Abstract
In addition to their capacity for template-directed 5′ → 3′ DNA synthesis at the polymerase (pol) site, DNA polymerases have a separate 3′ → 5′ exonuclease (exo) editing activity that is involved in assuring the fidelity of DNA replication. Upon misincorporation of an incorrect nucleotide residue, the 3′ terminus of the primer strand at the primer-template (P/T) junction is preferentially transferred to the exo site, where the faulty residue is excised, allowing the shortened primer to rebind to the template strand at the pol site and incorporate the correct dNTP. Here we describe the conformational changes that occur in the primer strand as it shuttles between the pol and exo sites of replication-competent Klenow and Klentaq DNA polymerase complexes in solution and use these conformational changes to measure the equilibrium distribution of the primer between these sites for P/T DNA constructs carrying both matched and mismatched primer termini. To this end, we have measured the fluorescence and circular dichroism spectra at wavelengths of >300 nm for conformational probes comprising pairs of 2-aminopurine bases site-specifically replacing adenine bases at various positions in the primer strand of P/T DNA constructs bound to DNA polymerases. Control experiments that compare primer conformations with available x-ray structures confirm the validity of this approach. These distributions and the conformational changes in the P/T DNA that occur during template-directed DNA synthesis in solution illuminate some of the mechanisms used by DNA polymerases to assure the fidelity of DNA synthesis.
Escherichia coli DNA polymerase (DNAP)2 I is a single subunit polymerase that is organized into three functional domains: an N-terminal domain that is associated with 5′ → 3′ exonuclease activity, an intermediate domain that carries the 3′ → 5′ proofreading activity, and a C-terminal domain that is associated with the 5′ → 3′ template-directed polymerization activity. An important role of DNAP I is to remove the RNA primers of the Okazaki fragments formed during lagging strand DNA synthesis in E. coli replication and to fill in the resulting gaps by template-directed DNA synthesis (1). An N-terminal deletion mutant of DNAP I, known as the “large fragment” or Klenow form of the enzyme, contains only the polymerase (pol) and the 3′ → 5′ exonuclease (exo) domains. The Klenow polymerase has served and continues to serve as an excellent model system for isolating and defining general structure-function relationships in polymerases and in the supporting machinery of DNA replication.
The main function of the 3′ → 5′ exonuclease activity of DNAP I is to remove misincorporated nucleotide residues from the 3′-end of the primer (2), thus contributing significantly to the overall fidelity of DNA replication (3). Contrary to initial expectations, crystallographic studies showed that the pol and exo active sites are quite far apart in replication polymerases, about 30 Å in Klenow (4). As a consequence, the ability of polymerases to “shuttle” the 3′-end of the primer strand efficiently between the pol and the exo sites in order to rectify misincorporation events during polymerization is critical to maintaining the overall accuracy of template-directed replication. Elucidation of the mechanisms of this shuttling and determination of the factors that control the rates (and equilibria) of the active site switching reaction will certainly increase our understanding of fidelity control by DNA polymerases.
An early crystallographic study of the Klenow polymerase complexed with fully paired primer-template (P/T) DNA revealed that 3–4 nt of the 3′-primer terminus had been unwound from the template stand and partitioned into the exo site and that an extended single-stranded DNA (ssDNA) binding pocket of the exo site appeared to make position-specific hydrophobic contacts with the unstacked bases at the 3′-end of the primer (4). A separate crystallographic study of an editing complex confirmed that an ssDNA fragment 4 nt in length was bound at the exo site in the same conformation as seen for the single-stranded 3′-primer sequence unwound from P/T DNA (5). A structure of Klenow polymerase with the DNA bound at the pol site has not yet been reported, although such structures have been obtained for other homologous polymerases, including Klentaq (the “large fragment” of Thermus aquaticus (Taq) DNAP), Bacillus stearothermophilus (Bst) “large fragment” polymerase, and the T7 DNAP (6–8), all of which are members of the polymerase family that includes Klenow.
The amino acid residues involved in the binding of DNA at the pol site in these polymerases (determined from co-crystal structures) and those of Klenow (determined by site-directed mutagenesis studies (9, 10)) are highly conserved, suggesting that a similar DNA binding mode at the pol site may apply to all of the DNAP I polymerases. The crystal structure of Klenow revealed that the polymerization domain has a shape reminiscent of a right hand in which the palm, fingers, and thumb domains form the DNA-binding crevice. Structural studies with various DNAP I polymerases in the presence of P/T DNA constructs yielded an “open” binary complex, whereas the addition of the next correct dNTP (as a chain-terminating dideoxy-NTP) resulted in the formation of a catalytically competent “closed” ternary complex (6–8). In the latter complex, the 3′-primer terminus was base-paired with the template DNA, and the templating base was poised for incorporation of the next correct nucleotide. These structures showed that the conformation of the DNA primer terminus bound at the pol site is markedly different from that of the “frayed open” primer observed at the exo site in Klenow (4, 5).
Although crystallographic studies have provided a wealth of information about the conformations of the DNA substrates bound at the active sites of DNAP, replication itself is a dynamic process (reviewed in Ref. 11), and it is critical to be able to distinguish between various forms of DNA-polymerase complexes in solution in order to fully understand the mechanistic details of the replication process. A solution approach used by Millar and co-workers (reviewed in Ref. 12) for studying the conformation of DNA in these complexes involved measuring the time-resolved fluorescence anisotropy properties of a dansyl fluorophore attached to a DNA base located 8 bp upstream of the P/T DNA junction. The changes in the lifetime of the fluorophore, which appeared to depend mostly on the local environment occupied by the probe within the protein (i.e. buried versus partially exposed), were correlated with specific binding conformations of the primer to provide an estimate of the fractional occupancy of the pol and the exo sites. Reha-Krantz and co-workers (13) more recently used a related approach, here involving the monitoring of changes in the fluorescent lifetimes of a single 2-aminopurine (2-AP) base (a fluorescent analogue of adenine) site-specifically substituted in the template strand at the P/T junction, to make similar fractional occupancy measurements. However, we note that structural interpretations of these fluorescence experiments relied heavily on the available crystal structures, and it remained to be shown directly that the 3′-end of the primer in P/T DNA constructs assumes the same distribution of conformations when bound to the protein in solution.
To get around this problem, as well as to directly investigate the conformations of the primer DNA in both active sites of the Klenow and Klentaq polymerases, we have used a novel CD spectroscopic approach to characterize the solution conformations of primer DNA bound to Klenow and Klentaq DNAPs. Previously, we had shown that CD spectroscopy, in conjunction with fluorescence measurements, can be used to examine changes in local DNA and RNA conformations at 2-AP dimer probes inserted at specified positions within the nucleic acid frameworks of a variety of macromolecular machines functioning in solution (14–16). 2-AP is a structural isomer of adenine that forms base pairs with thymine in DNA (and uridine in RNA), and the substitution of 2-AP for adenine in such bp does not significantly perturb the structure or stability of the resultant double helix. Furthermore, when these probes are used as dimer pairs, the CD spectrum primarily reflects the interaction of the transition dipoles of the two probes themselves and thus the local conformation of the DNA at those positions within the P/T DNA. The characteristic CD and fluorescence signals for 2-AP probes in nucleic acids occur at wavelengths of >300 nm, a spectral region in which the protein and the canonical nucleic acid components of the “macromolecular machines of gene expression” are otherwise transparent. In this study, we have examined the binding of Klenow and Klentaq polymerases to P/T DNA constructs that were designed to be comparable with the nucleic acid components of functioning replication complexes. By examining the low energy CD spectra of site-specifically placed 2-AP probes, we have been able to characterize base conformations at defined positions within the DNA to reveal conformational features of specific DNA bases bound at and near both the pol and the exo active sites of these polymerases. These measurements, in that they directly reflect the actual conformations of the DNA chains bound within the active sites of the functioning polymerase, have also provided a direct means to estimate the equilibrium distributions of primer ends between the two active sites for various P/T DNA constructs.
EXPERIMENTAL PROCEDURES
Materials
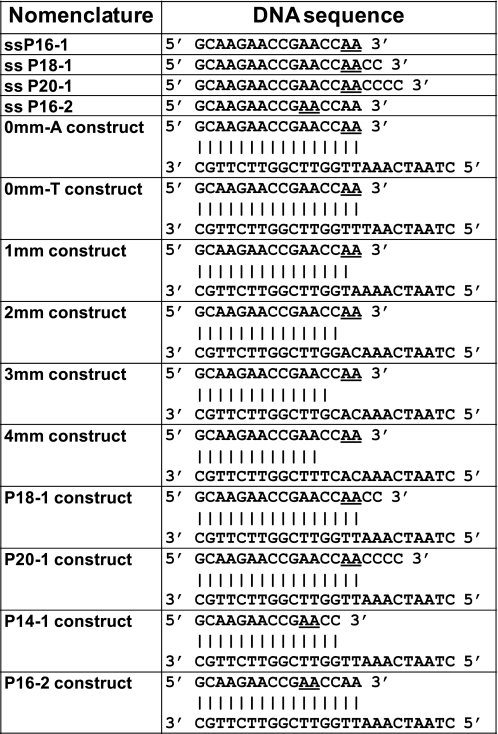
Unlabeled and 2-AP-labeled DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and from Operon (Huntsville, AL). Oligonucleotide concentrations were determined using absorbance measurements at 260 nm and extinction coefficients furnished by the manufacturer. The P/T constructs were annealed by heating equimolar concentrations of primer and template DNA strands at 92 °C for 4 min and then gradually cooling to room temperature over a period of 2 h. The sequences and nomenclatures of the DNA constructs examined in this study are shown in Fig. 1.
FIGURE 1.
Nomenclature and sequences for the DNA constructs used in this study. The underlined bases represent nucleotide positions substituted with 2-AP.
The clone (plasmid pXS106) for the exo− D424A derivative of Klenow fragment (a mutation that eliminates the 3′ → 5′ exonuclease activity of the polymerase but retains the binding affinity of the exo site (17)) and the host (CJ 376) cells were gifts from Catherine Joyce (Yale University). The protein was expressed and purified as described previously (18). Stock concentrations of Klenow were determined using an extinction coefficient (ϵ280) of 6.32 × 104 m−1 cm−1 (19). The Klentaq clone was obtained from the ATCC (Manassas, VA). Klentaq polymerase was purified in the presence of Tween 20 (protein grade from Calbiochem), following procedures described previously (20). Klentaq concentrations were determined by absorbance, using ϵ280 = 6.96 × 104 m−1 cm−1 (21).
Unless otherwise stated, all experiments with Klenow were performed at 25 °C in buffer containing 20 mm Hepes (pH 7.9), 50 mm sodium acetate, either 5 mm Ca(OAc)2 or 5 mm Mg(OAc)2, and 1 mm dithiothreitol. Experiments with Klentaq DNAP were performed at 25 °C in the same buffer used for Klenow plus 0.05% Tween 20. Except for the fluorescence titration shown in Fig. 3F and for velocity sedimentation experiments shown in Fig. S4, the DNA construct concentrations were 3 μm in all experiments. Concentrations of the dNTP substrates used were 500 μm, and those of α,β-methyleno-dATP were 200 μm.
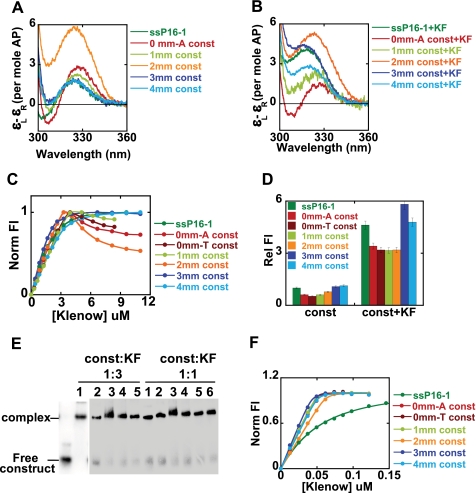
FIGURE 3.
Conformations of bases at the 3′-primer terminus of P/T DNA complexed with Klenow. A and B, low energy CD spectra for the free DNA constructs and the construct-Klenow complexes (see Fig. 1 for sequences and nomenclature of the constructs). C and F, fluorescence titrations for binding of the various constructs to Klenow at 3 μm and 50 nm DNA construct concentrations, respectively. D, relative fluorescence intensities for the same constructs without and with Klenow. E, gel shift assays for the various DNA constructs at equimolar and 1:3 DNA/Klenow concentrations. Lanes 1–6, constructs 4mm, 3mm, 2mm, 1mm, 0mm-T, and 0mm-A, respectively. Color coding is the same for all of the panels. Dark green, ssP16-1; red, 0mm-A; dark red, 0mm-T; light green, 1mm; orange, 2mm; dark blue, 3mm; light blue, 4mm.
Exonuclease and Polymerase Activity of Klenow in Mg2+ and Ca2+-containing Buffers
Some residual 3′ → 5′ exonuclease activity was observed with solutions of Klenow during data acquisition in the CD experiments, although an exo− mutant was used. Such residual activity has been seen by others (17), and appears to be due either to a slightly leaky exo− mutant or to a very low level contamination with wild-type Klenow. We were concerned that this activity could present a problem for the integrity of our DNA constructs, especially during the CD experiments that sometimes required data acquisition over a period of hours. Since Ca2+ (substituted for Mg2+) does not effectively catalyze either the exonuclease or the polymerase activities of DNAP I over the time scales of our experiments, we used Ca2+-containing solutions in our spectroscopic studies to eliminate any residual exonuclease activity of the polymerase and also to reduce polymerization activity in the presence of various dNTPs. Such use of Ca2+ for Mg2+ replacement had been reported previously in structural studies with RB69 DNA polymerase (22).
In addition, while we have shown that the primers for matched P/T DNA (Fig. 1, the 0mm-A and 0mm-T constructs) can be extended to full-length products by Klenow and Klentaq polymerases in the presence of dNTP substrates in buffers containing Mg2+ (Fig. S1), the efficiency of primer extension was significantly reduced in buffers containing Ca2+ instead (Fig. S1). In the presence of Ca2+, the Klenow polymerase only extended the DNA primer strand by 2–3 nt residues under our experimental conditions, showing that Ca2+ is not an effective replacement for Mg2+ in template-directed polymerization. This is consistent with a recent study that also showed that the rate of nucleotide incorporation by Klenow polymerase was significantly reduced when Ca2+ replaced Mg2+ (23). In addition, no significant elongation of the primer strand by either polymerase was observed for any of our mismatched primer-template DNA constructs in the presence of dNTP substrates in either Ca2+ or Mg2+ buffers, as expected from previous studies that had shown that these polymerases exhibit significantly reduced efficiency in extending mismatched primer termini, relative to their activities with fully template-matched P/T DNA constructs (2, 24, 25). We emphasize that experiments in either Ca2+ and Mg2+ buffers yielded very similar CD spectra for the complexes in the absence of substrate dNTPs. Fig. S2 shows CD spectra for ssP16-1 DNA and the P16-2 construct (green and red curves, respectively) with each of these two divalent metal ion co-factors. The changes (relative to the respective free constructs) in the shape and amplitude of the low energy CD with the addition of Klenow (pink curves) were very similar for both ions, suggesting that the conformational changes obtained for polymerase upon binding to these DNAs in the presence of either ion cofactor are also closely similar. This point is considered further under “Results.”
Fluorescence and CD Measurements
Fluorescence spectra were measured using a Jobin-Yvon Fluorolog spectrofluorimeter. Samples were excited at 315 nm, and emission spectra were recorded from 310 to 420 nm. The fluorescence intensities presented in the figures represent values measured at 370 nm. Enzyme titrations were performed by adding increasing amounts of protein to 3 μm concentrations of 2-AP-labeled DNA construct in order to determine the enzyme/DNA ratio needed to produce maximal fluorescence changes. DNA construct concentrations of 50 nm were used in the fluorescence titrations of Fig. 3F. The fluorescence intensities plotted for the titration curves were normalized by dividing the fluorescence at each polymerase concentration by the maximum fluorescence change. In experiments in which fluorescence changes were used to determine local base conformations under various conditions, the measured emission intensities at 370 nm were normalized with respect to the intensities of corresponding ssDNA.
CD spectra were measured at 300–400 nm using a Jasco model J-720 CD spectrophotometer equipped with a temperature-controlled cell holder. The CD spectra shown represent the average of 8–10 measurements and are reported as (ϵL–ϵR) per mol of 2-AP in units of m−1 cm−1. All CD results were obtained with 3 μm (equimolar) concentrations of polymerase and 2-AP-labeled DNA constructs. For both fluorescence and CD, the spectra of the unlabeled DNA and the polymerases over the wavelength range used were identical to those of the buffer base line; hence, only buffer base line spectra were subtracted from all of the experimental spectra shown.
Gel Shift and Activity Assays
These assays were performed under the same conditions as the fluorescence and CD titrations. For detection, the primer strands of the DNA constructs were labeled at the 5′ terminus with [γ-32P]ATP. DNAP stocks were added to 3 μm concentrations of construct and incubated at 25 °C for 30 min. For gel shift assays, the complexes were then loaded onto 4–20% TBE polyacrylamide Redigels (from Bio-Rad) that had been pre-equilibrated with 1× TBE running buffer (89 mm Tris, 89 mm boric acid, 2 mm EDTA, pH 8.3) containing 8 mm CaCl2. Gels were run at room temperature at 60 V. For activity assays, the DNAP-construct complexes were further incubated with dNTP substrate(s) for an additional 30 min, and the DNA products were separated on 12% sequencing gel in 7 m urea. All gels were visualized with a Storm scanner and quantified using ImageQuant software (GE Health Sciences) after exposure on a PhosphorImager screen.
Analytical Ultracentrifugation Experiments
Sedimentation velocity experiments were performed in a Beckman Optima XL-I analytical ultracentrifuge under the same solution conditions used for the spectroscopic assays and at equimolar (6 μm each) concentrations of Klenow and DNA constructs. The reference and sample sectors of double-sector cells were loaded with 425 μl of buffer and 400 μl of sample solution, respectively. All velocity runs were performed at 50,000 rpm in an An-60 Ti rotor for 8 h at 25 °C and were scanned continuously at 280 nm. The data were analyzed using the Sedfit program (26, 27).
Deconvoluting the CD Spectra of Klenow-DNA Complexes
In deconvoluting the spectra of these complexes into their exo and pol complex components, we have treated the spectra of the complexes representing the two “end states” as being additive, meaning that the experimental spectra obtained for the DNAP-construct mixtures can be represented as the sums of the spectra of the constituent complexes measured separately. In deconvoluting the spectra, the difference between the vector products for the sample complex and the individual component complexes was determined at 0.1-nm wavelength increments and then summed. The values obtained at each wavelength were reduced by the minimization routines of the Mathematica program (Mathematica 6), resulting in a best fit resolution of the given spectra under the restriction that the sum of the coefficients must add up to 1.
RESULTS
Nucleic Acid Construct Sequences and Nomenclature
The sequences and nomenclature of the single-stranded DNA primers and the annealed constructs used in this study are listed in Fig. 1. P16, P18, and P20 correspond to primer strands 16, 18, and 20 nt in length, respectively, whereas “mm” indicates the number of mismatched bases present at the 3′ terminus of the primer in the duplex DNA constructs. 0mm-A and 0mm-T designate matched P/T constructs that differ only in the identity of the base (A or T) at the respective templating positions. The 0mm-T construct was designed to permit the study of the effect of the addition of α,β-methyleno-dATP (a nonhydrolyzable analog of dATP) to the polymerase-DNA complex as a complementary (to the templating base) substrate nucleotide.
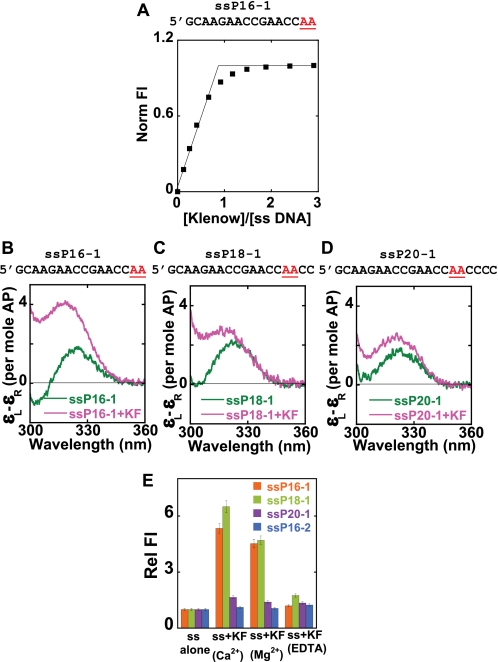
Interaction of ssDNA with the exo Site of Klenow in Solution
Fig. 2 documents the interactions of Klenow DNAP with ssDNA. Fig. 2A shows the binding curve obtained on adding Klenow to ssP16-1 DNA (see Fig. 1 for nomenclature) in Ca2+ buffer (see “Experimental Procedures” on using Ca2+ instead of Mg2+ in these experiments). The binding of Klenow to ssDNA at these concentrations is clearly stoichiometric, with the titration curve showing a breakpoint at equimolar concentrations of DNA and Klenow. In contrast, the addition of Klentaq DNAP to ssDNA under identical conditions did not result in any change in fluorescence intensity, and no complex formation was observed in gel shift assays (data not shown), suggesting that Klentaq does not bind significantly to ssDNA under the experimental conditions used here. This is not unexpected, because TaqDNAP lacks both the 3′ → 5′ exonuclease activity with ssDNA substrates (28) and a ssDNA binding site.
FIGURE 2.
Binding of Klenow to ssDNA. 2-AP bases at defined positions within the ssDNA sequences are shown in red. A, fluorescence titration curve for the binding of Klenow to ssP16-1. B–D, the low energy CD spectra of the three ssDNA chains used, in the presence and absence of Klenow fragment (KF). E, relative fluorescence intensities of the ssDNA chains in the absence and presence of Klenow. Color coding for the CD spectra is as follows. Dark green, free ssDNA; pink, ssDNA-Klenow complex at equimolar concentrations. Color coding for the fluorescence data is as follows. Orange, ssP16-1; light green, ssP18-1; purple, ssP20-1; blue, ssP16-2.
Comparison of crystal structures of Klenow and Klentaq reveals a very similar architecture of the N-terminal domain (which contains the exo site in Klenow). However, several of the key protein residues that are important for DNA binding, divalent metal binding, and catalysis in the exo site of Klenow are either missing or truncated in Klentaq (29, 30), consistent with the absence of measurable ssDNA binding to the latter DNAP. The CD spectra (Fig. 2, B–D) of the 2-AP dimer probes of the various ssDNA chains free in solution are quite similar (green curves), all showing peaks at about 325 nm, as expected (14, 31).3 This suggests that the local conformations of the ssDNA chains are likely to be similar as well.
The addition of equimolar concentrations of Klenow to the ssP16-1 and ssP18-1 DNA strands resulted in a blue shift of the low energy CD peak to 320 nm and a concomitant increase in peak amplitude (compare pink and green spectra in Fig. 2, B–D). These changes are characteristic of the unstacking of the 2-AP bases beyond the ssDNA level (14). However, the addition of Klenow to the ssP20-1 construct produced only a small increase in CD amplitude at 325 nm and no blue shift. These CD results suggest that the 4 nt at the 3′ terminus of the ssDNA primer adopt a significantly unstacked conformation upon binding to Klenow DNAP but that residues 5 and 6 are relatively unperturbed by this binding. Fig. 2E shows the fluorescence intensities at 370 nm of the 2-AP probes of the ssDNA chains alone and in the presence of equimolar concentrations of Klenow in Ca2+, Mg2+, and “EDTA buffer.” (The EDTA buffer samples contained no added divalent metal ions and 2 mm EDTA to scavenge any trace divalent ion contaminants.)
In the presence of either Ca2+ or Mg2+ ions, the fluorescence intensities of ssP16-1 and ssP18-1 showed a dramatic increase (5–6-fold) upon the addition of Klenow, consistent with a major unstacking of the 2-AP probes upon binding. However, the ssP20-1-Klenow complex showed very little increase in fluorescence intensity in either the Ca2+ or Mg2+ buffers, consistent with the finding that the conformations of 2-AP dimer probes located more than 4 nt from the 3′-primer end do not differ significantly from those of probes located in the “interior” of ssDNA chains. This suggests that the extent of propagation of conformational change down the length of the primer strand is a consequence of specific binding interactions of the DNA with the protein and is not contingent on how many base pairs the polymerase is able to unwind. Fluorescence data for ssP16-2 (a shorter ssDNA chain in which the positions of the 2-AP probes relative to the 3′ terminus are the same as those of ssP20-1) are also included for comparison.
Taken together, the fluorescence results are totally consistent with our CD experiments and demonstrate again that four 3′-terminal bases of the ssDNA are significantly unstacked as a consequence of binding in the exo site of the Klenow. These findings are also consistent with crystallographic studies of ssDNA-Klenow complexes that showed the 3′-terminal nucleotides of the ssDNA bound in the exo site in a significantly extended conformation (5).
We note that the addition of Klenow in EDTA buffer did not result in the production of a hyperfluorescent state (Fig. 2E) or in any change in the CD signal for the ssDNA constructs examined (data not shown), suggesting that divalent metal ions are required for proper binding of the 3′-end of the DNA at the exo site. This is consistent with previous site-directed mutagenesis studies that showed that at least one of the two exo site-bound catalytic divalent metal ions appeared to be important for DNA binding (32).
Interaction of P/T DNA Constructs with Klenow in Solution
P/T DNA constructs, each with different numbers of mismatched (mm) bases and all carrying a 2-AP dimer probe at the 3′ terminus of the primer strand (the 0mm-A, 0mm-T, 1mm, 2mm, 3mm, and 4mm constructs), were prepared by annealing the ssP16-1 chain with different template strands (see Fig. 1). Fig. 3, A and B, show the low energy CD spectra for the free P/T DNA constructs and the Klenow-DNA construct complexes in Ca2+ buffer, respectively.
The CD spectra (Fig. 3A) of the free 0mm-A P/T DNA construct showed a peak at 330 nm and a trough at 305 nm, corresponding to the expected B-form exciton coupling for 2-AP dimer probes paired with the template strand. The free 1mm P/T DNA construct, which carries a single mismatch at the 3′-primer terminus, showed a small decrease in the CD peak at 330 nm relative to the 0mm-A (fully base-paired) construct (Fig. 3A), whereas the spectra for the 3mm and 4mm constructs almost overlapped that of free ssP16-1. This demonstrates that the primer terminus reverts to a normal ssDNA conformation beyond 2 nucleotides from the P/T junction. In contrast, the free 2mm construct (two mismatches at the primer terminus directly adjacent to the P/T junction) exhibited a dramatic increase in CD intensity at 330 nm. This observed increase in CD amplitude most likely reflects the constrained conformation of an unpaired 2-AP dimer probe located directly at the P/T junction that we have demonstrated previously (33) for bases located directly vicinal to a replication fork. The above results also suggest that although these probes were only partially (the 1mm construct) or not (the 3mm and 4mm constructs) base-paired with the template strand, the terminal 2-AP dimer retained sufficient B-form character to display exciton character, as observed also for ssP16-1.
The addition of Klenow to the above constructs resulted in a blue shift of the CD peak relative to the peaks of the respective free DNA constructs (Fig. 3B), indicating that the bases at the primer terminus are significantly more unstacked in all of these complexes than in the equivalent free constructs. The 3mm construct with Klenow exhibited a spectrum that almost overlapped that of the ssP16-1-Klenow complex, whereas the spectrum for the 4mm construct complex showed significantly reduced amplitude. This suggests that the primer strand of the 3mm construct binds to Klenow in the same conformation as the 3′-end of the ssP16-1 chain binding in isolation. We speculate that four unpaired residues at the 3′-primer end of the 4mm construct represent too much ssDNA in a P/T DNA context to permit optimal primer contacts with the protein residues that make position-specific interactions with the primer bases at the exo binding site (4). This also suggests that a specific binding interaction occurs between the protein and the 3′-terminal nucleotide. No such length restriction would be expected in the absence of such an interaction, since otherwise the differing lengths of primer strand should be able to slide indefinitely through the exo site.
Both of the fully matched P/T DNA constructs (0mm-A and 0mm-T) examined in this study showed identical CD signals with the addition of Klenow (data shown for the 0mm-A construct only). We note that, in these perfectly matched DNA constructs, a significant proportion of the bases at the 3′-end of the primer appeared to be unstacked when bound to Klenow, as reflected in lowered CD (compare red spectra of Fig. 3, A and B) and increased fluorescence intensities (Fig. 3D). Significant unstacking of the primer terminus was also observed upon the addition of Klenow to the 1-mm construct (Fig. 3B). We believe that this spectrum represents a mixed population, with the primer terminus distributed in a dynamic equilibrium between the two active sites (the quantitation of this distribution is developed below). As discussed above, the complex of the 2mm P/T construct with Klenow also showed the constrained behavior of the bases immediately adjacent to the ssDNA-dsDNA junction that we observed for the corresponding free construct.
Fig. 3C shows the fluorescent titration curves obtained as Klenow was added to 3 μm concentrations of the various constructs in Ca2+ buffer. The 3mm and 4mm constructs showed titration curves similar to that obtained with ssP16-1, displaying a clear breakpoint at equimolar concentrations of protein and DNA. For the binding of the 0mm-A, 0mm-T, 1mm, and 2mm constructs, we observed a similar linear increase in fluorescence up to the equimolar concentration point. However, in the presence of excess Klenow, the fluorescence intensities for these constructs showed a gradual decrease (this is discussed below).
Fig. 3D presents the fluorescence intensities of the 2-AP dimer probes (normalized to the ssP16-1 free primer strand) for the free DNA constructs and for the Klenow-construct complexes at 3 μm equimolar Klenow-DNA concentrations. The fluorescence intensities of the free constructs showed a gradual transition from the duplex (fluorescence highly quenched) to a typical ssDNA (increased fluorescence) conformation as the number of mismatched nucleotides at the primer end increased from 0 to 4. This is consistent with the CD results (except for the 2mm construct, for which (unlike with fluorescence) we had previously noted anomalous behavior for the CD spectra; see above).
We observed an increase in fluorescence (relative to the fluorescence of the respective free constructs) upon the addition of Klenow for all of the constructs, suggesting that the 2-AP dimer probes become unstacked upon complex formation, consistent with the CD results. This increase in fluorescence was most dramatic (and almost equivalent to the amplitude observed for ssP16-1-Klenow complex) for the 3mm and 4mm construct-polymerase complexes, whereas the 0mm-A, 0mm-T, 1mm, and 2mm construct complexes displayed intermediate fluorescence intensities.
The binding of Klenow to P/T constructs was characterized further by fluorescence measurements at lower DNA concentration. At 50 nm DNA construct concentrations (60-fold lower than the concentration used in our other assays), Klenow binding still showed typical 1:1 binding stoichiometry but no apparent fluorescence decreases at higher protein concentrations (Fig. 3F). Binding affinities of both matched and mismatched primer-template constructs to Klenow were shown to be tight (Fig. 3F), with estimated Kd values in the nanomolar range or lower. Such tight binding is consistent with previous studies (34, 35). In contrast the binding of ssDNA to Klenow was relatively weak, with an apparent Kd of about 40 nm. This is very close to previously reported estimates of Kd for ssDNA binding to Klenow in similar buffer conditions (36). We conclude that a significant component of the binding free energy of P/T junction DNA to Klenow is contributed by interactions involving the double-stranded portion of the construct.
To further investigate the anomalous behavior of the titration curves for the 0mm-A (and 0mm-T), 1mm, and 2mm constructs at higher than stoichiometric concentrations of Klenow (Fig. 3C), we performed gel shift assays with radioactively labeled constructs (the 5′-end of the primer was labeled with γ-32P) at equimolar concentrations as well as at a 3-fold excess of Klenow relative to the construct (Fig. 3E). Under both conditions, we observed a single complex band for the various constructs that migrated at similar positions in the gel (Fig. 3E), suggesting that any additional complexes that might have formed at high Klenow concentrations were not sufficiently stable to be resolved in a gel mobility shift assay. The 2mm complex showed a slight smearing, but the majority of that complex migrated in the same position as the complexes with the other constructs. Thus, although a recent study by Millar and co-workers has suggested that Klenow can form 2:1 (Klenow/DNA) complexes with matched (and 1:1 complexes with mismatched) P/T DNA (37), we failed to observe stable higher order complexes for any of the constructs in our gel mobility shift assays. Taken together, our results suggest that the fluorescence anomaly observed in the binding titrations for 0mm-A (and 0mm-T), 1mm, and 2mm P/T constructs at 3 μm DNA with higher than saturating Klenow concentrations probably represents weak nonspecific aggregation of the complexes at higher concentrations.
Conformations of DNA Bases at or near the P/T Junction for Constructs Bound to Klenow
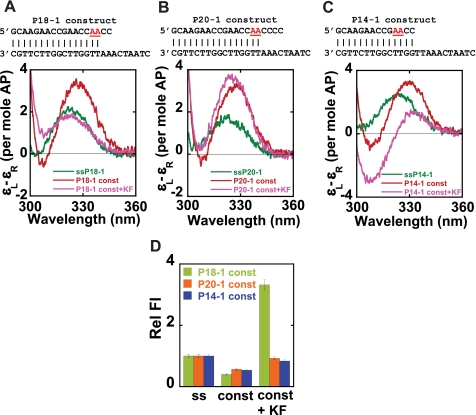
We next used our 2-AP probe approaches to determine the local conformations of bases at P/T junctions for both matched and mismatched DNA constructs bound to Klenow polymerase in order to establish both the total number of bases that the polymerase can actively unwind from the P/T junction in these constructs and to examine the local conformations of the bases bound at both the exo and the pol active sites. To this end, we examined constructs P18-1 (two mismatches at the primer terminus) and P20-1 (four mismatches at the primer terminus), with the 2-AP dimer probe in the first two base-paired positions of the P/T junction in both constructs. We also examined Klenow bound to the P14-1 construct, which has no mismatches, two GC bp immediately adjacent to the P/T junction, and the 2-AP dimer probes in the third and fourth positions upstream of the ssDNA-dsDNA junction.
The low energy CD spectra for these constructs are shown in Fig. 4, A–C. As expected, probes positioned in the ssDNA sequences of the free primer chains showed a peak at about 325 nm, whereas pairs of 2-AP base probes in the dsDNA portions of the free P/T DNA constructs exhibited characteristic exciton spectra with peaks at ∼330 nm. The addition of Klenow polymerase to the P18-1 construct resulted in a significant decrease in CD intensity relative to the spectrum of the free construct as well as a blue shift of the peak. The resulting spectrum almost overlapped that of the free ssP18-1 chain, suggesting that complex formation had resulted in the unwinding of at least the first two bp of the matched portion of this P/T DNA construct that carries two 3′-terminal mismatches.
FIGURE 4.
Klenow binding unwinds the 3′-end of the primer strand from matched P/T junctions. 2-AP bases at defined positions within the DNA constructs are shown in red. A–C, low energy CD spectra for the free and Klenow-bound P18-1, P20-1, and P14-1 constructs. D, relative fluorescence intensities for the same constructs without and with Klenow. Color coding for CD spectra is as follows. Dark green, ssDNA; red, free construct; pink, construct-Klenow complex. For fluorescence intensities, color coding is as follows. Light green, P18-1; orange, P20-1; blue, P14-1, either without or with Klenow.
However, for the P20-1 construct, we observed a very small blue shift of the exciton peak upon the addition of Klenow, indicating that the conformations of the 2-AP probes in this construct (with four mismatched bases in the primer terminus beyond the paired 2-AP dimer probe) are not significantly altered upon binding to Klenow. These results suggest that previously paired 2-AP probes located in the primer strand are unwound by Klenow binding if they are within 4 bases of the 3′-end of the primer but not unwound if they are flanked by four or more primer residues, whether or not these residues were paired in the original construct. Furthermore, our results with the P20-1 construct also show that the duplex DNA located at a fork next to four mismatched bases, with the 3′-primer terminus presumably bound in the exo site, maintains a B-form conformation.
Quite different behavior was seen for the fully matched P14-1 construct with base-paired 2-AP dimer probes located at positions 3 and 4 from the end of the primer (Fig. 4C). For this construct, the addition of Klenow resulted in a significant decrease in the intensity of the peak at 330 nm, with a concomitant increase in the depth of the trough. As previously shown, these spectral properties are characteristic of A-form DNA (14), suggesting that the base-paired probe region may not be significantly unwound in this complex but rather that the dsDNA portion of the P/T terminus tilts into a double-stranded A-like conformation as a consequence of Klenow binding.
As expected, the fluorescence intensities of our dimer probes in P/T DNA constructs were all quenched relative to ssDNA (Fig. 4D). The addition of Klenow polymerase resulted in a significant increase of the fluorescence of the P18-1 construct, whereas only a very small increase in fluorescence intensity was seen on complex formation with the P20-1 and P14-1 constructs. The fluorescence results with P18-1 and P20-1 constructs were consistent with the CD results and further confirm that a total of 3–4 bases at the 3′ terminus are unstacked (and not base-paired) in complexes formed with the various mismatched constructs.
We note that only a very small increase in fluorescence intensity was observed upon binding of Klenow to the P14-1 construct, whereas the CD spectrum of this complex resembles that of A-form DNA. Taken together, the results suggest that Klenow binding results in no significant unwinding of bases located 2 nt upstream of the matched P/T junction in the P14-1 construct. It is important to note that, unlike the 0mm-A (or 0mm-T) construct that has two 2-AP-T base pairs at the P/T junction, the P14-1 construct contains two GC base pairs at these positions, suggesting that Klenow may be less effective in opening GC-rich base-paired sequences at the P/T junction than it is in opening AT-rich sequences. This conclusion is consistent with earlier findings by Hariharan and Reha-Krantz (38) and others on the sequence dependence of the distribution of the primer ends between the exo and the pol active sites.
In addition, our results strongly suggest that the 2-AP bases at the defined positions in the P14-1 DNA construct adopt an A-form conformation when bound to Klenow. This is consistent with structural studies that have shown that 2–4 bp of the dsDNA located just adjacent to the primer-template junction adopt a distinct A-like conformation for P/T DNA constructs bound at the pol site of Klentaq, T7, or BstDNA polymerases (6–8). We note that these results also confirm that the primer strand of the P14-1 construct is bound as a duplex at the pol site of Klenow polymerase.
Conformation of Bases in the Duplex Region of Matched P/T DNA Constructs Bound to Klenow and Klentaq Polymerase
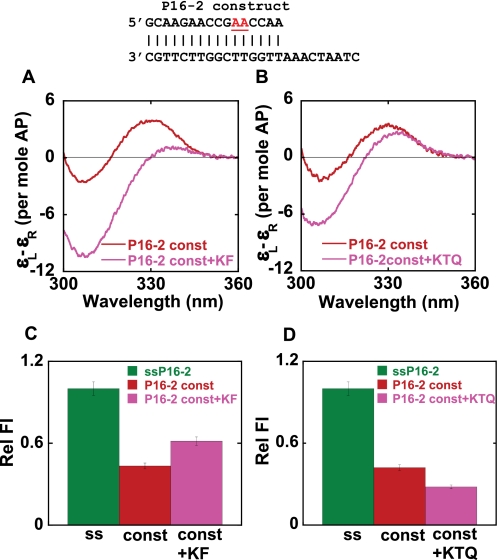
A matched P/T DNA construct (P16-2), carrying a dimer 2-AP probe 5 and 6 bp upstream of the matched P/T junction, was used to examine the local conformation of bases at these positions within polymerase-DNA construct complexes. Fig. 5, A and B, shows the low energy CD spectra of the P16-2 construct in the presence of Klenow and Klentaq polymerases, respectively. The free construct exhibited a characteristic exciton-coupled low energy CD spectrum, suggesting that the probe-containing base pairs were in a B-form DNA conformation. We note that this exciton coupling appears much larger than that seen with the 0mm-A or P14-1 constructs, perhaps reflecting some “end fraying” in these latter constructs in which the 2-AP probes are closer to or at the P/T junction (33).
FIGURE 5.
DNA base conformations 4 bp upstream from the P/T junction for matched P/T DNA constructs in the presence of Klenow and Klentaq. 2-AP bases at defined positions within the DNA construct are shown in red. A and B, low energy CD spectra of DNA constructs, without and with Klenow (KF) and Klentaq (KTQ) polymerases, respectively. C and D, relative fluorescence intensities for constructs without and with KF and KTQ. Color coding is the same for all panels. Dark green, ssDNA; red, free construct; pink, DNA construct-polymerase complexes.
The addition of Klenow to the P16-2 construct resulted in a large increase in the depth of the trough at 305 nm and a significant decrease in the peak intensity. Such CD spectral changes are characteristic of the shift of a dimer 2-AP probe from a B-form to an A-form DNA conformation. The addition of Klentaq resulted in a similar increase in the depth of the trough and a decrease in the peak amplitude, but the overall extent of the spectral changes was much smaller than those induced by Klenow. Nevertheless, the results with Klentaq also suggested that the 2-AP probes at these positions in the complex remained stacked upon binding and that this region of the DNA construct was probably also driven into (or at least toward) an A-like conformation when complexed with Klentaq. Relative to the free ssP16-2 DNA, the fluorescence intensities (Fig. 5, C and D) of the construct were significantly quenched on binding to both polymerases. Looking more carefully, however, we see that the fluorescence intensity of this construct increased slightly upon the addition of Klenow, whereas when Klentaq was added, the emission intensity of the binary complex was quenched somewhat further relative to the free construct.
The P16-2 construct is similar in sequence to the 0mm-A construct, differing only in the position of the 2-AP probes. As seen with the 0mm-A construct, the 3′-end of the primer strand of the P16-2 construct is probably distributed between the two active sites of Klenow. We believe that the spectrum of the 2-AP probes of this construct bound to Klenow reflects such a partitioning of the 3′-end of the primer between the pol active site, where it probably binds (with the template strand) in an A-form base-paired conformation, and the exo site, where it binds in an extended single-stranded form.
A structural comparison of the positions of the duplex DNA region in the pol site-bound complex of Klentaq (8) and the exo site-bound complex of Klenow (4) show that although these polymerases share a common duplex DNA binding site, the dsDNA regions themselves seem to be translated (relative to one another) by several Å along their helical axes, resulting in different backbone interactions with the respective polymerases. This may also account for the differences in the CD spectra observed for Klenow and Klentaq complexes with P16-2 construct, since in Klenow we observe a distribution between the two binding sites, whereas in Klentaq (which has no exo binding site), we expect to see interactions only with the pol site, with the P/T DNA bound in an A-form conformation.
Probing the Partitioning of the 3′-Primer Ends of Matched P/T DNA Constructs between pol and exo Sites for Klenow and Klentaq
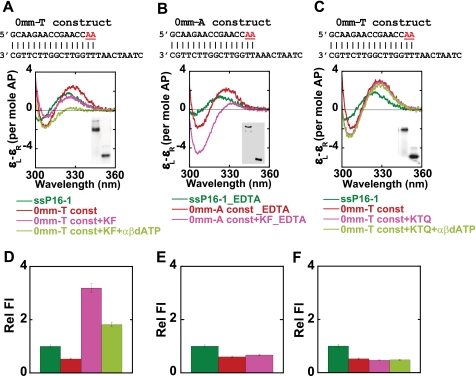
To further explore the binding conformations and fractional occupancies of primer termini in matched P/T DNA constructs between pol and exo sites, we examined the binding of Klentaq and Klenow polymerases to the 0mm-A and the 0mm-T constructs. Both of these constructs contain similar matched P/T DNA sequences (the ssP16-1 primer strand is identical for both) and differ only in the identity of the templating base. The binary complexes (containing only DNA and polymerase) for these two constructs behave identically.
The low energy CD spectra for the binding of these constructs to Klenow and Klentaq DNAP are shown in Fig. 6, A–C. The free ssP16-1 and the 0mm-T constructs exhibited probe CD spectra characteristic of ssDNA and B-form dsDNA, respectively. The addition of Klenow to the 0mm-T construct in the presence of Ca2+ yielded a CD spectrum similar to that observed previously with the 0mm-A construct-Klenow complex (Fig. 3B), showing a significant decrease of the CD amplitude at 330 nm accompanied by a blue shift of the spectrum (Fig. 6A). The addition of α,β-methyleno-dATP (a nonhydrolyzable analog of dATP) to form a ternary complex led to the disappearance of the CD amplitude at 330 nm, yielding a spectrum for the Klenow-0mm-T complex that resembles that obtained with 2-AP dimer probes in A-form DNA (Fig. 6A). In this experiment, we were not able to increase the concentration of α,β-methyleno-dATP to saturation levels, and as a consequence, we were unable to drive ternary complex formation to completion in either the CD or the fluorescence (Fig. 6D) measurement. Nevertheless, these results strongly suggest that the base-paired 3′-primer terminus adopts an A-like conformation on ternary complex formation.
FIGURE 6.
Klenow and Klentaq DNA polymerases bound to matched P/T DNA without and with nonhydrolyzable dATP. 2-AP bases at defined positions within the DNA are shown in red. A–C, low energy CD spectra of the 0mm-T without or with Klenow and with α,β-methyleno-dATP in Ca2+ buffer (A); 0mm-A without or with Klenow in EDTA buffer (B); and 0mm-T without or with Klentaq (KTQ) and with α,β-methyleno-dATP in Ca2+ buffer (C). Insets show the gels for the free DNA construct (lower bands) and DNAP-bound constructs (upper bands). D–F, plots of the fluorescence intensities for the same constructs and complexes. Color coding for all panels is as follows. Dark green, ssDNA; red, free construct; pink, construct-DNAP complex; light green, construct-DNAP complex plus α,β-methyleno-dATP.
We note also that the binary complex of Klenow with an end-matched P/T DNA construct in EDTA buffer (where Ca2+ is replaced with 2 mm EDTA) exhibited a very distinct spectrum that is also characteristic of an A-form conformation of the 2-AP dimer within the duplex region of the P/T junction (Fig. 6B). This spectrum is very similar to that obtained for a ternary complex in the presence of α,β-methyleno-dATP, where the major fraction of the (base-paired) 3′-primer end is expected to occupy the pol site.
Under other divalent ion concentration conditions, for example, in the absence of any additional divalent ions (labeled no Mg2+) and in the presence of 5 mm Mg2+ and 4 mm EDTA (labeled Mg2+ + EDTA), we also obtained CD spectra for the binary complex similar to that observed in 2 mm EDTA (see Fig. S3A). We note here that a Klenow complex in 5 mm Mg2+ and 4 mm EDTA is also active in extending the primer strand in presence of the next correct dNTP (here dTTP; data not shown), and that this ternary complex exhibits a CD signal that is almost identical to that obtained for the binary complex in 2 mm EDTA buffer (Fig. S3B). These results show the importance of divalent metal ions in modulating the binding conformation of DNA constructs with Klenow polymerase and also strongly suggest that the binary complex of Klenow in 2 mm EDTA represents a largely pol site-bound conformation.
In contrast, for Klentaq DNAP in Ca2+- or Mg2+-containing buffers, the CD spectra of the binary and the ternary complexes almost overlap with that of the free 0mm-T construct, suggesting that the primer termini retain a duplex B-form conformation in these complexes (Fig. 6C). The stacked conformation observed for the primer-template junction in Klentaq-DNA complex is not unexpected, since Klentaq DNAP lacks 3′ → 5′ exonuclease activity (28) and presumably also an exo binding site (29, 30). However, the observed B-form conformation of these base pairs is not consistent with observations from crystal structures, where this region has been seen to form an A-like conformation (see below, where this point is discussed further). For Klentaq, in the presence of 2 mm EDTA or in the absence of any additional divalent ions, we observed a similar B-form conformation of the primer end, suggesting that, unlike for Klenow, the presence or absence of divalent ions does not affect the binding conformation of the 3′-primer end in binary complexes with Klentaq polymerase (Fig. S3C). We also provide a gel shift assay (Fig. 6, A–C, inset) for the complexes shown in Fig. 6, A–C, to show that DNA-polymerase complexes were indeed formed under all of the buffer conditions examined.
The fluorescence intensities of the probes in the 0mm-T construct were, as expected, quenched relative to the ssP16-1 DNA (Fig. 6, D–F). The fluorescence increased significantly upon formation of the binary complex with Klenow in Ca2+-containing buffer (Fig. 6A), suggesting that the bases in these positions are unstacked. We observed an ∼1.6-fold decrease in fluorescence intensity, relative to the binary complex, upon forming the ternary complex with α,β-methyleno-dATP. This is consistent with the restacking of the 2-AP probes upon interaction with the next required NTP (Fig. 6D). In keeping with previous studies (35), this result also confirms that such ternary complexes can form, even in the presence of noncatalytic divalent metal ions. The binary complex of Klenow with the 0mm-A construct in EDTA showed no change in fluorescence compared with the free construct (Fig. 6E). In addition, there was no further change in fluorescence upon binary or ternary complex formation with Klentaq DNAP (Fig. 6F), which is consistent with our CD results.
Identifying the pol and exo Site-bound Conformations and Measuring the Fractional Occupancy of the Two Sites for Various Constructs
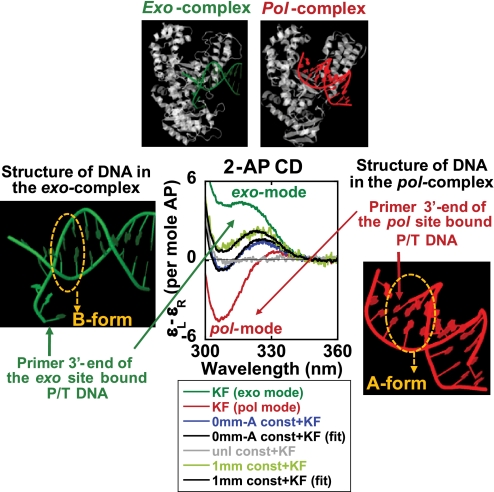
To permit us to estimate the fractional occupancy of the two active polymerase sites by the 3′-primer terminal nucleotides for Klenow bound to various constructs, it was necessary first to define “end point” spectra representing the fully bound exo and pol site conformations. Deconvolution of the mixed spectra obtained for various constructs into these conformational components should then permit us to establish the equilibrium partitioning of primer ends between the two sites in intermediate cases.
Our results with 2-AP-substituted ssP16-1 and 3mm constructs showed similar conformations of the primer end in complexes with Klenow. Previous studies have shown that the 3′ → 5′ exo site of Klenow shows a strong preference for ssDNA substrates (2, 5). Taken together, we believe that the CD spectra obtained for ssP16-1 and the 3mm construct correspond to a conformation in which the primer terminus is bound exclusively at the exo site (Fig. 3B). To obtain a spectrum corresponding to the fully pol site-bound conformation of the primer, we compared the CD spectra of 0mm-A and/or 0mm-T constructs with Klenow and Klentaq polymerases under various conditions. Our results strongly suggest that the Klenow complex with the 0mm-A (or 0mm-T) construct in 2 mm EDTA corresponds to a complex in which the 3′-primer end is totally base-paired and totally bound in the pol site of the polymerase.
To check that the above end point spectra represent fully homogeneous and polymerase-bound complexes, we also performed analytical velocity sedimentation experiments with Klenow complexed with the 3mm construct in Ca2+ (exo mode complex), 0mm construct in EDTA (pol mode complex), and 0mm construct in Ca2+, respectively, at 1:1 Klenow/DNA construct concentration ratios. The sedimentation velocity traces obtained in these experiments clearly showed the formation of stoichiometric complexes with Klenow for each of the above DNA constructs, with >90% of the population present in the complex form (Fig. S4).
A schematic of the deconvolution of the spectra for 0mm-A and 1mm constructs in Ca2+-containing buffer is presented in Fig. 7. The central set of CD spectra for Klenow-bound constructs show the fully exo site-bound extended ssDNA form spectrum (in dark green), and the fully pol site-bound A-form duplex spectrum (in red). Fitting the intermediate spectra to these end point spectra reveals that, in the binary complex of Klenow with matched probe base pairs at the P/T junction, a significant fraction (∼43%) of the primer termini are bound at the exo site. In the presence of a single mismatch at the 3′-primer end, the equilibrium distribution of the primer termini shifts significantly further (∼61%) toward exo site binding. Although, as noted earlier, the fractions partitioning between the two active sites will most likely vary depending on the nature (AT versus GC) of the base pairs at the P/T junction, our results demonstrate that this spectroscopic approach can be used as a direct “structure-based” method for measuring the partitioning of the primer ends between the two polymerase active sites for P/T DNA constructs of varying sequence.
FIGURE 7.
Deconvolution of the CD spectra of the different complexes into exo-bound and pol-bound components. The central panel represents the 2-AP CD spectra of the various DNA construct-Klenow complexes. Unless otherwise stated, all of the spectra were measured in Ca2+ buffer. Dark green, the Klenow-bound 3mm construct corresponding to total exo mode binding; red, the Klenow-bound 0mm-A construct in EDTA, corresponding to total pol mode binding; light green, the Klenow-bound 1mm construct; blue, the Klenow-bound 0mm-A construct; gray, the Klenow-bound unlabeled (no 2-AP probes) 0mm-A construct. The black curves represent the best fit for the resolved spectra. The panels at the top display the x-ray structures of the exo- and pol-bound modes of the polymerases. The exo complex is that of the Klenow “editing” complex (Protein Data Bank code 1KLN) (4), and the pol complex is that of the Klentaq-DNA complex (Protein Data Bank code 4KTQ) (8). The panels on the two sides show only the P/T DNA components of the above structures. The structure figures were made using the jmol program (available on the World Wide Web).
DISCUSSION
We have used the low energy CD and fluorescence spectra of 2-AP probes placed site-specifically in various DNA constructs to determine the local conformations of bases at and near the 3′-primer end of the various DNA structural motifs that can act as substrates for DNA polymerases. Using this technique we have (i) defined the structures of primer and P/T DNA bound at the pol and exo sites of Klenow polymerase, (ii) characterized the conformations of DNA bases bound at defined positions within the active sites, and (iii) determined the relative partitioning of the 3′-primer ends between the pol and exo sites of Klenow for various DNA constructs. Our results also shed light on the role of divalent metal ions in regulating DNA binding and exonuclease activity and in controlling the distribution of the primer termini between the active sites.
In some experiments, we used Ca2+ (instead of the canonical Mg2+) as the divalent metal ion cofactor to eliminate the residual exonuclease activity observed in the presence of Mg2+, since low levels of exo activity have been reported for the exo− mutant examined here (17). Wherever we could use Mg2+, we have shown that Mg2+ and Ca2+ behave essentially the same in controlling the binding of DNA constructs to Klenow. Substitution of Ca2+ for Mg2+ has previously been used to prevent exonuclease reactions and to trap ternary complexes in structural studies of RB69 DNAP complexes (22, 39). Also, an earlier and a very recent study with Klenow has shown that both binary and ternary complexes can form in the presence of Ca2+ but that the DNA synthesis is significantly inhibited in Ca2+-containing buffers (23, 35). Thus, we conclude that the conformations of the DNA constructs bound in the various complexes studied here probably do represent the physiologically relevant states defined above.
Probing Local Conformations at Defined Positions in the Primer-Template DNA
Based on structural studies, the DNA binding sites of the “large fragment” of DNAP I (Klenow polymerase) can be identified as (i) an exo site that binds ssDNA, (ii) a pol site that binds duplex P/T DNA, (iii) an extended “cleft” that lies between the exo and the pol sites and “clamps” (at somewhat different distances from the primer end) the dsDNA region located just upstream of the P/T junction when the 3′-end of the primer is bound in either the exo or the pol site, and (iv) a binding site that interacts with the single-stranded template downstream of the P/T junction. The protein residues interacting with the DNA in these sites are clearly different for the pol and exo mode-bound complexes (36, 40). In this study of polymerase-DNA complexes in solution, we have characterized the local conformations of bases interacting with the first three polymerase binding sites listed above. Elsewhere, we have used a different spectroscopic probe to characterize binding site (iv) as well.4
Our results with various mismatched P/T constructs binding to Klenow polymerase show that three unpaired nucleotide residues at the 3′-primer end are optimal for primer binding at the exo site. Consistent with earlier crystallographic findings (Fig. 7), we have shown that in solution the 3′-terminal residues of the primer strand of free ssDNA or mismatched P/T junctions adopt a significantly unstacked conformation when bound to the exo site. We have also shown that the P/T DNA duplex adopts an A-like conformation when bound to Klenow at the pol site but not when bound to Klentaq. A B-form to A-form transition for the 2–4 bp of duplex DNA adjacent to the P/T junction upon binding of this DNA region to the pol site has been reported for most DNAP I co-crystals (6–8). It is not clear why such a polymerase-induced conformational change does not occur in equivalent DNA constructs bound to Klentaq, but perhaps this may reflect the weaker binding of our DNA constructs to the latter polymerase, especially for complexes that are free in solution (i.e. not subject to the additional stabilizing interactions that may be present in crystals).
We also used our spectroscopic methods to probe the conformation of dsDNA portions of the bound constructs located further upstream of the P/T junction. For Klenow complexes with DNA constructs with a GC bp at the P/T junction (e.g. the P14-1 construct), we observed an A-like conformation for probes located at positions 2 bp upstream from the junction, indicating that these residues are probably bound to a pol site. For Klenow complexed with matched P/T DNA containing an AT-rich junction (the P16-2 construct) and 2-AP probes located further upstream (4 bp) from the P/T junction, the spectra also showed a stacked conformation at the site of the probes. However, this latter conformation deviated significantly from B-form, and most likely reflects a weighted average of conformations at these upstream dsDNA binding sites involved in holding the duplex DNA when the primer termini are partitioned between the pol and the exo binding modes. In contrast, for Klentaq (which contains no exo site) complexed with the P16-2 construct, the 2-AP probes bound in this region exhibited a much smaller divergence from the canonical B-form. Consistent with these findings, structural studies have suggested that when the primer end is in the exo site of Klenow, this duplex DNA sequence is significantly distorted and bent (4) relative to the more B-form-like conformation observed in the same region when the primer is bound in the pol site (8).
Probing of the dsDNA sequences at the P/T junction for pol and exo mode complexes with Klenow showed that this region of the DNA construct also adopts distinctly different conformations for the two binding modes. When the primer terminus is bound at the pol site (0mm-A construct in EDTA; Fig. 6B), the dsDNA at the P/T junction takes up a DNA A-like conformation, whereas the duplex P/T junction maintains a B-form structure when the primer end is bound at the exo site (P20-1 construct; Fig. 4B). We speculate that such A-form to B-form conversion of the dsDNA at the P/T junction could serve a mechanistic role that facilitates proper shuttling of the primer terminus between the two active sites in the DNAP I family of polymerases.
Partitioning of the Primer between the Two Active Sites of Klenow
For several of the constructs examined in this study, the CD spectra suggested that the 3′-primer ends are partitioned between the pol and the exo active sites. Such partitioning is expected to depend on the sequence and extent of mispairing at the P/T junction region of the DNA constructs. Previous studies of Klenow complexes with DNA, using time-resolved fluorescence anisotropy and a fluorescent probe attached to a base located 8 bp from the P/T junction, had suggested that the presence of 3–4 mismatched bases at the 3′-end of the primer strand at the P/T junction resulted in pushing the equilibrium distribution of the primer >80% toward binding to the exo site, whereas the primer strand of matched P/T DNA constructs seemed to be bound primarily (>85%) at the pol site (12, 41). In our study, we have used similar matched and mismatched DNA constructs but with fluorescent nucleotide analogue probes within the primer strand that interact directly with the relevant binding sites to permit us to identify and directly measure the DNA bound in the two active sites.
We have demonstrated that the primer terminus of P/T DNA carrying three mismatched residues at the 3′-primer end binds mostly at the exo site, consistent with previous measurements (12). However, in contrast to this earlier study, in which the presence of four terminal mismatches seemed to shift the equilibrium further toward the exo site, our results with 2-AP probes and CD spectra suggest that more than three terminal mismatched base pairs at the P/T DNA junction result in a primer end that is too extended to fit properly into the Klenow exo site. This difference may reflect the fact that the previous workers (12) relied on detecting apparent differences in the extent of burial within the protein of a fluorophore attached at a dsDNA site further removed from the P/T junction when the 3′-primer terminus was bound either in the pol or in the exo site of Klenow. Thus, these studies could not directly monitor the conformations of the primer strand bound at the two sites nor conclusively establish a linear relation between binding and probe fluorescence intensity. Nevertheless, our quantitative results (see below) are in general accord with the conclusions of these earlier workers. Our results also indicate that in the absence of divalent metal ions, the primer terminus of a matched P/T DNA is bound to the pol site of Klenow in duplex form.
Using CD spectra obtained with the 3mm construct in Ca2+ and the 0mm-A construct in EDTA to serve as reference end points for complete exo- and pol site binding, respectively, we deconvoluted the CD spectra obtained with the Klenow-bound 1mm and 0mm-A constructs to obtain the distribution of the primer ends into the respective active sites. On this basis, we calculated an ∼43% occupancy of the exo site by the 3′-primer end for the 0mm-A construct in Ca2+ buffer, indicating that Klenow can efficiently unwind the fully matched P/T DNA duplex in these constructs. We note here that the three terminal base pairs at the P/T junction of this construct were 5′-C(2-AP)(2-AP)-3′ (i.e. two AT-pairs at the 3′ terminus; see Fig. 1). In contrast, for the primer DNA of the fully matched constructs used by Millar and co-workers (41, 42), where the corresponding bases were 5′-ATG-3′ (a GC pair at the 3′ terminus), they estimated ∼14% of the primer end to be at the exo site. Making their P/T junction more GC-rich (5′-GGG-3′) reduced the apparent partitioning into the exo site to ∼7%. Taken together, these results are consistent with the notion that the positional distribution of the primer end between the two active sites is strongly dependent on the thermodynamic stability of the base pairs at the P/T junction and on the efficiency with which the polymerase involved unwinds the P/T junction.
In this context, we note that the CD (and fluorescence) results for Klenow binding to our P14-1-matched construct with a GC-rich P/T junction (5′-(2-AP)CC-3′) suggests that the 2-AP probes located upstream of the two terminal C residues are mostly stacked and base-paired. Although we have not probed the 3′-primer end of this construct, if we assume that ∼3 terminal primer nucleotides are unwound to an extent comparable with that of DNA constructs with AT base pairs at the P/T junction (0mm-A or T constructs), the 2-AP dimer probes in the P14-1 construct would be expected to show some unstacking. However, no significant unstacking of the probes in the P14-1 construct was seen, further supporting the suggestion that the extent of unwinding of the primer strand from matched P/T DNA constructs is dependent on the thermodynamic stability of the P/T junction.
We observed that ∼61% of the primer ends of the 1mm construct were bound at the exo site when the three terminal bases in our study were 5′-C(2-AP)(2-AP)-3′ (the underlined base here represents the mismatch). In contrast, the 1mm constructs used by Millar and co-workers (41, 42) showed exo site distributions of ∼18 and ∼40% for GC-rich (5′-GGG-3′) and AT-rich (5′-ATG-3′) P/T junctions, respectively. Our results agree with the previous observation that the fraction of primer ends located in the exo site increases with the increasing number of mismatches (up to three unpaired nucleotides). However, we note that the actual fraction of primer ends in the exo site in our studies is significantly higher than reported previously. Our results, showing a higher partitioning of the primer terminus into the exo site for matched DNA, are consistent with structural studies of Klenow and another proofreading-proficient polymerase (RB69 from the pol-α family). In these co-crystals, involving fully matched P/T DNA, the primer end exhibited an exo site-bound conformation (4, 39), suggesting this to be the favored equilibrium primer position for a matched P/T DNA construct bound to these polymerases.
The Roles of Divalent Metal Ions in DNA Binding and Partitioning of the Primer Terminus between the Two Active Sites in Klenow
The involvement of divalent metal ion co-factors in the catalytic activities of DNAP I polymerases have been extensively characterized. It is well established that two divalent metal ions, coordinated by amino acid residues and the DNA substrate, are essential for the 3′ → 5′ exonuclease activity of Klenow (43). Based on structural data from other homologous polymerases, phosphodiester bond formation at the pol site is also believed to involve a two-metal ion mechanism (44, 45). However, the role of metal cofactors in DNA binding at these active sites is less well understood.
Structural studies of various exonuclease mutants of Klenow (involving changes in catalytically conserved metal ion-coordinating residues at the exo site for mutants that retain normal pol activity but exhibit significantly reduced exo activity) have provided valuable information on the geometric arrangement of the essential divalent cations at the exo site (43). These studies demonstrated that the D424A mutant (the one used in our study) eliminates binding of one of the two catalytic metal ions at the exo active site without disrupting proper DNA binding at that site (17), whereas exo− double mutants (D355A/E357A) that eliminate the binding of both of the required divalent ions at the exo site abolished both exo function and the binding of dNMP (the product of exo activity that binds at the same position as the 3′-terminal nucleotide residue of the primer strand) (17). In addition, the structure of wild type Klenow in the absence of any substrate DNA or product showed a firmly bound divalent cation only at one divalent metal binding site, whereas the second metal site was observed only in Klenow-dNMP complexes (43, 46). Taken together, these results link the first metal ion binding site to both substrate binding and catalysis while suggesting that the second bound metal cation may be involved only in catalysis (17, 43).
Consistent with this, our studies with the D424A mutant (lacking the second metal binding site) polymerase bound to ssDNA and P/T DNA in the presence of EDTA showed no productive binding of the primer terminus at the exo site (we define “productive binding” as the distinct unstacked and “frayed” conformation of the primer end at the exo site that we observed in the presence of divalent ions (). We suggest that EDTA removes the first bound metal cation and possibly other protein-bound divalent cations as well, thereby eliminating binding of the primer terminus at the exo site. We note in this context that we do observe overall binding of P/T DNA to Klenow in the presence of EDTA (Fig. 6B, inset, and Fig. S4C), probably because other divalent metal ion-independent protein-mediated contacts involved in DNA binding are maintained. We have assigned this bound state as the pol site-bound conformation of Klenow in our deconvolution calculations, as discussed above.
A crystal structure of the Bst large fragment complexed with P/T DNA in an “open” binary complex (where the 3′-primer end was bound at the pol site) showed one Mg2+ ion coordinated with two catalytically conserved aspartate residues, Asp830 (Klenow residue Asp882) and Asp653 (Klenow residue Asp705), and also that the 3′-OH of the primer terminus formed a hydrogen bond with Asp830. A second bound divalent metal was observed only in the “closed” ternary complex in the presence of dNTP substrate (45), supporting the notion that the second metal ion is required for dNTP binding and catalysis. Thus, on the basis of these structural studies, it appears that two metal ions are required for dNTP binding and polymerase activity (6, 8, 45), although it has not been established whether metal ions are required for proper binding of the primer end at the pol site.
EPR and proton relaxation rate studies of the binding of Mn2+ to wild-type Klenow have revealed the presence of one tight (Kd in the micromolar range) and eight weak (Kd in the millimolar range) Mn2+ binding sites (47). Comparison of Mn2+ binding of wild type Klenow with the exonuclease mutants (D424A and D355A/E357A) in the absence and presence of dNMP (serving as a model of the binding of the 3′-end of ssDNA) and/or dGTP (the polymerase substrate) indicates that the tight binding site corresponds to the metal cation bound to the exo site in the crystallographic model. Of the eight weak binding sites, four are probably located in the exo domain, whereas the remaining four appear to be in the pol domain (47). A very tight Mn2+ binding site was created at the pol site only in the presence of dGTP, consistent with structural studies. This study thus shows that, unlike the exo domain, the pol domain does not exhibit a tight divalent metal binding site in the absence of substrate dNTP. We propose that the weakly bound metal ions, although they may affect the overall binding affinity of DNA, do not contribute significantly to determining the active site occupancy of the primer end, as long as the tightly bound divalent cation is present in the exo site. However, the removal of this cation from the exo domain by EDTA abolishes exo site binding, creating a condition in which the 3′-primer terminus can only bind at the pol site.
A previous study with T4 DNAP has also investigated the role of divalent metals in establishing the equilibrium distribution of the 3′-primer end of a P/T junction between the pol and the exo sites. In contrast to our findings with Klenow, the exo site-bound conformation dominated in the absence of Mg2+ for the T4 enzyme, whereas increasing the Mg2+ concentration increased the partitioning of the primer end into the pol site (48). This is clearly contrary to what we have observed for Klenow, where the absence of divalent metal ions resulted in more pol site binding, whereas the presence of these ions enhanced exo site binding. It may be that the divalent metal ion requirement for partitioning of the primer ends differs from one polymerase to another.
Supplementary Material
Acknowledgments
We are grateful to Walt Baase for advice on CD spectroscopy and other issues and to members of our laboratory for helpful discussions. We would also like to thank Casey Wilson and Eric Zaneveld for technical assistance with some of the experiments reported here.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-15792 (to P. H. v. H.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
We note that the green spectrum in Fig. 2B has more exciton character than do the green spectra in Fig. 2, C and D. This may reflect the fact that a pair of 2-AP probes located at the end of a free ssDNA strand can stack in a B-form conformation like free 2-AP duplexes (see Fig. 2 in Ref. 31), whereas interior pairs of 2-APs may be more constrained.
K. Datta et al., manuscript in preparation.
- DNAP
- DNA polymerase
- pol
- polymerase
- exo
- exonuclease
- P/T
- primer-template
- nt
- nucleotide(s)
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- 2-AP
- 2-aminopurine
- CD
- circular dichroism.
REFERENCES
- 1.De Lucia P., Cairns J. ( 1969) Nature 224, 1164– 1166 [DOI] [PubMed] [Google Scholar]
- 2.Brutlag D., Kornberg A. ( 1972) J. Biol. Chem. 247, 241– 248 [PubMed] [Google Scholar]
- 3.Kunkel T. A. ( 1988) Cell 53, 837– 840 [DOI] [PubMed] [Google Scholar]
- 4.Beese L. S., Derbyshire V., Steitz T. A. ( 1993) Science 260, 352– 355 [DOI] [PubMed] [Google Scholar]
- 5.Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. ( 1988) Proc. Natl. Acad. Sci. U.S.A. 85, 8924– 8928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doublié S., Tabor S., Long A. M., Richardson C. C., Ellenberger T. ( 1998) Nature 391, 251– 258 [DOI] [PubMed] [Google Scholar]
- 7.Kiefer J. R., Mao C., Braman J. C., Beese L. S. ( 1998) Nature 391, 304– 307 [DOI] [PubMed] [Google Scholar]
- 8.Li Y., Korolev S., Waksman G. ( 1998) EMBO J. 17, 7514– 7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polesky A. H., Steitz T. A., Grindley N. D., Joyce C. M. ( 1990) J. Biol. Chem. 265, 14579– 14591 [PubMed] [Google Scholar]
- 10.Polesky A. H., Dahlberg M. E., Benkovic S. J., Grindley N. D., Joyce C. M. ( 1992) J. Biol. Chem. 267, 8417– 8428 [PubMed] [Google Scholar]
- 11.Joyce C. M., Benkovic S. J. ( 2004) Biochemistry 43, 14317– 14324 [DOI] [PubMed] [Google Scholar]
- 12.Bailey M. F., Thompson E. H., Millar D. P. ( 2001) Methods 25, 62– 77 [DOI] [PubMed] [Google Scholar]
- 13.Tleugabulova D., Reha-Krantz L. J. ( 2007) Biochemistry 46, 6559– 6569 [DOI] [PubMed] [Google Scholar]
- 14.Datta K., Johnson N. P., von Hippel P. H. ( 2006) J. Mol. Biol. 360, 800– 813 [DOI] [PubMed] [Google Scholar]
- 15.Datta K., von Hippel P. H. ( 2008) J. Biol. Chem. 283, 3537– 3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson N. P., Baase W. A., von Hippel P. H. ( 2005) J. Biol. Chem. 280, 32177– 32183 [DOI] [PubMed] [Google Scholar]
- 17.Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. ( 1988) Science 240, 199– 201 [DOI] [PubMed] [Google Scholar]
- 18.Joyce C. M., Derbyshire V. ( 1995) Methods Enzymol. 262, 3– 13 [DOI] [PubMed] [Google Scholar]
- 19.Setlow P., Brutlag D., Kornberg A. ( 1972) J. Biol. Chem. 247, 224– 231 [PubMed] [Google Scholar]
- 20.Datta K., LiCata V. J. ( 2003) J. Biol. Chem. 278, 5694– 5701 [DOI] [PubMed] [Google Scholar]
- 21.Allen W. J., Rothwell P. J., Waksman G. ( 2008) Protein Sci. 17, 401– 408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franklin M. C., Wang J., Steitz T. A. ( 2001) Cell 105, 657– 667 [DOI] [PubMed] [Google Scholar]
- 23.Joyce C. M., Potapova O., Delucia A. M., Huang X., Basu V. P., Grindley N. D. ( 2008) Biochemistry 47, 6103– 6116 [DOI] [PubMed] [Google Scholar]
- 24.Huang M. M., Arnheim N., Goodman M. F. ( 1992) Nucleic Acids Res. 20, 4567– 4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuchta R. D., Benkovic P., Benkovic S. J. ( 1988) Biochemistry 27, 6716– 6725 [DOI] [PubMed] [Google Scholar]
- 26.Dam J., Schuck P. ( 2005) Biophys. J. 89, 651– 666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dam J., Velikovsky C. A., Mariuzza R. A., Urbanke C., Schuck P. ( 2005) Biophys. J. 89, 619– 634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tindall K. R., Kunkel T. A. ( 1988) Biochemistry 27, 6008– 6013 [DOI] [PubMed] [Google Scholar]
- 29.Kim Y., Eom S. H., Wang J., Lee D. S., Suh S. W., Steitz T. A. ( 1995) Nature 376, 612– 616 [DOI] [PubMed] [Google Scholar]
- 30.Korolev S., Nayal M., Barnes W. M., Di Cera E., Waksman G. ( 1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9264– 9268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson N. P., Baase W. A., Von Hippel P. H. ( 2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3426– 3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam W. C., Van der Schans E. J., Joyce C. M., Millar D. P. ( 1998) Biochemistry 37, 1513– 1522 [DOI] [PubMed] [Google Scholar]
- 33.Jose D., Datta K., Johnson N. P., von Hippel P. H. ( 2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4231– 4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Astatke M., Grindley N. D., Joyce C. M. ( 1995) J. Biol. Chem. 270, 1945– 1954 [DOI] [PubMed] [Google Scholar]
- 35.Gangurde R., Modak M. J. ( 2002) Biochemistry 41, 14552– 14559 [DOI] [PubMed] [Google Scholar]
- 36.Kukreti P., Singh K., Ketkar A., Modak M. J. ( 2008) J. Biol. Chem. 283, 17979– 17990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey M. F., Van der Schans E. J., Millar D. P. ( 2007) Biochemistry 46, 8085– 8099 [DOI] [PubMed] [Google Scholar]
- 38.Hariharan C., Reha-Krantz L. J. ( 2005) Biochemistry 44, 15674– 15684 [DOI] [PubMed] [Google Scholar]
- 39.Shamoo Y., Steitz T. A. ( 1999) Cell 99, 155– 166 [DOI] [PubMed] [Google Scholar]
- 40.Turner R. M., Jr., Grindley N. D., Joyce C. M. ( 2003) Biochemistry 42, 2373– 2385 [DOI] [PubMed] [Google Scholar]
- 41.Thompson E. H., Bailey M. F., van der Schans E. J., Joyce C. M., Millar D. P. ( 2002) Biochemistry 41, 713– 722 [DOI] [PubMed] [Google Scholar]
- 42.Carver T. E., Jr., Hochstrasser R. A., Millar D. P. ( 1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10670– 10674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beese L. S., Steitz T. A. ( 1991) EMBO J. 10, 25– 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brautigam C. A., Steitz T. A. ( 1998) Curr. Opin. Struct. Biol. 8, 54– 63 [DOI] [PubMed] [Google Scholar]
- 45.Johnson S. J., Taylor J. S., Beese L. S. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3895– 3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. ( 1985) Nature 313, 762– 766 [DOI] [PubMed] [Google Scholar]
- 47.Mullen G. P., Serpersu E. H., Ferrin L. J., Loeb L. A., Mildvan A. S. ( 1990) J. Biol. Chem. 265, 14327– 14334 [PubMed] [Google Scholar]
- 48.Beechem J. M., Otto M. R., Bloom L. B., Eritja R., Reha-Krantz L. J., Goodman M. F. ( 1998) Biochemistry 37, 10144– 10155 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.