Abstract
The human SLC28 family of integral membrane CNT (concentrative nucleoside transporter) proteins has three members, hCNT1, hCNT2, and hCNT3. Na+-coupled hCNT1 and hCNT2 transport pyrimidine and purine nucleosides, respectively, whereas hCNT3 mediates transport of both pyrimidine and purine nucleosides utilizing Na+ and/or H+ electrochemical gradients. These and other eukaryote CNTs are currently defined by a putative 13-transmembrane helix (TM) topology model with an intracellular N terminus and a glycosylated extracellular C terminus. Recent mutagenesis studies, however, have provided evidence supporting an alternative 15-TM membrane architecture. In the absence of CNT crystal structures, valuable information can be gained about residue localization and function using substituted cysteine accessibility method analysis with thiol-reactive reagents, such as p-chloromercuribenzene sulfonate. Using heterologous expression in Xenopus oocytes and the cysteineless hCNT3 protein hCNT3C−, substituted cysteine accessibility method analysis with p-chloromercuribenzene sulfonate was performed on the TM 11–13 region, including bridging extramembranous loops. The results identified residues of functional importance and, consistent with a new revised 15-TM CNT membrane architecture, suggest a novel membrane-associated topology for a region of the protein (TM 11A) that includes the highly conserved CNT family motif (G/A)XKX3NEFVA(Y/M/F).
Specialized nucleoside transporter proteins are required for passage of nucleosides and hydrophilic nucleoside analogs across biological membranes. Physiologically, nucleosides serve as nucleotide precursors in salvage pathways, and pharmacologically nucleoside analogs are used as chemotherapeutic agents in the treatment of cancer and antiviral diseases (1, 2). Additionally, adenosine modulates numerous cellular events via purino-receptor cell signaling pathways, including neurotransmission, vascular tone, immune responses, and other physiological processes (3, 4).
Human nucleoside transporter proteins are divided into two families: the SLC29 ENT (equilibrative nucleoside transporter) family and the SLC28 CNT (concentrative nucleoside transporter) family (3, 5–7). hENTs3 mediate bidirectional fluxes of purine and pyrimidine nucleosides down their concentration gradients and are ubiquitously found in most, possibly all, cell types (8). Additionally, the hENT2 isoform is capable of nucleobase transport (9). hCNTs, in contrast, are inwardly directed Na+-dependent nucleoside transporters found predominantly in intestinal and renal epithelial and other specialized cell types (10, 11). hCNT1 and hCNT2 are pyrimidine and purine nucleoside-selective, respectively, and couple Na+/nucleoside cotransport with 1:1 stoichiometry (12–18). In contrast, hCNT3 is broadly selective for both pyrimidine and purine nucleosides and couples Na+/nucleoside cotransport with 2:1 stoichiometry (10, 18, 19). hCNT3 is also capable of H+/nucleoside cotransport with a coupling stoichiometry of 1:1, whereby one of the two Na+ binding sites also functionally interacts with H+ (18, 19).
Current models of CNT topology have 13 putative transmembrane helices (TMs) (10, 14, 16, 20). Two additional TMs (designated 5A and 11A) are weakly predicted by computer algorithms (20), and immunocytochemical experiments with site-specific antibodies and studies of native and introduced glycosylation sites have confirmed an intracellular N terminus and an extracellular C terminus (20). Chimeric studies involving hCNTs and hfCNT, a CNT from the ancient marine prevertebrate, the Pacific hagfish Eptatretus stouti, have revealed that the functional domains responsible for CNT nucleoside selectivity and cation coupling reside within the C-terminal TM 7–13 half of the protein (19, 21). NupC, an H+-coupled CNT family member from Escherichia coli, lacks TMs 1–3 but otherwise shares a topology similar to that of its eukaryote counterparts (22, 23).
A functional cysteineless version of hCNT3 has been generated by mutagenesis of endogenous cysteine residues to serine, resulting in the cysteineless construct hCNT3C− employed originally in a yeast expression system for substituted cysteine accessibility method (SCAM) analysis of TMs 11, 12, and 13 using methanethiosulfonate (MTS) reagents (24). Subsequently, we have also characterized hCNT3C− in the Xenopus oocyte expression system (25) and have initiated SCAM analyses with the alternative thiol-specific reagent p-chloromercuribenzene sulfonate (PCMBS) (26). Measured by transport inhibition, reactivity of introduced cysteine residues with PCMBS, which is both membrane-impermeant and hydrophilic, indicates pore-lining status and access from the extracellular medium; the ability of a permeant to protect against this inhibition denotes location within, or closely adjacent to, the permeant-binding pocket (27, 28). Continuing the investigation of hCNT3 C-terminal membrane topology and function, the present study reports results of PCMBS SCAM analyses of TMs 11–13, including loop regions linking the putative TMs not previously studied using MTS reagents.
In earlier structure/function studies of hCNT3, we identified a cluster of conformationally sensitive residue positions in TM 12 (Ile554, Tyr558, and Cys561) that exhibit H+-activated inhibition by PCMBS, with uridine protection evident for Tyr558 and Cys561 (26). Located deeper within the plane of the membrane, other uridine-protectable residue positions in TM 12 were PCMBS-sensitive in both H+- and Na+-containing media (26). hCNT3 Glu519 and the corresponding residue in hCNT1 (Glu498) in region TM 11A were also identified as having key roles in permeant and cation binding and translocation (29, 30), and hCNT3 E519C showed inhibition of uridine uptake by PCMBS (30). Centrally positioned within the highly conserved CNT family motif (G/A)XKX3NEFVA(Y/M/F), residue 519 is proposed to be a direct participant in cation coupling via the common hCNT3 Na+/H+-binding site that, in other CNTs, is either Na+-specific (e.g. hCNT1) or H+-specific (e.g. NupC) (30).
Building upon the prior work with MTS reagents and other structure/function studies of hCNT3, the present study identified new residues of functional importance in the C-terminal one-third of hCNT3, established the orientations and α-helical structures of TMs 11–13, and determined a novel membrane-associated topology for the TM 11A region of the protein. A revised CNT membrane architecture is proposed.
EXPERIMENTAL PROCEDURES
Construction of Cysteineless hCNT3C− cDNA
hCNT3 cDNA (GenBankTM accession number AF305210) provided the template for the construction of a cysteineless version of hCNT3 (hCNT3C−). Using site-directed mutagenesis, individual cysteine residues were converted to serine (24). hCNT3C− was transferred from the yeast E. coli shuttle vector pYPGE15 (24) into the Xenopus oocyte expression vector pGEM-HE, which provides additional 5′- and 3′-untranslated regions from the Xenopus β-globin gene flanking the multiple cloning site and gives enhanced production and functional activity of proteins expressed in Xenopus oocytes (31). The hCNT3C− construct subcloned into pGEM-HE was sequenced in both directions by Taq dyedeoxy-terminator cycle sequencing to confirm that the correct base substitutions had been made.
Site-directed Mutagenesis and Expression in X. laevis Oocytes
In hCNT3C−, residues were individually converted into cysteine using the QuikChange® site-directed mutagenesis kit (Stratagene). Constructs were sequenced in both directions to confirm that the anticipated mutation had been introduced. Plasmid DNA was linearized and transcribed with T7 polymerase using the mMESSAGE mMACHINETM (Ambion) transcription system. Defolliculated stage VI Xenopus oocytes were microinjected with 20 nl of water or 20 nl of water containing capped RNA transcripts (20 ng) and incubated in modified Barth's medium (changed daily) at 18 °C for 72 h prior to assay of transport activity.
Flux Assays and Transport Inhibition
Transport assays were performed as described previously (26, 32). Groups of 12 oocytes were incubated at room temperature (20 °C) in 200 μl of transport medium containing either 100 mm NaCl or choline chloride (ChCl), and 2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, and 10 mm HEPES (pH 8.5) or MES (pH 5.5). Uptake was traced with 14C- or 3H-labeled uridine (1 or 2 μCi/ml, respectively) (GE Healthcare) at a concentration of 10 μm. One-min uptake intervals were used to measure initial rates of transport (influx), with the exception of mutants F482C(C−), E483C(C−), Y558C(C−), A601C(C−), and A609C(C−), which showed low rates of influx and thus were subjected to 5-min uptake intervals. At the end of the incubation period, extracellular label was removed by seven rapid washes in ice-cold Na+-free (choline chloride) transport medium (100 mm ChCl, pH 7.5), and individual oocytes were dissolved in 1% (w/v) SDS for quantitation of oocyte-associated radioactivity by liquid scintillation counting (LS 6000 IC; Beckman). Also in a volume of 200 μl, oocytes were treated on ice for 10 min with 200 μm PCMBS. Excess PCMBS was removed by three washes with ice-cold transport medium before the assay of transport activity. In protection experiments, unlabeled uridine (20 mm) was included along with PCMBS (26, 33, 34). The flux values shown represent mediated transport, corrected for basal uridine uptake measured in control water-injected oocytes, and are the means ± S.E. of 10–12 oocytes. For ease of comparison, all uptake values are reported in units of pmol/oocyte·min−1.
Cell Surface Expression
The presence of recombinant hCNT3C− and hCNT3C− mutant proteins at oocyte cell surfaces was determined by labeling intact oocytes with EZ-Link sulfo-NHS-LC-biotin (Pierce) followed by isolation of the resulting biotinylated plasma membrane proteins using immobilized streptavidin resin (Pierce) according to the manufacturer's instructions. For immunoblotting, solubilized proteins (one oocyte/lane) were resolved on NuPAGE® Novex® BisTris minigels (Invitrogen). The electrophoresed proteins were transferred to polyvinylidene difluoride membranes (GE Healthcare) and probed with affinity-purified anti-hCNT3-(45–69) polyclonal antibodies (35). Blots were then incubated with horseradish peroxidase-conjugated anti-rabbit antibodies (Pierce) and developed with enhanced chemiluminescence reagents (Pierce).
RESULTS
All 14 endogenous cysteine residues of hCNT3 were replaced with serine to produce hCNT3C−, a cysteineless hCNT3 construct (24, 25). hCNT3C− retained wild-type hCNT3 functional activity but with an increased K50 value for Na+ activation (25). In parallel with previous studies (24, 26), hCNT3C− was used as a template for the construction of single cysteine mutants prior to scanning for functional activity and inhibition by PCMBS. The 133 residues spanning a region between and including TMs 11–13 that were investigated in the present study are highlighted in Fig. 1.
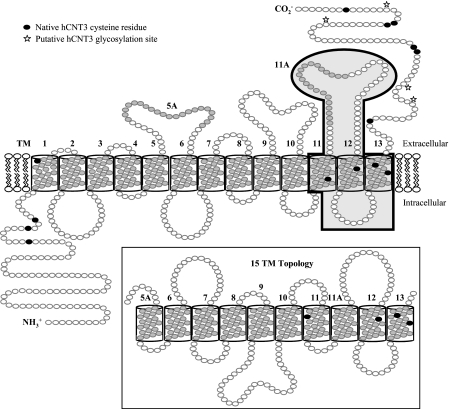
FIGURE 1.
Alternative models of hCNT3 topology. Shown is a schematic of proposed hCNT3 (GenBankTM accession number AF305210) topology with either 13 or 15 TMs. Insertion of TMs 5A and 11A into the membrane, resulting in a 15-TM membrane architecture and opposite orientations of TMs 6–11, is depicted in the inset. The positions of endogenous cysteine residues are indicated as black residues, and putative glycosylation sites are highlighted with a star. Residues studied by SCAM analysis are highlighted with a gray box.
Functional Activity of Single Cysteine Mutants
hCNT3 transports nucleosides using both Na+ and H+ electrochemical gradients (18, 19). Therefore, to examine the functional activity of single cysteine mutants, uptake of 10 μm radiolabeled uridine was determined in both Na+-containing, H+-reduced medium (100 mm NaCl, pH 8.5) and Na+-free, acidified medium (100 mm ChCl, pH 5.5, respectively). The Na+-containing medium was buffered at a pH of 8.5 to avoid the small but significant H+ activation of hCNT3 that occurs at pH 7.5 (18, 19). Previously, we have verified that Na+-coupled uridine transport by hCNT3 at pH 8.5 is kinetically indistinguishable from that at pH 7.5 (25). Initial rates of transport ± S.E. for each mutant, in units of pmol/oocyte·min−1, are given in supplemental Table 1. The uptake of 10 μm radiolabeled uridine (100 mm NaCl, pH 8.5) by oocytes expressing hCNT3C− varied between experiments, ranging from 2–4 pmol/oocyte·min−1 (data not shown). The flux values reported in supplemental Table 1 and elsewhere throughout, depict mediated transport activity, defined as the difference in uptake between RNA transcript-injected and control water-injected oocytes, and are from representative experiments. In all studies reported here, uridine uptake in water-injected oocytes was <0.02 pmol/oocyte·min−1 (data not shown).
TABLE 1.
Effect of PCMBS on uridine uptake in Xenopus oocytes expressing hCNT3C− single cysteine mutants. Influx of 10 μm [3H]uridine was measured in both Na+-containing, H+-reduced and Na+-free, acidified media (100 mm NaCl, pH 8.5, or 100 mm ChCl, pH 5.5, respectively) following a 10-min incubation on ice in the absence or presence of 200 μm PCMBS or 200 μm PCMBS + 20 mm uridine in media of the same composition used to determine uptake. Values are corrected for basal nonmediated uptake in control water-injected oocytes and are presented as a percentage of mediated uridine influx in the absence of inhibitor for each individual mutant. Each value is the mean ± S.E. of 10–12 oocytes.
| TM | Na+ (100 mm NaCl, pH 8.5) |
H+ (100 mm ChCl, pH 5.5) |
||
|---|---|---|---|---|
| With PCMBSa | With PCMBS + uridine | With PCMBSa | With PCMBS + uridine | |
| % | % | % | % | |
| 11 | ||||
| L480C(C−)b | 110 ± 20 | 90 ± 20 | 53 ± 8 | 91 ± 10 |
| F482C(C−) | 39 ± 7 | 91 ± 20 | 59 ± 8 | 98 ± 10 |
| E483C(C−) | 52 ± 6 | 42 ± 10 | 94 ± 20 | 85 ± 10 |
| L484C(C−) | 48 ± 5 | 57 ± 8 | 95 ± 10 | 86 ± 7 |
| S487C(C−)b | 47 ± 7 | 45 ± 5 | 48 ± 10 | 68 ± 5 |
| M491C(C−) | 38 ± 4 | 70 ± 7 | 60 ± 3 | 65 ± 7 |
| 11–12 loop | ||||
| M506C(C−) | 21 ± 5 | 21 ± 5 | 37 ± 3 | 37 ± 3 |
| A508C(C−) | 86 ± 10 | 78 ± 10 | 37 ± 6 | 94 ± 20 |
| G512C(C−) | 85 ± 10 | 92 ± 20 | 54 ± 10 | 83 ± 10 |
| Y513C(C−) | 45 ± 4 | 44 ± 6 | 60 ± 9 | 62 ± 6 |
| F516C(C−) | 9 ± 1 | 100 ± 10 | 11 ± 2 | 73 ± 7 |
| F517C(C−) | 12 ± 3 | 10 ± 3 | 16 ± 3 | 20 ± 3 |
| N518C(C−) | 6 ± 1 | 9 ± 1 | 5 ± 1 | 7 ± 1 |
| F520C(C−) | 26 ± 3 | 17 ± 3 | 5 ± 1 | 12 ± 2 |
| V521C(C−) | 26 ± 3 | 18 ± 2 | 35 ± 6 | 27 ± 3 |
| A522C(C−) | 30 ± 5 | 100 ± 10 | 90 ± 9 | 110 ± 10 |
| Y523C(C−) | 10 ± 1 | 77 ± 20 | 3 ± 2 | 3 ± 1 |
| L526C(C−) | 26 ± 4 | 120 ± 10 | 10 ± 2 | 35 ± 6 |
| Q545C(C−) | 83 ± 8 | 81 ± 7 | 47 ± 3 | 40 ± 3 |
| 12 | ||||
| I554C(C−) | 98 ± 20 | 93 ± 8 | 52 ± 6 | 67 ± 9 |
| T557C(C−)b | 7 ± 1 | 16 ± 2 | 22 ± 2 | 33 ± 2 |
| Y558C(C−) | 92 ± 20 | 100 ± 20 | 19 ± 3 | 71 ± 10 |
| S561C(C−) | 100 ± 20 | 99 ± 9 | 54 ± 5 | 94 ± 8 |
| N565C(C−)b | 37 ± 5 | 110 ± 10 | 46 ± 9 | 110 ± 20 |
| G567C(C−)b | 17 ± 4 | 97 ± 20 | 14 ± 5 | 94 ± 20 |
| I571C(C−)b | 44 ± 8 | 85 ± 9 | 30 ± 3 | 90 ± 8 |
| 13 | ||||
| A601C(C−) | 57 ± 7 | 43 ± 8 | 29 ± 5 | 29 ± 3 |
| A609C(C−) | 55 ± 5 | 69 ± 9 | 60 ± 6 | 76 ± 10 |
| L612C(C−) | 48 ± 5 | 100 ± 9 | 59 ± 3 | 86 ± 5 |
a Mediated uridine influx in the absence of inhibitor is given in pmol/oocytes.min−1 in supplemental Table I for each of the individual mutants.
b Previously identified as MTS-sensitive residues (24).
Mutants exhibiting uridine uptake values of <0.1 pmol/oocyte·min−1 were excluded from further analysis (supplemental Table 1). Only 9 of the 133 residues investigated fell into this category (6.8%), and in every case, the mutation to cysteine resulted in a protein with low functional activity in both Na+-containing, H+-reduced and Na+-free, acidified media (100 mm NaCl, pH 8.5, and 100 mm ChCl, pH 5.5, respectively). The nine residues were as follows: Met496 and Gly498 in TM 11; Glu519 in TM 11A; Phe563 and Ser568 in TM 12; and Arg593, Ala594, Ala606, and Gly610 in TM 13. Cell surface labeling with sulfo-NHS-LC-biotin and immobilized streptavidin resin were used to distinguish cell surface proteins from those associated with total (plasma + intracellular) membranes. Three of the nine mutants (residues Gly498, Phe563, and Ser568) that exhibited low levels of functional activity were present at cell surfaces in amounts and with electrophoretic mobilities similar to hCNT3C− (data not shown). The others were truncated (residue Met496) or present at cell surfaces in reduced amounts (residues Glu519, Arg593, Ala594, Ala606, and Gly610) (data not shown).
To facilitate comparisons between the remaining 124 mutants, the uridine transport activity of each construct is presented as the flux ratio of Na+-mediated to H+-mediated uptake (Na+/H+) in supplemental Table 1. The corresponding Na+/H+ ratios of uridine uptake (10 μm) for wild-type hCNT3 and cysteineless hCNT3C− were ∼1.7 and 1.0, respectively (averaged results from multiple experiments; data not shown), and were in good agreement with results of previous studies (18, 19, 25). Residue mutations that resulted in Na+/H+ ratios of uridine uptake of <0.5 and >2.5 (supplemental Table 1) are highlighted in the hCNT3 topology schematic shown in Fig. 2.
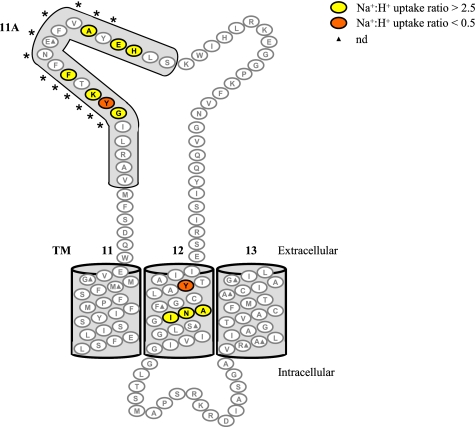
FIGURE 2.
hCNT3 TMs 11–13 depicting residues with altered Na+/H+ uridine uptake ratios. hCNT3C− mutants exhibiting Na+/H+ uridine uptake ratios of >2.5 are shown in yellow, and those with uptake ratios of <0.5 are shown in orange (supplemental Table 1). *, residues that form the conserved CNT family (G/A)XKX3NEFVA(Y/M/F) motif. Low activity mutants with uridine transport rates of <0.1 pmol/oocyte·min−1 in both Na+-containing, H+-reduced and Na+-free, acidified media (100 mm NaCl, pH 8.5, and ChCl, pH 5.5, respectively) are indicated by solid triangles. Corresponding numerical values are given in supplemental Table 1.
All mutants in TM 11, TM 13, and the loop between TMs 12 and 13 exhibited Na+/H+ uptake ratios similar to that of hCNT3/hCNT3C−. In contrast, mutant Y558C(C−) in TM 12 exhibited a ratio of 0.2, and three other TM 12 mutants exhibited ratios of >2.5 (A564C(C−), N565C(C−), and I566C(C−)) with values of 5.0, 3.3, and 2.6, respectively.
A cluster of residues with altered cation coupling characteristics was evident in the loop between TMs 11 and 12. Mutation of residue Tyr513 to cysteine resulted in a Na+/H+ ratio of 0.4, and mutation of six other residues resulted in Na+/H+ ratios of >2.5 (G512C(C−), 3.9; K514C(C−), 7.9; F516C(C−), 3.1; A522C(C−), 7.0; E524C(C−), 2.6; H525C(C−), 2.7). All seven of these residues lie within predicted TM 11A, with five located within the conserved CNT family (G/A)XKX3NEFVA(Y/M/F) motif. In hCNT3, this motif corresponds to residues GYKTFFNEFVAY, which span the region from Gly512 to Tyr523. Although the transport activity of E519C(C−) was <0.1 pmol/oocyte·min−1 and therefore not analyzed in the present study, we have previously characterized the transport activities of two Glu519 mutants (E519D and E519C) in an hCNT3 wild-type background (30). Both of these mutants displayed robust transport of uridine in the presence of Na+ but only low (E519D) or not detectable (E519C) levels of transport activity in Na+-free, acidified medium (30).
PCMBS Inhibition of Single Cysteine Mutants
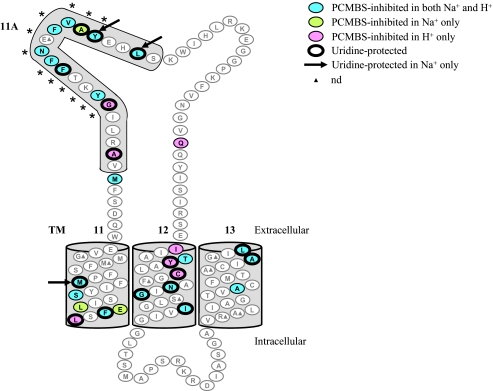
Wild-type hCNT3 and mutants bearing single cysteine residues at any of three TM 12 positions (Ile554, Tyr558, and Cys561) were previously reported to be sensitive to inhibition by PCMBS under acidic conditions only (i.e. in Na+-free, acidified medium) (26). Previous control experiments have also established the lack of inhibition by PCMBS for transport mediated by hCNT3C− in both Na+-containing, H+-reduced and Na+-free, acidified media (26). Therefore, single cysteine mutants of hCNT3C− were tested for inhibition by PCMBS both in Na+-containing, H+-reduced medium (100 mm NaCl, pH 8.5) and in Na+-free, acidified medium (100 mm ChCl, pH 5.5). After 10-min exposures to 200 μm PCMBS, uptake of 10 μm radiolabeled uridine was assayed in medium of the same composition. Exposure to PCMBS was performed on ice to minimize any possibility of diffusion or other movement across oocyte plasma membranes (26, 33). Results for each mutant calculated as a percentage of mediated uridine uptake in the absence of PCMBS are presented in Fig. 3(TM regions) and Fig. 4 (loop regions). For screening purposes, a residue was considered PCMBS-inhibitable upon exhibiting >20% inhibition of uridine uptake following incubation with PCMBS. Residues that were PCMBS-inhibitable are highlighted in Figs. 3 and 4, and the corresponding numeric values are presented in Table 1. A schematic of the locations of these PCMBS-inhibitable residues is presented in Fig. 5.
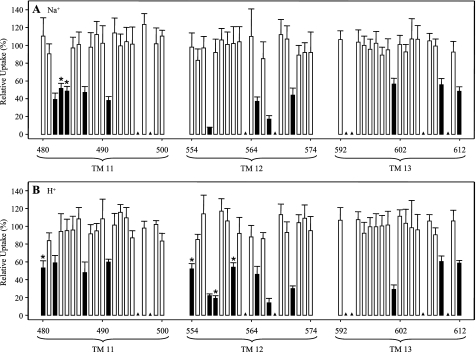
FIGURE 3.
PCMBS inhibition of residues in TMs 11–13 of hCNT3C−. Mediated influx of 10 μm radiolabeled uridine in Na+-containing, H+-reduced (A) or Na+-free, acidified (B) medium (100 mm NaCl, pH 8.5, or 100 mm ChCl, pH 5.5, respectively) was measured following 10-min incubations on ice in the same medium (A or B, respectively) in the presence of 200 μm PCMBS. Solid columns, residue positions inhibited by PCMBS; *, those residues that exhibited differential inhibition by PCMBS in the two media. Low activity mutants for which inhibition was not determined are indicated by solid triangles. Data are presented as mediated transport, calculated as uptake in RNA-injected oocytes minus uptake in water-injected oocytes and are normalized to the influx of uridine in the absence of inhibitor. Each value is the mean ± S.E. of 10–12 oocytes.
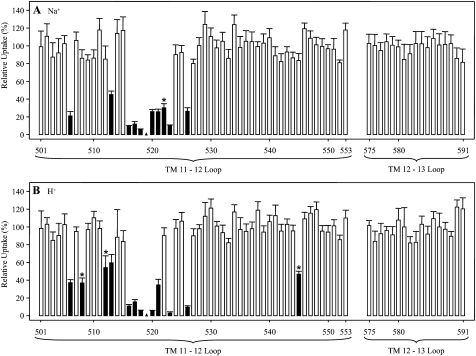
FIGURE 4.
PCMBS inhibition of residues in hCNT3C− loop regions between TMs 11 and 12 and TMs 12 and 13. Mediated influx of 10 μm radiolabeled uridine in Na+-containing, H+-reduced (A) or Na+-free, acidified (B) medium (100 mm NaCl, pH 8.5, or 100 mm ChCl, pH 5.5, respectively) was measured following 10-min incubations on ice in the same medium (A or B, respectively) in the presence of 200 μm PCMBS. Solid columns, residue positions inhibited by PCMBS; *, residues that exhibited differential inhibition to PCMBS in the two media. Low activity mutants for which inhibition was not determined are indicated by solid triangles. Data are presented as mediated transport, calculated as uptake in RNA-injected oocytes minus uptake in water-injected oocytes, and are normalized to the respective influx of uridine in the absence of inhibitor. Each value is the mean ± S.E. of 10–12 oocytes.
FIGURE 5.
hCNT3 TMs 11–13 depicting PCMBS-inhibited and uridine-protected residues. hCNT3C− mutants exhibiting inhibition of uridine uptake following incubation with PCMBS in both Na+-containing, H+-reduced and Na+-free, acidified media are indicated in blue; those that were inhibited only in Na+-containing, H+-reduced medium are indicated in green; and those that were inhibited only in Na+-free, acidified medium are indicated in red. Residues protected from PCMBS inhibition by excess unlabeled uridine are outlined in black. The three residues, Met491 in TM 11 and Tyr523 and Leu526 in TM 11A, that were inhibited by PCMBS in both media but protected from that inhibition only in the presence of Na+-containing, H+-reduced medium are indicated by a black arrow. *, residues that form the conserved CNT family (G/A)XKX3NEFVA(Y/M/F) motif. Low activity mutants are indicated by filled triangles. Corresponding numerical values are given in Table 1.
In TM 11, three residues, Phe482, Ser487, and Met491, were PCMBS-inhibitable in both Na+-containing, H+-reduced and Na+-free, acidified medium upon mutation to cysteine. Flanking Phe482, mutants E483C(C−) and L484C(C−) were PCMBS-inhibitable only in Na+-containing, H+-reduced medium, whereas L480C(C−) was PCMBS-inhibitable only in Na+-free, acidified medium. As described previously (26) and as also shown here, TM 12 contains four residues (Thr557, Asn565, Gly567, and Ile571) that, upon mutation to cysteine, were PCMBS-inhibitable in both cation conditions and three (Ile554, Tyr558, and Ser561) that were only PCMBS-inhibitable when tested in Na+-free, acidified medium. In TM 13, three mutants (A601C(C−), A609C(C−), and L612C(C−)) were PCMBS-inhibitable in both cation conditions.
Inhibition of transport by PCMBS was previously described for E519C in hCNT3 TM 11A (30). Scanning of the loop between TMs 11 and 12, which includes TM 11A, revealed additional PCMBS-inhibitable residues. Upon conversion to cysteine, residues Met506, Tyr513, Phe516, Phe517, Asn518, Phe520, Val521, Tyr523, and Leu526 were PCMBS-inhibitable under both cation conditions. Additionally, PCMBS inhibition was observed for Ala522 only in Na+-containing, H+-reduced medium, and for Ala508, Gly512, and Gln545 it was observed only in Na+-free, acidified medium. Of these 13 residues, 11 lie within the TM 11A region, and nine lie within the CNT family (G/A)XKX3NEFVA(Y/M/F) motif. In contrast, none of the residues in the TM 12–13 loop, when converted to cysteine in hCNT3C−, were inhibitable by PCMBS under either cation condition.
Uridine Protection of PCMBS Inhibition
Subsequent experiments investigated the ability of extracellular uridine (20 mm) to protect against inhibition by PCMBS for residues that were PCMBS-inhibitable in either or both cation conditions. Results for each individual mutant are presented in Table 1 as a percentage of mediated uridine uptake in the absence of PCMBS, and uridine-protectable residues are highlighted in the hCNT3 topology schematic of Fig. 5.
In TM 11, three residues exhibited uridine protection from PCMBS inhibition. L480C(C−), which was PCMBS-inhibitable only in Na+-free, acidified medium, demonstrated full protection against that inhibition, and F482C(C−), which was PCMBS-inhibitable in both Na+-containing, H+-reduced and Na+-free, acidified media was also fully protectable under both cation conditions. In contrast, M491C(C−), which was also PCMBS-inhibitable under both cation conditions, was partially protectable only in Na+-containing, H+-reduced medium. As previously described (26), five of the seven PCMBS-inhibitable mutants in TM 12 exhibited uridine protection. These were N565C(C−), G567C(C−), and I571C(C−), which were PCMBS-inhibitable under both cation conditions, and Y558C(C−) and S561C(C−), which were PCMBS-inhibitable only in Na+-free, acidified medium. Except for Y558C(C−), the four other TM 12 mutants exhibited full protection against PCMBS inhibition. Of the three mutants in TM 13 that were PCMBS-inhibitable under both cation conditions, L612C(C−) and A609C(C−) were fully and partially protectable, respectively, by extracellular uridine under both conditions.
In the TM 11–12 loop, only one mutant inhibitable by PCMBS under both cation conditions, F516C(C−), also exhibited uridine protection under both conditions. Y523C(C−) and L526C(C−), which were PCMBS-inhibitable under both cation conditions, were only protectable in Na+-containing, H+-reduced medium. A508C(C−) and G512C(C−), which were PCMBS-inhibitable only in Na+-free, acidified medium, and A522C(C−), which was PCMBS-inhibitable only in Na+-containing, H+-reduced medium, also showed uridine protection but only under the same cation conditions. All six of the uridine-protectable residues in the TM 11–12 loop are located in TM 11A, and three of these reside within the CNT family (G/A)XKX3NEFVA(Y/M/F) motif.
DISCUSSION
Studies from our laboratory have previously used PCMBS to investigate individual topological features and residues of hCNT1 and hCNT3 (26, 29, 30, 34). Incorporating results from TM 12 (26), the present investigation reports a SCAM analysis of the entire C-terminal one-third of the latter protein (hCNT3C− TMs 11–13, inclusive) (Fig. 1). In so doing, we have systematically analyzed functional and structural relationships among 133 consecutive residues in the C-terminal region of hCNT3.
Functional Activity of hCNT3C− Mutants
Initial characterization of the hCNT3C− single cysteine mutants in the present study measured uridine uptake in both Na+-containing, H+-reduced and Na+-free, acidified media (supplemental Table 1). Of the 133 mutants examined, nine exhibited uridine uptake values of <0.1 pmol/oocyte·min−1 under both cation conditions (supplemental Table 1). Two of these mutants were in TM 11 (M496C(C−) and G498C(C−)), one in TM 11A (E519C(C−)), two in TM 12 (F563C(C−) and S568C(C−)), and four in TM 13 (R593C(C−), A594C(C−), A606C(C−), and G610C(C−)). M496C(C−), G498C(C−), F563C(C−), and S568C(C−) were present at cell surfaces, suggesting loss of intrinsic transport capability. M496C(C−) was truncated, consistent with proteolytic cleavage to a lower molecular weight species. The other five exhibited reduced quantities in plasma membranes. All nine mutants were excluded from further functional analysis.
The previous MTS study that examined TMs 11–13 in yeast also identified the majority of these same residues, upon mutation to cysteine, to be nonfunctional (Met496, Gly498, Phe563, Ala594, and Ala606) or to exhibit low levels of transport activity (Ser568, Arg593, and Gly610) (24). In addition, G598C(C−) was nonfunctional, and I485C(C−), L595C(C−), and F603C(C−) exhibited low transport activities in yeast (24). Cell surface processing experiments by immunofluorescence and confocal microscopy found all of these mutants, except G498C(C−) and L595C(C−), to be present in yeast plasma membranes in similar abundance to the wild-type protein (24). Common to both studies, therefore, and excluding Met496 in TM 11, the following amino acids have potentially important roles in hCNT3 structure and/or function: Phe563 and Ser568 in TM 12 and Arg593, Ala594, Ala606, and Gly610 in TM 13. In the yeast expression system, mutants M496C(C−) and G598C(C−) were rescued by conversion to alanine (M496A(C−) and G598A(C−), respectively), whereas G498C(C−) and F563C(C−) were not (24). Although broadly similar in overall profile, the observed differences in the functional activities of some mutants indicate the existence of variations in processing, expression, and/or conformation of the mature proteins at cell surfaces in the Xenopus oocyte and yeast expression systems.
hCNT3 couples nucleoside transport to both Na+ and H+ electrochemical gradients and exhibits a Na+/H+ ratio of uridine uptake (10 μm) of ∼1.7 (18, 19). Similarly, hCNT3C− also mediates both Na+- and H+-coupled uridine transport, although the apparent K50 for Na+ is increased ∼11-fold, decreasing the Na+/H+ ratio of uridine uptake (10 μm) to ∼1.0 (25). Supplemental Table 1 and Fig. 2 highlight those residues for which mutation to cysteine in hCNT3C− resulted in Na+/H+ uridine uptake ratios of either <0.5 (i.e. H+-preferring) or >2.5 (i.e. Na+-preferring). Such mutants identified residues likely to be involved directly or indirectly in interactions with the coupling cation. The locations of these residues corresponded well with regions of the protein shown in other studies to be important for hCNT cation interactions.
In a previous investigation, mutation of hCNT3 Cys561, a conformationally sensitive TM 12 residue located at the Na+/H+ boundary of extracellularly accessible residues, influenced interactions with the two coupling cations (Na+ and H+) differently (26). In good agreement with a role for TM 12 in cation interactions, mutation to cysteine in hCNT3C− resulted in Na+/H+ ratios of uridine uptake of <0.5 for residue Tyr558 (i.e. H+-preferring) and >2.5 for residues Ala564, Asn565, and Ile566 (i.e. Na+-preferring). Also in a previous study, the hCNT3 TM 11A residue Glu519 was demonstrated to be an important determinant of Na+/H+ coupling to the conserved CNT cation binding site that in other family members interacts exclusively with either Na+ or H+ (30). In the present investigation, mutation to cysteine in hCNT3C− resulted in Na+/H+ ratios of uridine uptake of <0.5 for residue Tyr513 and >2.5 for residues Gly512, Lys514, Phe516, Ala522, Glu524, and His525 in the TM 11–12 loop, all of which also reside within TM 11A. Thus, the present results expand the role(s) of hCNT3 E519 (TM 11A) and C561 (TM 12) in cation binding and/or translocation to other adjacent residues in the same regions of the protein. Further analysis of the roles of these newly characterized amino acids in cation coupling is in progress.
PCMBS Inhibition of hCNT3C− Mutants; Transmembrane Architecture and Orientation of TMs 11–13
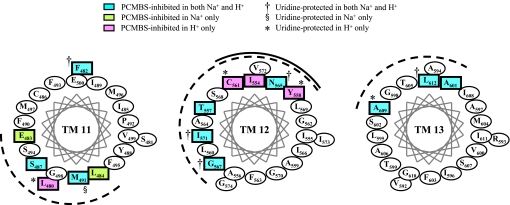
To summarize the data presented in Table 1 and Figs. 3 and 4, the schematic in Fig. 5 highlights those residues identified in hCNT3C− as PCMBS-sensitive and uridine-protectable. In agreement with previous predictions (20, 24), the pattern of PCMBS inhibition evident within TMs 11–13 supports a conventional membrane-spanning α-helical architecture for these regions, because affected residues, in general, cluster on one face of each of the helices. To more clearly demonstrate this, α-helical wheel projections for each of the TMs are presented in Fig. 6, although it is appreciated that the true structures of these regions of the protein may differ from the perfect α-helices illustrated (see also Ref. 26 for a space-filling representation of TM 12).
FIGURE 6.
Helical wheel projections of hCNT3 TMs 11–13. The helical wheel projections, as viewed from the extracellular side of the membrane, highlight in blue the locations of hCNT3 residues that were inhibited by PCMBS in both Na+-containing, H+-reduced and Na+-free, acidified media, in green those residues that were inhibited only in Na+-containing, H+-reduced medium, and in red those that were inhibited only in Na+-free, acidified medium. Those residues for which uridine protection was evident in both media or in either medium only are indicated by the †, §, and *, respectively. Corresponding numerical values are given in Table 1.
In TM 11, PCMBS inhibition is evident for residues Glu483 and Leu484 in Na+-containing, H+-reduced medium only; Leu480 in Na+-free, acidified medium only; and Phe482, Ser487, and Met491 in both cation conditions. Of these, Leu480 (Na+-free, acidified only), Phe482 (both cation conditions), and Met491 (Na+-containing, H+-reduced only) showed uridine protection. In the previous study in yeast, inhibition by MTS reagents for Leu480 and Ser487 in Na+-containing, H+-reduced medium was partially protected by uridine (24). In the current 13 TM model of hCNT3 topology, the six PCMBS-inhibitable residues in TM 11 span a central to deep region within the helix, including the last putative residue (Leu480) of the helix (Fig. 5). Uridine protection was evident for residues located at both ends of this region (Met491 and Leu480), a pattern of inhibition supporting the reversed orientation of this helix. As such, the majority of PCMBS-inhibitable residues would lie exofacially within the helix in a position more likely to be accessible to the extracellular medium and available for PCMBS binding. Five of the six PCMBS-inhibitable residues cluster to one face of the helix (Fig. 6), consistent with an α-helical structure. Residue Phe482, however, resides on a separate face of the helix. A reversed orientation of TM 11, as predicted by the 15-TM model of hCNT3 membrane topology (Fig. 1, inset), would place residue Phe482 at the exofacial boundary of the TM in a position potentially in contact with the external medium and accessible to PCMBS despite its location in a different face within the helix relative to the other PCMBS-inhibitable residues.
As previously reported (26), three residues in TM 12 (Ile554, Tyr558, and Ser561) showed PCMBS inhibition only in Na+-free, acidified medium and four additional residues (Thr557, Asn565, Gly567, and Ile571) showed PCMBS inhibition under both cation conditions (Fig. 5). Of the seven PCMBS-sensitive residues identified in TM 12, five (Tyr558, Ser561, Asn565, Gly567, and Ile571) showed uridine protection. In good agreement with these results, the yeast study also identified Thr557, Asn565, Gly567, and Ile571 as MTS-sensitive in Na+-containing, H+-reduced medium, and these four residues exhibited varying degrees of uridine protection (24). In addition to residing in the extracellular facing, exofacial half of the helix, the residues inhibitable by PCMBS only in Na+-free, acidified medium cluster to one quadrant of the helix surface, whereas those that were reactive under both cation conditions span a region that is deeper within the helix and extends over a wider aspect of the helix face (). Consistent with a conventional α-helical structure for TM 12, all of the PCMBS-accessible residues nevertheless reside within one-half of the helix surface. TM 12 Cys561 is the residue responsible for PCMBS inhibition of wild-type hCNT3 (26), which occurred under acidified conditions only, and therefore reported a specific conformational change associated with H+ binding (26). Adjacent residues Ile554 and Tyr558 reported the same H+-mediated conformational transition. Unlike TM 11, the longitudinal dispersal of PCMBS-sensitive residues shown in Fig. 5 for TM 12 suggests that the helix is presented in its correct transmembrane orientation. Nevertheless, the finding that residue Ile571 showed clear evidence of PCMBS inhibition and uridine protection (Table 1) indicates that the hCNT3 translocation pore penetrates deep within the membrane.
Although no TM 13 residues were previously reported as MTS-sensitive (24), the present study identified Ala601, Ala609, and Leu612 as PCMBS-sensitive under both cation conditions. Uridine protection from PCMBS inhibition was evident for the two most exofacially located of these residues, Ala609 and Leu612 (Fig. 5). Supporting an α-helical structure for TM 13, these three residues clustered to one face of the helix (Fig. 6). Similar to TM 12, but unlike TM 11, the predicted locations of Ala601, Ala609, and Leu612 within the extracellular half of the membrane (Fig. 5) provided evidence that TM 13 is correctly oriented in both the 13 and 15 TM topology models of hCNT3 membrane architecture (Fig. 1). Consistent with previous findings for rat CNT1 (20), our recent demonstration that all hCNT3 glycosylation sites are confined to the C-terminal end affords additional supporting evidence in this regard (25).
PCMBS Inhibition of hCNT3C− Mutants; Transmembrane Architecture and Orientation of TM 11A
The TM 11–12 loop, encompassing TM 11A, revealed an unexpected and novel pattern of reactivity to PCMBS (Table 1 and ). Fourteen of the 52 residues in this region showed inhibition by PCMBS upon conversion to cysteine in hCNT3C−, including residue Ala522 only in Na+-containing, H+-reduced medium; Ala508, Gly512, and Gln545 only in Na+-free, acidified medium; and Met506, Tyr513, Phe516, Phe517, Asn518, Phe520, Val521, Tyr523, and Leu526 under both cation conditions. Partial or complete uridine protection from PCMBS inhibition was evident for six of the residues: Ala522 and Tyr523 only in Na+-containing, H+-reduced medium; Ala508 and Gly512 only in Na+-free, acidified medium; and Phe516 and Leu526 under both cation conditions. Together, the results suggested that this region is critically involved in the transport mechanism of hCNT3 and predicted that at least a portion is membrane-associated, either in the form of a re-entrant loop or as part of a transmembrane domain. The majority of the PCMBS-sensitive residues reside within the previously predicted membrane domain TM 11A. One exception is M506, for which the corresponding hCNT3C- mutant showed PCMBS inhibition under both cation conditions and is positioned immediately adjacent to the TM 11A region. It is possible that the putative boundary of TM 11A includes Met506 within the membrane-associated region. The other exception is Gln545, which is separated from putative TMs 11A and 12 by 17 and 8 residues, respectively (Fig. 5). Q545C(C−) showed robust PCMBS inhibition only in Na+-free, acidified medium and was not uridine-protected. Centrally positioned in the putative exofacial loop region linking TMs 11A and 12 (Figs. 1 (inset) and 5), residue Gln545 is unlikely to be membrane-associated but may nevertheless reside in sufficiently close proximity to the exofacial aspect of the translocation pore that binding of PCMBS interferes with hCNT3 cotransport activity. Since Q545C(C−) was inhibited by PCMBS only in Na+-free, acidified medium, it may be similar to Cys561, reporting a specific H+-induced conformation of the transporter.
The overall pattern of PCMBS reactivity in putative TM 11A measured by transport inhibition suggested that this region, or at least part of it, differs from the traditional α-helical structure of membrane-associated TMs. Within the conserved CNT family (G/A)XKX3NEFVA(Y/M/F) motif of TM 11A, a sequence of eight consecutive residues, extending from Phe516 to Tyr523, were PCMBS-sensitive upon conversion to cysteine under both cation conditions, the only exceptions being Glu519, which showed low levels of uridine transport activity, and Ala522, which showed PCMBS inhibition only in Na+-containing, H+-reduced medium. In the wild-type hCNT3 background, mutant E519C exhibited Na+-specific nucleoside transport activity and was PCMBS-sensitive in the presence of Na+ (30). Within this region of eight PCMBS-inhibitable residues, three (Phe516, Ala522, and Tyr523) were uridine-protectable. Flanking either end of this block of PCMBS-sensitive residues, and especially noticeable in the first part of the TM adjacent to Met506, the pattern of PCMBS inhibition showed evidence of periodicity consistent with small segments of α-helical content. TM 11A therefore has characteristics of a pore-lining discontinuous helix in which the majority of the residues comprising the central conserved (G/A)XKX3NEFVA(Y/M/F) motif, including the key glutamate residue Glu519, adopt a relaxed, extended, and possibly mobile conformation within the translocation pore, which allows PCMBS binding to most of the residues within the motif. Block patterns of PCMBS reactivity also occur in TMs 7 and 8.4
As such, the pattern of PCMBS inhibition reported here for TM 11A (and apparent also in TMs 7 and 8)4 provides functional evidence of extended structures resembling the discontinuous membrane helices evident in crystal structures of the recently solved Na+-coupled bacterial leucine, galactose, and hydantoin membrane transport proteins Aquifex aeolicus LeuTAa (36), Vibrio parahemeolyticus SGLT (37), and Microbacterium liquefaciens NCS1 (38). In LeuTAa, nontraditional transmembrane α-helices are disrupted by the insertion of extended regions of polypeptide that comprise the Na+ binding sites of the protein and, upon Na+ binding, favor high affinity binding of the permeant amino acid leucine (36). A similar feature is also apparent in TM 7 of the glutamate transporter homologue GltPh from Pyrococcus horikoshii (39). Reviewed by Screpanti and Hunte (40), such discontinuous membrane helices are proposed to play important mechanistic roles in ion and permeant recognition, binding, and translocation in secondary active transporters. In the case of TM 11A in hCNT3, the centrally positioned glutamate residue Glu519, which resides in the conserved (G/A)XKX3NEFVA(Y/M/F) motif, plays a critical and possibly direct role in cation binding/coupling that is probably common to all CNT family members (30). Separated only by 26 amino acids, membrane-associated TM 11A is likely to lie within the translocation pore in close proximity to TM 12, which we have shown to undergo cation-dependent conformational changes (26).
Overall Topological and Mechanistic Implications for hCNT3
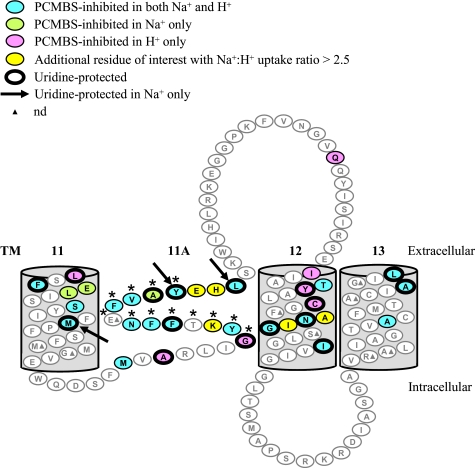
Although the pattern of PCMBS-inhibitable and uridine-protectable residues in TMs 12 and 13 support the current α-helical orientation of these regions, a reversed orientation is more plausible in TM 11. In this scenario, membrane-associated TM 11A would be membrane-spanning rather than a re-entrant loop (see schematic in Fig. 7). This is in good agreement with earlier predictions for TMs 5A and 11A as potentially membrane-spanning (20), as shown in the inset of Fig. 1. Previously, SCAM analysis of TMs 11–13 revealed MTS-sensitive residues buried deep within the TM 11 helix (24). Thus, the present study expands upon these earlier findings and provides solid experimental evidence to support the original hypothesis of an additional TM 11A. As shown in Fig. 1 (inset), insertion of both TMs 5A and 11A through the membrane bilayer would result in opposite orientation for TMs 6–11. In agreement with this, a recent structure-function study of negatively charged residues in hCNT1 provided evidence for an opposite orientation for TM 7 (29). Full PCMBS SCAM analysis of hCNT3C− TMs 7 and 8 is in progress and confirms their opposite orientation.4 Therefore, current evidence favors a revised model of hCNT3 membrane architecture (Fig. 1, inset). In parallel studies of E. coli NupC, the transmembrane nature of the TM 11A region has been unambiguously established by examining the susceptibilities of single cysteine mutants, in intact and lysed bacteria, to modification by membrane-impermeant Oregon green maleimide and ethyltrimethylammonium methanethiosulfonate.5
FIGURE 7.
Revised topology of hCNT3 TM 11–13 depicting TM 11A as membrane-spanning. This schematic depicts the insertion of TM 11A as membrane-spanning, thus reversing the orientation of TM 11. For illustrative purposes, the endofacial boundary of helix 11A, as illustrated in Figs. 1, 2 and 5, has been shifted by one residue to include the residue Met506. PCMBS-inhibited and uridine-protected residues, as well as additional residues of interest with Na+/H+ uridine uptake ratios of >2.5 but not inhibited by PCMBS, are highlighted according to Figs. 5 and 2, respectively. Two residues, Tyr513 in TM 11A and Tyr558 in TM 12, which exhibited Na+/H+ uridine uptake ratios of <0.5, were also PCMBS-inhibited and are only indicated as such. The three residues, Met491 in TM 11 plus Tyr523 and Leu526 in TM 11A, that were inhibited by PCMBS in both media but were protected from that inhibition only in Na+-containing, H+-reduced medium are indicated by a black arrow. *, residues that form the conserved CNT family (G/A)XKX3NEFVA(Y/M/F) motif. Low activity mutants are indicated by the filled triangles. Corresponding numerical values are given in supplemental Table 1 and Table 1.
PCMBS-inhibitable and uridine-protectable residues were identified in TMs 11, 11A, 12, and 13, thereby placing aspects of all four regions within or in close proximity to the permeant translocation pathway of hCNT3. At the very least, all of the residues inhibited by PCMBS must be solvent-accessible from the extracellular medium and in a location where PCMBS binding compromises transport activity. Serving as a control for the present experiments, the putative intracellular loop between TMs 12 and 13 showed no inhibition by PCMBS despite robust functional activity of all of the mutants. Quantitatively, the residues showing the most severe inhibition by PCMBS were located in TMs 11A and 12 and not in TMs 11 and 13 (Table 1). For example, TMs 11 and 13 contained no residues for which uptake was inhibited by PCMBS by >80%, whereas TM 11A contained six (Phe516, Phe517, Asn518, Phe520, Tyr523, and Leu526) and TM 12 contained three (Thr557, Tyr558, and Gly567). Furthermore, the five residues that were uridine-protectable (Phe516, Tyr523, Leu526, Tyr558, and Gly567) were fully protected by extracellular uridine (Table 1). This strongly implicated these two TMs in formation of key functional regions of the translocation pore and supports results of previous studies identifying important residues in TMs 11A (29, 30) and 12 (26). Additionally, residues that altered the Na+/H+ ratio of uridine uptake were also located in TMs 11A and 12 (Fig. 7). In contrast, residues in TMs 11 and 13 were inhibited by PCMBS to a lesser extent, and of the five residues that were uridine-protectable, two (Met491 in TM 11 and Ala609 in TM 13) were only partially protected by extracellular uridine (Table 1). Similarly, only two hCNT3C− residues in TM 11 (Leu480 and Ser487) and no residues in TM 13 were identified as inhibitable by MTS reagents in the yeast study, and those mutants also displayed only partial inhibition and protection (24). Together with the observation that PCMBS-accessible residues were more exofacially located in TMs 11 and 13 than in TMs 11A and 12 (Fig. 7), these findings suggested that TMs 11 and 13 may be less directly involved in formation of the translocation pore than TMs 11A and 12. Some cases of PCMBS inhibition and/or uridine protection may, for example, be secondary indirect effects of cation- or permeant-induced conformational changes rather than indicators of close proximity interaction with cation or nucleoside binding domains.
The present study also contributed two further insights into hCNT3 structure and function. The first relates to cation-dependent conformations adopted by the exofacially facing form of the protein. In contrast to Na+-specific hCNT1 and hCNT2, hCNT3 mediates both Na+- and H+-coupled nucleoside cotransport (10, 14, 16–19). The cation/nucleoside stoichiometry for hCNT3 H+-coupled transport is 1:1, compared with 2:1 for Na+, and when both cations are present, charge/uptake experiments suggest that hCNT3 binds one Na+ and one H+ (18, 19). The nucleoside and nucleoside drug selectivity pattern of hCNT3 in the presence of H+ also differs from that in the presence of Na+ (10, 19). Previously, mutation of hCNT3 Cys561 in TM 12 was reported to alter Na+ and H+ kinetics and, together with Tyr558 and Ile554, form a face of the helix that becomes extracellularly accessible to PCMBS only in the presence of H+ (Fig. 6), thus reporting a H+-dependent conformation of the protein (25, 26). Building upon these observations, the different patterns of residues exhibiting PCMBS inhibition and uridine protection in Na+-containing, H+-reduced medium only versus Na+-free, acidified medium versus both media provided strong additional support for the existence of multiple Na+- and/or H+-induced conformational states of hCNT3 (Table 1 and Fig. 5), some of which, like the H+-specific TM 12 I554/Y558/C561 cluster, involve subdomains within TMs. Other potential conformational differences are even more subtle. The arrows in Figs. 5 and 7, for example, identify residues in TMs 11 and 11A that were PCMBS-sensitive in both media but were uridine-protected only in Na+- containing, H+-reduced medium.
Second, the present studies support the concept of central closely adjacent cation-nucleoside binding domains within a common cation/nucleoside translocation pore. Similar to TM 12, TM 11A contains residues deep within the membrane that were PCMBS-inhibitable and uridine-protectable. Met491, a centrally positioned residue in TM 11, also shares this phenotype. In good agreement with this, other cation transporters for which high resolution molecular structures have been solved, including LeuTAa (36) and GltPh (41), also exhibit central cation and permeant binding domains.
Conclusions
Our SCAM analyses of the C-terminal one-third of hCNT3 highlight the functional importance of residues in TMs 11A and 12 in key cation and nucleoside binding and/or translocation events. The results also support a revised topology model for hCNTs, with the insertion of TM 11A as a membrane-spanning discontinuous helix. Confirmation of this new membrane architecture will come from on-going studies of TMs 7 and 8 as well as other remaining portions of the C-terminal half of hCNT3 and by extension of SCAM analysis to TMs in the N-terminal half of the transporter, including TM 5A.
This work was supported in part by the National Cancer Institute of Canada, with funds from the Canadian Cancer Society and the Alberta Cancer Board, Canada.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
R. Mulinta, A. M. L. Ng, S. Y. M. Yao, M. D. Slugoski, C. E. Cass, S. A. Baldwin, and J. D. Young, unpublished results.
H. Xie, L. Sun, D. A. Hadden, R. Mulinta, S. K. Loewen, J. C. Ingram, G. J. Litherland, M. P. Gallagher, P. J. F. Henderson, C. E. Cass, J. D. Young, and S. A. Baldwin, unpublished results.
- hENT
- human ENT
- hCNT
- human CNT
- TM
- putative transmembrane helix
- SCAM
- substituted cysteine accessibility method
- MTS
- methanethiosulfonate
- PCMBS
- p-chloromercuribenzene sulfonate
- MES
- 2-(N-morpholino)ethanesulfonic acid
- BisTris
- 2-[bis(2- hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Cass C. E. ( 1995) in Drug Transport in Antimicrobial and Anticancer Chemotherapy ( Georgopapadakou N. H. ed) pp. 403– 451, Marcel Dekker, New York [Google Scholar]
- 2.Griffith D. A., Jarvis S. M. ( 1996) Biochim. Biophys. Acta 1286, 153– 181 [DOI] [PubMed] [Google Scholar]
- 3.King A. E., Ackley M. A., Cass C. E., Young J. D., Baldwin S. A. ( 2006) Trends Pharmacol. Sci. 27, 416– 425 [DOI] [PubMed] [Google Scholar]
- 4.Latini S., Pedata F. ( 2001) J. Neurochem. 79, 463– 484 [DOI] [PubMed] [Google Scholar]
- 5.Young J. D., Cheeseman C. I., Mackey J. R., Cass C. E., Baldwin S. A. ( 2000) Curr. Top. Membr. 50, 329– 378 [Google Scholar]
- 6.Baldwin S. A., Beal P. R., Yao S. Y., King A. E., Cass C. E., Young J. D. ( 2004) Pflugers Arch. 447, 735– 743 [DOI] [PubMed] [Google Scholar]
- 7.Gray J. H., Owen R. P., Giacomini K. M. ( 2004) Pflugers Arch. 447, 728– 734 [DOI] [PubMed] [Google Scholar]
- 8.Young J. D., Yao S. Y., Sun L., Cass C. E., Baldwin S. A. ( 2008) Xenobiotica 38, 995– 1021 [DOI] [PubMed] [Google Scholar]
- 9.Yao S. Y., Ng A. M., Vickers M. F., Sundaram M., Cass C. E., Baldwin S. A., Young J. D. ( 2002) J. Biol. Chem. 277, 24938– 24948 [DOI] [PubMed] [Google Scholar]
- 10.Ritzel M. W., Ng A. M., Yao S. Y., Graham K., Loewen S. K., Smith K. M., Ritzel R. G., Mowles D. A., Carpenter P., Chen X. Z., Karpinski E., Hyde R. J., Baldwin S. A., Cass C. E., Young J. D. ( 2001) J. Biol. Chem. 276, 2914– 2927 [DOI] [PubMed] [Google Scholar]
- 11.Elwi A. N., Damaraju V. L., Baldwin S. A., Young J. D., Sawyer M. B., Cass C. E. ( 2006) Biochem. Cell Biol. 84, 844– 858 [DOI] [PubMed] [Google Scholar]
- 12.Huang Q. Q., Yao S. Y., Ritzel M. W., Paterson A. R., Cass C. E., Young J. D. ( 1994) J. Biol. Chem. 269, 17757– 17760 [PubMed] [Google Scholar]
- 13.Che M., Ortiz D. F., Arias I. M. ( 1995) J. Biol. Chem. 270, 13596– 13599 [DOI] [PubMed] [Google Scholar]
- 14.Ritzel M. W., Yao S. Y., Huang M. Y., Elliott J. F., Cass C. E., Young J. D. ( 1997) Am. J. Physiol. 272, C707– C714 [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Su S. F., Dresser M. J., Schaner M. E., Washington C. B., Giacomini K. M. ( 1997) Am. J. Physiol. 273, F1058– F1065 [DOI] [PubMed] [Google Scholar]
- 16.Ritzel M. W., Yao S. Y., Ng A. M., Mackey J. R., Cass C. E., Young J. D. ( 1998) Mol. Membr. Biol. 15, 203– 211 [DOI] [PubMed] [Google Scholar]
- 17.Smith K. M., Ng A. M., Yao S. Y., Labedz K. A., Knaus E. E., Wiebe L. I., Cass C. E., Baldwin S. A., Chen X. Z., Karpinski E., Young J. D. ( 2004) J. Physiol. 558, 807– 823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith K. M., Slugoski M. D., Cass C. E., Baldwin S. A., Karpinski E., Young J. D. ( 2007) Mol. Membr. Biol. 24, 53– 64 [DOI] [PubMed] [Google Scholar]
- 19.Smith K. M., Slugoski M. D., Loewen S. K., Ng A. M., Yao S. Y., Chen X. Z., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2005) J. Biol. Chem. 280, 25436– 25449 [DOI] [PubMed] [Google Scholar]
- 20.Hamilton S. R., Yao S. Y., Ingram J. C., Hadden D. A., Ritzel M. W., Gallagher M. P., Henderson P. J., Cass C. E., Young J. D., Baldwin S. A. ( 2001) J. Biol. Chem. 276, 27981– 27988 [DOI] [PubMed] [Google Scholar]
- 21.Yao S. Y., Ng A. M., Loewen S. K., Cass C. E., Baldwin S. A., Young J. D. ( 2002) Am. J. Physiol. Cell Physiol. 283, C155– 168 [DOI] [PubMed] [Google Scholar]
- 22.Craig J. E., Zhang Y., Gallagher M. P. ( 1994) Mol. Microbiol. 11, 1159– 1168 [DOI] [PubMed] [Google Scholar]
- 23.Loewen S. K., Yao S. Y., Slugoski M. D., Mohabir N. N., Turner R. J., Mackey J. R., Weiner J. H., Gallagher M. P., Henderson P. J., Baldwin S. A., Cass C. E., Young J. D. ( 2004) Mol. Membr. Biol. 21, 1– 10 [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., Tackaberry T., Ritzel M. W., Raborn T., Barron G., Baldwin S. A., Young J. D., Cass C. E. ( 2006) Biochem. J. 394, 389– 398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slugoski M. D., Smith K. M., Mulinta R., Ng A. M., Yao S. Y., Morrison E. L., Lee Q. O., Zhang J., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2008) J. Biol. Chem. 283, 24922– 24934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slugoski M. D., Ng A. M., Yao S. Y., Smith K. M., Lin C. C., Zhang J., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2008) J. Biol. Chem. 283, 8496– 8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan R. T., Maloney P. C. ( 1993) Cell 75, 37– 44 [PubMed] [Google Scholar]
- 28.Yan R. T., Maloney P. C. ( 1995) Proc. Natl. Acad. Sci. U. S. A. 92, 5973– 5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao S. Y., Ng A. M., Slugoski M. D., Smith K. M., Mulinta R., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2007) J. Biol. Chem. 282, 30607– 30617 [DOI] [PubMed] [Google Scholar]
- 30.Slugoski M. D., Smith K. M., Ng A. M. L., Yao S. Y. M., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 20April2009) J. Biol. Chem. 284, 17266– 17280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liman E. R., Tytgat J., Hess P. ( 1992) Neuron 9, 861– 871 [DOI] [PubMed] [Google Scholar]
- 32.Yao S. Y. M., Cass C. E., Young J. D. ( 2000) in Membrane Transport: A Practical Approach ( Baldwin S. A. ed) pp. 47– 78, Oxford University Press, Oxford [Google Scholar]
- 33.Yao S. Y., Sundaram M., Chomey E. G., Cass C. E., Baldwin S. A., Young J. D. ( 2001) Biochem. J. 353, 387– 393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slugoski M. D., Loewen S. K., Ng A. M., Smith K. M., Yao S. Y., Karpinski E., Cass C. E., Baldwin S. A., Young J. D. ( 2007) Biochemistry 46, 1684– 1693 [DOI] [PubMed] [Google Scholar]
- 35.Damaraju V. L., Elwi A. N., Hunter C., Carpenter P., Santos C., Barron G. M., Sun X., Baldwin S. A., Young J. D., Mackey J. R., Sawyer M. B., Cass C. E. ( 2007) Am. J. Physiol. Renal Physiol. 293, F200– 211 [DOI] [PubMed] [Google Scholar]
- 36.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. ( 2005) Nature 437, 215– 223 [DOI] [PubMed] [Google Scholar]
- 37.Faham S., Watanabe A., Besserer G. M., Cascio D., Specht A., Hirayama B. A., Wright E. M., Abramson J. ( 2008) Science 321, 810– 814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E. P., Rutherford N. G., Hadden J. M., O'Reilly J., Ma P., Saidijam M., Patching S. G., Hope R. J., Norbertczak H. T., Roach P. C., Iwata S., Henderson P. J., Cameron A. D. ( 2008) Science 322, 709– 713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yernool D., Boudker O., Jin Y., Gouaux E. ( 2004) Nature 431, 811– 818 [DOI] [PubMed] [Google Scholar]
- 40.Screpanti E., Hunte C. ( 2007) J. Struct. Biol. 159, 261– 267 [DOI] [PubMed] [Google Scholar]
- 41.Boudker O., Ryan R. M., Yernool D., Shimamoto K., Gouaux E. ( 2007) Nature 445, 387– 393 [DOI] [PubMed] [Google Scholar]