Abstract
Tumor suppressor p53 regulates the expression of p53-induced genes (PIG) that trigger apoptosis. PIG3 or TP53I3 is the only known member of the medium chain dehydrogenase/reductase superfamily induced by p53 and is used as a proapoptotic marker. Although the participation of PIG3 in the apoptotic pathway is proven, the protein and its mechanism of action were never characterized. We analyzed human PIG3 enzymatic function and found NADPH-dependent reductase activity with ortho-quinones, which is consistent with the classification of PIG3 in the quinone oxidoreductase family. However, the activity is much lower than that of ζ-crystallin, a better known quinone oxidoreductase. In addition, we report the crystallographic structure of PIG3, which allowed the identification of substrate- and cofactor-binding sites, with residues fully conserved from bacteria to human. Tyr-59 in ζ-crystallin (Tyr-51 in PIG3) was suggested to participate in the catalysis of quinone reduction. However, kinetics of Tyr/Phe and Tyr/Ala mutants of both enzymes demonstrated that the active site Tyr is not catalytic but may participate in substrate binding, consistent with a mechanism based on propinquity effects. It has been proposed that PIG3 contribution to apoptosis would be through oxidative stress generation. We found that in vitro activity and in vivo overexpression of PIG3 accumulate reactive oxygen species. Accordingly, an inactive PIG3 mutant (S151V) did not produce reactive oxygen species in cells, indicating that enzymatically active protein is necessary for this function. This supports that PIG3 action is through oxidative stress produced by its enzymatic activity and provides essential knowledge for eventual control of apoptosis.
p53 is an essential transcription factor in the control of the cell cycle, inducing cell cycle arrest or apoptosis, through the control of the expression of distinct genes (1). TP53I3 (tumor protein p53-inducible protein 3), also called PIG3 (p53-inducible gene 3), is one of the p53 protein targets and is used as a long lived proapoptotic marker. PIG3 was discovered along with 12 other proteins in a serial analysis of gene expression studies designed to determine genes induced by p53 before the apoptosis onset (2). In addition, PIG3 expression can also be elicited by p63 and p73 (3), which are able to induce apoptosis in a p53-independent manner (4) and which are implicated in the induction of PIG3 through flavonoid exposure (5). PIG3 expression is largely regulated through its promoter (6), and polymorphisms within this site have been associated with invasive bladder cancer (7) and leukemia (8). Some p53 mutants, which are able to induce cell cycle arrest but not apoptosis, could not induce PIG3 expression (9). Moreover, it has been recently reported that PIG3 expression is also regulated by a new transcription factor (hCAS/CSE1L), which associates with a subset of p53 target genes increasing apoptosis. The factor binds to the PIG3 promoter region, whereas hCAS/CSE1L silencing leads to decreased PIG3 transcription and to increased methylation of histone H3Lys-27 found within the PIG3 gene, finally resulting in decreased apoptosis (10). Despite its clear relationship with apoptosis, the mechanism of the PIG3 action has not been established. The protein shares significant sequence identity with the quinone oxidoreductase (QOR)2 family, which led us to propose a possible role in apoptosis through a putative QOR function. QORs catalyze quinone reduction, generating hydroxyquinone (11–13), which is reoxidized in the presence of O2 leading to the formation of reactive oxygen species (ROS) (Scheme 1). On the basis of this mechanism, it has been proposed that PIG3 may induce oxidative stress, like other p53 targets (14). In support of this notion, oxidative stress has been proposed as a proapoptotic mechanism (15), and PIG3 expression precedes the appearance of ROS in p53-induced apoptosis. However, evidence that supports an apoptotic role of PIG3 by increasing oxidative stress through a QOR function is only indirect because its enzymatic activity has never been reported, and only preliminary information exists on the role of PIG3 in ROS production in vivo (2, 16).
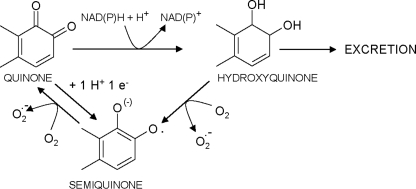
SCHEME. 1.
Quinone reduction and redox cycling. Quinone reduction by one-electron transfer yields the semiquinone radical. Two-electron reduction of a quinone by NAD(P)H-dependent quinone-reducing enzymes yields hydroxyquinone, which can be further conjugated by phase II enzymes and excreted. Alternatively, the hydroxyquinone can be oxidized to the semiquinone radical. Both the hydroxyquinone and semiquinone states can be oxidized in the presence of molecular oxygen, in a redox cycling process, yielding superoxide anion, thereby sustaining ROS generation. Taken in a modified form from Oppermann (13).
QOR is a poorly studied protein family within the medium chain dehydrogenases/reductases (MDR). There is no report on the involvement of any QOR in ROS increase or in vivo quinone oxidation. In QORs the enzymatic activity has been only described in the mammalian ζ-crystallin subfamily and related proteins in yeast (17) and bacteria (18). In fact, ζ-crystallins were described initially as proteins found in high amount in guinea pig and camel lenses (19), and recently their function as RNA- and DNA-binding proteins has been reported (17, 20, 21). However, the physiological role of any QOR has not been clearly demonstrated, and those roles proposed for ζ-crystallins, i.e. protection against UV light in lenses, and regulation of blood pH by destabilization/stabilization of certain RNAs in kidney cells (22), have no relationship with their enzymatic activity.
In this study, we have characterized the x-ray structure and analyzed the enzymatic properties of PIG3 and its role in ROS generation. The study has been supported by the side-by-side characterization of ζ-crystallin, a more active protein which served as a model and which allowed the recognition of general properties and the enzymatic mechanism of QORs. The identification of PIG3 substrates and active-site structure provides a target for the pharmacological control of apoptosis.
EXPERIMENTAL PROCEDURES
Sequence Analysis and Phylogeny of the PIG3 Subfamily
Orthologous PIG3 genes were identified in annotated eukaryotic genomes using the Ensembl Genome Browser version 38, and both their coding sequences and 5′-untranslated regions were retrieved. In addition, the Xenopus laevis PIG3 protein sequence was obtained by searching the UniProt data base, which also contained the complete Xenopus tropicalis, bovine, and human sequences. PIG3 proteins from nonvertebrate species, including Arabidopsis thaliana, Dictyostelium discoideum, and Paenibacillus larvae, were identified using BLASTP 2.0 (23) in a homology search against nonredundant PIG3 sequences at the NCBI protein data base. Sequence analysis and manipulation were carried out using BioEdit Sequence Alignment Editor, version 7.0.4.1 (24). Multiple amino acid sequence alignments were performed using the ClustalW (25) executable version 1.4 distributed with BioEdit and were manually revised.
Cloning of PIG3 cDNA into Bacterial Expression Vectors pGEX-4T-2 and pET-30 Xa/LIC and Mammalian Expression Vector pCMV-HA
Within this study several expression constructs were generated. PIG3 cDNA was obtained from Mammalian Gene Collection (MGC) clones (Open Biosystems, MGC:8642 IMAGE:2961571). Initially, PIG3 cDNA was cloned into pET-30 Xa/LIC (Novagen) by using two primers containing the ligation-independent cloning (LIC) tags (supplemental data). For PIG3 bacterial expression (orthorhombic crystal form) or transient expression in human cells, PIG3 cDNA was amplified by using two primers containing restriction sites for EcoRI, in the forward primer, and SalI, in the reverse primer (supplemental data). The resulting PCR product was inserted into pGEX-4T-2 (GE Healthcare) or pCMV-HA (Clontech). Alternatively, for PIG3 expression in the tetragonal crystal form, PIG3 cDNA was cloned by PCR-LIC into a LIC-adapted, isopropyl 1-thio-β-d-galactopyranoside-inducible (Roche Applied Science), bacterial expression vector (pNIC28-Bsa4, Structural Genomics Consortium, GenBankTM accession number EF198106) with an N-terminal His6 tag and an integrated tobacco etch virus protease site.
Site-directed Mutagenesis
PIG3 mutants were obtained by using the wild-type PIG3 cDNA cloned into pET30-Xa/LIC as a template. Based on the QuickChangeTM site-directed mutagenesis kit method (Stratagene), we designed two primers for each mutation as follows: Y51F, Y51A, and S151V (supplemental data).
The ζ-crystallin variants were obtained using ζ-crystallin cDNA cloned into pET30-Xa/LIC as a template (17) and two primers for each mutation (supplemental data). All reactions were performed in a DNA thermal cycler (MJ Research) with Pfu Turbo DNA polymerase (Stratagene). PCR products were incubated with DpnI at 37 °C for 60 min to select against the dam-methylated parental strand. The resulting nicked-circular mutagenic strands were transformed into Escherichia coli BL21. Prior to expression, DNA was completely sequenced to ensure that unwanted mutations were absent.
Protein Expression and Purification
For kinetic studies, PIG3 and ζ-crystallin were cloned into pET-30 Xa/LIC as described above and by Fernández et al. (17). Briefly, E. coli BL21, transformed with pET-30 Xa/LIC, was grown in 2× YT medium at 24 °C for 8 h. Protein expression was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside, and cells were further incubated for 16 h. After cell lysis by sonication, the homogenate was applied onto a nickel-charged chelating SepharoseTM fast flow column (5 ml) (GE Healthcare). After washing with 60 mm imidazole in 50 mm NaH2PO4, 0.5 m NaCl, pH 8.0, the enzyme was eluted with a 0.06–1.0 m imidazole gradient in the same buffer. Protein fractions were concentrated, and imidazole was removed with an Amicon ultra device (Millipore). The protein was stored at 4 °C in 50 mm NaH2PO4, 0.5 m NaCl, pH 8.0.
For PIG3 production (orthorhombic crystals), E. coli BL21 strain was transformed with pGEX-4T-2, containing a PIG3 cDNA insert, and grown in 2× YT medium at 24 °C for 8 h. Protein expression was induced by 0.1 mm isopropyl 1-thio-β-d-galactopyranoside and further incubated for 16 h. PIG3 was purified batchwise by using glutathione-SepharoseTM 4B (GE Healthcare). After washing with phosphate-buffered saline (PBS, 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4), the protein was eluted by thrombin digestion (GE Healthcare, 50 units/liter of culture) for 15 h at room temperature. Active enzyme was further purified with a Reactive Red 120-agarose (Sigma) resin and eluted by a 0–2 m NaCl gradient in PBS. Buffer was exchanged to 2.5 mm NADP+ in PBS using a PD-10 exchange buffer column (GE Healthcare). For PIG3 production (tetragonal crystals), expression and purification of uncleaved protein were achieved by using nickel-affinity chromatography (GE Healthcare) and size-exclusion chromatography (Hiload 16/60 Superdex 200, GE Healthcare). Pure protein was concentrated using Centricon (Millipore) up to 10–15 mg/ml and stored at −80 °C. Protein homogeneity was checked by SDS-PAGE followed by Coomassie® Brilliant Blue (Sigma) staining, as well as liquid chromatography-mass spectrometry using an Agilent electrospray ionization-time of flight instrument. Protein concentration was determined by a dye binding assay (Bio-Rad) using bovine serum albumin as a standard (26) or using a Nanodrop UV spectrophotometer.
Crystallization, Structure Determination, and Docking
The structure of human PIG3 was determined by x-ray crystallography using a tetragonal and an orthorhombic crystal form (Table 1). Tetragonal crystals were grown by vapor diffusion at 20 °C using sitting drops of 300 nl from a protein solution (15.1 mg/ml) and 300 nl from the reservoir solution containing 0.22 m lithium sulfate, 0.1 m sodium acetate, pH 4.0, 45% polyethylene glycol 300. Data were collected at the Swiss Light Source synchrotron beamline SLS-X10 from liquid nitrogen flash-cooled crystals. Orthorhombic crystals were grown also at 20 °C by vapor diffusion using hanging drops of 1 μl of a protein solution (10 mg/ml) mixed with 1 μl of the precipitant solution containing 30% polyethylene glycol 4000, 0.1 m Tris, and 0.2 m sodium acetate, pH 8.5. Data were collected at the automated crystallography platform (Parc Científic de Barcelona) using a Rigaku rotating anode generator with flash-cooled crystals. Diffraction data were integrated and scaled using DENZO and SCALEPACK (27). An initial solution was first obtained by molecular replacement (28) for the tetragonal crystal, with one protein subunit in the asymmetric unit, using as a search model the coordinates from Thermus thermophilus QOR (PDB code 1IYZ), a protein having 31% sequence identity with PIG3 (Table 2). However, completion of this tetragonal structure presented some difficulties that were only overcome when solving the orthorhombic crystal by molecular replacement using as a searching model the partial PIG3 structure then available. Refinement was carried out using REFMAC (29) and manual rebuilding with the graphic program O (30). During refinement noncrystallographic restraints were maintained between the subunits in the orthorhombic crystal. The resulting refined PIG3 models were deposited in the PDB with codes 2J8Z and 2OBY for the tetragonal and orthorhombic crystals, respectively (Table 1). The initial docking of 1,2-naphthoquinone into the PIG3 model was done manually and then regularized using REFMAC. Figures were prepared with PyMOL.
TABLE 1.
PIG3 data collection and refinement statistics
| Crystal |
||
|---|---|---|
| Orthorhombic (2OBY) | Tetragonal (2J8Z) | |
| Data collection | ||
| Space group | C2221 | P4322 |
| Cell parameters | ||
| a, b, c | 68.3, 184.4, 318.0 Å | 46.9, 46.9, 315.9 Å |
| α, β, γ | 90, 90, 90° | 90, 90, 90° |
| Subunits per asymmetric unit | 5 | 1 |
| No. of observed reflections | 34,254 (6514) | 13,313 (881) |
| Completeness (%) | 88.6 (65.0) | 99.8 (99.6) |
| Average 1/σ (I) | 6.9 (2.5) | 12.1 (4.4) |
| Rsym | 10.9% (42.4%) | 6.2% (31.1%) |
| Crystallographic refinement | ||
| Resolution range | 19.94 to 3 Å (3.14 to 3.0 Å) | 30 to 2.5 Å (2.56 to 2.5 Å) |
| Rworka | 22% (41.8%) | 23% (30.7%) |
| Rfreeb | 26% (45.2%) | 27% (37.7%) |
| No. of observed reflections | 34,254 (6514) | 13,313 (881) |
| Ramachandran plot statistics | ||
| Residues in most favored regions | 88.8% | 90.3% |
| Residues in additional allowed regions | 10.9% | 9.7% |
| Residues in generously allowed regions | 0.4% | 0% |
| Residues in disallowed regions | 0% | 0% |
| Final model parameters | ||
| No. of monomers | 5 | 1 |
| Residues (each monomer) | 334 | 330 |
| Hetero groups (each monomer) | 1 NADP+ | 1 NADP+ |
| No. of water molecules | 3 | 11 |
| Average B-factor protein | 40.6 Å2 | 53.8 Å2 |
| Covalent bond lengths | 0.005 Å | 0.010 Å |
| Bond angles | 0.935° | 1.302° |
aRwork = Σhkl‖Fobs|− |Fcalc‖/Σhkl|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
b Rfree has the same definition as Rwork for a cross-validation set of about 5% of the reflections. The last resolution shell is shown in parentheses.
TABLE 2.
PIG3-related proteins
A homology search with PIG3 amino acid sequence as the query using BLASTP 2.0 (23) was done against nonredundant protein sequences at NCBI protein data base. The nearest proteins and nonredundant Zn2+-lacking MDR proteins with reported kinetic constants or three-dimensional structure were selected. Swiss-Prot entry and Protein Data Bank code are shown in parentheses. All proteins are classified in the quinone oxidoreductase family, except trans-2-enoyl-CoA reductase, leukotriene B4 12-hydroxydehydrogenase, and NADP-dependent oxidoreductase P1 (previously named P1-ζ-crystallin).
| Protein | Organism | Amino acid sequence homology |
Reaction | Best substrates | ||
|---|---|---|---|---|---|---|
| Identity | Similarity | Gaps | ||||
| % | % | % | ||||
| PIG3 (Q53FA7) | Homo sapiens | 100 | 100 | 0 | ||
| Probable_QOR (Q8L3C8) (1IYZ) | T. thermophilus | 31 | 47 | 16 | ||
| TED2 (Q41724) | Zinnia elegans | 28 | 48 | 5 | ||
| QOR (P28304) (1QOR) | E. coli | 28 | 44 | 11 | ||
| Zta1p (P38230) | S. cerevisiae | 28 | 47 | 12 | Quinone → semiquinonea | 9,10-Phenanthrenequinone |
| ζ-Crystallin (Q08257) (1YB5) | H. sapiens | 27 | 47 | 3 | Quinone → semiquinonea | 9,10-Phenanthrenequinone |
| SA1989 protein (Q7A492) | S. aureus | 24 | 40 | 21 | Quinone → semiquinoneb | 9,10-Phenanthrenequinone |
| trans-2-Enoyl-CoA reductase (Q9BV79) (1ZSY) | H. SAPIENS | 22 | 37 | 20 | trans-2-Enoyl-CoA → acyl-CoAc | (2E)-Hexadecenoyl-CoA |
| Leukotriene B4 12-hydroxydehydrogenase (Q9EQZ5) (2DM6) | Cavia porcellus | 22 | 36 | 24 | Alk-2-enal → n-alkanald | 15-Keto prostaglandin E2 |
| NADP-dependent oxidoreductase P1 (Q39172) (2J3K) | A. thaliana | 21 | 37 | 27 | Quinone → semiquinoneeAlk-2-enal → n-alkanalf Azodicarbonyl → imidee | 9,10-Phenanthrenequinone trans-2-nonenal, diamide |
Determination of Coenzyme Binding by Fluorescence Quenching
NAD(P)H binding to PIG3 was followed by measuring the quenching in protein fluorescence by using a 650-40 spectrofluorimeter (PerkinElmer Life Sciences). Relative fluorescence intensity was determined at 25 °C in 100 mm sodium phosphate buffer, pH 7.0. The assay contained 50 μg of enzyme in 1-ml total volume, to which 5-μl aliquots from concentrated cofactor solution were added up to 2 μm NADPH with PIG3 and up of 20 μm for the control assays. Fluorescence was obtained by tryptophan excitation (280 nm) and by measuring the emission intensity (340 nm). Binding curves were reported as fluorescence quenching. The dissociation constant (KD) for NADPH binding to PIG3 was calculated as described previously (31, 32).
Kinetic Characterization
Activities were determined in 0.1 m sodium phosphate, pH 7.0, in the presence of 0.15 mm NADPH, with freshly prepared substrate solutions. Stock quinone solutions were prepared in ethanol, resulting in final concentrations lower than 3% (v/v) ethanol in the assay mixture. 4-Hydroxyestrone-o-quinone was a generous gift from Dr. A.R. de Lera, Dr. R. Alvarez, and P. García (University of Vigo, Spain). The enzymatic activity was measured in a Cary 400 Bio (Varian) spectrophotometer by following the consumption of NADPH at 340 nm (ϵNAD(P)H = 6200 m−1·cm−1). The activity toward 2,6-dichloroindophenol (DCIP) was determined by following the absorbance decrease at 600 nm (ϵDCIP = 4010 m−1·cm−1). The enzymatic assay toward retinaldehyde was performed as described (33). Controls, lacking either substrate or enzyme, were run routinely. The reactions started with the addition of the substrate. For the kinetic studies, initial velocities were measured in duplicate with five different substrate concentrations, and the kinetic constants were calculated using the nonlinear regression program Grafit 5.0 (Eritacus Software Ltd.). All reported values are expressed as the mean ± S.E. of at least three independent experiments.
Detection of Hydrogen Peroxide Production by Peroxyoxalate Chemiluminescent Reaction
H2O2 was measured as a product of the PIG3-catalyzed reaction. The reaction mixtures contained 50 μm 1,2-naphthoquinone or 100 μm diamide with 150 μm NADPH and 28 μg of PIG3. After 2 min at room temperature, a 2-μl aliquot of the 1,2-naphthoquinone reaction or a 5-μl aliquot of the diamide reaction was added to the H2O2 reaction assay. The detection of H2O2 was performed with 50 μl of Amplex Red reagent (Invitrogen), according to the manufacturer's instructions in a final volume of 100 μl. Amplex Red reagent reacts with H2O2 to produce highly fluorescent resorufin. The H2O2 reactions were performed on 96-well plates, and fluorescence was measured using a Victor3 spectrofluorimeter (PerkinElmer Life Sciences) by exciting at 530 nm and measuring emission at 590 nm. Positive controls were performed by adding 50 μl of 0.5, 2, and 10 μm hydrogen peroxide solutions.
Cell Culture and Transfection
HCT-116 cells were grown on 24-well plates in Dulbecco's modified Eagle's medium/F-12 (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Invitrogen). Incubation was performed at 37 °C in a humidified atmosphere containing 5% CO2, 95% air. For transfection, cells were plated and, after 24 h, transfected with LipofectamineTM Plus (Invitrogen) according to the manufacturer's instructions. After 4 h, fresh medium was added, and cells were incubated overnight.
Intracellular Measurement of ROS
The assay is based on 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA, Fluka), a fluorogenic marker for ROS in living cells (34, 35). DCFH-DA is deacetylated by nonspecific intracellular esterases, generating a fluorescein product, which could be oxidized by ROS and thus emitting bright green fluorescence. Briefly, cells were transiently transfected with pCMV-HA and PIG3/pCMV-HA for 24 h. Cells that had been treated with phorbol 12-myristate 13-acetate for 6 h were used as a positive control of ROS production. The cells were then incubated in 1 ml of Dulbecco's modified Eagle's medium/F-12 with 20 μm DCFH-DA at 37 °C for 30 min in the dark. After incubation, cells were washed with PBS, trypsinized, and finally resuspended in PBS. Cells, which should be protected from light, were analyzed by fluorescence-activated cell sorting (Caliber) within 30 min, using the excitation peak of fluorescein at 495 nm and the fluorescence emission at 529 nm. Data were expressed as means ± S.D. Statistical significance was assessed by the Student's t test using SigmaStat 3.5 version.
Western Blot Analysis
Polyclonal antiserum-specific against PIG3 was from OncogeneTM. Goat anti-rabbit IgG (H+L) peroxidase-conjugated antibody was from Bio-Rad.
RESULTS
Sequence Analysis and Phylogeny of the PIG3 Subfamily
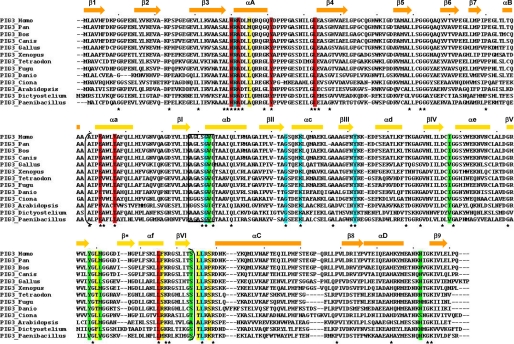
In vertebrates, the PIG3 gene is found in most of the genomes examined ranging from primitive chordates to humans (Fig. 1). Specifically, it is detected in ascidians (Ciona intestinalis), fish (Danio rerio, Fugu rubripes, and Tetraodon nigroviridis), amphibians (X. laevis), birds (Gallus gallus), mammals (Bos taurus and Canis familiaris), non-human primates (Pan troglodytes), and humans. At this point, a PIG3 gene, including a full-length coding sequence, has not been found in rodents (Rattus norvegicus, Mus musculus, Cavia porcellus, Spermophilus tridecemlineatus, and Dipodomys ordii). In contrast, it is detected in rabbit (Oryctolagus cuniculus). Promoter analysis reveals the presence of the p53-responsive element (TGYCC)n only in primate PIG3 promoters, confirming that p53 responsiveness was acquired during primate evolution (36). In addition, PIG3 orthologs are found in bacteria (Paenibacillus larvae), plants (A. thaliana), amoebozoa (D. discoideum), and other bacteria and fungi, indicating a wide distribution in living organisms. Comparison of the amino acid sequences from members of the PIG3 subfamily (Fig. 1) shows an overall 13% identity, increasing to 27, 35, and 60% within chordate, vertebrate, and tetrapod sequences, respectively. In all cases the identity is higher than 40% in pairwise comparisons with the human PIG3.
FIGURE 1.
Alignment of PIG3 amino acid sequences. Conserved residues from the substrate-binding pocket, including Thr-127 that interacts with NADP+ are shaded in red, and those not conserved are shaded in yellow. Residues interacting with NADP+ and conserved are shaded in blue, whereas those not conserved are shaded in green. The box indicates the (A/G)XXSXXG motif. The sequence segment in parentheses corresponds to the cofactor-binding domain. Asterisks indicate conserved residues. Secondary structure elements, obtained from the human PIG3 crystal (Fig. 3B), are shown above the sequences; bars indicate α-helices, and arrows denote β-sheets and are colored orange and light orange for the catalytic and cofactor-binding domains, respectively.
Substrate Analysis of PIG3 and Cofactor Specificity
Table 2 presents the proteins structurally closest to PIG3, from which three-dimensional and/or functional information is known. All of them are MDR proteins lacking Zn2+ as metal cofactor, and most of them can be classified as QORs, except trans-2-enoyl-CoA reductase, included in the enoyl thioester reductase family, and leukotriene B4 12-hydroxydehydrogenase and Arabidopsis NADP-dependent oxidoreductase P1, both included in the alkenal/one reductase family. Only mammalian ζ-crystallins, as well as the yeast and Staphylococcus aureus-related proteins, are known to possess enzymatic activity within the QOR family.
We have tested PIG3 NADPH-dependent reductase activity with reported ζ-crystallin substrates, with substrates for close MDR families (Table 2), and with related compounds. PIG3 exhibits enzymatic activity with naphthoquinones (Table 3), with a strong preference for the ortho-quinone isomer (1,2-naphthoquinone) over the para isomer (1,4-naphthoquinone). From the compounds tested, 1,2-naphthoquinone is the only substrate able to saturate the enzyme (Table 4). A similar but larger compound, 9,10-phenanthrenequinone, the best substrate for ζ-crystallins in terms of kcat/Km (Tables 2 and 4), is inactive with PIG3. Interestingly, activity was also detected for some non-quinone compounds. Thus, diamide [1,1′-azobis(N,N-dimethylformamide)] exhibited the highest rate with PIG3, although it did not saturate the enzyme. In contrast, diamide was not a substrate for ζ-crystallin. DCIP, a compound used as electron acceptor in several oxidoreductase reactions (37), showed low activity. Other compounds such as the para-quinone 1,4-benzoquinone, the physiological ortho-quinone 4-hydroxyestrone-o-quinone (a product of catechol estrogen metabolism), 2-trans-nonenal, and retinaldehyde were not active. Cytochrome c exhibited some activity with ζ-crystallin but was inactive toward PIG3.
TABLE 3.
NADPH-dependent reductase activities of PIG3 and ζ -crystallin with different compounds
Activities were determined in 0.1 m sodium phosphate, pH 7.0, with 0.15 mm NADPH, at 25 °C. Retinaldehyde reduction was measured as described in Ref. (33). NA means not active; ND means not determined.
| Concentration | Specific activity |
||
|---|---|---|---|
| PIG3 | ζ-Crystallin | ||
| mm | nmol/min·mg | ||
| 1,2-Naphthoquinone | 0.1 | 146 | 5500 |
| 1,4-Naphthoquinone | 0.1 | 3 | 165 |
| 1,4-Benzoquinone | 0.1–0.4 | NA | ND |
| 9,10-Phenanthrenequinone | 0.1 | NA | 4250 |
| 4-Hydroxyestrone-o-quinone | 0.05–0.1 | NA | NA |
| Diamide | 0.15 | 500 | NA |
| DCIP | 0.1 | 20 | 330 |
| 2-trans-Nonenal | 0.2–0.5 | NA | NAa |
| All-trans-retinaldehyde | 0.005–0.01 | NA | NAa |
| Cytochrome c | 0.1 | NA | 500 |
a Data are taken from Fernández et al. (17).
TABLE 4.
Kinetic analysis of PIG3, human ζ -crystallin, and their variants
Activities were determined in 0.1 m sodium phosphate, pH 7.0, with 0.15 mm NADPH, at 25 °C. NS means not saturable.
| PIG3 | PIG3 Y51F | PIG3 Y51A | |
|---|---|---|---|
| 1,2-Naphthoquinone | |||
| Km (μm) | 215 ± 3 | 150 ± 50 | NS |
| kcat (min−1) | 15.7 ± 1.6 | 192 ± 35 | NS |
| v (nmol·min−1·mg−1)a | 146 ± 18 | 2000 ± 350 | 290 ± 50 |
| kcat/Km (mm−1·min−1) | 73 ± 13 | 1270 ± 479 | NS |
| ζ-Crystallin | ζ-Crystallin Y59F | ζ-Crystallin Y59A | |
|---|---|---|---|
| 1,2-Naphthoquinone | |||
| Km (μm) | 29 ± 1b | 11 ± 3 | 26 ± 7 |
| kcat (min−1) | 170 ± 3b | 103 ± 14 | 244 ± 30 |
| kcat/Km (mm−1·min−1) | 5900 ± 320b | 9500 ± 3000 | 9260 ± 2700 |
| 9,10-Phenanthrenequinone | |||
| Km (μm) | 1.9 ± 0.1 | 0.25 ± 0.01 | 2 ± 0.4 |
| kcat (min−1) | 195 ± 4 | 90 ± 9 | 102 ± 5 |
| kcat/Km (mm−1·min−1) | 102600 ± 8000 | 371500 ± 137000 | 50800 ± 9400 |
a Activity was performed using 0.2 mm 1,2-naphthoquinone.
b Data are taken from Fernández et al. (17).
The kinetic constants could be calculated for 1,2-naphthoquinone (Table 4). Both the kinetic constants and the specific activities indicate that PIG3 has a considerably lower activity than ζ-crystallin with the substrates assayed, except for diamide.
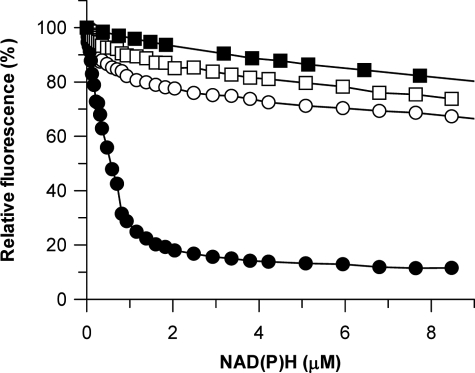
Regarding cofactor preference, no activity was found when the best substrates were assayed with NADH instead of NADPH. Fluorescence quenching experiments showed that PIG3 binds NADPH with high affinity (KD = 0.7 μm), whereas NADH binding is weak, similar to the binding to bovine serum albumin, used as the control for nonspecific interactions (Fig. 2). Therefore, kinetic and binding approaches demonstrate a strict specificity for NADPH.
FIGURE 2.
Fluorescence quenching on cofactor binding to PIG3 and to the S151V mutant. Fluorescence change on adding NADPH (solid circles) or NADH (solid squares) to PIG3, and the effect of adding NADPH to PIG3 S151V mutant (empty circles) and bovine serum albumin (BSA) (empty squares) are shown. Protein concentration was 2.1 μm.
PIG3 Overall Crystallographic Structure
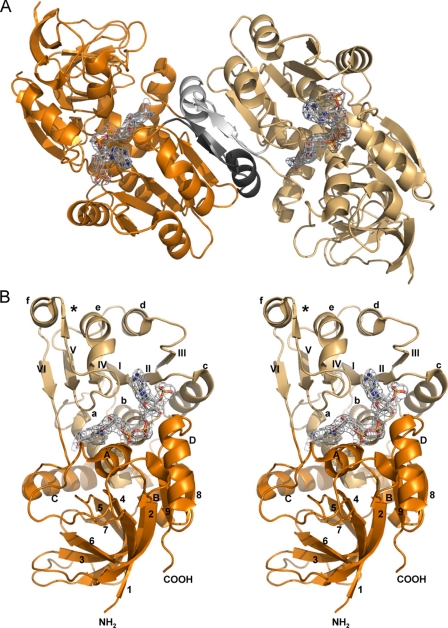
Two different constructs from the human PIG3 enzyme were crystallized in the presence of NADP+, giving tetragonal and orthorhombic crystal forms with space groups P4322 and C2221, and resolution limits of 2.5 and 3.0 Å, respectively (Table 1). The asymmetric units of the tetragonal and orthorhombic crystal forms contained one and five protein subunits, respectively (Table 1). Size-exclusion chromatography analysis yielded an apparent Mr of 70,300, whereas the calculated Mr of the subunit is 36,000. Therefore, PIG3 behaves in solution as a stable dimer. This result is consistent with the crystal structures determined (Fig. 3, A and B). The final refined models exhibit the typical MDR structural fold and include 333 residues (with 79 and 293–295 missing) for the subunit in the tetragonal crystal (PDB code 2J8Z) and 334 residues (with 293–295 missing) for each of the five subunits in the orthorhombic crystal (PDB code 2OBY). One NADP+ molecule was found bound to each of the subunits.
FIGURE 3.
Structure of PIG3. A, quaternary structure. The molecule consists of two identical subunits, colored orange and light orange. The dimer interface is highlighted in gray and black. B, stereo view image of the tertiary structure of the monomer. The catalytic domain is colored orange, and the cofactor binding domain is colored light orange. The secondary structure nomenclature is the same as that used in Fig. 1. The β-strands and α-helices of the cofactor-binding domain are indicated by roman numerals and lowercase letters, respectively. Strand β* is not part of the classical Rossmann fold topology. Nomenclature of the catalytic domain is in arabic numerals and capital letters. A and B, NADP+ molecule is displayed with the electron density map in gray.
The superimposition of C-α atoms from the PIG3 subunits shows root mean square deviations close to 0.6 Å between subunits from the tetragonal and the orthorhombic crystals. As it has been described for other MDR members (38), the PIG3 monomer is composed of two domains as follows: the catalytic domain (Met-1 to Ala-119 and Leu-265 to Gln-332) and the cofactor-binding domain (Ala-120 to Ser-264) (Fig. 3B). These domains are separated by a deep cleft, which accommodates NADP+. The catalytic domain is dominated by a seven-stranded antiparallel β-sheet, whereas the cofactor-binding domain consists of two mononucleotide binding βαβαβ motifs, which are assembled into the classical Rossmann fold. However, there is an additional β-strand (β*), which was also described in the human enoyl thioester reductase (39).
Two β-strands and an α-helix of β*-αf-βVI of the cofactor-binding domain are involved in the dimer interface between monomers (Fig. 3A). These β-strands are bonded through anti-parallel hydrogen bonds with the corresponding sheets of the neighboring monomer. In the dimer interface the buried accessible surface area per monomer is 1200 Å2.
NADP+ Binding
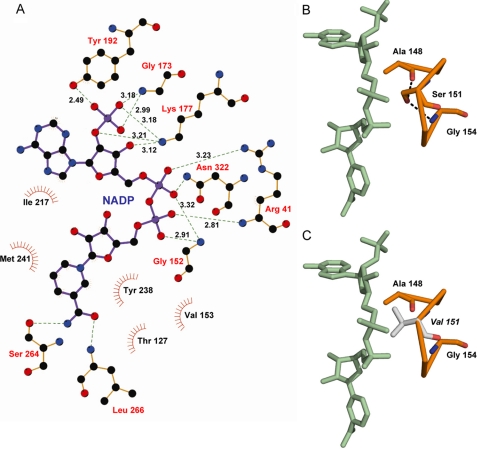
For all the subunits the NADP+ molecule was located in the cleft between the catalytic and the nucleotide-binding domains (Figs. 3 and 4). NADP+ is bound in an extended conformation with an averaged distance of 13.7 Å between the nicotinamide C-2 and the adenine C-6 reference atoms (40). Out of 13 residues directly implicated in NADP+ binding (Fig. 4A), 7 are fully conserved among the PIG3 sequences (Fig. 1). The binding of the dinucleotide cofactor to MDR enzymes has been described to require a GXGXX(G/A) sequence motif. In PIG3, the corresponding binding motif is modified as (A/G)XXSXXG, which is closer to the AXXGXXG and GXXGXXG binding motifs found in other QORs (41). The Ser-151 hydroxyl group from the PIG3 motif makes two hydrogen bonds with the main chain oxygen and nitrogen atoms from the motif Ala-148 and Gly-154, respectively (Fig. 4B). When a Gly is in that motif position, as in most MDR structures including other QOR subfamilies, a solvent molecule occupies the position corresponding to the Ser-151 hydroxyl group in PIG3, establishing similar interactions.
FIGURE 4.
NADP-binding site of PIG3. A, LIGPLOT (38) describing interactions in the NADP-binding site. Residues Gly-173, Lys-177, and Tyr-192 are interacting with the 2′-phosphate of NADP+. B, (A/G)XXSXXG motif in orange, showing Ser-151 hydrogen bonds with the main chain oxygen and nitrogen atoms from the motif residues Ala-148 and Gly-154. C, model of the same region as in B but using the S151V inactive mutant, which apparently presents steric hindrances between Val-151 side chain (gray) and NADP+ that may result in the inability of the protein to bind the cofactor.
For NAD(H)-dependent MDRs, the presence of an Asp or a Glu residue at the C-terminal end of the second β-sheet, in the nucleotide-binding domain, is characteristic (42). This acidic residue is replaced by a small neutral residue, predominantly Gly, in the NADP(H)-dependent enzymes (43). Therefore, this position is thought to have a key role in determining cofactor specificity. In PIG3 the corresponding residue is Gly-173, which appears fully conserved among PIG3 members and is also present in the E. coli QOR (Gly-173) and human ζ-crystallin (Gly-181). Cofactor binding and NADPH specificity in PIG3 is achieved through interactions of the 2′-phosphate group with the main chain of Gly-173, the Lys-177 amino group, and the Tyr-192 hydroxyl group (Fig. 4), also conserved residues. The presence of at least one basic residue interacting with this terminal phosphate of the NADP(H) molecule has also been proposed as an important contribution to cofactor specificity (44). Therefore, the presence of both Gly-173 and Lys-177 strongly suggests that PIG3 should be an NADP(H)-dependent enzyme, in full agreement with the kinetic and cofactor-binding experiments. Moreover, the NADP specificity may be reinforced by the proximity of Lys-193 increasing the positively charged environment. Arg-41 (equivalent to Arg/His-47 in alcohol dehydrogenases) interacts with the pyrophosphate bridge, whereas Ile-217, Tyr-221, and Met-241 define the adenine-binding hydrophobic pocket.
Active Site
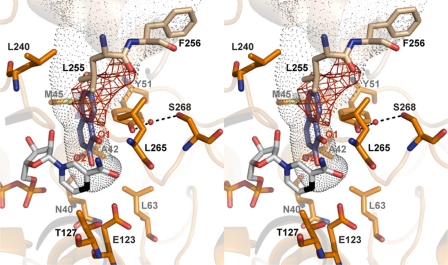
The pocket between the NADPH molecule and the catalytic domain, where the C-4 atom from the nicotinamide ring becomes accessible to the solvent, appears well suited as a substrate-binding site (Fig. 5). In fact, some unassigned electron density was found in this pocket and appears to reflect the binding tendency of this site toward small molecules. The pocket, predominantly polar around the nicotinamide ring but with several hydrophobic features, is formed by residues Asn-40, Ala-42, Met-45, Tyr-51, Leu-63, Glu-123, Thr-127, Leu-240 and Leu-265 from one subunit and Leu-255* and Phe-256* from the second subunit. Among these residues only Met-45, Leu-240, Leu-265, and Phe-256* present some variability (Fig. 1). The carboxylate group of Glu-123, a residue conserved in all PIG3 proteins but absent in all other QOR enzymes (Table 5), is about 3.0 Å from the C-4 atom and surrounded by a strongly hydrophobic environment with Leu residues 63, 89, and 266. In addition, side chains from Tyr-51 and Ser-268 appear to be tightly bound to each other through a solvent molecule (Fig. 5).
FIGURE 5.
Representation of the substrate-binding pocket in PIG3. A stereo view image of main amino acid residues that conform the substrate-binding cavity, which is shown in dots, and a computer model of the ternary complex with 1,2-naphthoquinone docked into the pocket. The NADP+ molecule is displayed with its C-4 atom in black. In this docking the O-1 atom of 1,2-naphthoquinone can establish a hydrogen bond with the hydroxyl group of Tyr-51, whereas the O-2 atom is in direct contact with the C-4 atom of NADP+. The nearest protein residues to the NADP+ C-4 atom are Glu-123, with the carboxyl group at 3.2 Å, and Thr-127, with the hydroxyl at 3.3 Å. Two additional electron densities are observed as follows: one between Tyr-51 and Ser-268, which has been attributed to a water molecule (red sphere) allowing interaction between these residues through hydrogen bonds (displayed in black dashed lines); and an uninterpreted extra electronic density in the substrate-binding pocket, highlighted in red. Mass spectrometry analysis suggests that the extra density corresponds to a fragment of ampicillin, the antibiotic used in the protein expression procedure. This ligand would not be present in the protein for kinetic studies, which was prepared using kanamycin.
TABLE 5.
Residues located at equivalent positions in the substrate-binding pocket of quinone oxidoreductases
| Human PIG3 | Human ζ-crystallin | E. coli QOR |
|---|---|---|
| Asn-40 | Asn-48 | Asn-41 |
| Ala-42 | Val-50 | Ile-43 |
| Met-45 | Tyr-53 | Tyr-46 |
| Tyr-51 | Tyr-59 | Tyr-52 |
| Leu-63 | Ser-71 | Thr-63 |
| Glu-123 | Ile-131 | Leu-123 |
| Thr-127 | Thr-135 | Thr-127 |
| Leu-240 | Ser-248 | –a |
| Leu-255 | Met-260 | Leu-253 |
| Phe-256 | Ala-261 | Asn-254 |
| Leu-265 | Thr-270 | Ser-265 |
a Dash means a gap is present at this position in the sequence alignment.
Docking of 1,2-naphthoquinone shows that the substrate fits well into the PIG3 putative substrate-binding pocket, with the ortho-carbonyl oxygen atoms in the appropriate orientations to interact with the Tyr-51 hydroxyl group and with the C-4 atom of NADP+ (Fig. 5A). In contrast, in the para-naphthoquinones the carbonyl oxygen atoms do not present the proper orientation, which would result in unfavorable binding. Similarly the bulkier 9,10-phenanthrenequinone could not be fitted into the PIG3 pocket, in particular due to the presence of Glu-123, in agreement with the lack of activity toward this substrate.
Site-directed Mutagenesis of the Active Site Tyr
In several Zn2+-independent MDRs, evidence supports a Tyr residue acting as an acid/base catalyst (39, 45–47). The crystallographic structures suggest that Tyr-51 in PIG3 and Tyr-59 in ζ-crystallin are candidates to perform this catalytic function. To evaluate the hypothesis, mutant variants were prepared, and kinetic experiments were conducted (Table 4). In both proteins the change of Tyr to Phe did not reduce activity, but in contrast, a significant increase of kcat/Km was observed. This conclusively demonstrates that the active site Tyr is not the catalytic acid/base of these two QORs. The increase in catalytic efficiency of the Tyr/Phe mutants indicates that, instead, the hydrophobic change is favorable to catalysis. The substitution of Tyr to Ala does not eliminate the activity either, although the change decreases the binding of PIG3 with 1,2-naphthoquinone, which could not saturate the enzyme. The ζ-crystallin Y59A mutant displays similar kinetic constants as those of the wild-type enzyme.
In Vitro PIG3 Activity and in Vivo PIG3 Overexpression Generate Reactive Oxygen Species
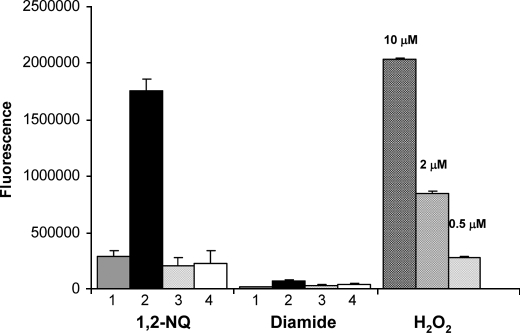
The activity toward quinones catalyzed by ζ-crystallin leads to the production of superoxide anion and hydrogen peroxide (11). Here we analyze ROS generation as a consequence of PIG3 activity (Fig. 6). Production of hydrogen peroxide is already observed in the 1,2-naphthoquinone/NADPH mixture, which is consistent with a significant background reaction in the absence of enzyme, demonstrating a chemical rate of electron transfer between NADPH and the quinone. However, ROS production is much higher in the presence of the enzyme. It can be concluded that the reduction of quinones catalyzed by PIG3 generates ROS, suggesting a similar reaction mechanism as found with ζ-crystallin. In contrast, PIG3 reduction of diamide does not yield ROS.
FIGURE 6.
Formation of hydrogen peroxide in the reaction catalyzed by human PIG3. Aliquots from the enzymatic reaction mixture containing 50 μm 1,2-naphthoquinone (1,2-NQ) or 100 μm diamide with 150 μm NADPH and 28 μg of PIG3 or 28 μg of PIG3 S151V were added to Amplex Red reagent, and fluorescence was measured. 1, no enzyme added; 2, wild-type PIG3; 3, no cofactor added; 4, PIG3 S151V mutant. Positive control using different hydrogen peroxide concentrations is indicated.
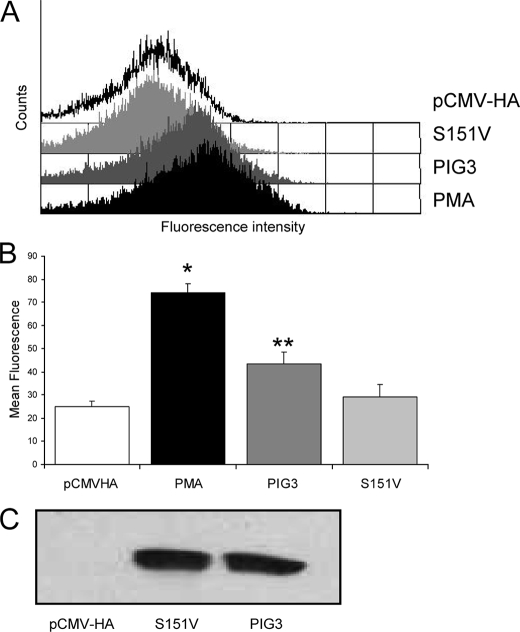
We have also investigated whether the overexpression of active PIG3 in eukaryotic cells (human colon carcinoma HCT-116 cells) influences the production of ROS in vivo (Fig. 7). As a control we prepared an inactive variant that contained the S151V mutation in the coenzyme-binding site. The PIG3 mutant was unable to bind coenzyme (Fig. 2) and was devoid of enzymatic activity. Otherwise, the mutant exhibited a normal expression and purification behavior, and size-exclusion chromatography demonstrated a dimeric structure (Mr = 71,000), indicating a stable native fold. HCT-116 cells were transfected with the corresponding vectors, and intracellular content of hydrogen peroxide was evaluated by a fluorescence method (Fig. 7). The mean fluorescence of the cells increased 24 h after transient transfection with the PIG3/pCMV-HA vector. The fluorescence was significantly higher than that of the cells transfected with the inactive mutant, which did not show differences toward mock transfection experiments. As expected, the inactive mutant did not generate ROS in vitro either (Fig. 6).
FIGURE 7.
Overexpression of PIG3-stimulated ROS production in living cells. HCT-116 cells were transfected with PIG3/pCMV-HA, PIG3 S151V/pCMV-HA, or empty pCMV-HA, treated with DCFH-DA, and analyzed by fluorescence-activated cell sorting, using phorbol 12-myristate 13-acetate (PMA) as a positive control of ROS production. A, cell counts versus relative fluorescence in transfected and phorbol 12-myristate 13-acetate-treated cells. B, mean fluorescence ± S.D. from six independent experiments plotted in a bar diagram. For PMA, single asterisk indicates p value of <0.05 versus pCMV-HA and versus PIG3 S151V, and for PIG3, two asterisks indicate p value of <0.05 versus pCMV-HA and versus PIG3 S151V. C, transfections were checked by Western blot analysis, using an anti-PIG3 polyclonal antibody.
Interaction of PIG3 with RNA
Using the electrophoretic mobility shift assay technique, we investigated whether PIG3 binds AU-rich elements in RNA. In contrast to ζ-crystallins, which exhibit significant interaction (17), PIG3 does not show any specific binding (supplemental data).
DISCUSSION
We here demonstrate that PIG3, found in most vertebrates, plants, protists, and bacteria, can reduce quinones. Kinetic constants could be determined for 1,2-naphthoquinone, the first substrate ever found for PIG3. Much lower activity is detected with the para isomer, which agrees with the specificity for ortho-quinones reported for other QORs (17) (Table 2). Interestingly, a related compound, 9,10-phenanthrenequinone, is inactive with PIG3, but it is the best substrate for the human and yeast ζ-crystallins, indicating a clear distinct specificity for the PIG3 subfamily.
In addition to its quinone reductase activity, PIG3 is also able to hydrogenate the N N double bond in diamide, or the N
N double bond in diamide, or the N C double bond in DCIP. The diamide reductase activity seems quite specific for PIG3, because mammalian ζ-crystallins do not use this compound as substrate. Remarkably, all substrates found for both PIG3 and ζ-crystallins exhibit nonenzymatic reduction by NADPH (and NADH). For PIG3, this background reaction may contribute up to 50% of the total rate measured after the addition of the enzyme. Whereas NADPH and NADH account for similar noncatalytic rates, the enzymatic reduction is exclusively NADPH-dependent.
C double bond in DCIP. The diamide reductase activity seems quite specific for PIG3, because mammalian ζ-crystallins do not use this compound as substrate. Remarkably, all substrates found for both PIG3 and ζ-crystallins exhibit nonenzymatic reduction by NADPH (and NADH). For PIG3, this background reaction may contribute up to 50% of the total rate measured after the addition of the enzyme. Whereas NADPH and NADH account for similar noncatalytic rates, the enzymatic reduction is exclusively NADPH-dependent.
The overall structure of human PIG3 exhibits a dimeric organization, where subunits present the expected MDR fold (Fig. 3). However, PIG3 differs from the best known MDR members by the absence of catalytic and structural Zn2+ and by lacking a large Zn2+-binding loop. These features, which are shared with other QOR enzymes (48, 49), suggested that PIG3 could act as a reductase using NADPH as a cofactor, a general property of non-Zn2+ MDRs. NADPH binding and reductase activity have been demonstrated in this study by fluorescence quenching and kinetic experiments, and fully supported by the cofactor binding site structure, which displays the key NADP-interacting residues (Gly-173, Lys-177, and Tyr-192) conserved in all PIG3 sequences.
The sequence motif (A/G)XXSXXG, which replaces the classical dinucleotide-binding motif GXGXX(G/A) found in MDR alcohol dehydrogenases, is also fully conserved. The importance of the motif in cofactor binding is corroborated by the fact that the PIG3 S151V mutant binds neither NADPH nor NADH (Fig. 2). This was predicted because Ser-151 (or the residue at the homologous position) has a key role in the motif conformation (50), and the replacement by a Val may also result in steric hindrance with the cofactor (Fig. 4). Residues lining the substrate pocket are highly conserved among PIG3 sequences (Fig. 1). Some of these residues are also conserved in ζ-crystallin and E. coli QOR. However, nonconservative substitutions, especially that of Glu-123 which changes to Ile and Leu in ζ-crystallin and E. coli QOR, respectively (Table 5), indicate critical differences in substrate specificity. Glu-123, a residue located close to the substrate and the cofactor (Fig. 5), is found uniquely in all PIG3 members and thus it may be used as a sequence signature to identify the subfamily. The PIG3 substrate-binding pocket is smaller than that of ζ-crystallin (PDB code 1YB5), which is consistent with the PIG3 specificity toward small quinones. The absence of the catalytic Zn2+, and the substitution of the catalytic residue Ser/Thr-48, found in most MDR alcohol dehydrogenases, by Ala-42 in PIG3 and Val-50 in ζ-crystallin, indicate a very different catalytic mechanism in QORs. In PIG3, the phenol oxygen from Tyr-51 sits in the vicinity of the reactive C-4 atom of the nicotinamide (Fig. 5). An equivalent Tyr residue is also present in most ζ-crystallins and in E. coli QOR. In contrast, in bovine ζ-crystallin, which shows minimal QOR activity, the corresponding residue is a His, suggesting that the Tyr is essential in the kinetic mechanism (51). In addition, mutagenesis and/or structural data support the participation of a Tyr in the mechanism of the related, Zn2+-lacking MDRs (Table 2) enoyl reductase (39, 47), Arabidopsis NADP-dependent oxidoreductase P1, and leukotriene B4 12-hydroxydehydrogenase (45, 46). Thus, it was appropriate to propose a catalytic role for PIG3 Tyr-51, the only Tyr residue in the active-site cavity, further supported by a docking model (Fig. 5). Remarkably, the PIG3 and ζ-crystallin mutants at the active site Tyr retained their enzymatic activity (Table 4). Therefore, Tyr-51 in PIG3 and Tyr-59 in ζ-crystallin do not appear to be essential for catalysis.
Other oxidoreductases using quinones may help to understand the kinetic mechanism of PIG3 and ζ-crystallin. A Tyr is also the proposed catalytic residue in enzymes that reduce quinones from other protein families: aldo-keto reductases (AKR), NADPH:quinone reductases (NQO1 or DT diaphorase), and short chain dehydrogenases/reductases (52). Several examples exist in which the Tyr/Phe exchange, or the absence of the Tyr, does not considerably affect the quinone reducing activity: NQO1 (53), a short chain dehydrogenases/reductase quinone reductase from E. coli (54), and the AKRs ρ-crystallin (55) and AKR1C9 (12). Interestingly, in AKR1C9, the exchange eliminates the 3-ketosteroid reductase activity but not the quinone reductase activity (12). In conclusion, the active site Tyr does not seem to be essential for catalysis with quinones. As indicated by Penning and co-workers (12), quinone reduction exhibits lower activation energy than other substrates to reach the transition state, which is supported by the high nonenzymatic reduction of the quinones in the presence of cofactor. Thus, it appears that many quinone reductases provide a scaffold to bring quinone and cofactor molecules in close proximity and with the right orientation, sufficient for catalysis, without the participation of a catalytic residue. In contrast, in enzymes with wide substrate specificity, such as AKRs, the Tyr would be necessary for catalysis of non-quinone compounds.
Regarding QORs, two possibilities could be proposed for their enzymatic function: 1) the substrates are endogenous and/or xenobiotic quinones, and the activity is through propinquity effects; or 2) the physiological substrates are as yet unidentified compounds that need a catalytic residue, probably a Tyr, whereas the activity with quinones has no functional relevance. Both possibilities can apply to the ζ-crystallin and PIG3 subfamilies. However, despite their common quinone reductase activity, clear functional differences exist between the two groups. ζ-Crystallin exhibits high activity, especially toward larger and bulkier substrates as compared with PIG3. Furthermore, ζ-crystallin has a demonstrated structural role in the eye lens of some mammals and may have additional functions through RNA binding (17). In contrast, PIG3 catalyzes the reduction of smaller substrates and has lower activity; it does not have a known structural function, and it does not bind RNA.
Strong evidence provided by this study and other studies (56, 57) suggests that PIG3 apoptotic action is through, or results in, oxidative stress. In this regard we have demonstrated that in vitro PIG3 activity generates ROS, and that PIG3 overexpression in cells leads to ROS generation, which is dependent on active enzyme or at least on protein able to bind cofactor. A PIG3 function, independent of its enzymatic activity, cannot be ruled out, but the strict conservation of the main active-site residues strongly supports a catalytic role. We have demonstrated two types of activity for PIG3, the reduction of quinones and the hydrogenation of an N N double bond such as in diamide. However, only the first type produces ROS. In conclusion, data suggest that PIG3 action on ROS generation and, probably, on apoptosis is through its enzymatic activity with an endogenous substrate of the quinone type.
N double bond such as in diamide. However, only the first type produces ROS. In conclusion, data suggest that PIG3 action on ROS generation and, probably, on apoptosis is through its enzymatic activity with an endogenous substrate of the quinone type.
Unbalanced apoptotic response mediated by PIG3 is an underlying principle behind many pathological situations. Thus, insufficient apoptosis may result in cancer or autoimmune disease, whereas accelerated cell death is evident in degenerative diseases, immunodeficiency, and infertility (58). Here we provide invaluable structural and functional information that will allow to design and test PIG3 inhibitor drugs for control of cell death.
Supplementary Material
Acknowledgments
We thank Dr. A. R. de Lera, Dr. R. Alvarez, and P. García (Universidade de Vigo, Spain) for the synthesis of 4-hydroxyestrone-o-quinone. We acknowledge Dr. Isidre Casals (Unitat de Tècniques Separatives, Serveis Cientificotècnics, Universitat de Barcelona, Spain) for mass spectrometry analysis. We thank Dr. Miguel Angel Peinado (Institut d'Investigació Biomèdica de Bellvitge, L'Hospitalet, Spain) for providing the HCT-116 cell line.
This work was supported by Spanish Dirección General de Investigación Grants BFU2005-02621, BFU2008-02945/BMC, and BFU2005-08686-C02-01 and the Generalitat de Catalunya Grant 2005 SGR 00112. The Structural Genomics Consortium is a registered charity (number 1097737) and receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada, through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck, the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” “Results,” Fig. S1, Table S1, and additional Ref. 1.
The atomic coordinates and structure factors (codes 2J8Z and 2OBY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- QOR
- quinone oxidoreductase
- ROS
- reactive oxygen species
- DCFH-DA
- 2′,7′-dichlorodihydrofluorescein diacetate
- PBS
- phosphate-buffered saline
- PDB
- Protein Data Bank
- LIC
- ligation independent cloning
- MDR
- medium chain dehydrogenases/reductase
- AKR
- aldo-keto reductase
- DCIP
- 2,6-dichloroindophenol.
REFERENCES
- 1.Levine A. J., Momand J., Finlay C. A. ( 1991) Nature 351, 453– 456 [DOI] [PubMed] [Google Scholar]
- 2.Polyak K., Xia Y., Zweier J. L., Kinzler K. W., Vogelstein B. ( 1997) Nature 389, 300– 305 [DOI] [PubMed] [Google Scholar]
- 3.Bergamaschi D., Samuels Y., Jin B., Duraisingham S., Crook T., Lu X. ( 2004) Mol. Cell. Biol. 24, 1341– 1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moll U. M., Slade N. ( 2004) Mol. Cancer Res. 2, 371– 386 [PubMed] [Google Scholar]
- 5.Zhang Q., Zhao X. H., Wang Z. J. ( 2008) Food Chem. Toxicol. 46, 2042– 2053 [DOI] [PubMed] [Google Scholar]
- 6.Contente A., Dittmer A., Koch M. C., Roth J., Dobbelstein M. ( 2002) Nat. Genet. 30, 315– 320 [DOI] [PubMed] [Google Scholar]
- 7.Ito M., Nishiyama H., Watanabe J., Kawanishi H., Takahashi T., Kamoto T., Habuchi T., Ogawa O. ( 2006) Jpn. J. Clin. Oncol. 36, 116– 120 [DOI] [PubMed] [Google Scholar]
- 8.Nomdedéu J. F., Perea G., Estivill C., Badell I., Lasa A., Aventín A. ( 2008) Leuk. Res. 32, 186– 188 [DOI] [PubMed] [Google Scholar]
- 9.Campomenosi P., Monti P., Aprile A., Abbondandolo A., Frebourg T., Gold B., Crook T., Inga A., Resnick M. A., Iggo R., Fronza G. ( 2001) Oncogene 20, 3573– 3579 [DOI] [PubMed] [Google Scholar]
- 10.Tanaka T., Ohkubo S., Tatsuno I., Prives C. ( 2007) Cell 130, 638– 650 [DOI] [PubMed] [Google Scholar]
- 11.Rao P. V., Krishna C. M., Zigler J. S., Jr. ( 1992) J. Biol. Chem. 267, 96– 102 [PubMed] [Google Scholar]
- 12.Schlegel B. P., Ratnam K., Penning T. M. ( 1998) Biochemistry 37, 11003– 11011 [DOI] [PubMed] [Google Scholar]
- 13.Oppermann U. ( 2007) Annu. Rev. Pharmacol. Toxicol. 47, 293– 322 [DOI] [PubMed] [Google Scholar]
- 14.Liang X. Q., Cao E. H., Zhang Y., Qin J. F. ( 2004) FEBS Lett. 569, 94– 98 [DOI] [PubMed] [Google Scholar]
- 15.Johnson T. M., Yu Z. X., Ferrans V. J., Lowenstein R. A., Finkel T. ( 1996) Proc. Natl. Acad. Sci. U. S. A. 93, 11848– 11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flatt P. M., Polyak K., Tang L. J., Scatena C. D., Westfall M. D., Rubinstein L. A., Yu J., Kinzler K. W., Vogelstein B., Hill D. E., Pietenpol J. A. ( 2000) Cancer Lett. 156, 63– 72 [DOI] [PubMed] [Google Scholar]
- 17.Fernández M. R., Porté S., Crosas E., Barberà N., Farrés J., Biosca J. A., Parés X. ( 2007) Cell. Mol. Life Sci. 64, 1419– 1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama A., Kumagai Y., Morikawa K., Taguchi K., Hayashi H., Ohta T. ( 2003) Microbiology 149, 389– 398 [DOI] [PubMed] [Google Scholar]
- 19.Huang Q. L., Russell P., Stone S. H., Zigler J. S., Jr. ( 1987) Curr. Eye Res. 6, 725– 732 [DOI] [PubMed] [Google Scholar]
- 20.Tang A., Curthoys N. P. ( 2001) J. Biol. Chem. 276, 21375– 21380 [DOI] [PubMed] [Google Scholar]
- 21.Kranthi B. V., Balasubramanian N., Rangarajan P. N. ( 2006) Nucleic Acids Res. 34, 4060– 4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibrahim H., Lee Y. J., Curthoys N. P. ( 2008) Kidney Int. 73, 11– 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. ( 1997) Nucleic Acids Res. 25, 3389– 3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall T. A. ( 1999) Nucleic Acids Symp. Ser. 41, 95– 98 [Google Scholar]
- 25.Thompson J. D., Higgins D. G., Gibson T. J. ( 1994) Nucleic Acids Res. 22, 4673– 4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford M. M. ( 1976) Anal. Biochem. 72, 248– 254 [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z., Minor W. ( 1997) Methods Enzymol. 276, 307– 326 [DOI] [PubMed] [Google Scholar]
- 28.Vagin A., Teplyakov A. ( 2000) Acta Crystallogr. Sect. D Biol. Crystallogr. 56, 1622– 1624 [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project ( 1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50, 760– 76315299374 [Google Scholar]
- 30.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. ( 1991) Acta Crystallogr. Sect. A 47, 110– 119 [DOI] [PubMed] [Google Scholar]
- 31.Stinson R. A., Holbrook J. J. ( 1973) Biochem. J. 131, 719– 728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valencia E., Larroy C., Ochoa W. F., Parés X., Fita I., Biosca J. A. ( 2004) J. Mol. Biol. 341, 1049– 1062 [DOI] [PubMed] [Google Scholar]
- 33.Gallego O., Belyaeva O. V., Porté S., Ruiz F. X., Stetsenko A. V., Shabrova E. V., Kostereva N. V., Farrés J., Parés X., Kedishvili N. Y. ( 2006) Biochem. J. 399, 101– 109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Royall J. A., Ischiropoulos H. ( 1993) Arch. Biochem. Biophys. 302, 348– 355 [DOI] [PubMed] [Google Scholar]
- 35.Delia D., Aiello A., Meroni L., Nicolini M., Reed J. C., Pierotti M. A. ( 1997) Carcinogenesis 18, 943– 948 [DOI] [PubMed] [Google Scholar]
- 36.Contente A., Zischler H., Einspanier A., Dobbelstein M. ( 2003) Cancer Res. 63, 1756– 1758 [PubMed] [Google Scholar]
- 37.Hiromi K., Kuwamoto C., Ohnishi M. ( 1980) Anal. Biochem. 101, 421– 426 [DOI] [PubMed] [Google Scholar]
- 38.Wallace A. C., Laskowski R. A., Thornton J. M. ( 1995) Protein Eng. 8, 127– 134 [DOI] [PubMed] [Google Scholar]
- 39.Chen Z. J., Pudas R., Sharma S., Smart O. S., Juffer A. H., Hiltunen J. K., Wierenga R. K., Haapalainen A. M. ( 2008) J. Mol. Biol. 379, 830– 844 [DOI] [PubMed] [Google Scholar]
- 40.Rossmann M. G., Liljas A., Brändén C. I., Banaszak L. J. ( 1975) in The Enzymes ( Boyer P. D. ed) Vol. XI, pp. 61– 102, Academic Press, New York [Google Scholar]
- 41.Edwards K. J., Barton J. D., Rossjohn J., Thorn J. M., Taylor G. L., Ollis D. L. ( 1996) Arch. Biochem. Biophys. 328, 173– 183 [DOI] [PubMed] [Google Scholar]
- 42.Baker P. J., Britton K. L., Rice D. W., Rob A., Stillman T. J. ( 1992) J. Mol. Biol. 228, 662– 671 [DOI] [PubMed] [Google Scholar]
- 43.Rosell A., Valencia E., Parés X., Fita I., Farrés J., Ochoa W. F. ( 2003) J. Mol. Biol. 330, 75– 85 [DOI] [PubMed] [Google Scholar]
- 44.Carugo O., Argos P. ( 1997) Proteins 28, 10– 28 [DOI] [PubMed] [Google Scholar]
- 45.Hori T., Yokomizo T., Ago H., Sugahara M., Ueno G., Yamamoto M., Kumasaka T., Shimizu T., Miyano M. ( 2004) J. Biol. Chem. 279, 22615– 22623 [DOI] [PubMed] [Google Scholar]
- 46.Youn B., Kim S. J., Moinuddin S. G., Lee C., Bedgar D. L., Harper A. R., Davin L. B., Lewis N. G., Kang C. ( 2006) J. Biol. Chem. 281, 40076– 40088 [DOI] [PubMed] [Google Scholar]
- 47.Airenne T. T., Torkko J. M., Van den Plas S., Sormunen R. T., Kastaniotis A. J., Wierenga R. K., Hiltunen J. K. ( 2003) J. Mol. Biol. 327, 47– 59 [DOI] [PubMed] [Google Scholar]
- 48.Jörnvall H. ( 2008) Cell. Mol. Life Sci. 65, 3873– 3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riveros-Rosas H., Julián-Sánchez A., Villalobos-Molina R., Pardo J. P., Piña E. ( 2003) Eur. J. Biochem. 270, 3309– 3334 [DOI] [PubMed] [Google Scholar]
- 50.Yokomizo T., Ogawa Y., Uozumi N., Kume K., Izumi T., Shimizu T. ( 1996) J. Biol. Chem. 271, 2844– 2850 [DOI] [PubMed] [Google Scholar]
- 51.Rao P. V., Gonzalez P., Persson B., Jörnvall H., Garland D., Zigler J. S., Jr. ( 1997) Biochemistry 36, 5353– 5362 [DOI] [PubMed] [Google Scholar]
- 52.Kavanagh K. L., Jörnvall H., Persson B., Oppermann U. ( 2008) Cell. Mol. Life Sci. 65, 3895– 3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma Q., Cui K., Xiao F., Lu A. Y., Yang C. S. ( 1992) J. Biol. Chem. 267, 22298– 22304 [PubMed] [Google Scholar]
- 54.Kim I. K., Yim H. S., Kim M. K., Kim D. W., Kim Y. M., Cha S. S., Kang S. O. ( 2008) J. Mol. Biol. 379, 372– 384 [DOI] [PubMed] [Google Scholar]
- 55.Fujii Y., Watanabe K., Hayashi H., Urade Y., Kuramitsu S., Kagamiyama H., Hayaishi O. ( 1990) J. Biol. Chem. 265, 9914– 9923 [PubMed] [Google Scholar]
- 56.Sablina A. A., Budanov A. V., Ilyinskaya G. V., Agapova L. S., Kravchenko J. E., Chumakov P. M. ( 2005) Nat. Med. 11, 1306– 1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naumann U., Huang H., Wolburg H., Wischhusen J., Weit S., Ohgaki H., Weller M. ( 2006) Cancer Gene Ther. 13, 469– 478 [DOI] [PubMed] [Google Scholar]
- 58.MacFarlane M., Williams A. C. ( 2004) EMBO Rep. 5, 674– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokomizo T., Izumi T., Takahashi T., Kasama T., Kobayashi Y., Sato F., Taketani Y., Shimizu T. ( 1993) J. Biol. Chem. 268, 18128– 18135 [PubMed] [Google Scholar]
- 60.Mano J., Babiychuk E., Belles-Boix E., Hiratake J., Kimura A., Inzé D., Kushnir S., Asada K. ( 2000) Eur. J. Biochem. 267, 3661– 3671 [DOI] [PubMed] [Google Scholar]
- 61.Mano J., Torii Y., Hayashi S., Takimoto K., Matsui K., Nakamura K., Inzé D., Babiychuk E., Kushnir S., Asada K. ( 2002) Plant Cell Physiol. 43, 1445– 1455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.