Abstract
Human concentrative nucleoside transporter 3 (hCNT3) utilizes electrochemical gradients of both Na+ and H+ to accumulate pyrimidine and purine nucleosides within cells. We have employed radioisotope flux and electrophysiological techniques in combination with site-directed mutagenesis and heterologous expression in Xenopus oocytes to identify two conserved pore-lining glutamate residues (Glu-343 and Glu-519) with essential roles in hCNT3 Na+/nucleoside and H+/nucleoside cotransport. Mutation of Glu-343 and Glu-519 to aspartate, glutamine, and cysteine severely compromised hCNT3 transport function, and changes included altered nucleoside and cation activation kinetics (all mutants), loss or impairment of H+ dependence (all mutants), shift in Na+:nucleoside stoichiometry from 2:1 to 1:1 (E519C), complete loss of catalytic activity (E519Q) and, similar to the corresponding mutant in Na+-specific hCNT1, uncoupled Na+ currents (E343Q). Consistent with close-proximity integration of cation/solute-binding sites within a common cation/permeant translocation pore, mutation of Glu-343 and Glu-519 also altered hCNT3 nucleoside transport selectivity. Both residues were accessible to the external medium and inhibited by p-chloromercuribenzene sulfonate when converted to cysteine.
Physiologic nucleosides and the majority of synthetic nucleoside analogs with antineoplastic and/or antiviral activity are hydrophilic molecules that require specialized plasma membrane nucleoside transporter (NT)3 proteins for transport into or out of cells (1–4). NT-mediated transport is required for nucleoside metabolism by salvage pathways and is a critical determinant of the pharmacologic actions of nucleoside drugs (3–6). By regulating adenosine availability to purinoreceptors, NTs also modulate a diverse array of physiological processes, including neurotransmission, immune responses, platelet aggregation, renal function, and coronary vasodilation (4, 6, 7). Two structurally unrelated NT families of integral membrane proteins exist in human and other mammalian cells and tissues as follows: the SLC28 concentrative nucleoside transporter (CNT) family and the SLC29 equilibrative nucleoside transporter (ENT) family (3, 4, 6, 8, 9). ENTs are normally present in most, possibly all, cell types (4, 6, 8). CNTs, in contrast, are found predominantly in intestinal and renal epithelia and other specialized cell types, where they have important roles in absorption, secretion, distribution, and elimination of nucleosides and nucleoside drugs (1–3, 5, 6, 9).
The CNT protein family in humans is represented by three members, hCNT1, hCNT2, and hCNT3. Belonging to a CNT subfamily phylogenetically distinct from hCNT1/2, hCNT3 utilizes electrochemical gradients of both Na+ and H+ to accumulate a broad range of pyrimidine and purine nucleosides and nucleoside drugs within cells (10, 11). hCNT1 and hCNT2, in contrast, are Na+-specific and transport pyrimidine and purine nucleosides, respectively (11–13). Together, hCNT1–3 account for the three major concentrative nucleoside transport processes of human and other mammalian cells. Nonmammalian members of the CNT protein family that have been characterized functionally include hfCNT, a second member of the CNT3 subfamily from the ancient marine prevertebrate the Pacific hagfish Eptatretus stouti (14), CeCNT3 from Caenorhabditis elegans (15), CaCNT from Candida albicans (16), and the bacterial nucleoside transporter NupC from Escherichia coli (17). hfCNT is Na+- but not H+-coupled, whereas CeCNT3, CaCNT, and NupC are exclusively H+-coupled. Na+:nucleoside coupling stoichiometries are 1:1 for hCNT1 and hCNT2 and 2:1 for hCNT3 and hfCNT3 (11, 14). H+:nucleoside coupling ratios for hCNT3 and CaCNT are 1:1 (11, 16).
Although much progress has been made in molecular studies of ENT proteins (4, 6, 8), studies of structurally and functionally important regions and residues within the CNT protein family are still at an early stage. Topological investigations suggest that hCNT1–3 and other eukaryote CNT family members have a 13 (or possibly 15)-transmembrane helix (TM) architecture, and multiple alignments reveal strong sequence similarities within the C-terminal half of the proteins (18). Prokaryotic CNTs lack the first three TMs of their eukaryotic counterparts, and functional expression of N-terminally truncated human and rat CNT1 in Xenopus oocytes has established that these three TMs are not required for Na+-dependent uridine transport activity (18). Consistent with this finding, chimeric studies involving hCNT1 and hfCNT (14) and hCNT1 and hCNT3 (19) have demonstrated that residues involved in Na+- and H+-coupling reside in the C-terminal half of the protein. Present in this region of the transporter, but of unknown function, is a highly conserved (G/A)XKX3NEFVA(Y/M/F) motif common to all eukaryote and prokaryote CNTs.
By virtue of their negative charge and consequent ability to interact directly with coupling cations and/or participate in cation-induced and other protein conformational transitions, glutamate and aspartate residues play key functional and structural roles in a broad spectrum of mammalian and bacterial cation-coupled transporters (20–30). Little, however, is known about their role in CNTs. This study builds upon a recent mutagenesis study of conserved glutamate and aspartate residues in hCNT1 (31) to undertake a parallel in depth investigation of corresponding residues in hCNT3. By employing the multifunctional capability of hCNT3 as a template for these studies, this study provides novel mechanistic insights into the molecular mechanism(s) of CNT-mediated cation/nucleoside cotransport, including the role of the (G/A)XKX3NEFVA(Y/M/F) motif.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis of hCNT3 and Expression in Xenopus Oocytes
hCNT3 mutants were constructed using the QuikChange® site-directed mutagenesis kit (Stratagene) with hCNT3 cDNA (GenBankTM accession number AF305210) in the pGEM-HE vector (32) as the template for mutant construction. Introduction of the correct mutation was confirmed by Taq DyeDeoxyTM terminator cycle sequencing of the constructs in both directions. Plasmid DNA was linearized with NheI and transcribed with T7 polymerase using the mMESSAGE mMACHINETM (Ambion) transcription system. Defolliculated stage VI Xenopus oocytes were microinjected with 20 nl of water or 20 nl of water containing capped RNA transcript (20 ng) and incubated in modified Barth's medium (changed daily) at 18 °C for either 4 days (radioisotope flux studies) or 4–7 days (electrophysiology experiments) prior to the assay of transport activity (33).
Radioisotope Flux Assays
Transport assays were performed as described previously (11, 31) on groups of 12 oocytes at room temperature (20 °C) using 14C- or 3H-labeled nucleosides (GE Healthcare) (1 or 2–4 μCi/ml, respectively) in 200 μl of transport medium containing either 100 mm NaCl or 100 mm choline chloride (ChCl) and 2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, and 10 mm HEPES, pH ≥ 7.0, or 10 mm MES, pH < 7.0. In experiments examining the H+ dependence of transport, 100 mm NaCl was replaced with 100 mm choline chloride (ChCl). ChCl also substituted for NaCl in Na+-activation experiments. Experiments replacing NaCl with equimolar ChCl included 10-min preincubation periods and several washes with ChCl-containing transport medium prior to addition of radiolabeled nucleoside to ensure complete removal of extracellular Na+. Except where otherwise indicated, nucleoside uptake was determined at a concentration of 20 μm using an incubation period of 1 or 3 min, depending on the catalytic activity of the mutant, to determine initial rates of transport. At the end of the incubation period, extracellular label was removed by six rapid washes in ice-cold 100 mm ChCl transport medium, pH 7.5, and individual oocytes were dissolved in 1% (w/v) SDS for quantitation of cell-associated radioactivity by liquid scintillation counting (LS 6000 IC, Beckman). Also in a volume of 200 μl, treatment of oocytes with p-chloromercuribenzene sulfonate (PCMBS) was performed on ice for 10 min. Excess organomercurial was removed by three washes with ice-cold transport medium before the assay of transport activity. In protection experiments, unlabeled uridine (20 mm) was included along with PCMBS (31, 34). Flux values represent mediated transport, corrected for basal uridine uptake measured in control water-injected oocytes, and are the means ± S.E. of 10–12 oocytes. Kinetic parameters (Km, K50, Vmax, and Hill coefficient) (±S.E.) were calculated using SigmaPlot software (Jandel Scientific Software). Each experiment was repeated at least twice on oocytes from different frogs.
Electrophysiology Current Measurements
Membrane currents were measured at room temperature (20 °C) using the whole-cell, two-electrode voltage clamp technique (GeneClamp 500B, Molecular Devices Corp.), as described previously (19, 35). The GeneClamp 500B was interfaced to an IBM-compatible PC via a Digidata 1322A A/D converter and controlled by pCLAMP software (version 9.0, Molecular Devices Corp.). The microelectrodes were filled with 3 m KCl and had resistances ranging from 0.5 to 1.5 megohms. Following microelectrode penetration, resting membrane potential was measured over a 10-min period prior to the start of the experiment. Oocytes exhibiting an unstable membrane potential or a potential more positive than −30 mV were discarded. Individual oocytes with good resting membrane potentials were clamped at a holding voltage (Vh) of −90 mV, and current measurements were sampled in transport media of the same composition used in the radioisotope transport assays. During the course of data collection, the transport medium perfusing the oocyte was changed to one containing uridine for ∼60 s, and then immediately washed and exchanged with fresh medium lacking the nucleoside permeant. Current signals were filtered at 2 kHz (four-pole Bessel filter) at a sampling interval of 50 ms. For data presentation, the signals were further filtered at 0.5 Hz by use of pCLAMP 9.0 software (Molecular Devices Corp.).
Data from individual electrophysiological experiments are presented as nucleoside-evoked currents from single representative cells or as mean values (±S.E.) from four or more oocytes from the same batch of oocytes used on the same day. Each experiment was repeated at least twice on oocytes from different frogs. No nucleoside-evoked currents were detected in oocytes injected with water alone, demonstrating that currents in transcript-injected oocytes were transporter-specific.
Current-voltage (I-V) curves were determined from differences in steady-state currents generated in the presence and absence of uridine during 300-ms voltage pulses to potentials between −110 and +60 mV (10 mV steps). Oocytes were voltage-clamped at −50 mV and exposed to 100 μm uridine.
Charge-to-Nucleoside Stoichiometry
Na+:nucleoside and H+:nucleoside coupling stoichiometries were determined by simultaneously measuring currents and [3H]uridine (200 μm; 2 μCi/ml) uptake under voltage clamp conditions, as described above. Experiments measuring the Na+:nucleoside and H+:nucleoside coupling ratios used media containing 100 mm NaCl, pH 8.5, and 100 mm ChCl, pH 5.5, respectively. Individual oocytes in nucleoside-free medium were voltage-clamped at Vh of −90 mV, and the base-line current was monitored for 5 min. When the base line was stable, perfusion was stopped, and medium of the same composition containing radiolabeled nucleoside was manually added to the perfusion chamber. Current was measured for 2 min, and nucleoside uptake was terminated by rapidly washing the oocyte with nucleoside-free medium until the current returned to base line. The oocyte was then transferred to a scintillation vial and solubilized with 1% (w/v) SDS for quantitation of oocyte-associated radioactivity. The nucleoside-induced current was obtained as the difference between base-line current and the inward nucleoside-induced current. The total charge translocated into the oocyte during the uptake period was calculated from the current-time integral and correlated with the measured radiolabeled flux for each oocyte to determine the charge:flux ratio. Basal [3H]uridine uptake was determined in control water-injected oocytes (from the same donor frog) under equivalent conditions and used to correct for endogenous nonmediated radiolabeled nucleoside uptake over the same incubation period. Coupling ratios (±S.E.) were calculated from slopes of least squares fits of nucleoside-dependent charge versus nucleoside accumulation for five or more individual oocytes. Uridine itself carries no charge over the pH range used in these and other electrophysiological experiments, pH 5.5–8.8.
Immunoblotting
Production of recombinant wild-type hCNT3 and hCNT3 Glu-343 and Glu-519 mutant proteins at the oocyte cell surface was determined by labeling intact cells with EZ-Link sulfosuccinimidyl-6-(biotinamido) hexanoate (Pierce) followed by isolation of the resultant biotinylated plasma membrane proteins using immobilized streptavidin resin (Pierce) according to the manufacturer's instructions. Protein was determined by the bicinchoninic acid protein assay (Pierce) using bovine serum albumin as standard. For immunoblotting, solubilized proteins from two oocytes/lane were resolved on 12% SDS-polyacrylamide gels (36). The electrophoresed proteins were transferred to polyvinylidene difluoride membranes and probed with affinity-purified anti-hCNT3-(45–69) polyclonal antibodies (37). Blots were then incubated with horseradish peroxidase-conjugated anti-rabbit antibodies (GE Healthcare) and developed with enhanced chemiluminescence reagents (GE Healthcare). Staining intensities were quantified using ImageJ software from the National Institutes of Health.
RESULTS
Residues Identified for Mutagenesis
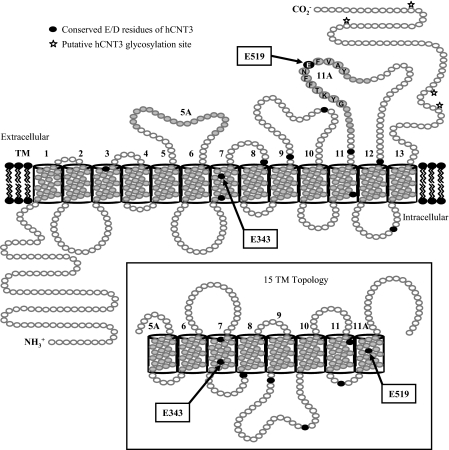
In this study, we employed site-directed mutagenesis and heterologous expression in Xenopus oocytes to analyze the roles of acidic amino acid residues in hCNT3-mediated cation/nucleoside cotransport. The locations of the residues selected for study are shown in Fig. 1.
FIGURE 1.
Putative hCNT3 topology model. Schematic of proposed hCNT3 (GenBankTM accession number AF305210) membrane architecture with 13 ± 2 TMs, as predicted by bioinformatic analyses of currently identified CNT family members. The location of a conserved (G/A)XKX3NEFVA(Y/M/F) motif in TM 11A is indicated. Insertion of TMs 5A and 11A into the membrane, resulting in a 15-TM topology and opposite orientations of TMs 6–11, is depicted in the inset. The positions of highly conserved negatively charged aspartate and glutamate residues are indicated in black.
hCNT3 contains 10 aspartate and glutamate residues that are conserved in other mammalian members of the CNT protein family (Asp-192, Glu-329, Glu-343, Glu-359, Glu-410, Glu-434, Asp-503, Glu-519, Glu-553, and Asp-586) and were included in this study for comparison with previous mutagenesis studies of the corresponding residues in hCNT1 (31). Also included was one additional glutamate residue unique to, and conserved within, the CNT3/hfCNT subfamily (Glu-483). All but one of the selected residues (Asp-192) were located in the C-terminal half of the protein. In initial mutagenesis experiments, the 11 hCNT3 aspartate and glutamate residues were individually replaced by asparagine or glutamine, respectively. All mutations were verified by sequencing the entire coding region of the double-stranded plasmid DNA in both directions. Except for the desired base changes, all sequences were identical to wild-type hCNT3.
Transport Activity of hCNT3 Mutants
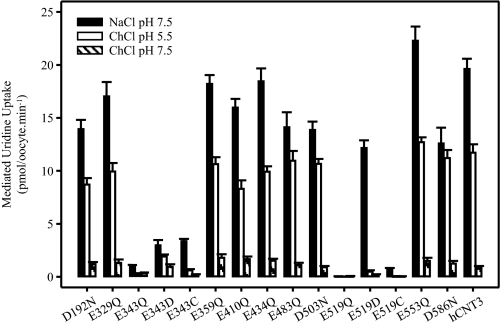
Different from hCNT1/2, hCNT3 transports both purine and pyrimidine nucleosides, and couples nucleoside transport to H+ as well as Na+ electrochemical gradients (10, 11, 19). Each of the 11 hCNT3 mutants was therefore produced in Xenopus oocytes and screened for altered permeant selectivity (Table 1) and changes in Na+ and/or H+ dependence (Fig. 2). Measured in Na+-containing, H+-reduced transport medium (100 mm NaCl, pH 7.5), Table 1 presents for each mutant initial rates of transport (1-min flux) of 20 μm radiolabeled uridine (a universal mammalian CNT permeant), inosine (a representative purine nucleoside), and thymidine (a representative pyrimidine nucleoside). Fig. 2 compares uridine uptake (20 μm; 1 min flux) in Na+-containing, H+-reduced (100 mm NaCl, pH 7.5), Na+-free, acidified (100 mm ChCl, pH 5.5), and Na+-free, H+-reduced (100 mm ChCl, pH 7.5) transport media. The flux values shown in this figure (and in all subsequent experiments) depict mediated transport activity, defined as the difference in uptake between RNA transcript-injected and control water-injected oocytes. Under all conditions tested, nucleoside uptake by control water-injected oocytes was ≤0.1 pmol/oocyte·min−1 (data not shown).
TABLE 1.
Mediated uptake of radiolabeled nucleosides by hCNT3 and mutants
Uptake of 20 μm radiolabeled uridine, inosine, and thymidine in 100 mm NaCl, pH 7.5, was measured in oocytes producing hCNT3 mutants or wild-type hCNT3. Values were corrected for basal nonmediated uptake in control water-injected oocytes and are the means ± S.E. of 10–12 oocytes.
| Mediated nucleoside uptake |
|||
|---|---|---|---|
| Uridine | Inosine | Thymidine | |
| pmol/oocyte·min−1 | |||
| D192N | 12.2 ± 0.7 | 15.3 ± 0.6 | 12.5 ± 0.4 |
| E329Q | 16.5 ± 0.4 | 19.4 ± 0.9 | 13.2 ± 0.6 |
| E343Q | 1.7 ± 0.1 | 1.3 ± 0.1 | 1.9 ± 0.1 |
| E343D | 4.5 ± 0.6 | 3.3 ± 0.3 | 4.4 ± 0.6 |
| E343C | 4.0 ± 0.2 | 1.1 ± 0.1 | 1.9 ± 0.3 |
| E359Q | 14.8 ± 0.7 | 18.8 ± 0.9 | 13.7 ± 0.7 |
| E410Q | 16.9 ± 1.1 | 17.4 ± 0.9 | 13.4 ± 0.6 |
| E434Q | 14.3 ± 0.6 | 18.5 ± 1.0 | 13.8 ± 0.5 |
| E483Q | 15.6 ± 0.9 | 18.8 ± 0.6 | 11.6 ± 0.4 |
| D503N | 11.7 ± 0.8 | 14.6 ± 0.9 | 10.4 ± 0.8 |
| E519Q | <0.1 | <0.1 | <0.1 |
| E519D | 20.7 ± 0.7 | 13.5 ± 0.9 | 9.6 ± 0.6 |
| E519C | 0.6 ± 0.1 | 0.1 ± 0.1 | 0.2 ± 0.1 |
| E553Q | 20.8 ± 1.1 | 24.4 ± 0.7 | 14.2 ± 0.6 |
| D586N | 19.7 ± 1.0 | 21.0 ± 0.8 | 13.4 ± 0.5 |
| hCNT3 | 17.4 ± 1.1 | 18.7 ± 1.0 | 15.6 ± 0.7 |
FIGURE 2.
Cation selectivity of hCNT3 and mutants. Mediated uptake of 20 μm radiolabeled uridine in transport medium containing 100 mm NaCl, pH 7.5 (black bars), ChCl, pH 5.5 (open bars), or ChCl, pH 7.5 (hatched bars), for oocytes producing hCNT3 mutants (as indicated) or wild-type hCNT3 is shown. Values were corrected for basal nonmediated uptake in control water-injected oocytes and are means ± S.E. of 10–12 oocytes.
Of the 11 hCNT3 mutants investigated, 9 displayed robust levels of nucleoside uptake (>10 pmol/oocyte·min−1) similar to that of wild-type hCNT3 (Table 1) and were capable of both Na+- and H+-coupled uridine uptake (Fig. 2). In marked contrast, mutant E343Q exhibited only ∼5% of wild-type hCNT3 uridine transport activity in the presence of Na+, and even lower rates of transport in the absence of Na+ under acidified or H+-reduced conditions. In Na+-containing, H+-reduced medium, E343Q-mediated fluxes of inosine and thymidine were decreased to similar extents as uridine. The final mutant, E519Q, was nonfunctional under all conditions tested. In case its pH profile was shifted to be more acidic, E519Q was also tested for H+-dependent uridine uptake at pH 4.0. Again, no functional activity was detected (data not shown).
Subsequent studies therefore focused on Glu-343 and Glu-519. Other residues whose mutation produced wild-type functional characteristics were not investigated further.
Additional hCNT3 Glu-343 and Glu-519 Mutants
To provide additional investigative tools for subsequent experiments, residues Glu-343 and Glu-519 were also mutated to aspartate (to restore the negative charge) and to cysteine (to test for inhibition by thiol-reactive reagents). Table 1 and Fig. 2 show their properties. Compared with E343Q, reintroduction of the negative charge (mutant E343D) increased both Na+- and H+-dependent nucleoside influx to ∼25% of wild-type levels. E343C, in contrast, showed a selective increase in Na+-dependent uridine transport activity relative to H+-dependent activity. For residue 519, although charge restoration (mutant E519D) returned Na+-dependent uridine uptake to wild-type levels, H+-dependent uridine uptake in the absence of Na+ remained very low. Relative to E519Q, E519C exhibited a small increase in Na+-dependent uridine transport activity, with no detectable H+-dependent transport. E343D and E519D exhibited wild-type nucleoside selectivity, whereas E343C and E519C preferentially transported uridine relative to inosine and thymidine.
Cell-surface Expression of hCNT3 Glu-343 and Glu-519 Mutants
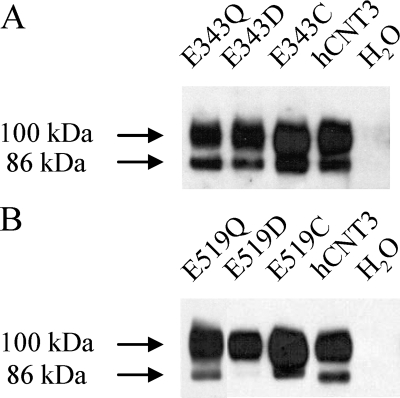
To verify that the observed decreases in transport activities were not secondary to altered cell-surface expression, oocytes producing Glu-343 or Glu-519 mutants were subjected to cell-surface labeling with sulfosuccinimidyl-6-(biotinamido) hexanoate using immobilized streptavidin resin to separate cell-surface protein from that associated with intracellular membranes. Immunoblots were probed with hCNT3 polyclonal antibodies (37) directed against amino acid residues 45–69 of the intramembranous N-terminal region of the protein. Similar to previous findings (38) and consistent with the presence of multiple C-terminal sites of N-linked glycosylation, cell-surface hCNT3 exhibited discrete immunostained bands at 100 and 86 kDa (Fig. 3). Both these immunostained bands correspond to glycosylated forms of the transporter, aglyco-hCNT3 having a lower molecular mass of 75 kDa (38). Antibody specificity was confirmed by lack of immunoreactivity in membranes from control, water-injected oocytes.
FIGURE 3.
Cell-surface expression of hCNT3 and mutants. Oocytes injected with RNA transcripts encoding E343Q, E343C, or E343D (A), E519Q, E519C, or E519D (B), or wild-type hCNT3 (A and B) and control water-injected oocytes (A and B) were assayed for the presence of plasma membrane recombinant protein by immunoblotting. Staining intensities relative to wild-type hCNT3 (100- and 86-kDa immunostained bands combined) were 0.77, 0.83, and 1.08 for E343Q, E343D, and E343C, respectively, and 0.98, 0.71, and 1.15 for E519Q, E519D, and E519C, respectively.
Patterns of immunoreactivity and staining intensity similar to the wild-type protein were apparent for E343Q, E343D, and E343C (Fig. 3A) and for E519Q and E519C (Fig. 3B). Also present in the plasma membrane, the majority of E519D immunoreactivity was associated with the more extensively glycosylated (100 kDa) form of the transporter (Fig. 3B). The robust transport activity exhibited by E519D in Table 1 and Fig. 2 indicates that the altered glycosylation status of this mutant had no effect on function. Because all Glu-343 and Glu-519 mutants were present at cell surfaces in amounts similar to wild-type hCNT3 (see Fig. 3 legend for numerical values), the impaired functional activities of E343Q, E343D, E343C, E519C, and E519D resulted from loss of intrinsic transport capability, rather than reduced quantities in plasma membranes.
Nucleoside Selectivity of hCNT3 Glu-343 and Glu-519 Mutants
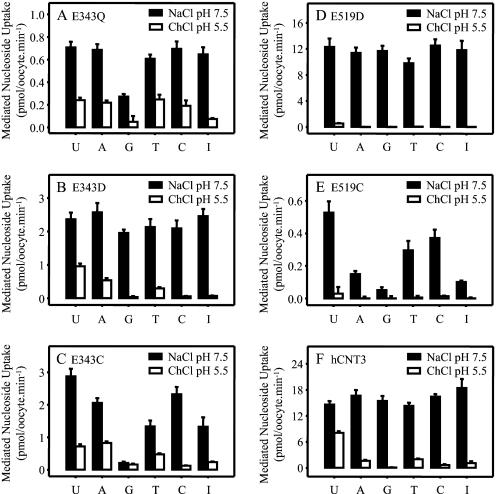
There are indications from Table 1 that some Glu-343 and Glu-519 mutants may have altered permeant selectivities. To further investigate the role of residues Glu-343 and Glu-519 in nucleoside binding and/or translocation, subsequent experiments investigated the uptake of a full panel of physiological radiolabeled purine and pyrimidine nucleosides (20 μm) measured in both Na+-containing, H+-reduced (100 mm NaCl, pH 7.5) and Na+-free acidified transport medium (100 mm ChCl, pH 5.5, respectively) (Fig. 4).
FIGURE 4.
Nucleoside selectivity of hCNT3 and mutants. Uptake of 20 μm radiolabeled uridine (U), adenosine (A), guanosine (G), thymidine (T), cytidine (C), and inosine (I) was measured in oocytes producing E343Q (A), E343D (B), E343C (C), E519D (D), E519Q (E), or hCNT3 (F) in 100 mm NaCl, pH 7.5 (black bars), or 100 mm ChCl, pH 5.5 (open bars). Values were corrected for basal nonmediated uptake in control water-injected oocytes and are means ± S.E. of 10–12 oocytes.
Na+-coupled hCNT3 exhibits broad selectivity for both pyrimidine and purine nucleosides, whereas H+-coupled hCNT3 has a different conformation of the permeant binding pocket and preferentially transports uridine > thymidine, adenosine > cytidine, and inosine > guanosine (19). These contrasting patterns of nucleoside selectivity were confirmed for wild-type hCNT3 in the present analysis (Fig. 4F). Extending the results presented in Table 1, a role for hCNT3 residue Glu-343 in nucleoside selectivity was apparent from the nucleoside uptake patterns of both E343Q and E343C. In Na+-containing, H+-reduced medium, mutant E343Q exhibited a selective decrease in guanosine influx compared with that for uridine (Fig. 4A). Guanosine transport was impaired to an even greater extent in the case of E343C, which also showed a reduction in thymidine and inosine transport (Fig. 4C). Relative to the pattern of nucleoside transport exhibited by wild-type hCNT3 in Na+-free, acidified medium (Fig. 4F), E343Q and E343C showed increased influx of adenosine, thymidine, and cytidine (E343Q) and adenosine and thymidine (E343C). In contrast, restoration of the negative charge (mutant E343D) resulted in wild-type Na+- and H+-coupled nucleoside selectivity (Fig. 4B).
A role in nucleoside selectivity was also apparent for residue Glu-519. In Na+-containing medium, mutant E519C exhibited reduced influx of all three purine nucleosides (adenosine, guanosine, and inosine) (Fig. 4E), whereas charge replacement (mutant E519D) restored wild-type transport selectivity (Fig. 4D). With the exception of very small fluxes of uridine evident for E519D (Fig. 4D) (see also Fig. 2), neither Glu-519 mutant transported nucleosides in Na+-free, acidified medium. Mutant E519Q could not be included in this (and subsequent) analysis because of a total lack of functional activity in either Na+-containing, H+-reduced, or Na+-free acidified medium (Table 1 and Fig. 2).
Uridine Transport Kinetics of hCNT3 Glu-343 and Glu-519 Mutants
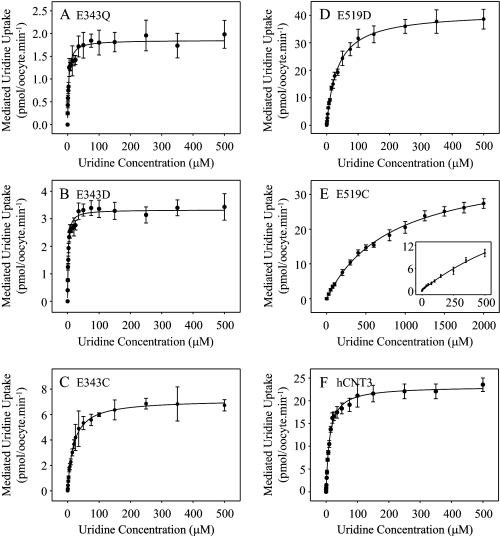
Fig. 5 presents representative concentration dependence curves for radiolabeled uridine influx in Na+-containing, H+-reduced medium (100 mm NaCl, pH 7.5) in oocytes producing E343Q, E343D, E343C, E519D, E519C, or wild-type hCNT3. Kinetic parameters derived from these curves are summarized in Table 2. In good agreement with previous studies (10, 19), the apparent Km value of hCNT3 for uridine was 11 μm, with a Vmax value of 23 pmol/oocyte·min−1 and a Vmax:Km ratio of 2.1. In comparison with hCNT3, the Glu-343 series of mutants exhibited markedly lower Vmax values (rank order E343Q < E343D < E343C), with apparent Km values that were either higher (E343C) or lower (E343Q and E343D) than that of wild-type hCNT3. The Vmax:Km ratio, a measure of transport efficiency, was lowest for E343Q and E343C (both 0.4), and intermediate for the charge-restored mutant E343D (1.3).
FIGURE 5.
Uridine kinetics of hCNT3 and mutants. Radiolabeled uridine uptake was measured under initial rate conditions in 100 mm NaCl transport medium, pH 7.5, in oocytes producing E343Q (A), E343D (B), E343C (C), E519D (D), E519C (E), or hCNT3 (F) for uridine concentrations of 0–500 μm (A–D, E inset only, and F) and 0–2 mm (E). Values were corrected for basal nonmediated uptake in control water-injected oocytes and are means ± S.E. of 10–12 oocytes. Kinetic parameters calculated from the data are presented in Table 2.
TABLE 2.
Kinetic parameters for uridine uptake mediated by hCNT3 and mutants
| Apparent Kma | Vmaxa,b | Vmax:Kmb ratio | |
|---|---|---|---|
| μm | pmol/oocytes·min−1 | ||
| E343Q | 3.9 ± 0.7 | 1.7 ± 0.1 (0.09) | 0.44 (0.27) |
| E343D | 2.4 ± 0.3 | 3.2 ± 0.1 (0.16) | 1.33 (0.75) |
| E343C | 18.9 ± 0.9 | 7.2 ± 0.1 (0.28) | 0.38 (0.16) |
| E519D | 35.0 ± 1.6 | 40.1 ± 0.5 (2.41) | 1.15 (0.75) |
| E519C | 876 ± 57 | 39.0 ± 0.9 (1.45) | 0.04 (0.02) |
| hCNT3 | 10.9 ± 0.7 | 23.4 ± 0.4 (1.00) | 2.15 (1.00) |
In contrast to the Glu-343 series of mutants, E519D and E519C exhibited Vmax values higher than that of wild-type hCNT3. For E519D, this was offset by an ∼3-fold increase in the apparent Km value, resulting in an intermediate Vmax:Km ratio (1.1) similar to that of E343D. In contrast, mutant E519C showed a dramatic ∼80-fold increase in the apparent Km value, resulting in the lowest Vmax:Km ratio (<0.1) of all the mutants studied. Vmax values and Vmax:Km ratios adjusted for differences in cell-surface expression (Fig. 3) showed similar trends (Table 2).
Guanosine Inhibition Kinetics of hCNT3 Glu-343 and Glu-519 Mutants
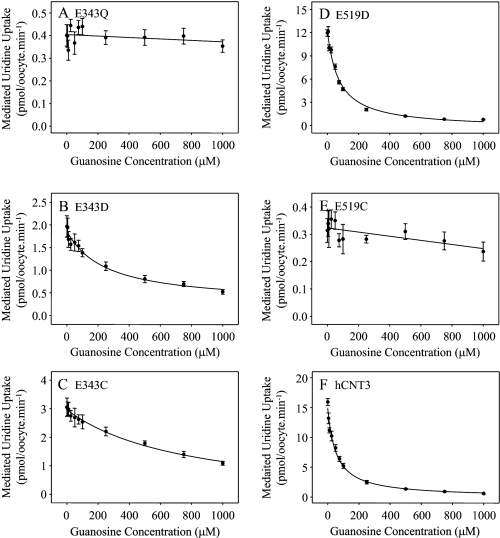
Competition experiments were undertaken to determine whether the decreased uptake of guanosine by E343Q, E343C, and E519C in Na+-containing medium (Fig. 4) was a consequence of altered nucleoside binding (Fig. 6). Influx of 20 μm radiolabeled uridine by the Glu-343 and Glu-519 series mutants and wild-type hCNT3 was measured in Na+-containing, H+-reduced medium (100 mm NaCl, pH 7.5) in the presence of increasing concentrations (0–1000 μm) of nonlabeled guanosine. Because of low solubility, guanosine concentrations greater than 1 mm could not be achieved. In good agreement with the previously reported apparent Km value of 43 μm for guanosine transport by hCNT3 (10), and consistent with the robust level of guanosine uptake shown by wild-type hCNT3 in Fig. 4F, the calculated IC50 value for guanosine inhibition of uridine transport by hCNT3 was 54 ± 9 μm. Corresponding IC50 values for guanosine transport-impaired mutants E343Q, E343C, and E519C were too high to determine, whereas values of 248 ± 78 and 73 ± 11 μm were obtained for E343D and E519D, respectively. Therefore, decreased guanosine transport reflects alterations to the nucleoside binding pocket, at least in part.
FIGURE 6.
Guanosine inhibition of hCNT3 and mutants. Uptake of 20 μm radiolabeled uridine was measured in the presence of 0–1000 μm nonlabeled guanosine in 100 mm NaCl transport medium, pH 7.5, in oocytes producing E343Q (A), E343D (B), E343C (C), E519D (D), E519C (E), or hCNT3 (F). Values were corrected for basal nonmediated uptake in control water-injected oocytes and are means ± S.E. of 10–12 oocytes. IC50 values calculated from the data are presented in the text.
Na+ Activation Kinetics of hCNT3 Glu-343 and Glu-519 Mutants
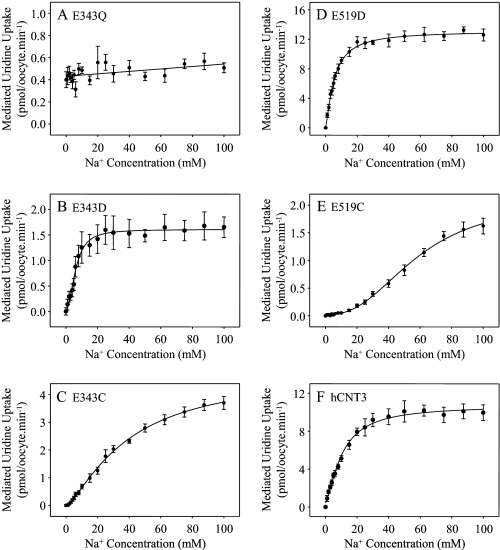
Na+ activation of the Glu-343 and Glu-519 mutants was investigated by measuring 20 μm radiolabeled uridine influx as a function of Na+ concentration (0–100 mm NaCl, pH 8.5) (Fig. 7). Kinetic parameters derived from the curves are summarized in Table 3. Na+-containing medium at pH 8.5 was used to avoid the small, but significant, amount of hCNT3 H+ activation that occurs at pH 7.5 (11, 19). Na+-coupled uridine transport by hCNT3 at pH 8.5 is kinetically indistinguishable from that at pH 7.5 (19, 38).
FIGURE 7.
Na+-activation kinetics of hCNT3 and mutants. Na+-activation curves for oocytes producing E343Q (A), E343D (B), E343C (C), E519D (D), E519C (E), or wild-type hCNT3 (F) were measured in transport medium containing 0–100 mm NaCl at pH 8.5 with isomolality maintained by addition of ChCl. A radiolabeled uridine concentration of 20 μm was used. Values were corrected for basal nonmediated uptake in control water-injected oocytes and are means ± S.E. of 10–12 oocytes. Kinetic parameters calculated from the data are presented in Table 3.
TABLE 3.
Na+- and H+-activation kinetic parameters for hCNT3 and mutants
| Na+-activation kinetic parameters |
H+-activation kinetic parameters |
|||||
|---|---|---|---|---|---|---|
| Apparent K50a | Vmaxa,b | Hill coefficienta | Apparent K50a | Vmaxa,b | Hill coefficienta | |
| mm | pmol/oocyte·min−1 | nm | pmol/oocyte·min−1 | |||
| E343Q | ND | ND | ND | ND | ||
| E343D | 6.3 ± 0.4 | 1.5 ± 0.1 (0.18) | 1.8 ± 0.1 | 46 ± 12 (pH 7.3) | 1.9 ± 0.1 (0.26) | 0.8 ± 0.1 |
| E343C | 40 ± 3 | 4.8 ± 0.3 (0.43) | 1.4 ± 0.1 | 231 ± 54 (pH 6.6) | 0.5 ± 0.1 (0.05) | 0.7 ± 0.1 |
| E519D | 5.5 ± 0.3 | 13.0 ± 0.4 (1.78) | 1.4 ± 0.1 | 42 ± 13 (pH 7.4) | 0.7 ± 0.1 (0.11) | 0.7 ± 0.1 |
| E519C | 58 ± 3 | 2.1 ± 0.1 (0.18) | 1.9 ± 0.1 | |||
| hCNT3 | 10.5 ± 0.7 | 10.3 ± 0.4 (1.00) | 1.5 ± 0.1 | 495 ± 106 (pH 6.3) | 8.7 ± 0.4 (1.00) | 0.7 ± 0.1 |
In good agreement with previous studies (11, 19, 38), Na+ activation of uridine uptake by wild-type hCNT3 occurred with an apparent K50 value of 11 mm and a Vmax value of 10 pmol/oocyte·min−1. In marked contrast, E343Q exhibited very low affinity for Na+, such that the curve appeared linear with concentration, although uridine uptake at 100 mm Na+ was marginally greater than that in the absence of Na+ (see also Fig. 2). Relative to wild-type, E343D showed a modest decrease in apparent K50 value for Na+ and a marked decrease in Vmax. In contrast, E343C exhibited a dramatic increase in the Na+-apparent K50 value, such that influx approached saturation only at the highest Na+ concentration used (100 mm). E519C showed a similar marked decrease in affinity for Na+, and the Vmax value was also greatly decreased. However, consistent with the robust transport activity shown in Fig. 2, E519D exhibited modest increases in apparent affinity and Vmax for Na+. Vmax values for all mutants adjusted for differences in cell-surface expression (Fig. 3) showed similar trends (Table 3). All curves were sigmoidal, and calculated Hill coefficients were ≥1.4 (Table 3).
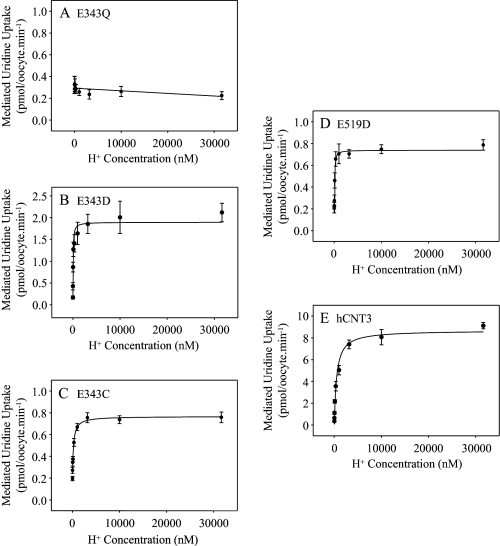
H+ Activation Kinetics of hCNT3 Glu-343 and Glu-519 Mutants
H+ activation of Glu-343 and Glu-519 mutants was investigated by measuring 20 μm radiolabeled uridine influx in 100 mm ChCl transport medium at pH values ranging from 4.5 to 8.5 (Fig. 8). Kinetic parameters derived from the curves are summarized alongside those for Na+ in Table 3. In good agreement with previous studies, H+-coupled uridine uptake by wild-type hCNT3 occurred with an apparent K50 value of 495 nm and a corresponding Vmax value of 9 pmol/oocyte·min−1 (11, 19, 38). Consistent with Fig. 2, the H+-activation curve for E343Q was linear and independent of H+ concentration. The corresponding H+-activation curves for E343D, E343C and E519D exhibited decreases in both apparent K50 and Vmax compared with wild-type hCNT3. Of these, E343D and E519D exhibited the most pronounced shift in H+ apparent affinity, whereas E343C and E519D had the lowest Vmax values. Similar to hCNT3, Hill coefficients were between 0.7 and 0.8. H+-activation kinetics were not determined for mutant E519C which lacked transport activity in Na+-free, acidified transport medium (Fig. 2). Vmax values adjusted for differences in cell-surface expression (Fig. 3) showed similar trends (Table 3).
FIGURE 8.
H+-activation kinetics of hCNT3 and mutants. H+-activation curves for oocytes producing E343Q (A), E343D (B), E343C (C), E519D (D), or hCNT3 (E) were measured in transport medium containing 100 mm ChCl transport medium with pH values ranging from 4.5 to 8.5 (H+ concentration 31.6 μm and 3.16 nm, respectively). The concentration of radiolabeled uridine was 20 μm. Values were corrected for basal nonmediated uptake in control water-injected oocytes and are means ± S.E. of 10–12 oocytes. Kinetic parameters calculated from the data are presented in Table 3.
Steady-state Currents of hCNT3 Glu-343 and Glu-519 Mutants
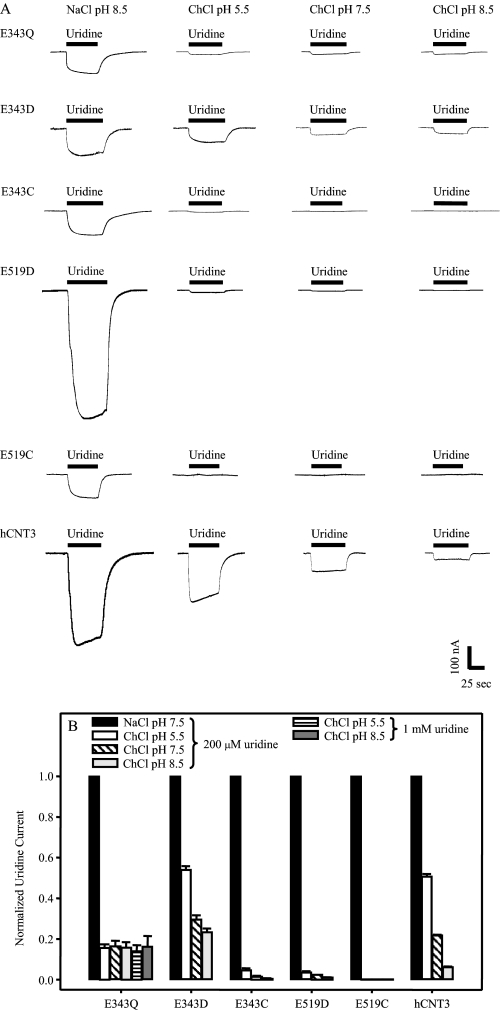
Complementary to the uridine flux studies of cation dependence shown in Fig. 2, steady-state electrophysiological experiments were undertaken to measure uridine-induced Na+ and H+ inward currents in oocytes producing E343Q, E343D, E343C, E519D, E519C or, as a control, hCNT3. Representative recordings comparing currents evoked by uridine (200 μm) in Na+-containing, H+-reduced (100 mm NaCl, pH 8.5), and Na+-free acidified transport media (100 mm ChCl, pH 5.5), as well as in Na+-free, H+-reduced transport media (100 mm ChCl at pH 7.5 and pH 8.5) are shown in Fig. 9A. Fig. 9B depicts mean currents for ≥4 oocytes normalized to that obtained in 100 mm NaCl, pH 8.5.
FIGURE 9.
Uridine-evoked steady-state currents of hCNT3 and mutants. Uridine-evoked currents in 100 mm NaCl, pH 8.5, or 100 mm ChCl, pH 5.5, pH 7.5, or pH 8.5 transport media (right to left columns, respectively) are shown for representative oocytes producing E343Q, E343D, E343C, E519D, E519C, or hCNT3 (A). No current was detected in control water-injected oocytes (data not shown). Bars indicate the duration of exposure to 200 μm uridine. The bar graph of B shows the same uridine-evoked steady-state currents from ≥4 oocytes each normalized to current in 100 mm NaCl, pH 8.5. Error bars are not shown where values were smaller than that represented by the symbols.
As demonstrated in previous studies (19), wild-type hCNT3 exhibited large inward currents for both Na+ and H+, the latter decreasing as a function of pH (5.5 > 7.5 > 8.5) indicating a strong dependence of H+. No currents were observed in control water-injected oocytes (data not shown). Smaller uridine-induced Na+ currents were apparent for all three Glu-343 mutants. Uridine-induced H+ currents were also evident for E343D, but were very small for E343C. Consistent with a lack of H+ dependence in radioisotope flux studies (Fig. 2 and Fig. 8A), mutant E343Q exhibited small uridine-induced currents in the absence of Na+ that were of similar magnitude irrespective of the H+ concentration in the medium. Increasing the uridine concentration to 1 mm did not influence the magnitude of these currents, either at pH 5.5 or pH 8.5 (Fig. 9B). Uridine-induced Na+ currents were also apparent for E519D and E519C, whereas H+ currents were very small (E519D) or not detectable (E519C).
Cation:Nucleoside Coupling Ratios of hCNT3 Glu-343 and Glu-519 Mutants
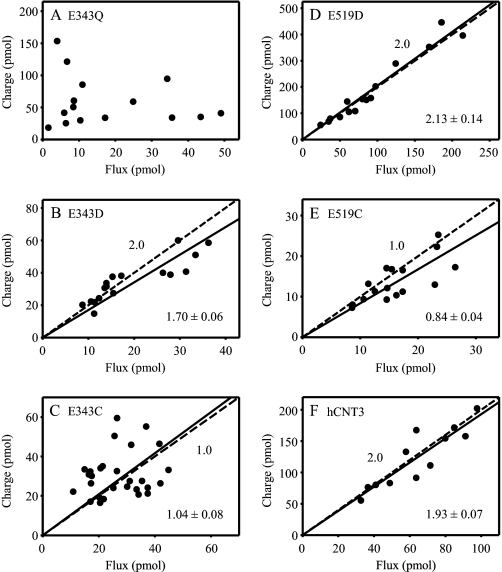
Na+:nucleoside and H+:nucleoside stoichiometries of E343C, E343Q, E343D, E519C, E519D and, as a control, wild-type hCNT3 were determined by simultaneously measuring cation currents and radiolabeled uridine uptake under voltage clamp conditions, as described previously (11, 19, 35). Na+ and H+ currents were determined in transport media containing 100 mm NaCl at pH 8.5 and 100 mm ChCl at pH 5.5, respectively. The uridine concentration was 200 μm. Charge versus flux plots for groups of individual oocytes producing each of the recombinant transporters are shown for Na+ in Fig. 10. Na+:uridine stoichiometries, derived from linear regression analyses of the data, together with corresponding H+:uridine stoichiometries (data plots not shown) are presented in Table 4.
FIGURE 10.
Na+:uridine stoichiometry of hCNT3 and mutants. Charge to radiolabeled uridine uptake ratio plots were generated with oocytes producing E343Q (A), E343D (B), E343C (C), E519D (D), E519C (E), or hCNT3 (F) in 100 mm NaCl transport medium at pH 8.5. A uridine concentration of 200 μm was used, and the oocytes were clamped at a holding potential of −90 mV. Integration of the uridine-evoked current was used to calculate the net cation influx (charge) and was correlated to the net [3H]uridine influx (flux). Linear fits passed through the origin, and linear regression analysis of the data for each plot is indicated by the solid line. The dashed line represents a theoretical 2:1 (B, D, and F) and 1:1 (C and E) charge:uptake ratio. Calculated Na+:uridine coupling ratios from these data are presented in Table 4.
TABLE 4.
Na+:uridine and H+:uridine stoichiometry for hCNT3 and mutants
| Na+:uridine stoichiometrya | H+:uridine stoichiometryb | |
|---|---|---|
| E343Q | ∼7.5 | ∼7.4 |
| E343D | 1.70 ± 0.06 | 1.15 ± 0.04 |
| E343C | 1.04 ± 0.09 | ND |
| E519D | 2.13 ± 0.14 | 0.61 ± 0.09 |
| E519C | 0.84 ± 0.04 | ND |
| hCNT3 | 1.93 ± 0.07 | 1.02 ± 0.04 |
a Data are from Fig. 10.
b Measured in transport medium containing 100 mm ChCl, pH 5.5; ND indicates not determined.
Consistent with kinetic indications of uncoupled transport, and different from the other mutants studied, there was no correlation between E343Q Na+ current and radiolabeled uridine uptake, and charge:flux ratios for individual E343Q-producing oocytes ranged from 0.8 to 39 (mean value ∼7.5) (Fig. 10A and Table 4). Uncoupling was also apparent for E343Q H+ currents (mean charge:flux ratio ∼7.4) (Table 4). Still somewhat variable, but with a statistically significant linear correlation between Na+ current and uridine flux (correlation coefficient (r) 0.589, p < 0.001, n = 28), regression analysis of data for individual E343C-producing oocytes gave a calculated Na+:uridine stoichiometry of 1.04 ± 0.09 (Fig. 10C and Table 4). Also consistent with cotransport of one Na+ ion per uridine molecule, the mean E343C Na+ charge:flux ratio in Fig. 10C was 1.24 ± 0.10 (range 0.6–2.2). H+:uridine coupling ratios could not be determined for E343C because of low levels of H+-coupled transport activity (Fig. 2 and Fig. 9A). In marked contrast, re-introduction of the negative charge at this position (mutant E343D) gave 2:1 and 1:1 Na+:uridine and H+:uridine stoichiometries, respectively, thus restoring wild-type characteristics (Fig. 10B and Table 4).
Similar to wild-type hCNT3, E519D also exhibited a Na+:uridine coupling ratio of 2:1 (Fig. 10D and Table 4). Different from this, however, and with less scatter than for E343C, mutant E519C gave a Na+:uridine stoichiometry of 0.84 ± 0.04, suggesting cotransport of only one Na+ ion per uridine molecule (Fig. 10E and Table 4). The corresponding H+:uridine coupling ratio for E519D was 0.61 ± 0.09 (Table 4). An H+:uridine coupling ratio could not be determined for E519C because of a lack of transport activity in the absence of Na+ (Fig. 2 and Fig. 9B).
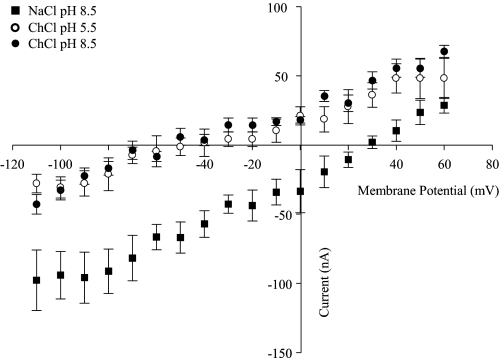
Current-Voltage (I-V) Relationships of hCNT3 Mutant E343Q
As a further test of the uncoupled behavior of mutant E343Q, Fig. 11 shows current-voltage (I-V) curves for E343Q-producing oocytes generated as the difference between steady-state currents recorded in the presence and absence of uridine. Measured in Na+-containing, H+-reduced (100 mm NaCl, pH 8.5), Na+-free, acidified (100 mm ChCl, pH 5.5), and Na+-free, H+-reduced (100 mm ChCl, pH 8.5) transport media, E343Q-mediated currents evoked by uridine (100 μm) at potentials between −110 and +60 mV were voltage-dependent and, in contrast to those of wild-type hCNT3 (19), reversed polarity at membrane potentials of approximately +30 and −50 mV in the presence and absence of Na+, respectively. Consistent with the lack of H+ dependence of E343Q shown by the radioisotope flux studies in Fig. 2 and the current data in Fig. 9, A and B, curves in Na+-free, acidified (100 mm ChCl, pH 5.5),and Na+-free, H+-reduced (100 mm ChCl, pH 8.5) transport media overlapped. No uridine-induced currents were observed in control water-injected oocytes (data not shown). Reported concentrations of Na+ inside the oocyte range from 4 to 22 mm (39). At an extracellular Na+ concentration of 100 mm, however, the calculated equilibrium potential for Na+ ranges respectively from +81 to +38 mV. This corresponds well with the reversal potential of +30 mV evident for E343Q-mediated currents in Na+-containing, H+-reduced medium (100 mm NaCl, pH 8.5) (Fig. 11). With a common reversal potential of −50 mV, the curves in Na+-free, acidified (100 mm ChCl, pH 5.5) and Na+-free, H+-reduced (100 mm ChCl, pH 8.5) transport media are incompatible with involvement of H+ (calculated equilibrium potentials of +116 and −58 mV, respectively, at an internal pH of 7.5). Assuming instead that the cation is Na+ (derived by leakage from intracellular pools), a reversal potential of −50 mV in Na+-free media implies a localized extracellular concentration of Na+ in the range 0.8–4 mm.
FIGURE 11.
Current-voltage relationships of E343Q-mediated uridine transport. Current-voltage (I-V) curves for representative E343Q-producing oocytes in 100 mm NaCl, pH 8.5 (solid squares), 100 mm ChCl, pH 5.5 (open circles), and 100 mm ChCl, pH 8.5 (solid circles) transport media were generated from the difference between steady-state currents recorded in the presence and absence of 100 μm uridine. Oocytes were clamped at a holding potential (Vh) of −50 mV and pulsed to test (Vt) ranging from −110 to +60 mV in 10-mV increments. Data points represent the mean ± S.E. from 6 to 8 oocytes. No uridine-induced currents were observed in control water-injected oocytes. Error bars are not shown where values were smaller than that represented by the symbols.
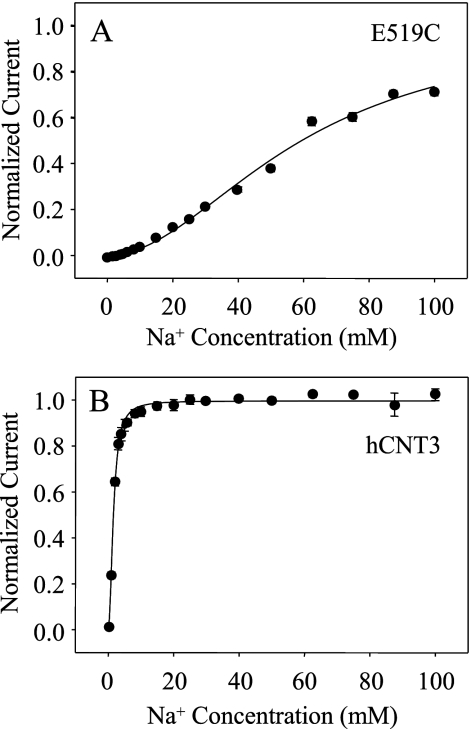
Na+ Activation Kinetics of hCNT3 Mutant E519C Determined by Electrophysiology
The Na+-activation kinetics of E519C were also investigated by electrophysiology. Fig. 12A depicts the relationship between Na+ concentration and 200 μm uridine-evoked current for oocytes producing E519C clamped at a membrane holding potential of −90 mV. For comparison, the corresponding Na+-activation curve for wild-type hCNT3 was also determined (Fig. 12B). Currents from individual oocytes were normalized to their predicted Imax values and subsequently averaged to produce mean kinetic parameters. Similar to the radioisotope flux data in Fig. 7E, E519C exhibited an apparent K50 value for Na+-coupled uridine uptake of 60 ± 4 mm and a corresponding Hill coefficient of 2.1 ± 0.4. In agreement with previous studies (11, 19, 38), the apparent K50 value for Na+-coupled uridine uptake by wild-type hCNT3 under these conditions was 1.6 ± 0.1 mm, and the corresponding Hill coefficient was 1.7 ± 0.6. As determined previously, the difference in apparent K50 value for hCNT3 when determined by electrophysiology (1.6 mm, Fig. 12B) compared with radioisotope flux analysis (10.3 mm, Fig. 7F and Table 3) reflects the difference in membrane potential under the two experimental conditions (−90 versus approximately −40 mV, respectively) (19, 38). The similar apparent affinity of E519C for Na+ in Fig. 7E (radioisotope flux assay) and Fig. 12A (electrophysiology) suggests a loss of voltage dependence of Na+ K50 for this mutant.
FIGURE 12.
Na+-activation kinetics of hCNT3 and E519C determined by electrophysiology. The Na+-activation curves for E519C (A) and wild-type hCNT3 (B) were determined at a holding potential of −90 mV in 100 mm NaCl, pH 8.5, transport medium. The uridine concentrations used were 200 and 20 μm for E519C and hCNT3, respectively. Uridine-evoked currents at each Na+ concentration were normalized to the respective fitted Imax value and are presented as the mean ± S.E. of 8 (E519C) or 5 (hCNT3) oocytes. Error bars are not shown where values were smaller than that represented by the symbols. No currents were detected in control water-injected oocytes (data not shown).
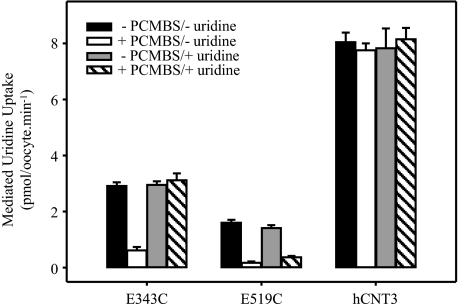
PCMBS Inhibition of hCNT3 Mutants E343C and E519C
Residues lining the translocation pore can be identified through the use of hydrophilic thiol-reactive reagents such as PCMBS. In a final series of experiments, uptake of 20 μm radiolabeled uridine was measured in oocytes producing hCNT3 mutants E343C and E519C or wild-type hCNT3 incubated in the presence and absence of 200 μm PCMBS and in the presence and absence of 20 mm extracellular uridine. Exposure to PCMBS and uridine was performed on ice to minimize passage across the oocyte plasma membrane. Because the experiment was performed in a wild-type background, and because wild-type hCNT3 contains a conformationally mobile cysteine residue (Cys-561) that becomes exposed to PCMBS in the H+-bound state of the transporter (34), reactivity to PCMBS was investigated in Na+-containing, H+-reduced transport medium (100 mm NaCl, pH 8.5) (Fig. 13). Both E343C and E519C showed marked inhibition by PCMBS consistent with pore-lining status. In the case of E343C, this inhibition was prevented by uridine, suggesting a location close to or within the nucleoside binding pocket. A small amount of uridine protection was also seen for E519C. Under the same conditions, and in agreement with previous results (34), there was no inhibition of the wild-type transporter.
FIGURE 13.
PCMBS inhibition of hCNT3-, E343C-, and E519C-mediated uridine transport. Radiolabeled uridine uptake (20 μm) was measured in oocytes producing E343C, E519C, or wild-type hCNT3 following incubation in the absence (solid and gray bars) or presence (open and hatched bars) of 500 μm PCMBS and in the absence (solid and open bars) or presence (gray and hatched bars) of 20 mm uridine. The medium for exposure to PCMBS and subsequent assay of transport activity was 100 mm NaCl, pH 8.5. Data are presented as mediated transport, calculated as uptake in RNA-injected oocytes minus uptake in water-injected oocytes. Each value represents the mean ± S.E. of 10–12 oocytes. Error bars are not shown where values were smaller than that represented by the symbols.
DISCUSSION
hCNT3 is the most functionally versatile of the three human members of the SLC28 (CNT) protein family. More widely distributed in cells and tissues than paralogs hCNT1 or hCNT2 (10), and with a central role in renal nucleoside and nucleoside drug transport (37, 40), the multifunctional capability of hCNT3 and robust heterologous expression in Xenopus oocytes makes it the protein of choice for molecular characterization by site-directed mutagenesis and other approaches. Here we report an investigation of hCNT3 glutamate and aspartate residues. With the foundation of a corresponding analysis of Na+-specific hCNT1 (31), this study revealed two glutamate residues with key roles in hCNT3 Na+/nucleoside and H+/nucleoside cotransport. A side-by-side comparison of hCNT1 and hCNT3 highlighting the new discoveries of this study is presented in Table 5.
TABLE 5.
Comparison of hCNT1 and hCNT3 mutations
✓ indicates transporter was detected immunologically and/or by function at the cell surface; x indicates transporter was immunologically absent from the cell surface; WT indicates wild type-like functional activity; ? indicates no measurable transport activity because of retention in intracellular membranes; ND indicates not determined.
| Mutation |
Plasma membrane expression |
Change in phenotype |
|||
|---|---|---|---|---|---|
| hCNT1a | hCNT3 | hCNT1 | hCNT3b | hCNT1 | hCNT3c |
| D172N | D192N | ✓ | ✓ | WT | WT |
| E308Q | E329Q | ✓ | ✓ | Na+ and uridine kinetics | WT |
| E308D | ✓ | Uridine kinetics only | |||
| E308C | ✓ | ND | |||
| E322Q | E343Q | ✓ | ✓ | Na+ and uridine kinetics, uncoupled transport | Na+ and uridine kinetics, no H+ dependence, uncoupled transport |
| E322D | E343D | ✓ | ✓ | Na+ and uridine kinetics | Na+, H+, and uridine kinetics |
| E322C | E343C | ✓ | ✓ | ND | Na+, H+, and uridine kinetics, Na+:uridine stoichiometry |
| E338Q | E359Q | ✓ | ✓ | WT | WT |
| E389Q | E410Q | ✓ | ✓ | WT | WT |
| E413Q | E434Q | ✓ | ✓ | WT | WT |
| E483Q | ✓ | WT | |||
| D482N | D503N | x | ✓ | ? | WT |
| D482E | ✓ | WT | |||
| D482C | ND | ? | |||
| E498Q | E519Q | x | ✓ | ? | No functional activity |
| E498D | E519D | ✓ | ✓ | WT | H+ kinetics |
| E498C | E519C | ND | ✓ | ? | Na+ and uridine kinetics, no H+-dependence, Na+:uridine stoichiometry |
| E532Q | E553Q | x | ✓ | WT | |
| E532D | x | ||||
| E532C | ND | ? | |||
| D565N | D586N | ✓ | ✓ | WT | WT |
Aspartate and Glutamate Residues of hCNT3
There are 11 highly conserved negatively charged residues in hCNT3. Ten of these are also present in hCNT1/2, whereas one is unique to CNT3/hfCNT subfamily members.
Substitution of 9 of 11 acidic amino acids in hCNT3 with the corresponding neutral amino acid was without effect. Two of these (D503N and E553Q) corresponded to mutants in hCNT1 (D482N and E532Q) that were retained in intracellular membranes (31), and therefore not previously amenable to testing for roles in CNT structure or function. Another (E329Q) corresponded to an hCNT1 mutant (E308Q) that was processed normally to the cell surface but exhibited impaired transport activity (31). Because hCNT3 E329Q exhibited wild-type transport function, this residue may have roles specific to hCNT1 or, perhaps, to the CNT1/CNT2 subfamily.
Subsequent studies focused exclusively on the remaining two hCNT3 acidic amino acids Glu-343 and Glu-519. In addition to insertion of glutamine at these positions, the residues were also mutated to aspartate and cysteine. Different from hCNT1 (Table 5), all constructs were targeted to the oocyte plasma membrane, enabling their characterization by both radioisotope flux and electrophysiological techniques.
Glu-343, Functional Considerations
Located in TM 7 (Fig. 1), mutation of Glu-343 to glutamine had marked effects on hCNT3 kinetics. Similar to the corresponding hCNT1 mutant E322Q (31), E343Q retained the ability to bind uridine, although the translocation capacity (i.e. transporter turnover number) was dramatically decreased. Also like hCNT1 E322Q (31), E343Q retained the ability to bind and transport Na+, but only with low affinity. Thus, uridine influx and uridine-induced currents were higher in Na+-containing medium than in Na+-free medium, and cation activation analysis revealed a linear relationship between Na+ concentration and uridine influx. No H+ dependence of transport was evident, and although small uridine-induced currents were observed in Na+-free media, the magnitudes of these currents did not respond to changes in external pH.
Extending the previous findings of Na+-leak currents for hCNT1 mutant E322Q (31), E343Q charge:flux experiments revealed uncoupled uridine-induced cation movement through the transporter in both Na+-containing, H+-reduced, and Na+-free acidified transport media. Because the range of apparent coupling ratios encountered in these experiments was similar in both Na+-containing, H+-reduced, and Na+-free acidified transport media, H+ movement did not seem to account for the small pH-independent currents seen under the latter condition. Instead, we hypothesize that small local concentrations of Na+ in the immediate vicinity of the oocyte cell surface, in conjunction with the uncoupled Na+ flux, may have been responsible for the minor E343Q currents seen under otherwise Na+-free conditions. The current-voltage (I-V) curves in Na+-containing, H+-reduced, and Na+-free acidified transport media and broad nucleoside selectivity and impaired transport of guanosine under both conditions support this possibility.
Providing important additional new evidence of uridine-gated uncoupled Na+ flux, hCNT3 mutant E343Q exhibited an ohmic relationship between uridine-induced current and membrane potential. Residues of other transporters for which mutation causes uncoupled behavior include Asp-204 of the human Na+-dependent glucose transporter SGLT1 and Asn-177 of the rat 5-hydroxytryptamine transporter (41, 42).
Restoring the negative charge at hCNT3 residue position Glu-343 by mutation to aspartate (E343D) regained the wild-type phenotype, although Vmax values for uridine transport and cation activation were lower than those for hCNT3. Thus, both the presence and positioning of the Glu-343 negative charge are critical for normal functional activity.
New evidence of the importance of hCNT3 Glu-343 also arose from its mutation to cysteine (E343C). Compromised kinetically with respect to both uridine transport and Na+/H+-activation, and with a marked decrease in Na+ binding affinity, E343C exhibited small uridine-evoked H+ currents relative to those of Na+. Additionally, E343C demonstrated Na+:uridine coupling features intermediate between those of E343Q (uncoupled) and E343D (fully coupled). Different from wild-type hCNT3, however, the mean Na+:uridine stoichiometry was 1:1. E343C nevertheless retained sigmoid Na+-activation kinetics, indicating retention of both Na+-binding sites. Similar to E343Q, E343C showed decreased transport of the purine nucleoside guanosine. E343 therefore functions in determination of both cation coupling and nucleoside specificity.
Glu-519, Functional Considerations
hCNT3 Glu-519 was also revealed to have key roles in cation/nucleoside cotransport. This residue resides in putative TM 11A (Fig. 1) and is centrally positioned in the highly conserved (G/A)XKX3NEFVA(Y/M/F) motif common to all eukaryote and prokaryote CNTs. Previous indications for a role of this residue in CNT function comes from studies of the corresponding residue in hCNT1, for which mutant E498D exhibited a modest reduction in Vmax value for Na+, and a small increase in apparent Km value for uridine (31). Anticipated to show more pronounced phenotypic changes, hCNT1 mutant E498Q was not processed to the cell surface in oocytes and therefore could not be analyzed functionally (31). hCNT1 E498C was also nonfunctional, presumably for the same reason (31). In the case of hCNT3, however, all three hCNT3 Glu-519 mutants (E519Q, E519D, and E519C) were processed to the oocyte cell surface similar to wild-type hCNT3 providing, for the first time, a means to assess the role of this glutamate residue in CNT Na+/nucleoside cotransport.
Despite its confirmed presence in the oocyte plasma membrane, hCNT3 mutant E519Q showed no measurable transport activity and therefore is the first reported example of a mutation in hCNT3 rendering the protein catalytically inactive. Mutation of Glu-519 to aspartate (E519D), which restored a negative charge at this position, also fully restored Na+-coupled functional activity, with only small kinetic differences from the wild-type transporter. Similar to E343Q and E343C, however, and to a greater extent than E343D, this protein showed severely reduced H+-dependent uridine uptake compared with hCNT3, and barely detectable uridine-evoked H+ currents. Thus, H+ coupling has more stringent requirements for the positioning of the Glu-519 negative charge than Na+ coupling.
Altered interaction with H+ was also evident for E519C with radiolabeled uptake assays and current measurements both reporting no detectable H+-mediated uridine uptake. Additionally, E519C showed preference for pyrimidine nucleosides compared with purine nucleosides (uridine > cytidine, thymidine > adenosine, inosine > guanosine) and, kinetically, exhibited large decreases in uridine and Na+ binding affinity and Na+ Vmax, with the change in Na+ binding resembling that for E343C. In contrast to wild-type hCNT3 (19), E519C also exhibited an apparent absence of voltage dependence of Na+ K50. As well, flux/charge experiments found a change in Na+:nucleoside stoichiometry from 2:1 to 1:1, a characteristic shared by the more variable Na+ coupling of E343C. Differences in the stringency of altered Na+ coupling between E519C and E343C nevertheless reinforce other indications of dissimilar mechanistic roles of the two residue positions.
Recently, a naturally occurring hCNT3 variant has been described in the Spanish population in which Cys-602 is substituted by arginine (43). This single amino acid replacement in TM 13 leads to a shift in Hill coefficient consistent with a possible change in Na+:nucleoside stoichiometry from 2:1 to 1:1. In contrast, both of the hCNT3 mutants with altered stoichiometry investigated in this study (E343C and E519C) retained sigmoidal Na+-activation curves. With calculated Hill coefficients of 1.4 (E343C) and 1.9 (E519C), it is possible that both of these constructs still bind two Na+ ions but that only one is translocated.
In studies of wild-type hCNT1 and hCNT3, we have established that Na+ binds to the transporter first, increasing the affinity for nucleoside, which then binds second (19, 35). To minimize the potential effects of altered Na+ apparent affinity on uridine kinetic parameters, experiments to investigate uridine transport kinetics were undertaken at the maximum possible Na+ concentration of 100 mm. It is nevertheless possible that the very large reduction in the uridine apparent Km value observed for E519C is, at least in part, secondary to the low apparent binding affinity for Na+ and the change in Na+:nucleoside coupling ratio from 2:1 to 1:1.
Glu-343 and Glu-519, Mechanistic Considerations
CNTs have previously been defined by 13 putative TMs (Fig. 1), with two additional TMs (5A and 11A) weakly predicted by computer algorithms (18). A number of topological and structure/function studies are consistent with both a 13- and 15-TM membrane architecture (18, 34, 44–46). However, a newly completed SCAM analysis of TMs 11, 11A, 12, and 13 of a functional cysteine-free version of hCNT3 (hCNT3C−) using the membrane-impermeant thiol reagent PCMBS (47) provides unequivocal evidence for the revised 15-TM membrane topology shown in Fig. 1 (inset). In progress SCAM analyses of TMs 7 and 8 support this alternative membrane architecture.4 Relevant to the current analysis, TM 7 (containing Glu-343) is now in an orientation opposite to that shown in the original 13-TM topology model,4 and TM 11A (containing Glu-519) is now transmembrane (47).
In this investigation, the introduced cysteine residues in mutants E343C and E519C in wild-type hCNT3 were PCMBS-accessible, resulting in marked inhibition of uridine transport activity, and suggesting that both residues are pore-lining. In the case of E343C (and to a lesser extent for E519C), this inhibition was prevented by externally applied uridine, implying a location within, or close to, the nucleoside binding pocket of the translocation pore. Similarly, the equivalent TM 7 mutant of hCNT1, E322C, showed uridine-protectable inhibition by both PCMBS and the methanethiosulfonate reagent 2-aminoethylmethanethiosulfonate hydrobromide, confirming that there are topological similarities between the two transporters (31). Supporting the pore-lining location of hCNT3 Glu-343, the corresponding residue in hCNT3C−, when converted to cysteine, also results in PCMBS inhibition of uridine uptake.4 The hCNT3C− mutant corresponding to E519C was nonfunctional and not therefore amenable to testing (47). However, in hCNT3C−, residues flanking Glu-519 were both PCMBS-inhibitable and uridine-protectable (47). From the pattern of PCMBS inhibition and uridine protection observed in SCAM analysis of hCNT3C− (47),4 and from electrophysiological studies of hCNT3 presteady-state currents,5 it is likely that the sites of uridine and Na+ binding are approximately half-way across the membrane. Glu-343 may be deeper within the translocation pore, whereas Glu-519 is likely to be centrally positioned within the membrane (Fig. 1, inset).
From a mechanistic standpoint, this positions Glu-343 in a location consistent with this residue stabilizing the internal gate of the transporter vestibule. Such a function is strongly suggested by the kinetic characteristics of Glu-343 mutants, where large reductions in Vmax relative to wild-type hCNT3 were a consistent feature and, diagnostically, by the partially uncoupled activity of mutant hCNT3 E343Q (and E343C). Parallel findings of altered Na+-coupling for hCNT1 mutant E322Q support this interpretation (31).
A paradigm for the role proposed here for hCNT3 Glu-343 can be found in the potential gating function of negatively charged residues within the common cation/solute translocation pore of the recently solved three-dimensional crystal structure of the Aquifex aeolicus LeuTAa Na+/Cl−-dependent leucine transporter (48). In this protein, negatively charged residues stabilize the transporter in a closed conformation that occludes closely associated Na+- and leucine-binding sites halfway across the membrane lipid bilayer. Similar to the mammalian GAT1 Na+/Cl−-dependent γ-aminobutyric acid transporter (49, 50), a member of the same protein family as LeuTAa, hCNT3 (and hCNT1) presteady-state currents largely reflect binding and potential occlusion of extracellular Na+.5 Consistent with a potential gating function for hCNT1 Glu-322, its mutation markedly decreased hCNT1 presteady-state currents (31). Examples of other transporters where glutamate and aspartate residues are proposed to stabilize conformational transitions within the transport cycle include the E. coli PutP Na+/proline transporter (24) and LacY H+-coupled lactose permease (21, 22, 51).
hCNT3 and the other CNT3 subfamily member hfCNT each have two cation-binding sites (11, 14, 19). One hCNT3 site is Na+-specific, whereas the second site may functionally interact with both Na+ and H+ (11, 19). In the case of hfCNT, both sites are specific for Na+ (14). hCNT1 and hCNT2, in contrast, have single Na+-specific sites (11, 35). C. albicans CaCNT (16) also has a single cation-binding site, in this case H+-specific. Other CNT family members that are exclusively H+-coupled, and presumed also to have single H+-specific binding sites, include E. coli NupC (17) and C. elegans CeCNT3 (15). A central question in CNT energetics is the structural, functional, and evolutionary relationship between these various cation-binding sites in different CNT family members. In the case of human and other mammalian CNTs, for example, which of the two cation-binding sites of hCNT3 corresponds to the single Na+-specific site of hCNT1/2? Another key question is the functional role of the (G/A)XKX3NEFVA(Y/M/F) motif.
The present findings with respect to hCNT3 Glu-519 provide insights into both of these issues. Because mutation of Glu-519 led to (i) complete abolition of transport function (mutant E519Q), (ii) selective reduction of H+ dependence (mutant E519D), and (iii) loss of H+ dependence in conjunction with a change in Na+:uridine stoichiometry from 2:1 to 1:1 (mutant E519C), this residue is shown to be of crucial importance to cation coupling perhaps, as suggested by the very low Na+-binding affinity of E519C, through direct electrostatic interaction with the coupling cation. The restored functionality of E519D in Na+-containing but not H+-enriched (acidified) medium supports this conclusion and points to subtly different positional requirements for this negative charge in Na+ and H+ coupling. Alternatively, Glu-519 may facilitate cation-induced conformational transitions. Furthermore, Glu-519 appears to be specifically linked to the hCNT3 cation-binding site common to Na+ and H+. This being the case, and given that Glu-519 and the (G/A)XKX3NEFVA(Y/M/F) motif to which it belongs are present in all CNTs, we hypothesize that the shared Na+/H+-binding site of hCNT3 is the equivalent of the Na+-specific site of hCNT1/2 and the H+-specific site of NupC, CeCNT3, and CaCNT. Therefore, in hCNT3, the Na+/H+-binding site can be viewed as the primary cation-binding site of the transporter, and the Na+-specific site as a secondary (auxiliary) site. If substitution of hCNT3 Glu-519 with glutamine totally abolishes cation binding to the primary site, then complete loss of transport would be anticipated. Indeed, E519Q was catalytically inactive. In contrast to E519Q, E519C retained some functional activity, possibly because the slight electronegative character of the sulfur atom was sufficient to allow low affinity binding of Na+ to the primary site, enabling the observed low efficiency (and sigmoidal) Na+-coupled nucleoside transport to occur.
Similar primary and auxiliary cation-binding sites have also been identified in GAT1 (50). Also similar to GAT1 (50), electrophysiological studies of hCNT3 presteady-state currents predict that a Na+-dependent occlusion step (or other slow conformational change) occurs prior to uridine binding and transport.5 In the case of GAT1, this occlusion step occurs following the binding of Na+ to the primary site, thereby resulting in the auxiliary Na+ site becoming available (50).
With properties intermediate between those of E343Q and E343D yet similar to E519C, hCNT3 mutant E343C also exhibited characteristics consistent with functional impairment of the primary cation-binding site. In addition to possibly forming part of the inward gate of the transporter, Glu-343 also directly or indirectly contributes to cation binding. Mutation of both glutamate residues influences nucleoside selectivity of the transporter, providing further evidence that TMs 7 and 11A have closely aligned roles in cation/nucleoside translocation.
TMs 7 and 11A as Discontinuous Membrane Helices
Within the conserved CNT family (G/A)XKX3NEFVA(Y/M/F) motif of hCNT3/hCNT3C-TM 11A, a sequence of eight consecutive residues, extending from Phe-516 to Tyr-523 were PCMBS-sensitive upon conversion to cysteine (Fig. 13) (47). Of these, three (Phe-516, Ala-522, and Tyr-523) were uridine protectable. Flanking either end of this region, the pattern of PCMBS inhibition showed evidence of periodicity consistent with small segments of α-helical content. TM 11A therefore has characteristics of a pore-lining discontinuous helix in which the majority of the residues comprising the central conserved (G/A)XKX3NEFVA(Y/M/F) motif, and including the key glutamate residue Glu-519, adopts a relaxed, extended, and possibly mobile conformation within the translocation pore, and which allows PCMBS binding to most of the residues within the motif. A similar feature also occurs in TM 7, where Glu-343 is the last of four sequential PCMBS-sensitive residues (the others being Gly-340, Gln-341, and Thr-342), all of which are uridine-protected.4
These patterns of PCMBS inhibition in TMs 7 and 11A provide functional evidence of extended structures resembling the discontinuous membrane helices evident in LeuTAa (48) and equivalent Vibrio parahemeolyticus SGLT (52) and Microbacterium liquefaciens NCS1 (53) crystal structures. In LeuTAa, nontraditional transmembrane α-helices are disrupted by the insertion of extended regions of polypeptide that include the Na+-binding sites of the protein and, upon Na+ binding, favor high affinity binding of the permeant amino acid leucine (36). A similar feature is also apparent in TM 7 of the unrelated glutamate transporter homolog GltPh from Pyrococcus horikoshii (54) and in the E. coli NhaA Na+/H+ antiporter (55). Such regions are proposed to play important roles in ion and permeant recognition, binding, and translocation (56), a conclusion supported by the present mutagenesis studies of hCNT3 Glu-343 and Glu-519.
Within the extended regions of hCNT3 TMs 7 and 11A, two of the other three residues in TM 7 and seven of the nine other residues in TM 11A exhibit altered Na+- and/or H+-mediated uridine uptake (47),4 providing evidence that Glu-343 and Glu-519 are central elements of larger linear groupings of amino acids involved in hCNT3 cation coupling.
Conclusions
This study provides important and novel insights into the mechanism(s) of cation/nucleoside cotransport by hCNT3 and other CNT family members. Located within PCMBS-accessible and extended nonhelical regions of TMs 7 and 11A, mutation of Glu-343 and Glu-519 had primary effects on Na+/H+ coupling and stoichiometry. Parallel changes in nucleoside selectivity support close proximity integration of cation and nucleoside binding and transport within a common hCNT translocation pore.
This work was supported in part by the National Cancer Institute of Canada with funds from the Canadian Cancer Society and the Alberta Cancer Board.
R. Mulinta, A. M. L. Ng, S. Y. M. Yao, M. D. Slugoski, C. E. Cass, S. A. Baldwin, and J. D. Young, unpublished observations.
K. M. Smith, C. E. Cass, S. A. Baldwin, E. Karpinski, and J. D. Young, unpublished observations.
- NT
- nucleoside transporter
- CNT
- concentrative nucleoside transporter
- hCNT
- human CNT
- ENT
- equilibrative nucleoside transporter
- TM
- putative transmembrane helix
- MES
- 2-(N-morpholino)ethanesulfonic acid
- PCMBS
- p-chloromercuribenzene sulfonate
- SCAM
- substituted cysteine accessibility method
- hf
- hagfish.
REFERENCES
- 1.Cass C. E. ( 1995) in Drug Transport in Antimicrobial and Anticancer Chemotherapy ( Georgopapadakou N. H. ed) pp. 403– 451, Marcel Dekker, Inc., New York [Google Scholar]
- 2.Griffith D. A., Jarvis S. M. ( 1996) Biochim. Biophys. Acta 1286, 153– 181 [DOI] [PubMed] [Google Scholar]
- 3.Young J. D., Cheeseman C. I., Mackey J. R., Cass C. E., Baldwin S. A. ( 2000) in Current Topics in Membranes ( Barrett K. E., Donowitz M. eds) Vol. 50, pp. 329– 378, Academic Press, San Diego [Google Scholar]
- 4.Young J. D., Yao S. Y., Sun L., Cass C. E., Baldwin S. A. ( 2008) Xenobiotica 38, 995– 1021 [DOI] [PubMed] [Google Scholar]
- 5.Damaraju V. L., Damaraju S., Young J. D., Baldwin S. A., Mackey J., Sawyer M. B., Cass C. E. ( 2003) Oncogene 22, 7524– 7536 [DOI] [PubMed] [Google Scholar]
- 6.King A. E., Ackley M. A., Cass C. E., Young J. D., Baldwin S. A. ( 2006) Trends Pharmacol. Sci. 27, 416– 425 [DOI] [PubMed] [Google Scholar]
- 7.Latini S., Pedata F. ( 2001) J. Neurochem. 79, 463– 484 [DOI] [PubMed] [Google Scholar]
- 8.Baldwin S. A., Beal P. R., Yao S. Y., King A. E., Cass C. E., Young J. D. ( 2004) Pflugers Arch. 447, 735– 743 [DOI] [PubMed] [Google Scholar]
- 9.Gray J. H., Owen R. P., Giacomini K. M. ( 2004) Pflugers Arch. 447, 728– 734 [DOI] [PubMed] [Google Scholar]
- 10.Ritzel M. W., Ng A. M., Yao S. Y., Graham K., Loewen S. K., Smith K. M., Ritzel R. G., Mowles D. A., Carpenter P., Chen X. Z., Karpinski E., Hyde R. J., Baldwin S. A., Cass C. E., Young J. D. ( 2001) J. Biol. Chem. 276, 2914– 2927 [DOI] [PubMed] [Google Scholar]
- 11.Smith K. M., Slugoski M. D., Cass C. E., Baldwin S. A., Karpinski E., Young J. D. ( 2007) Mol. Membr. Biol. 24, 53– 64 [DOI] [PubMed] [Google Scholar]
- 12.Ritzel M. W., Yao S. Y., Huang M. Y., Elliott J. F., Cass C. E., Young J. D. ( 1997) Am. J. Physiol. 272, C707– C714 [DOI] [PubMed] [Google Scholar]
- 13.Ritzel M. W., Yao S. Y., Ng A. M., Mackey J. R., Cass C. E., Young J. D. ( 1998) Mol. Membr. Biol. 15, 203– 211 [DOI] [PubMed] [Google Scholar]
- 14.Yao S. Y., Ng A. M., Loewen S. K., Cass C. E., Baldwin S. A., Young J. D. ( 2002) Am. J. Physiol. Cell Physiol. 283, C155– 168 [DOI] [PubMed] [Google Scholar]
- 15.Xiao G., Wang J., Tangen T., Giacomini K. M. ( 2001) Mol. Pharmacol. 59, 339– 348 [DOI] [PubMed] [Google Scholar]
- 16.Loewen S. K., Ng A. M., Mohabir N. N., Baldwin S. A., Cass C. E., Young J. D. ( 2003) Yeast 20, 661– 675 [DOI] [PubMed] [Google Scholar]
- 17.Loewen S. K., Yao S. Y., Slugoski M. D., Mohabir N. N., Turner R. J., Mackey J. R., Weiner J. H., Gallagher M. P., Henderson P. J., Baldwin S. A., Cass C. E., Young J. D. ( 2004) Mol. Membr. Biol. 21, 1– 10 [DOI] [PubMed] [Google Scholar]
- 18.Hamilton S. R., Yao S. Y., Ingram J. C., Hadden D. A., Ritzel M. W., Gallagher M. P., Henderson P. J., Cass C. E., Young J. D., Baldwin S. A. ( 2001) J. Biol. Chem. 276, 27981– 27988 [DOI] [PubMed] [Google Scholar]
- 19.Smith K. M., Slugoski M. D., Loewen S. K., Ng A. M., Yao S. Y., Chen X. Z., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2005) J. Biol. Chem. 280, 25436– 25449 [DOI] [PubMed] [Google Scholar]
- 20.Pourcher T., Zani M. L., Leblanc G. ( 1993) J. Biol. Chem. 268, 3209– 3215 [PubMed] [Google Scholar]
- 21.Franco P. J., Brooker R. J. ( 1994) J. Biol. Chem. 269, 7379– 7386 [PubMed] [Google Scholar]
- 22.Karlin A. ( 1997) Proc. Natl. Acad. Sci. U. S. A. 94, 5508– 5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quick M., Jung H. ( 1997) Biochemistry 36, 4631– 4636 [DOI] [PubMed] [Google Scholar]
- 24.Quick M., Jung H. ( 1998) Biochemistry 37, 13800– 13806 [DOI] [PubMed] [Google Scholar]
- 25.Griffith D. A., Pajor A. M. ( 1999) Biochemistry 38, 7524– 7531 [DOI] [PubMed] [Google Scholar]
- 26.Pirch T., Quick M., Nietschke M., Langkamp M., Jung H. ( 2002) J. Biol. Chem. 277, 8790– 8796 [DOI] [PubMed] [Google Scholar]
- 27.Grewer C., Watzke N., Rauen T., Bicho A. ( 2003) J. Biol. Chem. 278, 2585– 2592 [DOI] [PubMed] [Google Scholar]
- 28.Noël J., Germain D., Vadnais J. ( 2003) Biochemistry 42, 15361– 15368 [DOI] [PubMed] [Google Scholar]
- 29.Chen N., Rickey J., Berfield J. L., Reith M. E. ( 2004) J. Biol. Chem. 279, 5508– 5519 [DOI] [PubMed] [Google Scholar]
- 30.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. ( 2003) Science 301, 610– 615 [DOI] [PubMed] [Google Scholar]
- 31.Yao S. Y., Ng A. M., Slugoski M. D., Smith K. M., Mulinta R., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2007) J. Biol. Chem. 282, 30607– 30617 [DOI] [PubMed] [Google Scholar]
- 32.Liman E. R., Tytgat J., Hess P. ( 1992) Neuron 9, 861– 871 [DOI] [PubMed] [Google Scholar]
- 33.Yao S. Y., Cass C. E., Young J. D. ( 2000) in Membrane Transport. A Practical Approach ( Baldwin S. A. ed) pp. 47– 78, Oxford University Press, Oxford [Google Scholar]
- 34.Slugoski M. D., Ng A. M., Yao S. Y., Smith K. M., Lin C. C., Zhang J., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2008) J. Biol. Chem. 283, 8496– 8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith K. M., Ng A. M., Yao S. Y., Labedz K. A., Knaus E. E., Wiebe L. I., Cass C. E., Baldwin S. A., Chen X. Z., Karpinski E., Young J. D. ( 2004) J. Physiol. 558, 807– 823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli U. K. ( 1970) Nature 227, 680– 685 [DOI] [PubMed] [Google Scholar]
- 37.Damaraju V. L., Elwi A. N., Hunter C., Carpenter P., Santos C., Barron G. M., Sun X., Baldwin S. A., Young J. D., Mackey J. R., Sawyer M. B., Cass C. E. ( 2007) Am. J. Physiol. Renal Physiol. 293, F200– 211 [DOI] [PubMed] [Google Scholar]
- 38.Slugoski M. D., Smith K. M., Mulinta R., Ng A. M., Yao S. Y., Morrison E. L., Lee Q. O., Zhang J., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2008) J. Biol. Chem. 283, 24922– 24934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dascal N. ( 1987) CRC Crit. Rev. Biochem. 22, 317– 387 [DOI] [PubMed] [Google Scholar]
- 40.Pastor-Anglada M., Errasti-Murugarren E., Aymerich I., Casado F. J. ( 2007) J. Physiol. Biochem. 63, 97– 110 [DOI] [PubMed] [Google Scholar]
- 41.Quick M., Loo D. D., Wright E. M. ( 2001) J. Biol. Chem. 276, 1728– 1734 [DOI] [PubMed] [Google Scholar]
- 42.Lin F., Lester H. A., Mager S. ( 1996) Biophys. J. 71, 3126– 3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Errasti-Murugarren E., Cano-Soldado P., Pastor-Anglada M., Casado F. J. ( 2008) Mol. Pharmacol. 73, 379– 386 [DOI] [PubMed] [Google Scholar]
- 44.Zhang J., Tackaberry T., Ritzel M. W., Raborn T., Barron G., Baldwin S. A., Young J. D., Cass C. E. ( 2006) Biochem. J. 394, 389– 398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loewen S. K., Ng A. M., Yao S. Y., Cass C. E., Baldwin S. A., Young J. D. ( 1999) J. Biol. Chem. 274, 24475– 24484 [DOI] [PubMed] [Google Scholar]
- 46.Slugoski M. D., Loewen S. K., Ng A. M., Smith K. M., Yao S. Y., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2007) Biochemistry 46, 1684– 1693 [DOI] [PubMed] [Google Scholar]
- 47.Slugoski M. D., Ng A. M., Yao S. Y., Lin C. C., Mulinta R., Cass C. E., Baldwin S. A., Young J. D. ( 2009) J. Biol. Chem. 284, 17281– 17292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. ( 2005) Nature 437, 215– 223 [DOI] [PubMed] [Google Scholar]
- 49.Lu C. C., Hilgemann D. W. ( 1999) J. Gen. Physiol. 114, 445– 457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hilgemann D. W., Lu C. C. ( 1999) J. Gen. Physiol. 114, 459– 475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaback H. R. ( 2005) C. R. Biologies 328, 557– 567 [DOI] [PubMed] [Google Scholar]
- 52.Faham S., Watanabe A., Besserer G. M., Cascio D., Specht A., Hirayama B. A., Wright E. M., Abramson J. ( 2008) Science 321, 810– 814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E. P., Rutherford N. G., Hadden J. M., O'Reilly J., Ma P., Saidijam M., Patching S. G., Hope R. J., Norbertczak H. T., Roach P. C., Iwata S., Henderson P. J., Cameron A. D. ( 2008) Science 322, 709– 713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yernool D., Boudker O., Jin Y., Gouaux E. ( 2004) Nature 431, 811– 818 [DOI] [PubMed] [Google Scholar]
- 55.Hunte C., Screpanti E., Venturi M., Rimon A., Padan E., Michel H. ( 2005) Nature 435, 1197– 1202 [DOI] [PubMed] [Google Scholar]
- 56.Screpanti E., Hunte C. ( 2007) J. Struct. Biol. 159, 261– 267 [DOI] [PubMed] [Google Scholar]