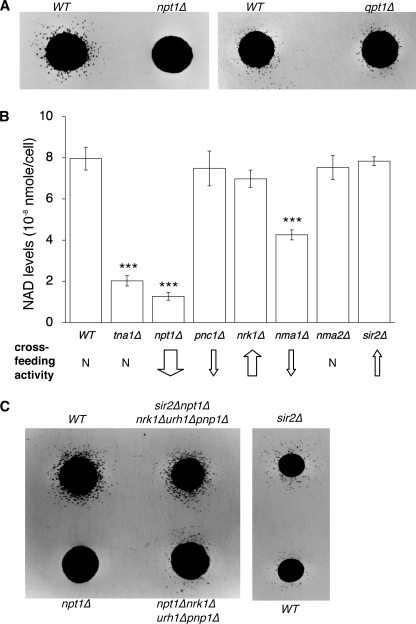
Analyses of NmR release and NAD+ levels in the NA/Nam salvage mutants. A, the level of NmR released by the npt1Δ mutant is extremely low. The qpt1Δ mutant releases WT-level NmR. B, comparisons of the extent of NmR release (cross-feeding activities) and NAD+ levels in the NA/Nam salvage mutants. Upward arrows, increased NmR release; downward arrows, decreased NmR release; N, normal. The width of the arrows indicates the extent of NmR release compared with WT. One set of representative experiments conducted in triplicate is shown. The error bars denote standard deviations. The p values are calculated using Student's t test (***: p < 0.005). C, inactivation of NmR assimilating enzymes partially restores NmR release in the npt1Δ mutant (left panel). Deleting Sir2 slightly increases NmR release in WT (right panel) and further increases NmR release in the npt1Δnrk1Δurh1Δpnp1Δ mutant (left panel). The npt1Δqpt1Δ mutant is used as recipient in all cross-feeding assays.
We next directly quantitated the levels of intracellular and released NmR by LC-MS. Cell extracts (intracellular fractions) and culture supernatants (extracellular fractions) of WT and the nrk1Δurh1Δpnp1Δ mutant were prepared and analyzed as described (28, 29). As shown in Fig. 2C, NmR was detected in the intracellular fractions of WT and the nrk1Δurh1Δpnp1Δ mutant, which confirmed that NmR was an endogenous metabolite. As expected, the concentrations of both intracellular and released NmR were higher in the nrk1Δurh1Δpnp1Δ mutant compared with WT (Fig. 2C, left panel). In both strains, extracellular concentrations of NmR were maintained at much lower levels compared with the intracellular fractions, indicating that NmR transporters Nrt1 and Fun26 efficiently retrieved NmR back to the cell. Fig. 2C (right panel) showed that the amount of NmR released by each nrk1Δurh1Δpnp1Δ cell was ∼40-fold higher than that of WT cell. Overall, results obtained by LC-MS analysis correlated well with results acquired by cross-feeding assays (Figs. 1E and 2A).
It has been reported that exogenous NaR could function as a NAD+ precursor, which also relied on Nrk1 and Urh1/Pnp1/Meu1 for assimilation (16). We therefore examined whether NaR was also present in the intracellular and the extracellular fractions. Interestingly, intracellular concentrations of NaR were higher (∼6-fold) than NmR in both WT and the nrk1Δurh1Δpnp1Δ mutant (Fig. 2D, left panel). However, extracellular NaR concentrations were extremely low in both strains. Consistent with a recent study (21), we found that NaR was a much less efficient (∼100-fold less) NAD+ precursor supplement compared with NmR (supplemental Fig. S2). This was likely due to inefficient transport of NaR across the cell membrane (21). Collectively, our in vivo cross-feeding data and LC-MS quantitative results provide evidence that both NmR and NaR are endogenous metabolites. Because most NaR is retained intracellularly, NmR is likely the key NAD+ precursor that rescues the growth of recipients. The constant release and re-uptake cycle of NmR may represent a novel extended pool of NAD+.
Perturbations of NmR Salvage Shorten Life Span and Increase Sensitivity to Heat Stress
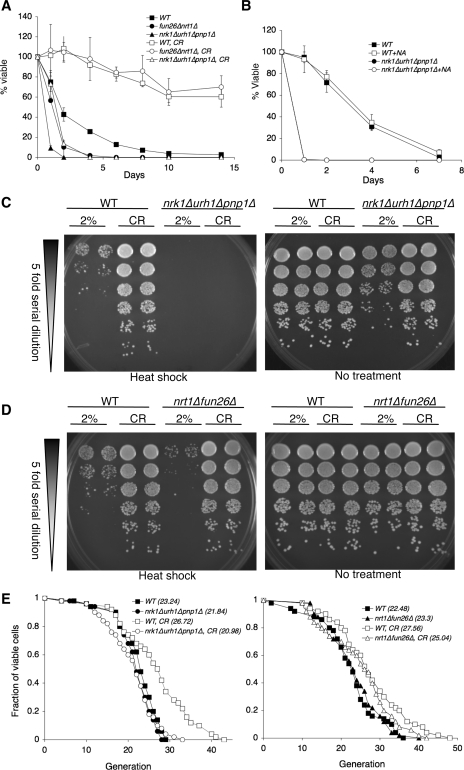
We next determined whether deficiencies in salvaging endogenous NmR would cause any growth defects. Because NmR was mainly produced during late log phase (Fig. 2B), NmR salvage might be central for cell survival in the stationary phase. Yeast chronological life span (CLS) is defined as the length of time cells remain viable in a nondividing state (stationary phase or post-diauxic shift) and is suggested to be a model for studying life span regulation of post-mitotic cells in metazoan (31). As shown in Fig. 3A, the nrk1Δurh1Δpnp1Δ mutant displayed short CLS. CR-induced CLS extension was largely abolished in the nrk1Δurh1Δpnp1Δ mutant. The nrt1Δfun26Δ mutant showed moderate decrease in CLS. However, CR-induced CLS extension was not affected in this mutant (Fig. 3A). NmR supplement has been shown to restore the NAD+ level and the life span of cells grown in media lacking NA (15). However, NA supplement failed to rescue the short CLS of the nrk1Δurh1Δpnp1Δ mutant (Fig. 3B), indicating that NmR salvage played a more important role than the classical NA/Nam salvage in CLS. Yeast CLS extension has been attributed to enhanced stress resistance (31, 34–36). In line with these studies, the nrk1Δurh1Δpnp1Δ and the nrt1Δfun26Δ mutants were sensitive to heat shock stress (Fig. 3, C and D). Also consistent with CLS shown in Fig. 3A, CR-induced heat resistance was abolished in the nrk1Δurh1Δpnp1Δ mutant (Fig. 3C) but was unaffected in the nrt1Δfun26Δ mutant (Fig. 3D). We next examined whether NmR salvage was also required for the CR-induced RLS (division potential of individual cells) extension. As shown in Fig. 3E, CR-induced RLS extension was completely abolished in the nrk1Δurh1Δpnp1Δ mutant and was only partially prevented in the nrt1Δfun26Δ mutant. Collectively, our data demonstrated that the severity of growth defects observed in the nrt1Δfun26Δ and nrk1Δurh1Δpnp1Δ mutants correlated with the amount of NmR released (Fig. 2A) and that NmR assimilation was essential for CR-induced benefits.
FIGURE 3.
Balanced NmR salvage is required for CR-induced life span extension and heat resistance. A, CLS analyses of the NmR salvage mutants grown under normal and CR conditions. CR fails to extend the CLS of the nrk1Δurh1Δpnp1Δ mutant. B, NA does not rescue the short CLS of the nrk1Δurh1Δpnp1Δ mutant. One representative set of three independent experiments, each conducted in quadruplicate, is shown. The error bars denote standard deviations. C, deleting Nrk1, Urh1 and Pnp1 abolishes CR-induced heat resistance. D, deleting Nrt1 and Fun26 has no effect on CR-induced heat resistance. E, RLS analyses of NmR salvage mutants grown under normal and CR conditions. CR requires NmR assimilation to confer RLS extension. Statistical analysis of RLS was carried out using the JMP statistics software, and the Wilcoxon rank-sum test p values are calculated for each pair of life spans. The p values for WT versus CR and nrk1Δurh1Δpnp1Δ versus nrk1Δurh1Δpnp1Δ, CR are both <0.05. One set of representative data is shown. For A–E, WT, wild type control; CR, 0.5% glucose.
NmR Production Requires Functional NA/Nam-mediated NAD+ Salvage
We next investigated the endogenous sources of NmR. Because the nicotinamide moiety of NmR is likely to originate from de novo synthesis or NA/Nam-mediated salvage (Fig. 1A), we compared the cross-feeding activities of the qpt1Δ and npt1Δ mutants. As shown in Fig. 4A, the npt1Δ mutant was unable to cross-feed the recipients, whereas the qpt1Δ mutant exhibited similar cross-feeding ability as the WT cells, suggesting that NA/Nam-mediated salvage was required for NmR production. However, it was also possible that in the npt1Δ mutant, NmR assimilation was activated, thereby resulting in decreased NmR levels. Deleting NmR assimilation enzymes in the npt1Δ mutant only slightly restored NmR release (Fig. 4C). Therefore, NmR production was indeed compromised in the npt1Δ mutant. Because the level of NAD+ in the npt1Δ mutant was significantly reduced compared with WT and the qpt1Δ mutant cells (Fig. 4B) (37), it appeared that the cross-feeding activities were determined by intracellular NAD+ levels. We therefore determined the contribution of each step of the NA/Nam salvage pathway to cross-feeding activities and NAD+ levels. Deleting TNA1 (the major NA transporter) (Fig. 1A) significantly decreased total cellular NAD+ levels (Fig. 4B); however, the cross-feeding activity of the tna1Δ mutant remained similar to that of WT cells (Fig. 4B). Deleting other components of the NA/Nam salvage pathway, such as PNC1, NMA1, and NMA2, only slightly reduced cross-feeding (Fig. 4B). These results demonstrated that the amount of NmR released did not simply reflect the levels of intracellular NAD+. Interestingly, unlike other components in the NA/Nam salvage pathway, deleting SIR2 increased cross-feeding (Fig. 4, B and C).
Sir2 Modulates the Flux of the NmR Salvage Cycle
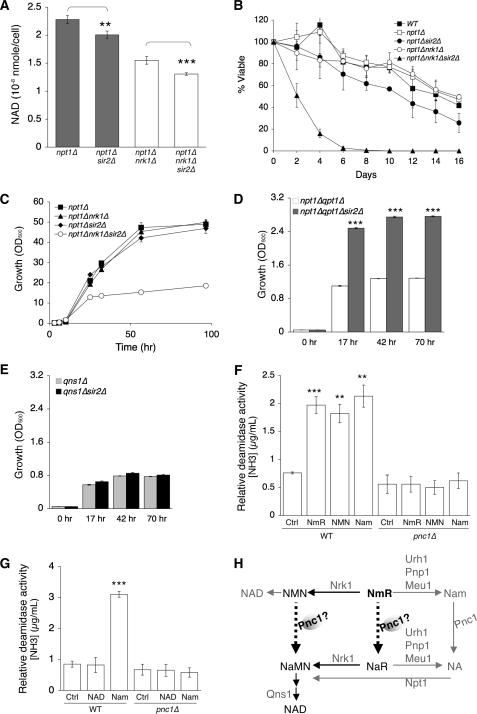
The NAD+-dependent Sir2 deacetylase family has been proposed to support the NA/Nam salvage pathway via producing Nam in deacetylation reactions (38) (Fig. 1A). However, the precise role of Sir2 in NAD+ metabolism remained unclear because deleting SIR2 did not significantly affect total NAD+ levels in WT cells (Fig. 4B). Increased NmR release observed in the sir2Δ mutant suggested a specific role of Sir2 in NmR salvage (Fig. 4C). This role of Sir2 was further uncovered in cells with low basal NAD+ levels (Fig. 4C). It was possible that Sir2 helped to preserve the NAD+ pool by decreasing the pyridine nucleotide flux into the NmR branch. Indeed, deleting SIR2 in these cells further decreased the size of the NAD+ pool (Fig. 5A), which might result in certain growth defects. As shown in Fig. 5B, deleting SIR2 significantly shortened CLS of the npt1Δnrk1Δ mutant. In addition, deletion of SIR2 also severely compromised the growth of the npt1Δnrk1Δ mutant in rich growth medium (Fig. 5C).
FIGURE 5.
Analyses of the role of Sir2 in NmR salvage. A, measurements of intracellular NAD+ levels. Deleting Sir2 decreases NAD+ levels in the npt1Δ and npt1Δnrk1Δ mutants. B, deleting Sir2 in combination with deleting Npt1 and Nrk1 shortens CLS. C, cells lacking Sir2, Npt1 and Nrk1 show severe growth defect in rich medium. D, NmR supplement supports growth of the npt1Δqpt1Δ mutant, which is further enhanced by deleting Sir2. E, deleting Sir2 does not further enhance growth of the qns1Δ mutant supported by NmR. F and G, relative deamidase activities toward various substrates. Cell extracts of WT show deamidase activities toward Nam, NmR, and NMN (F) but not NAD+ (G). Deleting Pnc1 abolishes most deamidase activities in cell extracts. H, a model showing additional routes for NmR assimilation. For A–G, one set of representative experiments conducted in triplicate is shown. The error bars denote standard deviations. The p values are calculated using Student's t test. **, p < 0.01; ***, p < 0.005. For D and E, NmR is supplemented at a final concentration of 0.1 μm.
To further confirm that Sir2 functioned in modulating the pyridine nucleotide flux, we examined the efficiency of NmR utilization of NAD+ auxotrophic mutants. As shown in Fig. 5D, SIR2 deletion enhanced growth of the npt1Δqpt1Δ mutant on NmR supplement, demonstrating the benefit of derepressing NmR flux when NmR salvage became essential. Interestingly, deleting SIR2 failed to enhance NmR utilization in the qns1Δ mutant (Fig. 5E), indicating the beneficial effects induced by sir2Δ required functional Qns1. Current model shows that NmR is converted to NMN or Nam, which are then assimilated into NAD+ (Fig. 1A). Because NMN assimilation does not require Qns1 and the ability to salvage Nam is blocked in the npt1Δqpt1Δ mutant, our results here suggest alternate routes for NmR and/or NMN assimilation. Pnc1 has been shown to function as a deamidase for Nam, which converts Nam to NA (32, 33). We therefore examined whether Pnc1 could also function as a deamidase for NMN and NmR, leading to NaMN and NaR production respectively (Fig. 5H), which would require Qns1 to enter the NAD+ pool. As shown in Fig. 5 (F and G), yeast cells indeed exhibited Pnc1-dependent deamidase activities toward NMN and NmR. In line with these results, NMN deamidase activity has also been reported in bacteria (39). In addition, we showed that NaR was an abundant endogenous NAD+ metabolite (Fig. 2D). Although our results suggested that Pnc1 is a potential NmR and NMN deamidase, further enzyme kinetic analysis is required to show the specificity of Pnc1 toward different pyridine nucleotides. Together, our studies have uncovered additional factors/paths of NAD+ salvage that further underscored the complexity of NAD+ metabolism.
Utilization of Extracellular NMN Requires Prior Conversion to NmR
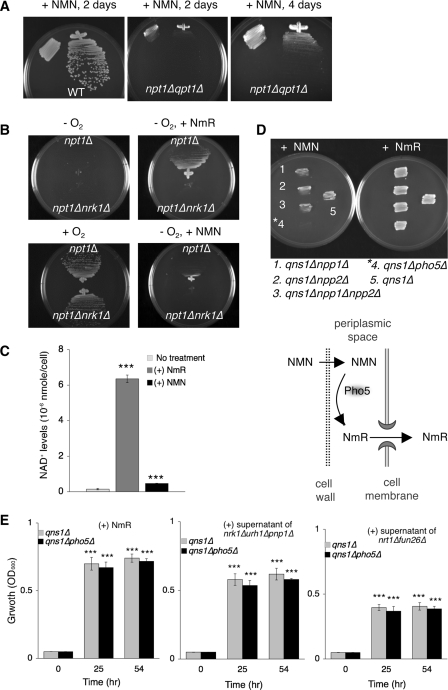
NMN supplement has also been shown to replenish the intracellular NAD+ pool (19, 40, 41). Because the structure of NMN is unfavorable for direct diffusion across the cell membrane, either specific NMN transporters or extracellular NMN catabolizing enzymes would be required to utilize NMN. It has been reported that some bacteria utilize exogenous NMN via conversion to NmR, which is then imported into cell (40, 41). To date, it remained unclear how eukaryotic cells utilized extracellular NMN. To address this question, we first demonstrated that yeast cells were able to utilize exogenous NMN (Fig. 6A). Interestingly, growth of yeast cells on NMN-supplemented medium required Nrk1 (Fig. 6B), indicating that conversion of NMN to NmR was necessary prior to entering cells. However, NMN supported cell growth less efficiently compared with NmR (Fig. 6B, right panels). This contrast between NMN and NmR in supporting cell growth was correlated with their abilities in replenishing the NAD+ pool (Fig. 6C).
FIGURE 6.
Utilization of exogenous NMN requires prior conversion to NmR. A, yeast cells can utilize exogenous NMN as NAD+ precursor. NMN rescues the lethality of the npt1Δqpt1Δ mutant. B, utilization of exogenous NMN requires Nrk1. NMN fails to support the growth of the npt1Δnrk1Δ mutant under anaerobic conditions, which are used to shut down de novo NAD+ synthesis to mimic a npt1Δqpt1Δnrk1Δ mutant. C, measurements of intracellular NAD+ levels of the qpt1Δ recipient grown in NA-free medium supplemented with 10 μm NmR or NMN. NMN is a less efficient NAD+ precursor compared with NmR. D, utilization of exogenous NMN requires Pho5. The qns1Δ recipient carrying deletions of Pho5 or Npp1/Npp2 are grown on media with NMN (left) or NmR (right) added to a final concentration of 10 μm. E, NMN is not a major cross-feeding metabolite. Utilization of cross-feeding metabolites does not require Pho5. The results show growth of the qns1Δ and qns1Δpho5Δ mutants supplemented with 50 μl of 10 μm NmR (left) or culture supernatant of the nrk1Δurh1Δpnp1Δ mutant (middle) or the nrt1Δfun26Δ mutant (right). For C and E, one set of representative experiments conducted in triplicate is shown. The error bars denote standard deviations. The p values are calculated using Student's t test. ***, p < 0.005.
We next determined how exogenous NMN was converted to NmR. In bacteria, NMN assimilation is facilitated by an acid phosphatase, which cleaves NMN to NmR within the periplasmic space (40). In yeast, there are four identified periplasmic acid phosphatases localized to the cell wall or the periplasmic space (42). Among those, Pho5 accounts for more than 90% of acid phosphatase activity in this compartment (43). In addition, Npp1 and Npp2 are two ecto-nucleotide pyrophophatases that exhibit overlapping function with Pho5 in salvaging extracellular phosphates from nucleotides (44). Here we showed that deleting PHO5 totally abolished growth of the qns1Δ mutant on NMN containing medium, whereas deleting NPP1 and NPP2 had no effect (Fig. 6D). These results indicated Pho5 played a major role in the utilization of exogenous NMN.
These results also raised the possibility that released NAD+ precursors might include NMN. However, deleting PHO5 did not significantly reduce the ability of recipients to utilize released metabolites (Fig. 6E), indicating that NMN was not a major cross-feeding molecule. We concluded that prior conversion of NMN to NmR by Pho5 and NmR salvage enzymes were central for assimilating exogenous NMN. Other organisms including mammals might utilize extracellular NMN by a similar mechanism.
DISCUSSION
In this study, we showed that yeast cells constitutively produce, release, and import NmR. The lethality of mutants defective in both de novo and salvage synthesis of NAD+ could be rescued by NAD+ precursors released by WT cells grown in proximity (Fig. 1B), which we defined as “cross-feeding.” We developed NmR-specific cross-feeding reporter systems to further study factors that regulate NmR/NAD+ metabolism. Through LC-MS (Fig. 2, C and D) and cross-feeding analyses (Fig. 6) of WT and the NmR utilization mutants, we demonstrated that NmR was the major NAD+ precursor released by NAD+ prototrophic feeders that rescued the lethality of the npt1Δqpt1Δ and qns1Δ recipients. We showed that functional NmR salvage was required for CR-induced life span extension and stress resistance (Fig. 3). We also provided evidence that endogenous NmR generation was largely dependent on NA/Nam-mediated salvage and was negatively regulated by Sir2 (Fig. 4). Deleting SIR2 in cells defective in NAD+ salvage further decreased intracellular NAD+ levels, resulting in reduced growth fitness and short life span (Fig. 5, A–C). We also demonstrated that utilization of exogenous NMN required prior conversion to NmR by a periplasmic acid phosphatase Pho5 (Fig. 6D). We proposed that similar mechanisms might take place in mammalian cells.
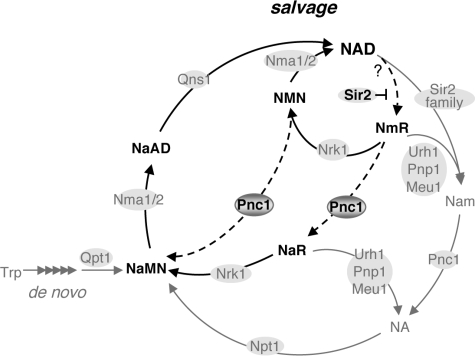
Our studies have uncovered novel paths for NmR-mediated NAD+ salvage (Figs. 5H and 7), in which both NmR and NMN were deamidated by Pnc1 (and perhaps other deamidaes), giving rise to NaR and NaMN respectively. Conversion of NmR to NaR might represent a mechanism to preserve the NAD+ pool because most NaR remained intracellular (Fig. 2D). Supporting this model, our LC-MS analysis showed that NaR was an abundant endogenous metabolite (Fig. 2D). Multiple NmR/NMN assimilation pathways might help confer metabolic flexibility in response to changes in growth conditions. It would be interesting to investigate whether this metabolic strategy could be extrapolated to mammals. Although there is no direct Pnc1 homolog in mammals, it is possible that other aminotransferases could mediate the deamidation of NmR and NMN.
FIGURE 7.
An updated model of NAD+ biosynthesis. NmR and NaR are produced as endogenous metabolites. Sir2 inhibits the flow of pyridine nucleotides into the NmR branch. Additional routes for assimilating NmR and NMN are mediated by Pnc1-like deamidases. Multiple NmR assimilation paths in conjunction with the ability of cycling NmR in and out of cell confer metabolic flexibility in response to various growth conditions.
Although both NA/Nam salvage and Sir2 have been demonstrated to play important roles in regulating RLS (3, 15), their roles in CLS remained unclear. Because both the npt1Δ and nrk1Δurh1Δpnp1Δ mutants had low basal NAD+ levels (15), it was unexpected that the nrk1Δurh1Δpnp1Δ mutant showed reduced CLS (Fig. 3A), whereas the npt1Δ mutant exhibited normal CLS (Fig. 5B). Moreover, NA supplement failed to rescue the short CLS of the nrk1Δurh1Δpnp1Δ mutant (Fig. 3B). These data reinforced the distinct role of NmR salvage in CLS. Interestingly, it was reported that sir2Δ extended the maximum CLS of certain long-lived mutants (45). Because deleting SIR2 enhanced the pyridine nucleotide flow into the NmR branch (Fig. 5D), it would be interesting to investigate whether NmR metabolism plays a role in sir2Δ-induced CLS extension. Our studies suggested that Sir2, as a metabolic sensor, not only senses the NAD+ levels but also modulates the pyridine nucleotide flux. This unique role of Sir2 might provide a feedback control mechanism for cells to respond to metabolic changes. Because deleting other Sir2 family members, such as Hst1 and Hst2, did not show similar effects (supplemental Fig. S3), the growth benefits conferred by sir2Δ was not likely due to decreased NAD+ consumption. It was possible that Sir2 directly regulated yet to be identified components in the NmR pathway. Our data showed that the increase in NmR release resulted from sir2Δ was not affected by deleting any known components of the NmR assimilation pathway (Fig. 4C and data not shown). Future studies to identify novel components in the NmR pathway will provide insights into the molecular basis underlying the roles of Sir2 and NAD+ metabolism in life span regulation.
It is intriguing that cells allow NmR to traffic between intra- and extracellular compartments, which poses the potential risk of losing NAD+. NAD+ participates in many biological processes (1, 46). It is possible that keeping a flexible NmR/NAD+ pool facilitates prompt adjustments of the activities of NAD+-dependent enzymes. In bacteria, it has been suggested that conversion of intracellular NMN to NmR and subsequent release of NmR relieve NMN inhibition toward the NAD+-dependent DNA ligase during active aerobic growth (40). NmR assimilation might also protect cells against stress because NmR assimilation mutant was sensitive to heat stress (Fig. 3C), and intracellular NmR level in WT yeast was decreased upon heat treatment (data not shown). Interestingly, recent studies on the role of NAD+ in protecting neuronal degeneration also showed that expression levels of most NAD+ biosynthetic enzymes were moderately increased after injury (19). In particular, Nrk2, the isoform of Nrk1 identified in mammals (14), was dramatically up-regulated in response to neuronal stresses (19). In addition, it has been suggested that NmR also circulated in the peripheral bloodstream in mammals (47). It will be of great interest to further investigate whether intra- and extracellular cycling of NmR identified in yeast represents a primordial design for metabolic flexibility that also functions in higher eukaryotes.
In mammals, extracellular NMN circulating in peripheral bloodstream has been shown to exert systemic function by regulating NAD+ requiring enzymes in response to altered physiological demand signaled by remote tissues (24, 48). In addition, NMN administration effectively elevated NAD+ level and SIRT1 function in old BESTO mice in which SIRT1 was overexpressed specifically in the pancreatic β cells (24, 48). The additions of NaMN, NMN, NmR, and NAD+ were also shown to delay axonal degeneration in vitro (18, 19). Therefore, administrating NAD+ precursors appeared to be a promising therapeutic or preventive strategy for certain age-associated diseases (17, 19, 48–50). Here we showed that yeast cells utilized exogenous NMN through conversion to NmR by the periplasmic acid phosphatase Pho5 (Fig. 6D). Although there seemed no direct homologs of Pho5 in mammals, CD73, an ecto-5′-nucleotidase, has been suggested to break down extracellular pyridine nucleotides to salvagable precursors (49). In summary, our results have uncovered novel components involved in NmR and NMN assimilation. These studies may also provide insights into the molecular basis of diseases associated with aberrant NAD+ metabolism and aging.
Supplementary Material
Acknowledgments
We thank Dr. A. Sauve and Dr. C. Brenner for discussions on NmR metabolism; Dr. J. Roth, Dr. J. Engebrecht, and members of the Roth and Lin laboratories for discussions and suggestions; and Dr. J. Sporty, Dr. G. Bench, Dr. V. Tolstikov, and Dr. W. Zou for assistance with metabolite extraction and LC-MS analysis.
This work was supported by the National Institute on Aging and the Ellison Medical Foundation.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1–S3.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1–S3.
Footnotes
- CR
- calorie restriction
- LC-MS
- liquid chromatography-mass spectrometry
- NmR
- nicotinamide riboside
- NaMN
- nicotinic acid mononucleotide
- NaR
- nicotinic acid riboside
- NMN
- nicotinamide mononucleotide
- Nam
- nicotinamide
- NA
- nicotinic acid
- WT
- wild type
- CLS
- chronological life span
- RLS
- replicative life span.
REFERENCES
- 1.Lin S. J., Guarente L. ( 2003) Curr Opin Cell Biol. 15, 241– 246 [DOI] [PubMed] [Google Scholar]
- 2.Sauve A. A., Wolberger C., Schramm V. L., Boeke J. D. ( 2006) Annu. Rev. Biochem. 75, 435– 465 [DOI] [PubMed] [Google Scholar]
- 3.Lin S. J., Defossez P. A., Guarente L. ( 2000) Science 289, 2126– 2128 [DOI] [PubMed] [Google Scholar]
- 4.Easlon E., Tsang F., Skinner C., Wang C., Lin S. J. ( 2008) Genes Dev. 22, 931– 944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weindruch W., Walford R. L. ( 1998) The Retardation of Aging and Diseases by Dietary Restriction, Charles C. Thomas, Springfield, IL [Google Scholar]
- 6.Roth G. S., Ingram D. K., Lane M. A. ( 2001) Ann. N.Y. Acad. Sci. 928, 305– 315 [DOI] [PubMed] [Google Scholar]
- 7.Easlon E., Tsang F., Dilova I., Wang C., Lu S. P., Skinner C., Lin S. J. ( 2007) J. Biol. Chem. 282, 6161– 6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dilova I., Easlon E., Lin S. J. ( 2007) Cell Mol. Life Sci. 64, 752– 767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guarente L. ( 2008) Cell 132, 171– 176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guarente L. ( 2006) Nature 444, 868– 874 [DOI] [PubMed] [Google Scholar]
- 11.Panozzo C., Nawara M., Suski C., Kucharczyka R., Skoneczny M., Bécam A. M., Rytka J., Herbert C. J. ( 2002) FEBS Lett. 517, 97– 102 [DOI] [PubMed] [Google Scholar]
- 12.Emanuelli M., Carnevali F., Lorenzi M., Raffaelli N., Amici A., Ruggieri S., Magni G. ( 1999) FEBS Lett. 455, 13– 17 [DOI] [PubMed] [Google Scholar]
- 13.Emanuelli M., Amici A., Carnevali F., Pierella F., Raffaelli N., Magni G. ( 2003) Protein Expr. Purif. 27, 357– 364 [DOI] [PubMed] [Google Scholar]
- 14.Bieganowski P., Brenner C. ( 2004) Cell 117, 495– 502 [DOI] [PubMed] [Google Scholar]
- 15.Belenky P., Racette F. G., Bogan K. L., McClure J. M., Smith J. S., Brenner C. ( 2007) Cell 129, 473– 484 [DOI] [PubMed] [Google Scholar]
- 16.Tempel W., Rabeh W. M., Bogan K. L., Belenky P., Wojcik M., Seidle H. F., Nedyalkova L., Yang T., Sauve A. A., Park H. W., Brenner C. ( 2007) PLoS Biol. 5, e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogan K. L., Brenner C. ( 2008) Annu. Rev. Nutr. 28, 115– 130 [DOI] [PubMed] [Google Scholar]
- 18.Araki T., Sasaki Y., Milbrandt J. ( 2004) Science 305, 1010– 1013 [DOI] [PubMed] [Google Scholar]
- 19.Sasaki Y., Araki T., Milbrandt J. ( 2006) J. Neurosci. 26, 8484– 8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma B., Pan S. J., Zupancic M. L., Cormack B. P. ( 2007) Mol. Microbiol. 66, 14– 25 [DOI] [PubMed] [Google Scholar]
- 21.Belenky P., Christensen K. C., Gazzaniga F., Pletnev A. A., Brenner C. ( 2009) J. Biol. Chem. 284, 158– 164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belenky P. A., Moga T. G., Brenner C. ( 2008) J. Biol. Chem. 283, 8075– 8079 [DOI] [PubMed] [Google Scholar]
- 23.McClure J. M., Gallo C. M., Smith D. L., Jr., Matecic M., Hontz R. D., Buck S. W., Racette F. G., Smith J. S. ( 2008) Genetics 180, 797– 810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Revollo J. R., Körner A., Mills K. F., Satoh A., Wang T., Garten A., Dasgupta B., Sasaki Y., Wolberger C., Townsend R. R., Milbrandt J., Kiess W., Imai S. ( 2007) Cell Metab 6, 363– 375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. ( 1998) Yeast 14, 115– 132 [DOI] [PubMed] [Google Scholar]
- 26.Burke D., Dawson D., Sterns T. ( 2000) in Methods in Yeast Genetics, pp. 171– 174, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27.Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. ( 1996) Nucleic Acids Res. 24, 2519– 2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen P., Wolf W. R. ( 2007) Anal. Bioanal. Chem. 387, 2441– 2448 [DOI] [PubMed] [Google Scholar]
- 29.Sporty J. L., Kabir M. M., Turteltaub K. W., Ognibene T., Lin S. J., Bench G. ( 2008) J. Sep. Sci. 31, 3202– 3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang T., Chan N. Y., Sauve A. A. ( 2007) J. Med. Chem. 50, 6458– 6461 [DOI] [PubMed] [Google Scholar]
- 31.Fabrizio P., Longo V. D. ( 2003) Aging Cell 2, 73– 81 [DOI] [PubMed] [Google Scholar]
- 32.Ghislain M., Talla E., François J. M. ( 2002) Yeast 19, 215– 224 [DOI] [PubMed] [Google Scholar]
- 33.Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. ( 2003) Nature 423, 181– 185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabrizio P., Pozza F., Pletcher S. D., Gendron C. M., Longo V. D. ( 2001) Science 292, 288– 290 [DOI] [PubMed] [Google Scholar]
- 35.Powers R. W., 3rd, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S. ( 2006) Genes Dev. 20, 174– 184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonawitz N. D., Chatenay-Lapointe M., Pan Y., Shadel G. S. ( 2007) Cell Metab. 5, 265– 277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandmeier J. J., Celic I., Boeke J. D., Smith J. S. ( 2002) Genetics 160, 877– 889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Cohen H., Lin S. S., Manchester J. K., Gordon J. I., Sinclair D. A. ( 2002) J. Biol. Chem. 277, 18881– 18890 [DOI] [PubMed] [Google Scholar]
- 39.Cheng W., Roth J. ( 1995) J. Bacteriol. 177, 6711– 6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grose J. H., Bergthorsson U., Xu Y., Sterneckert J., Khodaverdian B., Roth J. R. ( 2005) J. Bacteriol. 187, 4521– 4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kemmer G., Reilly T. J., Schmidt-Brauns J., Zlotnik G. W., Green B. A., Fiske M. J., Herbert M., Kraiss A., Schlör S., Smith A., Reidl J. ( 2001) J. Bacteriol. 183, 3974– 3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oshima Y. ( 1997) Genes Genet. Syst. 72, 323– 334 [DOI] [PubMed] [Google Scholar]
- 43.Svaren J., Hörz W. ( 1997) Trends Biochem. Sci. 22, 93– 97 [DOI] [PubMed] [Google Scholar]
- 44.Kennedy E. J., Pillus L., Ghosh G. ( 2005) Eukaryot. Cell 4, 1892– 1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fabrizio P., Gattazzo C., Battistella L., Wei M., Cheng C., McGrew K., Longo V. D. ( 2005) Cell 123, 655– 667 [DOI] [PubMed] [Google Scholar]
- 46.Sauve A. A. ( 2008) J. Pharmacol. Exp. Ther. 324, 883– 893 [DOI] [PubMed] [Google Scholar]
- 47.Schmidt-Brauns J., Herbert M., Kemmer G., Kraiss A., Schlör S., Reidl J. ( 2001) Int. J. Med. Microbiol. 291, 219– 225 [DOI] [PubMed] [Google Scholar]
- 48.Ramsey K. M., Mills K. F., Satoh A., Imai S. ( 2008) Aging Cell 7, 78– 88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belenky P., Bogan K. L., Brenner C. ( 2007) Trends Biochem. Sci. 32, 12– 19 [DOI] [PubMed] [Google Scholar]
- 50.Spronck J. C., Nickerson J. L., Kirkland J. B. ( 2007) Nutr. Cancer 57, 88– 99 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.