Abstract
Activation of the phosphatase calcineurin and its downstream targets, transcription factors of the NFAT family, results in cardiomyocyte hypertrophy. Recently, it has been shown that the dual specificity tyrosine (Y) phosphorylation-regulated kinase 1A (DYRK1A) is able to antagonize calcineurin signaling by directly phosphorylating NFATs. We thus hypothesized that DYRK1A might modulate the hypertrophic response of cardiomyocytes. In a model of phenylephrine-induced hypertrophy, adenovirus-mediated overexpression of DYKR1A completely abrogated the hypertrophic response and significantly reduced the expression of the natriuretic peptides ANF and BNP. Furthermore, DYRK1A blunted cardiomyocyte hypertrophy induced by overexpression of constitutively active calcineurin and attenuated the induction of the hypertrophic gene program. Conversely, knockdown of DYRK1A, utilizing adenoviruses encoding for a specific synthetic miRNA, resulted in an increase in cell surface area accompanied by up-regulation of ANF- mRNA. Similarly, treatment of cardiomyocytes with harmine, a specific inhibitor of DYRK1A, revealed cardiomyocyte hypertrophy on morphological and molecular level. Moreover, constitutively active calcineurin led to robust induction of an NFAT-dependent luciferase reporter, whereas DYRK1A attenuated calcineurin-induced reporter activation in cardiomyocytes. Conversely, both knockdown and pharmacological inhibition of DYRK1A significantly augmented the effect of calcineurin in this assay. In summary, we identified DYRK1A as a novel negative regulator of cardiomyocyte hypertrophy. Mechanistically, this effect appears to be mediated via inhibition of NFAT transcription factors.
Cardiac hypertrophy accompanies a variety of heart diseases and is an independent predictor of cardiovascular morbidity and mortality (1). Hypertrophy may develop in response to a variety of pathological stimuli, e.g. arterial hypertension or heart valve disease (2–4). While numerous molecular pathways have been implicated in the development of cardiomyocyte hypertrophy, the phosphatase calcineurin and its downstream targets, transcription factors of the nuclear factor of activated T-cells (NFAT)2 family, appear to play a central role. Activation of calcineurin leads to dephosphorylation of NFATs, subsequent nuclear translocation, and initiation of the hypertrophic gene program as well as marked cardiomyocyte hypertrophy (5). In contrast, genetic ablation of calcineurin Aβ renders the heart unresponsive to hypertrophic stimuli (6). In addition, inhibitors of calcineurin signaling have been shown to attenuate cardiac hypertrophy (7–9).
Recently, the dual specificity tyrosine (Y) phosphorylation-regulated kinase 1A (DYRK1A) has been identified as a novel modifier of NFAT transcription factors in Drosophila (10) and vertebrate neurons (11). DYRK1A directly phosphorylates NFATs, thereby promoting their nuclear export followed by a reduction of transcriptional activity. DYRK1A, which belongs to a family of dual specificity kinases, is ubiquitously expressed with high levels in the developing nervous system and the heart (12). All isoforms share the ability to autocatalyze their own phosphorylation at an YXY motif (13), while all other substrates are phosphorylated at Ser/Thr residues. DYRK1A, DYRK1B, and the Drosophila homolog, minibrain, are predominantly localized to the nucleus. In addition to NFAT, DYRK1A is capable of phosphorylating other transcription factors, including FKHR/Foxo1 (14), STAT3 (15), and Gli1 (16). Furthermore DYRK1A stimulates the phosphorylation of CREB (17). Because the DYRK1A gene is localized to the Down syndrome critical region and DYRK1A is overexpressed in Down syndrome fetal brain, it was proposed to contribute to the phenotype of trisomy 21. Consistent with this notion, transgenic mouse models with overexpression of DYRK1A reveal learning impairment and hyperactivity (18, 19).
In contrast, little is known about the function of DYRK1A in the heart. Because DYRKs were recently recognized as kinases that phosphorylate NFATs, we hypothesized that the cardiac-enriched isoform DYRK1A might modulate cardiac hypertrophy. Here we show for the first time that DYRK1A overexpression potently inhibits cardiomyocyte hypertrophy. Conversely, miRNA-mediated knockdown or pharmacological inhibition of DYRK1A causes a hypertrophic phenotype accompanied by induction of the hypertrophic gene program and increased NFAT activity.
EXPERIMENTAL PROCEDURES
Cloning and Generation of Recombinant Adenoviruses Encoding for DYRK1A
Cloning of rat DYRK1A into a pDonRTM201 gateway vector (Invitrogen) was carried out using standard PCR techniques. An adenovirus (AdV) encoding for full-length rat DYRK1A cDNA with a C-terminal V5 tag was generated using the ViraPowerTM Adenoviral Expression System (Invitrogen) according to the manufacturer's protocol. A β-galactosidase V5-encoding adenovirus served as control (Invitrogen).
Cloning and Generation of Recombinant Adenoviruses Encoding for Synthetic microRNAs Targeting Rat DYRK1A
Oligonucleotides encoding for microRNAs specifically targeting rat DYRK1A were designed using the Invitrogen oligo perfect software. Sequences of the inserts, which were subsequently cloned into the pcDNATM6.2-GW/EmGFP-miR vector: miR-544: 5′-TGC TGT TTA ATG GCG ACC CAT TCT TGG TTT TGG CCA CTG ACT GAC CAA GAA TGT CGC CAT TAA A-3′, miR-1331: 5′-TGC TGT CAA GTA GTC AGC TAC AGT GTG TTT TGG CCA CTG ACT GAC ACA CTG TAT GAC TAC TTG A-3′, miR-1732: 5′-TGC TGA ACA GGA TGA GTT TCA ACA GTG TTT TGG CCA CTG ACT GAC ACT GTT GAC TCA TCC TGT T-3′.
As negative control, we used the pcDNATM6.2-GW/ EmGFP-miR plasmid, which can form a hairpin structure and is consecutively processed into a mature miRNA, yet is predicted not to target any known mammalian gene (sequence of the insert: 5′-GAA ATG TAC TGC GCG TGG AGA CGT TTT GGC CAC TGA CTG ACG TCT CCA CGC AGT ACA TTT-3′). Adenoviruses encoding for synthetic microRNAs were generated using the ViraPowerTM Adenoviral Expression System (Invitrogen) according to the manufacturer's protocol.
Isolation and Culture of Neonatal Rat Ventricular Cardiomyocytes (NRVMs)
Hearts from 1–2-day-old Wistar rats (Charles River) were excised and minced in ADS buffer (120 mmol/liter NaCl, 20 mmol/liter HEPES, 8 mmol/liter NaH2PO4, 6 mmol/liter glucose, 5 mmol/liter KCl, 0.8 mmol/liter MgSO4, pH 7.4). A series of digestion steps was carried out with an enzymatic solution containing collagenase type II (0.5 mg/ml, Worthington) and pancreatin (0.6 mg/ml, Sigma-Aldrich) in sterile ADS buffer. A Percoll (GE Healthcare) gradient centrifugation step was applied to remove contaminating fibroblasts from cardiomyocytes. NRVMs were resuspended and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum, penicillin/streptomycin, and l-glutamine (PAA) for 24 h. After 24 h, cells were either infected in serum-free medium or serum-starved for 24 h before applying stimulants or inhibitors for the indicated time.
RNA Isolation and Purification
Total RNA from NRVMs was isolated using the TRIzol method (Invitrogen) according to the manufacturer's protocol. DNaseI digestion and purification of RNA was carried out on RNeasy columns (Qiagen).
Quantitative Real-time PCR
DNaseI-digested total RNA of each condition was transcribed into cDNA using the Superscript III first strand kit (Invitrogen). Real-time PCR primer sequences are shown in supplemental Table S1. 18S rRNA served as an internal standard. For quantitative real-time PCR, the Platinum SYBR Green qPCR SuperMix-UDG system (Invitrogen) was used in the ABI Prism 7000 Sequence Detection System (Applied Biosystems).
Immunoblotting
NRVMs were harvested in a lysis buffer containing 20 mmol/liter Tris, pH 7.5, 12.5% (v/v) glycerol, 10 mmol/liter dithiothreitol, 500 mmol/liter NaCl, 1% (v/v) Nonidet P-40 (Sigma-Aldrich), and protease inhibitor mixture tablets (Roche Diagnostics). After up to three brief freeze-and-thaw cycles and a centrifugation step, whole cell lysate was obtained. SDS-PAGE Western blotting was carried out using the WestranTM polyvinylidene difluoride membrane (Schleicher & Schuell). Application of the primary antibody (monoclonal anti-V5 (1:1000, Invitrogen), anti-DYRK1A (1:50, Santa Cruz Biotechnology), polyclonal anti-RCAN1 (1:1000), monoclonal anti-α-tubulin (1:10,000, Sigma-Aldrich)) was followed by incubation with a horseradish peroxidase-coupled secondary antibody (1:10,000, Santa Cruz Biotechnology). Visualization was achieved using a chemiluminescence kit (GE Healthcare).
Immunofluorescence Experiments
NRVMs were fixed in 4% paraformaldehyde (Sigma-Aldrich) in phosphate-buffered saline (PBS), permeabilized with 0.3% Triton X-100 (Sigma-Aldrich), and blocked for 1 h with 2.5% bovine serum albumin (Sigma-Aldrich) in PBS. The monoclonal antibody against sarcomeric α-actinin (Sigma-Aldrich) was used at a dilution of 1:200. The secondary antibody (Cy3-coupled antimouse antibody, Dianova) was used at 1:200 for 1 h. Nuclei were labeled by DAPI. Cell surface areas of cardiomyocytes were determined applying AxioVision Release 4.4 (Carl Zeiss Vision).
Reporter Gene Assays
24 h after plating, neonatal cardiomyocytes were infected by an adenovirus (AdNFAT-luc, 20 moi) carrying a firefly luciferase construct under the transcriptional control of three consensus NFAT binding sites (Biomyx) and serum-starved for another 24 h. Simultaneous to the infection with the reporter virus, cells were infected by adenoviruses as indicated. To apply an identical virus load per condition, we used an adenovirus encoding for luciferase without NFAT binding sites (Adluc), which had been tested to not activate the reporter.
Luciferase assays were performed according to the manufacturer's instructions (Promega). A β-galactosidase assay was performed in a 0.1 mol/liter sodium phosphate buffer (pH 7.3) with 1 mmol/liter MgCl2 as an internal control of infection. 2-Nitrophenyl-β-d-galactopyranoside (30 μg/ml) served as substrate. After incubation for 30 min, a photometric analysis was performed.
Inhibitor Assays
Inhibitor assays were carried out using the DYRK1A inhibitor harmine (Fluka) dissolved in ethanol. Control cells received the vehicle only.
Statistical Analyses
All results are shown as the means ± standard error of the mean (S.E.) unless stated otherwise. Real-time PCR data analyses were carried out using the ΔΔct method. Statistical analyses of the data were carried out using one-way analysis of variance (ANOVA) followed by Student-Newman-Keuls post-hoc tests. If appropriate, the Student's t test (two sided) was employed. p values <0.05 were considered statistically significant.
RESULTS
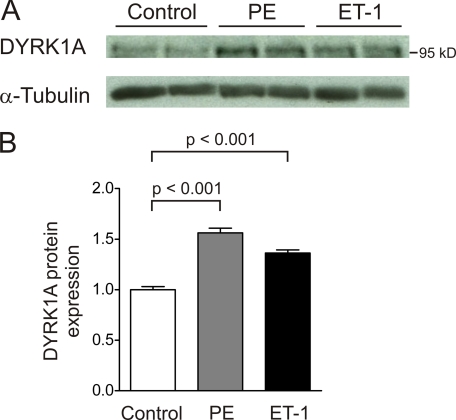
Recently it has been shown that DYRK1A inhibits the activation of NFAT transcription factors by phosphorylation (10, 11). We thus hypothesized that DYRK1A might have an influence on cardiomyocyte hypertrophy. First, we asked whether the expression level of DYRK1A is modulated in established models of cardiomyocyte hypertrophy. Neonatal rat ventricular cardiomyocytes (NRVM) were stimulated with ET-1 (100 nmol/liter) or phenylephrine (PE, 50 μmol/liter) for 24 h, and DYRK1A protein expression was found to be significantly induced by either hypertrophic agent (PE: 1.6 ± 0.05-fold, ET-1: 1.4 ± 0.03-fold, p < 0.001, n = 6, Fig. 1, A and B).
FIGURE 1.
DYRK1A is induced by hypertrophic agents in vitro. A, treatment of cardiomyocytes with PE (50 μmol/liter) and endothelin-1 (ET-1, 100 nmol/liter) for 24 h induces DYRK1A protein expression. B, PE as well as ET-1 lead to a significant induction of DYRK1A (PE: 1.6 ± 0.05-fold, ET-1: 1.4 ± 0.03-fold, p < 0.001, n = 6).
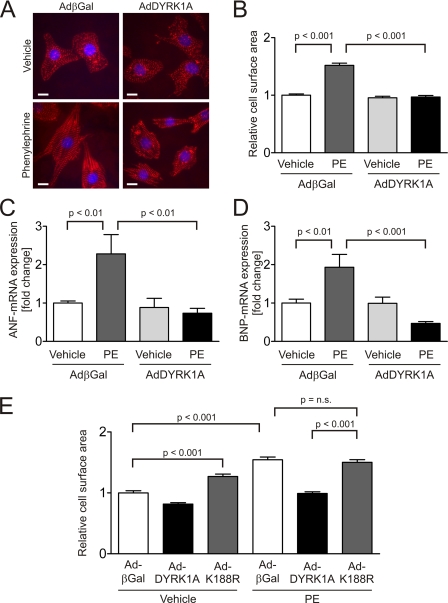
To further elucidate the role of DYRK1A in cardiomyocyte hypertrophy, we utilized adenoviral gene transfer for overexpression of DYRK1A (AdDYRK1A) in NRVMs treated with prohypertrophic agents. Infection of NRVM led to a dose-dependent overexpression of DYRK1A on mRNA and protein level (data not shown). NRVMs were infected by AdDYRK1A or by a control virus overexpressing β-galactosidase (AdβGal) and treated with the hypertrophic agonist phenylephrine. While stimulation with PE (100 μmol/liter, 24 h) led to a robust 1.5 ± 0.04-fold increase in cardiomyocyte surface area in control cells (p < 0.001, 50 cells per condition, n = 3), AdDYRK1A treatment completely abrogated cardiomyocyte hypertrophy induced by PE (p < 0.001, Fig. 2, A and B). Because cellular hypertrophy is accompanied by activation of specific genes (e.g. Nppa/ANF/ANP, Nppb/BNP, β-MyHC), we searched for expression changes of these members of the hypertrophic gene program by DYRK1A. DYRK1A blunted PE-mediated up-regulation of the mRNAs encoding for the natriuretic peptides ANF by 67.8% (p < 0.01, n = 6, Fig. 2C) and BNP by 75.8% (p < 0.001, n = 6, Fig. 2D).
FIGURE 2.
DYRK1A inhibits cardiomyocyte hypertrophy induced by phenylephrine. A, representative images of β-galactosidase (25 moi) and DYRK1A (25 moi)-overexpressing cardiomyocytes stained with an α-actinin antibody reveal the resistance of DYRK1A-overexpressing NRVMs to PE-induced hypertrophy. DAPI is used as counterstain for nuclei. B, while stimulation with PE (100 μmol/liter) for 24 h leads to a 1.5 ± 0.04-fold increase in cardiomyocyte surface area in control cells (p < 0.001), AdDYRK1A treatment abrogates cardiomyocyte hypertrophy induced by PE (p < 0.001, 50 cells per condition, n = 3). C, AdDYRK1A (25 moi) reverses the induction of ANF- (−67.8%, p < 0.01, n = 6) and D, BNP-mRNA (−75.8%, p < 0.001, n = 6) caused by PE (100 μmol/liter, 24 h). E, catalytically inactive point mutant of DYRK1A (AdK188R) causes cellular hypertrophy (1.3 ± 0.04-fold, p < 0.001) and fails to inhibit PE-induced hypertrophy (100 μmol/liter, 24 h). The cell surface area of PE-stimulated cardiomyocytes overexpressing DYRK1A-K188R did not differ from PE-treated cells overexpressing the control virus (50 cells per condition, n = 3). Scale bars, 10 μm.
Interestingly, a catalytically inactive mutant (20) of DYRK1A (AdK188R) failed to inhibit PE-induced hypertrophy (50 cells per condition, n = 3, Fig. 2E). The cell surface area of PE-stimulated cardiomyocytes overexpressing DYRK1A-K188R did not differ from PE-treated cells overexpressing the control virus, suggesting that DYRK1A′s kinase activity is required for its antihypertrophic effect. Moreover, the overexpression of DYRK1A-K188R led to significant cellular hypertrophy (1.3 ± 0.04-fold, p < 0.001).
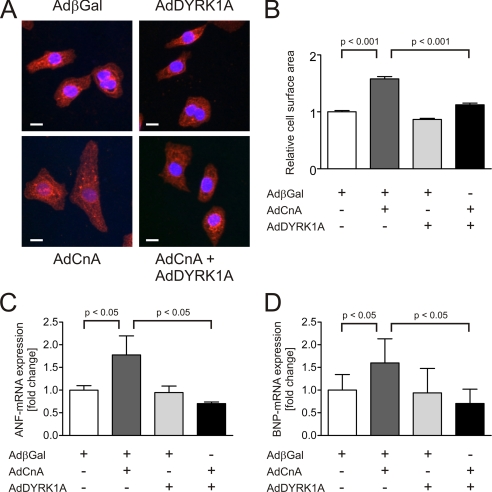
To dissect the underlying pathway we utilized a constitutively active mutant of calcineurin A (CnA), a well known inducer of cardiomyocyte hypertrophy (21). Whereas overexpression of CnA led to a significant increase in cardiomyocyte surface area (1.6 ± 0.04-fold, p < 0.001, 100 cells per condition, n = 3, Fig. 3, A and B), DYRK1A inhibited the CnA-induced increase by 78% (p < 0.001). This difference in cell size was accompanied by an attenuated induction of the natriuretic peptides ANF and BNP. Both members of the hypertrophic gene program have been shown to be responsive to the calcineurin/NFAT pathway (5, 21, 22). DYRK1A caused a reduction of CnA-induced ANF-mRNA by −60% (p < 0.05, n = 3, Fig. 3C) and BNP-mRNA by −56% (p < 0.05, n = 3, Fig. 3D). Taken together, these data suggest that DYRK1A is sufficient to inhibit cardiomyocyte hypertrophy as well as the hypertrophic gene program.
FIGURE 3.
DYRK1A reverses cardiomyocyte hypertrophy induced by calcineurin. A, representative α-actinin immunofluorescence images of cardiomyocytes overexpressing a constitutively active mutant of CnA and DYRK1A. B, while the overexpression of CnA causes a significant increase in cardiomyocyte surface area (1.6 ± 0.04-fold, p < 0.001, 100 cells per condition, n = 3), DYRK1A inhibits the CnA-induced increase in cell surface area by 78% (p < 0.001). The antihypertrophic effect of DYRK1A is accompanied by a reduction of CnA-induced (C) ANF-mRNA by −60% (p < 0.05, n = 3) and (D) BNP-mRNA by −56% (p < 0.05, n = 3). Scale bars, 10 μm.
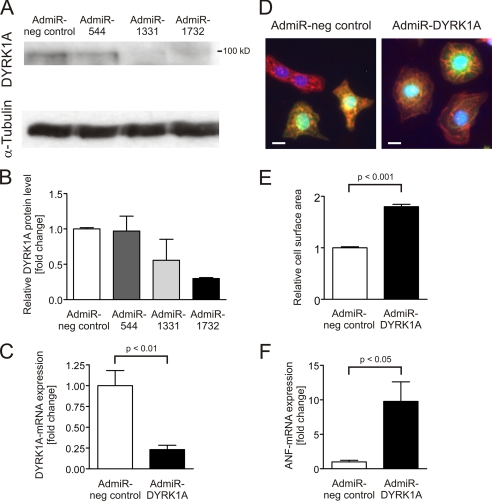
Next, we evaluated whether down-regulation of DYRK1A might in turn facilitate the development of cardiomyocyte hypertrophy. Thus, adenoviruses encoding for three different synthetic miRNAs targeting DYRK1A (AdmiR-544, -1331, -1732) were generated, and the most potent one was chosen for further analyses (AdmiR-1732/AdmiR-DYRK1A: −70.1% DYRK1A protein, Fig. 4, A and B). Treatment with 10 moi AdmiR-DYRK1A led to a reduction of −76.9% DYRK1A mRNA (p < 0.01, Fig. 4C). Knockdown of DYRK1A resulted in a significant increase of cardiomyocyte surface area (1.8 ± 0.05-fold, p < 0.001, 75 cells per condition, n = 3, Fig. 4, D and E), accompanied by a significant up-regulation of ANF expression (9.8 ± 2.8-fold, p < 0.05, n = 8, Fig. 4F), suggesting that DYRK1A is not only sufficient but also required for the inhibition of cardiomyocyte hypertrophy.
FIGURE 4.
Knockdown of DYRK1A causes cardiomyocyte hypertrophy. A, evaluation of the knockdown efficiency of three adenovirally encoded synthetic miRNAs (AdmiR-544, -1331, -1732; 25 moi each) against DYRK1A reveals (B) AdmiR-1732 as the most potent miRNA with −70.1% down-regulation of DYRK1A protein expression (n = 2). C, likewise, this synthetic miRNA reduces DYRK1A mRNA levels by −76.9% (p < 0.01, n = 3). D, representative cardiomyocytes stained with an α-actinin antibody after adenovirally delivered miRNA against DYRK1A (10 moi). The adenovirally delivered knockdown construct carries a GFP cassette. E, only double-labeled cells were measured, revealing a significant increase in cell surface area (1.8 ± 0.05-fold, p < 0.001, 75 cells per condition, n = 3) in response to DYRK1A down-regulation. F, hypertrophic phenotype is accompanied by a 9.8 ± 2.8-fold induction of ANF-mRNA (p < 0.05, n = 8). Scale bars, 10 μm.
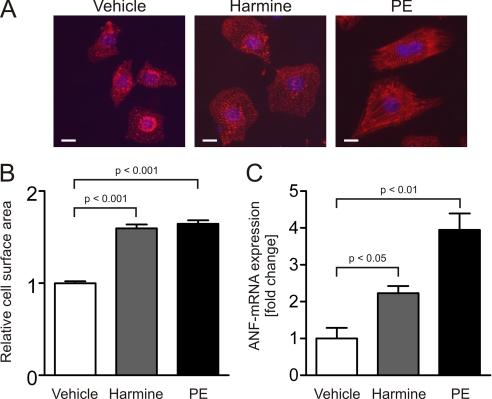
To further confirm the antihypertrophic properties of DYRK1A, we utilized harmine, a specific inhibitor of DYRK1A (23–25). Consistent with the knockdown data, treatment of NRVMs with 1 μmol/liter harmine for 24 h resulted in a significant increase in cell size (1.6 ± 0.04-fold, p < 0.001, 100 cells per condition, n = 3, Fig. 5, A and B) compared with cardiomyocytes treated with vehicle only. Furthermore, harmine caused an induction of ANF expression up to 2.2 ± 0.2-fold (p < 0.05, Fig. 5C).
FIGURE 5.
Pharmacological inhibition of DYRK1A results in cardiomyocyte hypertrophy. A, representative α-actinin immunofluorescence images of NRVMs treated with vehicle, DYRK1A inhibitor harmine (1 μmol/liter) or PE (50 μmol/liter) for 24 h. B, treatment of NRVMs with harmine causes an increased cell surface area (1.6 ± 0.04-fold, p < 0.001, 100 cells per condition, n = 3). C, hypertrophic phenotype is accompanied by an induction of ANF-mRNA up to 2.2 ± 0.2-fold (p < 0.05, n = 3). Scale bars, 10 μm.
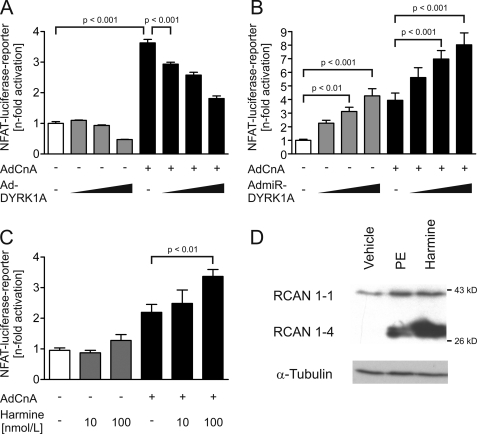
To answer the question whether DYRK1A modulates the activity of NFAT transcription factors in NRVMs we performed luciferase reporter assays employing a reporter under transcriptional control of three NFAT binding sites. Infection of NRVMs by a constitutively active variant of calcineurin A (AdCnA; 30 moi) led to marked induction of the reporter (3.6 ± 0.1-fold, p < 0.001), whereas DYRK1A significantly and dose-dependently (AdDYRK1A; 30, 60, 90 moi) attenuated AdCnA-induced reporter activity by up to 50.1% (n = 3, Fig. 6A), indicating a negative regulation of NFAT activity by DYRK1A. Conversely, miRNA-mediated knockdown of DYRK1A (5, 10, 25 moi) resulted in an up to 4.3 ± 0.5-fold increase of luciferase activity (p < 0.001, Fig. 6B). Interestingly, DYRK1A knockdown further augmented NFAT induction by calcineurin (30 moi) up to 2.0-fold (p < 0.001, n = 12–18, Fig. 6B). Likewise, the inhibition of DYRK1A by 0.1 μmol/liter harmine markedly exacerbated the effect of calcineurin (+53%, p < 0.01, n = 9, Fig. 6C).
FIGURE 6.
DYRK1A attenuates NFAT-activity in cardiomyocytes. A, AdDYRK1A reduces calcineurin (AdCnA)-induced NFAT luciferase reporter activity dose dependently up to −50.1% (n = 3). B, conversely, AdmiR-DYRK1A increases reporter activity up to 4.3 ± 0.5-fold and exaggerates the calcineurin effect up to 2-fold (n = 12–18). C, likewise, the inhibition of DYRK1A by harmine (100 nmol/liter) exacerbates the effect of calcineurin up to 1.5-fold (p < 0.01, n = 9). D, expression of RCAN 1–4, a direct target gene of NFAT, is found markedly up-regulated in harmine-treated cardiomyocytes (1 μmol/liter, 24 h, n = 3).
Finally, we examined the regulation of the RCAN isoform 1–4 which is expressed from the RCAN1 locus by an alternative promoter (22, 26). This promoter is activated via binding of transcription factors of the NFAT family, whereas the promoter of RCAN1–1 is not sensitive to calcineurin/NFAT signaling. RCAN1–4 was significantly up-regulated in harmine-treated cardiomyocytes (Fig. 6D), again indicating that inhibition of DYRK1A leads to increased calcineurin/NFAT activity. Taken together, these data suggest that the antihypertrophic effect of DYRK1A is at least in part mediated via inhibition of NFAT activity.
DISCUSSION
Here we show for the first time that DYRK1A plays a significant role in the regulation of cardiomyocyte hypertrophy. Our results suggest that DYRK1A is sufficient to completely inhibit agonist- as well as calcineurin-induced hypertrophy. Conversely, both down-regulation and pharmacological inhibition of DYRK1A leads to a hypertrophic phenotype in cardiomyocytes.
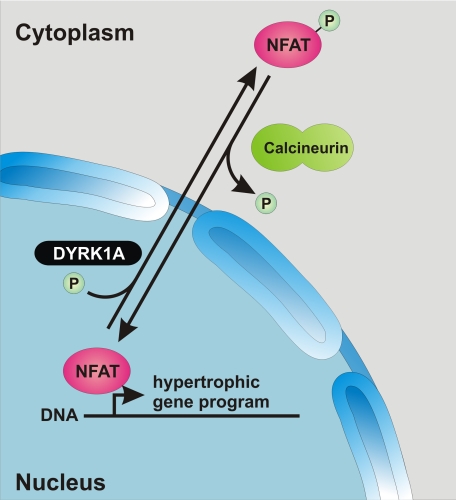
It is well established that agonists of Gq/11-protein-coupled receptors, such as PE, mediate their effect through activation of the calcineurin/NFAT cascade (27). Activated calcineurin dephosphorylates NFATs in the cytoplasm which results in their nuclear translocation and subsequent activation of prohypertrophic target genes. In contrast, DYRK1A is capable of phosphorylating transcription factors of the NFAT family leading to their nuclear export (10, 11). Thus, the underlying mechanism of the antihypertrophic properties of DYRK1A is likely to be a reduction of NFAT activity (Fig. 7). Consistent with this notion DYRK1A directly reduces calcineurin-induced NFAT reporter activity in cardiomyocytes, while its baseline activity is markedly augmented by knockdown or inhibition of DYRK1A.
FIGURE 7.
Proposed role of DYRK1A in cardiomyocyte hypertrophy. DYRK1A directly phosphorylates NFAT thereby mediating its nuclear export which leads to a reduced transcriptional activity of prohypertrophic target genes.
NFAT transcription factors serve as integrators of multiple signaling pathways. Several kinases have been identified that can antagonize calcineurin-signaling by phosphorylating NFATs, including glycogen synthase kinase 3 (GSK3), c-Jun N-terminal kinase (JNK), and p38 MAP kinases (28). Interestingly, GSK3 phosphorylates Ser/Thr residues only after a priming phosphorylation reaction (29). Because DYRK1A targets a serine in NFATc4 in close proximity to the GSK3 phosphorylation site and DYRK1A synergizes with GSK3 to inhibit NFAT activity (11), DYRK1A represents a good candidate for a physiological priming kinase. Of note, adenovirally overexpressed GSK3β blocks the hypertrophic response to pharmacological inducers of hypertrophy such as PE or endothelin-1 (30). Likewise, cardiac hypertrophy induced by adrenergic stimulation or pressure overload is diminished by overexpression of constitutively active GSK3β (31). It will thus be interesting to see whether GSK3β is also required for the antihypertrophic properties of DYKR1A. Finally, we cannot exclude that phosphorylation of other targets than NFATs by DYRK1A may contribute to the antihypertrophic effects of DYRK1A.
In summary, we identified DYRK1A as a novel regulator of cardiomyocyte hypertrophy. Whereas DYRK1A inhibited pharmacological agonist- and calcineurin-induced hypertrophy, the absence or inhibition of DYRK1A caused a hypertrophic phenotype. Mechanistically, the antihypertrophic effect of DYRK1A is likely mediated via increased phosphorylation of NFAT transcription factors.
Supplementary Material
Acknowledgments
We thank Walter Becker for DYRK1A expression constructs and Ulrike Oehl and Jutta Krebs for excellent technical assistance.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG, FR1289/5-1) as well as by a grant from the Bundesministerium für Bildung und Forschung (Nationales Genomforschungsnetz: NGFNplus, Germany) (to N. F. and H. A. K.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
- NFAT
- nuclear factor of activated T-cells
- ANF
- atrial natriuretic factor
- ANP
- atrial natriuretic peptide
- BNP
- B-type natriuretic peptide
- CnA
- calcineurin A
- DYRK1A
- dual specificity tyrosine(Y) phosphorylation-regulated kinase 1A
- ET-1
- endothelin-1
- moi
- multiplicity of infections
- NRVM
- neonatal rat ventricular cardiomyocytes
- PE
- phenylephrine
- RCAN
- regulator of calcineurin
- DAPI
- 4′,6-diamidino-2-phenylindole.
REFERENCES
- 1.Levy D., Garrison R. J., Savage D. D., Kannel W. B., Castelli W. P. ( 1990) N. Engl. J. Med. 322, 1561– 1566 [DOI] [PubMed] [Google Scholar]
- 2.Frey N., Olson E. N. ( 2003) Annu. Rev. Physiol. 65, 45– 79 [DOI] [PubMed] [Google Scholar]
- 3.Hill J. A., Olson E. N. ( 2008) N. Engl. J. Med. 358, 1370– 1380 [DOI] [PubMed] [Google Scholar]
- 4.Frey N., Katus H. A., Olson E. N., Hill J. A. ( 2004) Circulation 109, 1580– 1589 [DOI] [PubMed] [Google Scholar]
- 5.Molkentin J. D., Lu J. R., Antos C. L., Markham B., Richardson J., Robbins J., Grant S. R., Olson E. N. ( 1998) Cell 93, 215– 228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bueno O. F., Wilkins B. J., Tymitz K. M., Glascock B. J., Kimball T. F., Lorenz J. N., Molkentin J. D. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 4586– 4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Windt L. J., Lim H. W., Bueno O. F., Liang Q., Delling U., Braz J. C., Glascock B. J., Kimball T. F., del Monte F., Hajjar R. J., Molkentin J. D. ( 2001) Proc. Natl. Acad. Sci. U. S. A. 98, 3322– 3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank D., Kuhn C., van Eickels M., Gehring D., Hanselmann C., Lippl S., Will R., Katus H. A., Frey N. ( 2007) Circulation 116, 2587– 2596 [DOI] [PubMed] [Google Scholar]
- 9.Rothermel B. A., McKinsey T. A., Vega R. B., Nicol R. L., Mammen P., Yang J., Antos C. L., Shelton J. M., Bassel-Duby R., Olson E. N., Williams R. S. ( 2001) Proc. Natl. Acad. Sci. U. S. A. 98, 3328– 3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gwack Y., Sharma S., Nardone J., Tanasa B., Iuga A., Srikanth S., Okamura H., Bolton D., Feske S., Hogan P. G., Rao A. ( 2006) Nature 441, 646– 650 [DOI] [PubMed] [Google Scholar]
- 11.Arron J. R., Winslow M. M., Polleri A., Chang C. P., Wu H., Gao X., Neilson J. R., Chen L., Heit J. J., Kim S. K., Yamasaki N., Miyakawa T., Francke U., Graef I. A., Crabtree G. R. ( 2006) Nature 441, 595– 600 [DOI] [PubMed] [Google Scholar]
- 12.Okui M., Ide T., Morita K., Funakoshi E., Ito F., Ogita K., Yoneda Y., Kudoh J., Shimizu N. ( 1999) Genomics 62, 165– 171 [DOI] [PubMed] [Google Scholar]
- 13.Becker W., Weber Y., Wetzel K., Eirmbter K., Tejedor F. J., Joost H. G. ( 1998) J. Biol. Chem. 273, 25893– 25902 [DOI] [PubMed] [Google Scholar]
- 14.Woods Y. L., Rena G., Morrice N., Barthel A., Becker W., Guo S., Unterman T. G., Cohen P. ( 2001) Biochem. J. 355, 597– 607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuo R., Ochiai W., Nakashima K., Taga T. ( 2001) J. Immunol. Methods 247, 141– 151 [DOI] [PubMed] [Google Scholar]
- 16.Mao J., Maye P., Kogerman P., Tejedor F. J., Toftgard R., Xie W., Wu G., Wu D. ( 2002) J. Biol. Chem. 277, 35156– 35161 [DOI] [PubMed] [Google Scholar]
- 17.Yang E. J., Ahn Y. S., Chung K. C. ( 2001) J. Biol. Chem. 276, 39819– 39824 [DOI] [PubMed] [Google Scholar]
- 18.Altafaj X., Dierssen M., Baamonde C., Martí E., Visa J., Guimerà J., Oset M., González J. R., Flórez J., Fillat C., Estivill X. ( 2001) Hum. Mol. Genet. 10, 1915– 1923 [DOI] [PubMed] [Google Scholar]
- 19.Branchi I., Bichler Z., Minghetti L., Delabar J. M., Malchiodi-Albedi F., Gonzalez M. C., Chettouh Z., Nicolini A., Chabert C., Smith D. J., Rubin E. M., Migliore-Samour D., Alleva E. ( 2004) J. Neuropathol. Exp. Neurol. 63, 429– 440 [DOI] [PubMed] [Google Scholar]
- 20.Kentrup H., Becker W., Heukelbach J., Wilmes A., Schürmann A., Huppertz C., Kainulainen H., Joost H. G. ( 1996) J. Biol. Chem. 271, 3488– 3495 [DOI] [PubMed] [Google Scholar]
- 21.De Windt L. J., Lim H. W., Taigen T., Wencker D., Condorelli G., Dorn G. W., 2nd, Kitsis R. N., Molkentin J. D. ( 2000) Circ. Res. 86, 255– 263 [DOI] [PubMed] [Google Scholar]
- 22.Yang J., Rothermel B., Vega R. B., Frey N., McKinsey T. A., Olson E. N., Bassel-Duby R., Williams R. S. ( 2000) Circ. Res. 87, E61– 68 [DOI] [PubMed] [Google Scholar]
- 23.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. ( 2007) Biochem. J. 408, 297– 315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sitz J. H., Baumgärtel K., Hämmerle B., Papadopoulos C., Hekerman P., Tejedor F. J., Becker W., Lutz B. ( 2008) Neuroscience 157, 596– 605 [DOI] [PubMed] [Google Scholar]
- 25.Seifert A., Allan L. A., Clarke P. R. ( 2008) Febs J. 275, 6268– 6280 [DOI] [PubMed] [Google Scholar]
- 26.Davies K. J., Ermak G., Rothermel B. A., Pritchard M., Heitman J., Ahnn J., Henrique-Silva F., Crawford D., Canaider S., Strippoli P., Carinci P., Min K. T., Fox D. S., Cunningham K. W., Bassel-Duby R., Olson E. N., Zhang Z., Williams R. S., Gerber H. P., Pérez-Riba M., Seo H., Cao X., Klee C. B., Redondo J. M., Maltais L. J., Bruford E. A., Povey S., Molkentin J. D., McKeon F. D., Duh E. J., Crabtree G. R., Cyert M. S., de la Luna S., Estivill X. ( 2007) Faseb J. 21, 3023– 3028 [DOI] [PubMed] [Google Scholar]
- 27.Taigen T., De Windt L. J., Lim H. W., Molkentin J. D. ( 2000) Proc. Natl. Acad. Sci. U. S. A. 97, 1196– 1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkins B. J., Molkentin J. D. ( 2004) Biochem. Biophys. Res. Commun. 322, 1178– 1191 [DOI] [PubMed] [Google Scholar]
- 29.Sugden P. H., Fuller S. J., Weiss S. C., Clerk A. ( 2008) Br. J. Pharmacol. 153, Suppl. 1, S137– S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haq S., Choukroun G., Kang Z. B., Ranu H., Matsui T., Rosenzweig A., Molkentin J. D., Alessandrini A., Woodgett J., Hajjar R., Michael A., Force T. ( 2000) J. Cell Biol. 151, 117– 130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antos C. L., McKinsey T. A., Frey N., Kutschke W., McAnally J., Shelton J. M., Richardson J. A., Hill J. A., Olson E. N. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 907– 912 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.