Abstract
Previously we showed that xenobiotic inducible cytochrome P450 (CYP) proteins are bimodally targeted to the endoplasmic reticulum and mitochondria. In this study, we investigated the mechanism of delivery of chimeric signal containing CYP proteins to the peripheral and channel-forming mitochondrial outer membrane translocases (TOMs). CYP+33/1A1 and CYP2B1 did not require peripheral TOM70, TOM20, or TOM22 for translocation through the channel-forming TOM40 protein. In contrast, CYP+5/1A1 and CYP2E1 were able to bypass TOM20 and TOM22 but required TOM70. CYP27, which contains a canonical cleavable mitochondrial signal, required all of the peripheral TOMs for its mitochondrial translocation. We investigated the underlying mechanisms of bypass of peripheral TOMs by CYPs with chimeric signals. The results suggested that interaction of CYPs with Hsp70, a cytosolic chaperone involved in the mitochondrial import, alone was sufficient for the recognition of chimeric signals by peripheral TOMs. However, sequential interaction of chimeric signal containing CYPs with Hsp70 and Hsp90 resulted in the bypass of peripheral TOMs, whereas CYP27A1 interacted only with Hsp70 and was not able to bypass peripheral TOMs. Our results also show that delivery of a chimeric signal containing client protein by Hsp90 required the cytosol-exposed NH2-terminal 143 amino acids of TOM40. TOM40 devoid of this domain was unable to import CYP proteins. These results suggest that compared with the unimodal mitochondrial targeting signals, the chimeric mitochondrial targeting signals are highly evolved and dynamic in nature.
Protein targeting to different subcellular compartments is directed by specific NH2-terminal or internal signals, which serve as destination-specific mail delivery codes (1, 2). The signal sequences of proteins targeted to different membrane compartments, such as the endoplasmic reticulum (ER),2 peroxisomes, and mitochondria, vary markedly in terms of amino acid sequence, hydrophobicity, and secondary structure and interact with distinctly different sets of carrier proteins, receptors, and protein translocator complexes (3–8). These observations have led to a widely accepted view that most protein targeting signals are unimodal in nature, which in turn restricts the destination of a given protein to a single subcellular compartment.
ER-targeted proteins contain a distinct NH2-terminal hydrophobic signal for binding to a signal recognition particle, which in turn targets the emerging nascent chains to the ER (9). With certain exceptions (1, 10), ER targeting is thought to be co-translational. Current models of protein targeting imply that the ER or mitochondrial destination of a protein is determined at the pre-translational level by virtue of the signal sequence that the protein carries. Many mitochondria- and ER-targeted proteins are encoded by a distinct set of genes. In a limited number of cases, characteristic mitochondrial targeting sequences are generated by differential expression of the gene using either alternate transcription/translation start sites or differential splicing of the primary transcripts (11–13). Thus, proteins targeted to different cytoplasmic organelles carry a specific signal that directs the protein to a specific organelle.
In contrast to the prevailing dogma, studies from our laboratory and other laboratories have shown that a number of proteins are bimodally targeted to different subcellular compartments (14–20). Our results showed that different CYPs, glutathione S-transferase (GST) isoforms, and the amyloid precursor protein (APP) are targeted to the mitochondria in addition to their well established subcellular locations in the ER, cytosol, and plasma membranes, respectively (21, 22). Bimodal targeting of CYPs and APP was facilitated by the NH2-terminal chimeric signal, which consists of a cryptic mitochondrial targeting signal, immediately flanking the NH2-terminal ER-targeting signal. By contrast, a COOH-terminal cryptic mitochondrial targeting signal was critical for the mitochondrial translocation of cytosolic glutathione S-transferases (21). We also showed that mitochondrial targeting of these proteins required the activation of a cryptic mitochondrial signal either by sequence-specific processing by a cytosolic endoprotease as in the case of CYP1A1 (23) or PKA-mediated phosphorylation of Ser residues at positions 128 or 129 as in the case of CYP2B1 and CYP2E1 (16, 18, 24). In contrast, phosphorylation of a tandem PKA/PKC target site close to the COOH terminus was involved in the activation of the mitochondrial targeting signal of GSTA4-4 (21).
Proteins with canonical mitochondrial targeting signals specifically bind to the outer membrane protein import complex, TOM, as the critical initial step in mitochondrial protein targeting (13, 25–28). The TOM complex consists of nine subunits, of which TOM70, TOM40, TOM22, and TOM20 are the major components. The COOH termini of TOM20 and TOM70 and the NH2 terminus of TOM22 are exposed to the cytosol and provide receptor sites for binding to client proteins. The small TOMs (TOM7, TOM6, and TOM5), on the other hand, are associated with the channel-forming TOM40 protein, which is embedded in the outer membrane (13, 25, 27). Current models suggest that the NH2-terminal signal regions of pre-proteins bind to the cytosol-exposed domains of TOM20 and TOM22 through both hydrophobic and ionic interactions. TOM20 and TOM70 exhibit overlapping substrate specificity. TOM22 is believed to regulate the gating activity of the TOM40 channel (29–32). A recent study suggests the involvement of Hsp70 in the targeting of phosphorylated CYP2E1 to TOM40 (18). However, the precise mode of transport of chimeric signal-containing proteins to mitochondria and the interaction of chimeric signals with members of the TOM complex remain unclear. A number of studies have observed low levels of mitochondrial protein import by mitochondria pretreated with proteases or those depleted of peripheral TOMs (29, 33–36) suggesting a distinct role for these peripheral TOMs in the mitochondrial import of proteins. However, the role of these peripheral TOMs in the mitochondrial import of proteins containing chimeric signals remains unclear.
In this study, we investigated the requirement for peripheral TOM proteins for the translocation of CYPMT2a (+5/1A1), CYPMT2b (+33/1A1) (the NH2-terminal truncated forms of CYP1A1), CYPMT4, and CYPMT5 (phosphorylated forms of CYP2B1 and CYP2E1, respectively) that contain NH2-terminal chimeric signals. We found that the activated chimeric signals of these proteins can bypass the peripheral TOM20 and TOM22 receptors. Our results also suggest that differential interaction of chimeric signals with Hsp70 and Hsp90 might be the key discriminating factor in the TOM20/TOM22 bypass mechanism of mitochondrial protein targeting.
EXPERIMENTAL PROCEDURES
Yeast Strains Used
Strains BY4741 (MATa and MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), tom70Δ (MATa Tom70::KAN his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), and tom20Δ (MATa Tom20::KAN his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) were obtained from Research Genetics, Inc. Strain tom22Δ (his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 tom22::HIS3 rho0) and the control haploid strain OL223 (his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 rho0) were a kind gift from N. Pfanner (Germany). Standard yeast genetic techniques and media were used (37).
Construction of Expression Plasmids
Rat CYP2E1, CYP2B1, CYP+5/1A1, CYP+33/1A1, and CYP27A1 cDNAs were generated by reverse transcriptase-PCR and cloned in pGEM7zf (Promega Biotech, Madison, WI) plasmid vectors. Full-length rat liver cDNA to TOM40 was purchased from ATCC (IMAGE clone). NH2-terminal truncated TOM40 cDNA (Δ143TOM40) was generated by PCR amplification using full-length TOM40 DNA as a template. Both full-length and truncated TOM40 constructs were cloned in pGEM7zf vector and pET vectors to carry out expression in vitro and bacterial cells, respectively.
Wild-type and tom70Δ, tom20Δ, and tom22Δ yeast strains were used for expressing rat CYP cDNAs driven by the elongation factor promoter on either 2-μm or centromeric URA3 plasmids. The cDNAs for full-length CYP2E1, CYP+5/1A1, CYP+33/1A1, and su9-DHFR were cloned into the 2-μm vector, pTEF-URA3, or the centromeric pTEF-URA3 plasmid (38). CYP27A1 cDNA was cloned in pYES2 (Invitrogen) vector.
Disruption of Outer Membrane Receptors by Protease Treatment
Rat liver mitochondria were isolated by differential centrifugation in H medium (5 mm HEPES (pH 7.4), 210 mm mannitol, 70 mm sucrose, 1 mm EDTA), as described previously (39), and washed three times with the H medium. Mitochondria were subjected to limited Pronase treatment (30 μg/mg protein) at 4 °C for 30 min. Following the addition of a 10-fold excess protease inhibitor, the mitochondrial pellet was re-isolated by passing through 1.2 m sucrose and used for protein import as described below.
In Vitro Import of Labeled Proteins into Mitochondria
In vitro translation products were generated in the transcription-linked translation RRL system (Promega, Madison, WI) in the presence of added [35S]Met (40 μCi/50 μl reaction, 1175 Ci/mmol; PerkinElmer Life Sciences), according to the manufacturer's recommended protocol. In the indicated experiments, CYP2B1 and CYP2E1 translation products were phosphorylated for 45 min at 30 °C by supplementing the translation mix with 10 units/100 μl PKA (Sigma) and 100 μm ATP before using for import assays. Import reactions were performed in 200-μl final volumes containing 200 μg of mitochondria at 28 °C for 60 min. In some experiments the reaction volume for import assays was increased to 500 μl using 500 μg of mitochondria. After import, the samples were treated with 200 μg/ml trypsin for 20 min at 4 °C. Trypsin-treated and untreated samples were mixed with trypsin inhibitor (10× molar excess), and mitochondria were recovered by sedimentation through 1 m sucrose as described previously (18). Mitochondrial proteins were solubilized in 2× Laemmli sample buffer (43) for 10 min at 75 °C and analyzed by SDS-PAGE and fluorography.
Effects of Immuno-inhibition of Membrane Translocases on Mitochondrial Import
Rat liver mitochondria in H medium containing 1 mm phenylmethylsulfonyl fluoride were incubated with antibodies to TOM20, TOM22, TOM70, and TOM40 (Santa Cruz Biotechnology), porin (Calbiochem), or preimmune serum at 30 °C for 30 min. The reaction mixtures were centrifuged at 5,000 × g for 5 min to remove excess antibody, and the resultant mitochondria were washed twice with H medium and used in import assays.
Immunodepletion of Hsp70 and Hsp90 from the Rabbit Reticulocyte Lysate
For immunodepletion, RRL (200-μl each) was incubated with 20 μg of Hsp70 and/or Hsp90 antibodies (Sigma) overnight (12–16 h) at 4 °C. Immune complexes were pulled down by incubation with protein A-agarose (1 h at 4 °C) followed by centrifugation at 5000 × g for 5 min. In parallel experiments, RRL was incubated with preimmune IgG (20 μg/200 μl RRL) as antibody controls. The depleted RRL were used for in vitro translation as described above.
Interaction of CYPs with Cytosolic Hsp70 and Hsp90
Interaction of CYP proteins with cytosolic Hsp70 under in vitro and in vivo conditions was tested by co-immunoprecipitation. For studying in vitro interactions, nascent proteins were synthesized in the presence (for CYP2E1 and CYP2B1) and absence (for CYP+/51A1, CYP+33/1A1, and CYP27A1) of PKA in the RRL system. Interaction of CYP proteins with cytosolic Hsp70 or Hsp90 present in the lysate under these conditions was estimated by co-immunoprecipitation. Reaction mixtures were immunoprecipitated with polyclonal antibodies against the respective CYP proteins. One portion of the immunoprecipitate was resolved by SDS-PAGE on a 10% gel and subjected to immunoblot analysis with anti-Hsp70 and -Hsp90 antibodies. The other portion was subjected to SDS-PAGE, and gels were subjected to autoradiography.
Bacterial Expression of TOM40 and Binding of CYPs to Reconstituted TOM40
Full-length and Δ143 TOM40 lacking the NH2-terminal cytosol-exposed Pro-rich domain (25) expressed in Escherichia coli BL21 cells were purified from inclusion bodies to near homogeneity and used for reconstitution in liposomes. Liposomes were prepared from azolectin (Sigma, type IVS) in MOPS/Tris (pH 6.9) buffer using a Branson sonifier as described previously (40). Liposomes were freeze-thawed three times and solubilized by the addition of n-octyl glucopyranoside (6% v/v) on ice for 30 min. Solubilized liposomes (10 mg) were mixed with 9–300 μg of TOM40 and diluted 2-fold with 10 mm MOPS/Tris (pH 6.9) containing 0.5 mm phenylmethylsulfonyl fluoride and 0.5 mm dithiothreitol. Reconstitution was performed by dialysis of the mixture at 4 °C for 24 h against the same buffer. 35S-Labeled and phosphorylated RRL translation products (80,000–100,000 cpm) were incubated for 1 h at 30 °C with vesicles containing full-length and truncated TOM40 in a buffer containing 20 mm KCl and 2% bovine serum albumin. Liposomes were re-isolated by sedimenting through a 0.4 m sucrose layer by centrifugation at 125,000 × g for 1 h. Client 35S-labeled proteins bound to TOM40 vesicles were eluted with 10 mm Tris (pH 7.4) containing 10–80 mm NaCl. Liposomes and eluted proteins were subjected to SDS-PAGE on a 12% gel, and the amount of radioactivity in the liposomal and post-liposomal fractions was determined by radiometric imaging through the STORM system (GE Healthcare).
Measurement of Pore Function of Liposome Reconstituted TOM40 Protein
The orientation and function of reconstituted TOM40 pore were investigated by measuring the release of [14C]sucrose and retention of [3H]dextran (70 kDa) from the proteoliposomes containing WT or NH2-terminal truncated Δ143 TOM40 proteins as described (25, 41). As a negative control purified P4501A1, which does not form a channel, was used following reconstitution in lipid vesicles. Reconstituted vesicles were suspended in 2 mm HEPES buffer (pH 7.4) containing 20 mm NaCl, 2 mm MgCl2, 0.3 mm NaN3, and 2 mm sucrose. Reaction mixtures were incubated with equal amounts (∼100,000 cpm) of [14C]sucrose (7.5 mCi/mmol; American Radiolabeled Chemicals Corp.) or [3H]dextran (55 mCi/mmol; American Radiolabeled Chemicals Corp.) at room temperature for 30 min. Following vortexing, tubes were placed in a bath-type sonic oscillator for 10 s. The tubes were incubated at 45 °C for 30 min and were allowed to cool down to room temperature. The reaction mixture was passed through a Sepharose 4B column, and the vesicles were eluted with 10 mm HEPES-KOH buffer (pH 7.4) containing 100 mm NaCl, 0.1 mm MgCl2, and 0.3 mm NaN3 and assayed for the amount of radioactivity retained by using a Beckman 5000 liquid scintillation counter.
Preparation of Mitochondria from TOM-deleted Yeast Strains
Mitochondria were isolated from both the control and tom-deleted yeast strains essentially as described previously (42). Steady-state levels of mitochondrial proteins were analyzed by separation via SDS-PAGE and immunoblotting.
SDS-PAGE and Immunoblot Analysis
Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes for immunoblot analysis as described previously (43). Polyclonal antibodies against CYP2E1, CYP2B1, CYP1A1, monoclonal antibody to CYP27A1, and monoclonal antibodies against cytosolic Hsp70 and Hsp90 (Sigma) were used. Immunoblots were developed using a chemiluminescence SuperSignal West Femto kit (Pierce), and the blots were imaged and quantitated using a Versadoc system (Bio-Rad).
RESULTS
Distinctive TOM Protein Requirements for the Import of Chimeric Signal-containing CYPs
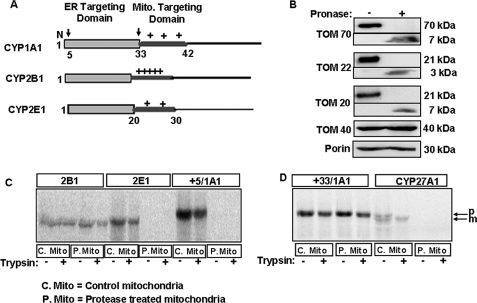
Fig. 1A shows the NH2-terminal chimeric signals of CYP1A1, CYP2B1, and CYP2E1 that encompass the NH2-terminal transmembrane domain and the cryptic mitochondrial targeting signal sequences. We have shown previously that mitochondrial import of these CYPs is dependent on transmembrane potential and energy (14, 16–18). In this study, we initially tested whether the major peripheral TOM receptors were needed for the mitochondrial import of these CYPs. For this purpose rat liver mitochondria were subjected to limited Pronase treatment, which is known to remove cytosol-exposed regions of the proteins (33). Fig. 1B shows that Pronase treatment degraded most of the cytosol-exposed regions of TOM70, TOM20, and TOM22. This treatment protected the 7-, 3-, and 7-kDa putative transmembrane segments that were detected by antibodies raised against full-length TOM70, TOM20, and TOM22, respectively. Under these treatment conditions nearly full-length 40-kDa TOM40 protein and the 30-kDa porin protein were protected. Although not shown, mitochondrial inner membrane protein cytochrome c oxidase subunit I (CcOI) and matrix protein Hsp70 were also fully protected. Using these protease-treated mitochondria, we tested the import efficiency of CYP proteins. CYP27A1 (44), which contains a canonical (NH2-terminal cleavable) mitochondrial targeting signal, was used as a positive control. The results of in vitro import showed that nearly intact CYP2B1 protein was imported and rendered resistant to trypsin treatment, although CYP2E1 and CYP+5/1A1 were not imported into Pronase-treated mitochondria (Fig. 1C). Fig. 1D shows that CYP+33/1A1 was imported efficiently into Pronase-treated mitochondria, whereas CYP27A1 was not. These results suggest that different chimeric signal-containing proteins have different requirements for peripheral TOM proteins.
FIGURE 1.
Import of CYP proteins into protease-treated mitochondria. A, schematic representation of chimeric signals of CYP1A1, CYP2B1, and CYP2E1 with NH2-terminal ER-targeting domain followed by the positively charged mitochondrial (Mito.) targeting domains. Arrows over the CYP1A1 schematic indicate processing sites at the 4th and 32nd residues. B, isolated rat liver mitochondria were treated with or without Pronase as indicated under “Experimental Procedures.” The TOM components (full-length and fragments) were detected by immunoblot analysis (30 μg of protein each) using subunit-specific antibodies. C and D, 35S-labeled CYPs translated in a RRL system were used for import into Pronase-treated (P-mito) and untreated control (C-mito) mitochondria. The extent of import was determined by resistance to treatment with trypsin (200 μg/ml); mitochondria were pelleted through 1 m sucrose, and 250 μg of protein in each case was analyzed by SDS-PAGE on a 14% gel and subjected to fluorography. p and m indicate the precursor and mature forms of CYP27A1, respectively.
In agreement with previously published results from our laboratory (14–18), all four chimeric signal-containing CYPs and also canonical signal-containing CYP27A1 protein imported in WT yeast mitochondria were resistant to digitonin plus trypsin treatment suggesting their localizations inside the inner membrane matrix compartment (see supplemental Fig. 1, A–E). Furthermore, in all cases mitochondria-imported CYPs were extractable with alkaline Na2CO3 indicating that they are mostly membrane extrinsic proteins (supplemental Fig. 1, A–E).
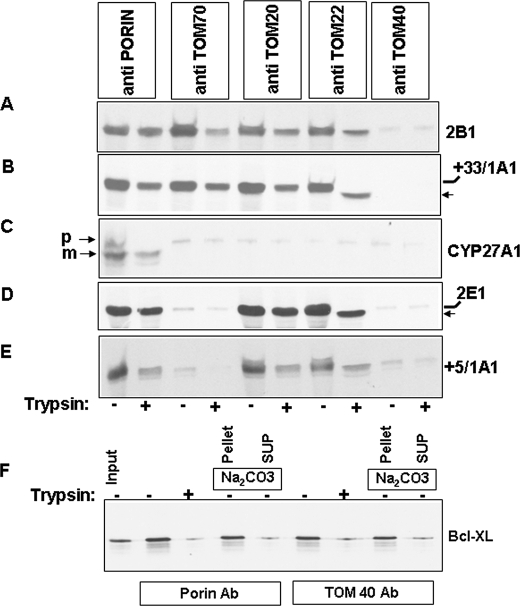
The results of import with protease-treated mitochondria were further ascertained using two parallel approaches. In the first approach, we used immuno-inhibition of individual TOM proteins using saturating levels of specific antibodies. As is seen in Fig. 2, A–E, the TOM40 antibody blocked the import of all five proteins in rat liver mitochondria. It is likely that binding of client proteins to peripheral TOMs is a cooperative process and requires functional TOM40. Import of CYP2B1 and CYP+33/1A1 was not blocked by antibodies to the peripheral TOM proteins, TOM70, TOM22, and TOM20 (Fig. 2, A and B). Import of CYP2E1 and CYP+5/1A1 was blocked by TOM70 antibody (Fig. 2, D and E) but not by antibodies to either TOM20 or TOM22. Also, the import of CYP+33/1A1 and CYP2E1 in the presence of TOM22 antibodies was incomplete as seen by relatively shorter protease-protected products (Fig. 2, B and D). Import of CYP27A1 was affected by all antibodies tested (Fig. 2C). Antibody to porin was used as a negative control and had no effect on the import of any of the proteins tested (Fig. 2, A–E).
FIGURE 2.
Effects of antibody blocking of peripheral TOMs on the mitochondrial import of proteins with different chimeric signals. Isolated rat liver mitochondria were preincubated with anti-rat TOM40, anti-rat TOM20, anti-rat TOM22, anti-rat porin, or anti-rat TOM70 antibodies at 30 °C for 30 min. Mitochondria were re-isolated and used for the import of 35S-labeled CYP2B1 (A), CYP+33/1A1 (B), CYP27A1 (C), CYP2E1 (D), and CYP+5/1A1 (E). F, mitochondria preincubated with antibodies (Ab) to porin and TOM40 were used of the import of 35S-labeled Bcl-XL protein. Mitochondria were extracted with alkaline Na2CO3 as described previously (14, 15). Details of import and protease treatment were as described under “Experimental Procedures.” Mitochondrial proteins (250 μg each) were resolved by SDS-PAGE, and the gels were subjected to fluorography. B and D, ← indicates the shorter protected fragment. SUP, supernatant.
The specificity of TOM40 antibody effect observed above was ascertained using Bcl-XL protein, which is targeted to the mitochondrial outer membrane in the absence of functional TOM40 protein (45). Fig. 2F shows that, as expected, TOM40 antibody and also porin antibody had no significant effect on the membrane association of Bcl-XL. The transmembrane orientation of the protein is apparent from its relative insolubility in Na2CO3. Furthermore, trypsin treatment caused substantial reduction in protein level as expected of a protein with most part exposed outside the outer membrane (Fig. 2F).
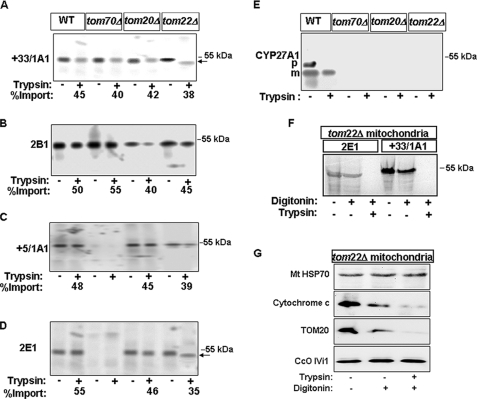
In the second approach, we used mitochondria from yeast strains that contained selectively deleted tom genes. As seen from Fig. 3A, CYP+33/1A1 was imported into yeast mitochondria that were devoid of TOM70, TOM20, and TOM22 proteins at the same level as in WT mitochondria. However, the protease-protected fragment in TOM22-deficient mitochondria was shorter by almost 3–4 kDa. Similarly, CYP2B1 was imported into mitochondria devoid of all three TOM proteins at the level similar to that in WT mitochondria (Fig. 3B). On the other hand, CYP+5/1A1 and CYP2E1 were not imported efficiently by mitochondria deficient in TOM70, suggesting a critical requirement for TOM70 in the import of these two proteins (Fig. 3, C and D). Additionally, the protease-protected fragment of CYP2E1 was shorter (∼3–4 kDa) in mitochondria deficient in TOM22 (Fig. 3D). Finally, CYP27A1 was not imported significantly in mitochondria deficient in any of the three TOMs (Fig. 3E).
FIGURE 3.
Import of CYP proteins into yeast mitochondria with deleted TOM subunits. A–E, mitochondria were isolated from wild-type, tom70Δ, tom20Δ, or tom22Δ yeast strains. 35S-Labeled proteins (CYP+33/CYP1A1 (A), CYP2B1 (B), CYP+5/1A1 (C), CYP2E1 (D), and CYP27A1 (E)) were incubated with mitochondria from either wild-type or TOM deletion strains as indicated. Following import, mitochondria were treated with trypsin, re-isolated, and analyzed by SDS-PAGE and fluorography. The radioactive proteins bound to mitochondria and the protease-protected fractions were quantified for calculations of the % import (values presented at the bottom of lanes). F, 35S-labeled CYP+331A1 and CYP2E1 were imported into tom22Δ mitochondria as described for A–E and resolved on a higher resolution gradient gel. G, trypsin- and digitonin (75 μg/mg protein)-treated fractions were also analyzed for marked proteins by immunoblot analysis using indicated antibodies. A and D, ← indicates the shorter protected fragment.
The topology of incompletely translocated CYP2E1 and CYP+33/1A1 in tom22Δ mitochondria was tested by treatment with digitonin prior to treatment with trypsin. The results shown in Fig. 3F reveal that digitonin treatment rendered both proteins completely sensitive to trypsin digestion, suggesting that imported CYP2E1 and CYP+33/1A1 in tom22Δ mitochondria are located in the intermembrane space. The intermembrane space location of CYP2E1 and CYP+33/1A1 in Fig. 3F was further confirmed in Fig. 3G showing that cytochrome c, an intermembrane space protein, is sensitive to both digitonin and digitonin plus trypsin treatment (Fig. 3G) in these mitochondria. However, mitochondrial inner membrane protein cytochrome c oxidase subunit IVi1 (CcOIVi1) and matrix protein Hsp70 were resistant to these treatments.
We tested whether the import of incompletely translocated CYP2E1 and CYP+33/1A1 in tom22Δ mitochondria is dependent on membrane potential (supplemental Fig. 1, F and G). Results show that the import of CYP+33/1A1 and CYP2E1 by WT mitochondria was inhibited by more than 75%, whereas the import of these proteins into tom22Δ mitochondria was moderately inhibited by CCCP. Furthermore, <60% of tom22Δ mitochondria-associated CYP+33/1A1 and CYP2E1 was extracted with alkaline Na2CO3 suggesting their membrane extrinsic topology. These results together with the results of digitonin plus trypsin treatment (Fig. 3F) suggest that the incompletely imported CYP+33/1A1 and CYP2E1 proteins in tom22Δ mitochondria are localized in the intermembrane space as opposed to the matrix compartment in the WT mitochondria (supplemental Fig. 1, A–E).
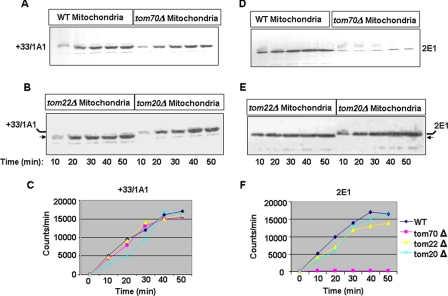
As shown in Fig. 4, A–C, the rate of in vitro import of CYP+33/1A1 in tom70Δ, tom20Δ, and tom22Δ mitochondria up to 50 min of incubation were nearly similar and compared well with the rate of import into WT yeast mitochondria. The rate of import of CYP2E1 was nearly comparable in the WT, tom22Δ, and tom20Δ mitochondria, although there was no significant import into tom70Δ mitochondria. In support of results in Figs. 2 and 3, the protected CYP+33/1A1 and CYP2E1 proteins in tom22Δ mitochondria were notably shorter. The results from Figs. 2–4 suggest that different chimeric signal-containing proteins have notably different requirements for peripheral TOMs. These results also show that TOM22 may play a significant role in the complete translocation of CYP+33/1A1 and CYP2E1 proteins through TOM40.
FIGURE 4.
Rates of in vitro import of CYP+33/1A1 and CYP2E1 proteins into mitochondria from Tom-deleted yeast strains. Import of 35S-labeled proteins was carried out as described for Fig. 3, for 0–50 min. Trypsin-treated mitochondria were subjected to SDS-PAGE analysis (A, B, D, and E), and the gels were imaged through a STORM radiometric imager. The rate of import as a function of time was plotted in C and F. B and E, ← indicates the shorter protected fragment.
Binding of Chimeric Signal-containing Proteins to Hsp70 Is a Prerequisite for Binding to Hsp90
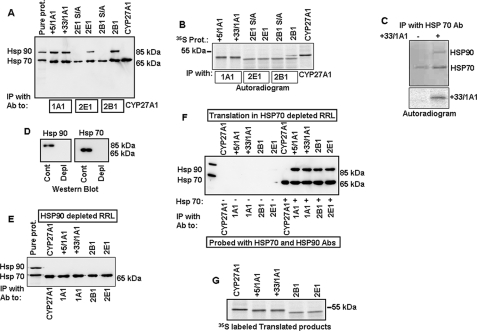
A marked difference in the requirement for different TOM proteins suggested a possible bypass mechanism for the import of certain chimeric signal-containing proteins. These results prompted an investigation into the role of other cytosolic factors on the ability of chimeric signals to bypass peripheral TOMs on their way to mitochondrial entry. Recent studies from our laboratory and others indicated a possible role for cytosolic Hsp70 in the import of mitochondria-targeted proteins (18). In this study, we investigated the ability of different proteins to bind Hsp70 and Hsp90 by a co-immunoprecipitation method. In vitro-translated CYP+5/1A1, CYP+33/1A1, CYP2B1, CYP2E1, and CYP27A1 in RRL were immunoprecipitated with CYP-specific antibodies, and the immunoprecipitates were then probed with Hsp70 and Hsp90 antibodies using an immunoblot method. The immunoblot shown in Fig. 5A reveals that CYP+5/1A1, CYP+33/1A1, CYP2E1, and CYP2B1 interacted with both Hsp70 and Hsp90, whereas CYP27A1 interacted only with Hsp70. Additionally, the S128A and S129A phosphorylation site mutants of CYP2B1 and CYP2E1 (S/A mutants), respectively, interacted weakly with Hsp70 but did not interact with Hsp90. These latter results are consistent with our observation that PKA-mediated phosphorylation is important for efficient mitochondrial import of CYP2B1 and CYP2E1 (16, 18, 24). The fluorogram presented in Fig. 5B shows the levels of 35S labeled client proteins in the immunoprecipitates shown in Fig. 5A.
FIGURE 5.
Interaction of chimeric and canonical import signal containing CYPs with Hsp70 and Hsp90 chaperones. A and B, 35S-labeled wild-type, CYP+5/1A1, CYP+33/1A1, CYP2E1, CYP2B1, and CYP27A1 proteins, and phosphorylation site mutant 2E1S/A (S129A CYP2E1), and 2B1S/A (S128A CYP2B1) proteins in RRL were subjected to immunoprecipitation (IP) with CYP-specific antibodies (Ab) as indicated. Half of the immunoprecipitate was analyzed by SDS-PAGE and probed with anti-Hsp70 and -Hsp90 antibodies (A). The other half of immunoprecipitate was analyzed by SDS-PAGE and fluorography (B). C, RRL with 35S-labeled CYP+33/1A1 and that incubated with empty vector were immunoprecipitated with Hsp70 antibody. Half of immunoprecipitate was subjected to immunoblot analysis using Hsp70 plus Hsp90 antibodies (upper panel), and the other half was resolved on the gel and subjected to fluorography (lower panel). D, levels of Hsp70 and Hsp90 in immunodepleted RRL. E, various CYP proteins were translated in Hsp90-depleted RRL and immunoprecipitated with CYP-specific antibodies. The immunoprecipitates were subjected to immunoblot analysis with anti-Hsp70 plus -Hsp90 antibodies. F, various CYP proteins were translated in Hsp70-depleted or Hsp70-replenished RRL and immunoprecipitated with CYP-specific antibodies. The immunoprecipitates were probed with a combination of anti-Hsp70 plus -Hsp90 antibodies. Immunodepletion of lysates was carried out as described under “Experimental Procedures.” G, autoradiogram representing the 35S-labeled products of CYPs translated in reticulocyte lysates depleted of Hsp70.
The possibility of a ternary complex formation between the client protein and Hsp70 and Hsp90 was tested by co-immunoprecipitation. Fig. 5C shows that immunoprecipitation of RRL containing 35S-labeled CYP+33/1A1 with Hsp70 antibody resulted in the pulldown of the client protein and Hsp90. Immunoprecipitation of RRL devoid of client protein with Hsp70 antibody, on the other hand, immunoprecipitated only Hsp70. These results confirm the formation of ternary complex consisting of the client protein and the two Hsp chaperones.
We next investigated whether the binding of various CYPs to Hsp70 and Hsp90 are interdependent or independent of each other. Various CYPs were translated in RRLs depleted of either Hsp90 or Hsp70 and subjected to immunoprecipitation with CYP-specific antibodies (Fig. 5, E and F). The immunoprecipitates were then probed with Hsp-specific antibodies by immunoblot analysis. Fig. 5D shows the levels of Hsp70 and Hsp90 in control and immunodepleted RRL used in this experiment. Fig. 5E shows that in the absence of Hsp90, all the CYPs tested were able to interact with Hsp70. Surprisingly, these CYPs failed to co-immunoprecipitate Hsp90 in Hsp70-depleted lysates (Fig. 5F). However, supplementation with purified Hsp70 restored CYP binding to Hsp90. The only exception was CYP27A1, which did not show any significant interactions with Hsp90 either in complete or Hsp70-depleted extracts. As seen in Fig. 5G, the level of translation of all CYP proteins was comparable in Hsp70-depleted RRL. Although not shown, Hsp90 depletion also did not adversely affect the translation of CYP proteins. These results suggest that the interaction between chimeric signal-containing proteins and Hsp90 is dependent on the presence of Hsp70, suggesting a degree of cooperativity between the two factors.
Roles of Hsp70 and Hsp90 in Modulating the TOM Bypass Mechanism and Translocation
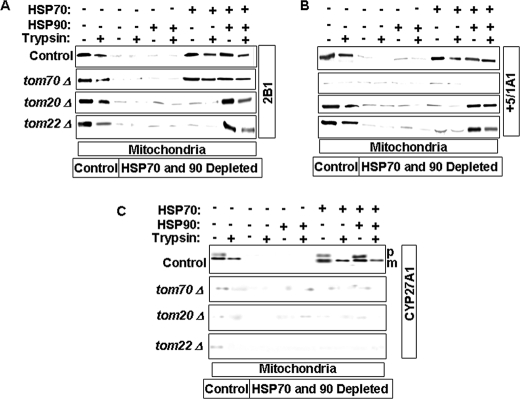
In a previous study, we showed that the binding of CYP2E1 to Hsp70 was important for the mitochondrial targeting of the protein (18). In an extension of this study, we have now investigated the roles of Hsp70 and Hsp90 chaperones in the mitochondrial targeting of different chimeric signal-containing proteins and their propensity to bypass peripheral TOM proteins. For this purpose, we translated CYP2B1, CYP+5/1A1, and CYP27A1 proteins in the RRL depleted of Hsp70, Hsp90, or both. The translated products were used for import into mitochondria isolated from various TOM protein-deleted yeast cells. In each case, the effects of added Hsp70 and/or Hsp90 were tested. The results of in vitro import assay (Fig. 6A) revealed that depletion of Hsp70 and Hsp90 completely abolished CYP2B1 import into WT as well as TOM70-, TOM20-, and TOM22-deleted mitochondria. The addition of Hsp90 alone did not rescue the import defect. However, the addition of Hsp70 alone was able to restore import of CYP2B1 into both WT and tom70Δ mitochondria. Supplementation with both Hsp70 and Hsp90 was required for restoration of the import of CYP2B1 into both tom20Δ and tom22Δ mitochondria. Although not shown, CYP+33/1A1 showed a similar pattern of import into TOM-deficient mitochondria.
FIGURE 6.
Role of Hsp70 and Hsp90 in the import of CYP proteins into mitochondria with deleted TOM subunits. CYP proteins were translated in RRL depleted of Hsp70, Hsp90, or both and used for in vitro import into wild-type or TOM subunit-deficient mitochondria. RRL were immunodepleted with anti-Hsp70 and -Hsp90 antibodies as described under “Experimental Procedures.” 35S-Labeled CYP2B1 (A), CYP+5/1A1 (B), and CYP27A1 (C) were used for import into mitochondria from wild-type, tom70Δ, tom20Δ, or tom22Δ yeast cells. The import assays were carried out with or without the addition of purified Hsp70 and Hsp90. The details of the import reaction and treatment with trypsin and protease inhibitor were described in the legend for Fig. 2. Proteins (250 μg each) were subjected to SDS-PAGE and fluorography. p, precursor and m, mature protein.
The import of CYP+5/1A1 into tom-deficient mitochondria was somewhat different in that the addition of Hsp70 and Hsp90 did not restore the import of this protein. Furthermore, both Hsp70 and Hsp90 were required for import of this protein into tom20Δ and tom22Δ mitochondria (Fig. 6B), whereas mitochondria from tom70Δ cells were unable to import CYP+5/1A1 under all conditions tested. Although not presented, CYP2E1 showed an identical requirement for TOM70 for import. CYP27A1 showed a distinctly different requirement (Fig. 6C). First, Hsp90 depletion did not have a significant effect on the import of CYP27A1 into WT mitochondria. Second, deletion of tom70, tom20, or tom22 almost completely inhibited the import of CYP27A1 (Fig. 6C). These results together suggest that Hsp70 and Hsp90 are important factors that facilitate the presentation of different chimeric signals to the TOM complex. They also play a major role in the ability of chimeric signals to bypass peripheral TOMs. The results also suggest different requirements for peripheral TOMs in terms of the import of the four chimeric signal-containing proteins tested.
Targeting of CYP Proteins to Mitochondria in Intact Cells Involves TOM Bypass Mechanism
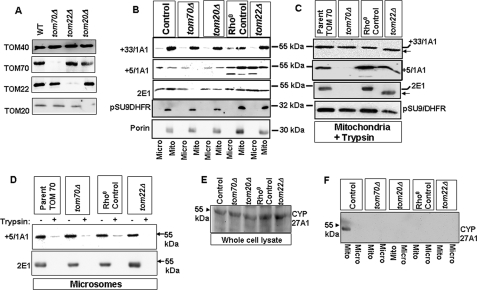
The occurrence of a TOM bypass mechanism for the mitochondrial targeting of chimeric signal-containing CYPs was confirmed by the expression of various CYPs in control (parent strains of tom70 and tom20 null mutant) and tom null mutant yeast cells. Immunoblot analysis of mitochondrial proteins in Fig. 7A shows that mutant strains deficient in a given peripheral TOM, for example tom70Δ cells, expressed intact peripheral TOM20 and TOM22 proteins at levels comparable with the wild-type yeast. Thus the deficiency in these strains was restricted to the specified gene product.
FIGURE 7.
Mitochondrial targeting of chimeric signal-containing CYPs in tom70Δ, tom20Δ, and tom22Δ yeast cells. Wild-type and tom null mutant strains were transformed with CYP+5/1A1, CYP+33/1A1, CYP2E1, or su9-DHFR. A, TOM40, TOM70, TOM20, and TOM22 levels of various deletion strains. Immunoblots were carried out using 40 μg of protein each. B, mitochondrial and microsomal fractions of cells expressing different CYPs or Su9-DHFR were subjected to immunoblot analysis (30 μg each) using CYP-specific antibodies as indicated. C, mitochondria from tom70, tom22, tom70, or tom22 null mutant cells expressing different CYPs or Su9-DHFR were subjected to trypsin treatment, and proteins were subjected to immunoblot analysis as described in A. D, microsomal fractions from tom70Δ, tom70Δ parent, tom22Δ, and tom22Δ parent strains expressing CYP+5/1A1 and CYP2E1 were treated with or without trypsin and subjected to immunoblot analysis using CYP-specific antibodies. E and F, proteins from total cell extract (E) and mitochondria (F) from WT and mutant yeast strains expressing CYP27A1 were subjected to immunoblot analysis (50 μg each) using CYP27A1 specific antibody. C, ← indicates the shorter protected fragment. Mito, mitochondrial; Micro, microsomal.
The tom22 null mutation is known to induce ρ0 status. To assess the effects of the ρ0 mutation on mitochondrial protein targeting, we expressed different CYP cDNAs in the respective parent strain made ρ0 by ethidium bromide treatment (ρ0 control). As shown in Fig. 7B, CYP+33/1A1 was efficiently targeted to mitochondria in both control and tom null mutant cells. The protein was inefficiently targeted to the ER as expected of a protein that lacks the signal recognition particle-binding signal domain. CYP+5/1A1, on the other hand, showed a distinct requirement for TOM70, as no detectable protein was found in the mitochondrial compartment of tom70Δ cells. These results essentially confirm the in vitro data (Fig. 6), which suggests the distinctive nature of chimeric signals of CYP2E1 and CYP+5/1A1 requiring TOM70 for mitochondrial translocation. Although not shown, CYP2B1 behaved similarly to CYP+33/1A1 and was targeted efficiently to mitochondria in tom70Δ cells. Furthermore, the mitochondrial targeting of all three proteins occurred efficiently in tom20Δ and tom22Δ cells, although the overall level of expression in both mitochondrial and microsomal compartments was relatively low in tom20Δ cells. Surprisingly, the level of expression of CYP+5/1A1 and CYP2E1 was generally higher in both ρ0 control and tom22Δ cells possibly reflecting strain-specific differences.
The topology of mitochondria- and microsome-associated CYPs in tom null mutants was tested by limited digestion with trypsin followed by immunoblot analysis. As shown in Fig. 7C, mitochondria-associated CYPs were resistant to trypsin digestion, although microsome-associated CYPs were sensitive to trypsin treatment (Fig. 7D). In this respect, mitochondria-associated CYPs exhibited resistance to trypsin similar to the matrix-associated protein pSU9-DHFR used as a control (Fig. 7C, bottom panel). Confirming the results in Figs. 3, 4, and 6, CYP2E1 and CYP+5/1A1 were not targeted to mitochondria significantly in tom70Δ cells. Also, the trypsin-protected fragments of CYP2E1 and CYP+33/1A1 were shorter in tom22Δ cells compared with ρ0 control cells. These results confirm that mitochondria-associated proteins are probably localized inside the inner membrane compartment, although the microsome-associated proteins are exposed externally to the ER in a protease-accessible orientation. These results together confirm the occurrence of the TOM20/TOM22 bypass protein-targeting mechanism in vivo.
We used pSU9/DHFR as a control for matrix-targeted protein because Stan et al. (46) showed that the protein is efficiently transported in tom70Δ cells. In contrast, results from other groups (47, 48) showed that deletion or depletion of TOM70, TOM20, and TOM22 affects mitochondrial import of pSU9/DHFR. However, we did not observe significant inhibition of mitochondrial import of this protein in tom70Δ and tom22Δ cells either under in vivo (Fig. 7C) or in vitro (not shown) conditions. Furthermore, CYP27A1, which required all three peripheral TOMs for mitochondrial transport (Figs. 1–3 and 6) was also not targeted to mitochondria in tom70Δ, tom22Δ, and tom20Δ yeast cells (Fig. 7F), although the protein is expressed at nearly comparable levels in all cells (Fig. 7E) and targeted efficiently to mitochondria in WT yeast cells. These results confirm the specificity of the mitochondrial transport in these mutant strains.
Association of Hsp90 with Reconstituted TOM40 during the Delivery of CYP+33/1A1 Protein
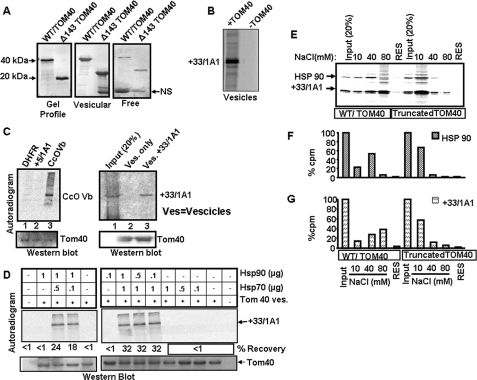
It is known that a number of mitochondria-destined proteins with canonical targeting signal require functional TOM70 receptor protein for import. In these cases Hsp90 has been shown to physically associate with TOM70 for delivering the client protein to the translocase complex (49, 50). The in vitro and in vivo experiments in this study (Figs. 2–7) suggested that in the presence of Hsp70 and Hsp90 and CYP+33/1A1 and CYP2B1 proteins are translocated to mitochondria in the absence of TOM20, TOM22, and TOM70. To understand the molecular basis of this bypass mechanism, we tested the ability of Hsp90 to bind to purified and reconstituted TOM40 under in vitro conditions. Our preliminary results with antibody pulldown assays showed that 35S-labeled CYP+33/1A1 efficiently bound to WT TOM40 but not with Δ143 TOM40 lacking the NH2-terminal cytosol-exposed Pro-rich domain (25). We have therefore used purified and reconstituted WT and Δ143 TOM40 proteins in lipid vesicles for binding in these studies. Fig. 8A (left panel) shows the Coomassie Blue-stained gel profiles of purified WT and Δ143 TOM40 proteins. The reconstituted lipid vesicles were recovered by sedimentation at 125,000 × g for 1 h, and both the vesicular fraction (Fig. 8A, middle panel) and supernatant representing “free” proteins (right panel) were analyzed by PAGE. The middle and right panels (Fig. 8A) show that a large fraction of proteins in both cases is associated with lipid vesicles, and a relatively small fraction is recovered in the supernatant fraction. Another control experiment in Fig. 8B shows that only vesicles containing TOM40 protein binds efficiently to CYP+33/1A1 in complete RRL, whereas vesicles devoid of TOM40 fail to bind to the protein indicating the specificity.
FIGURE 8.
Delivery of CYP+33/1A1 client protein by Hsp70 and Hsp90 to intact TOM40 but not the Δ143 TOM40. A, Coomassie Blue-stained patterns of WT and Δ143 TOM40 proteins (left panel), liposome-integrated proteins (125,000 × g pellet, middle panel), and the free proteins (125,000 × g supernatant, right panel). B, 35S-labeled CYP+33/1A1 protein (40,000 cpm) was incubated with lipid vesicles (5 mg each) containing TOM40 or that lacking TOM40 for 20 min at 30 °C. The vesicles were recovered by pelleting through 0.4 m sucrose as described in C and subjected to SDS-PAGE and fluorography. C, 35S-labeled client proteins, dihydrofolate reductase (DHFR), CcOVb (Vb), CYP+5/1A1 (left panel), and CYP+33/1A1 (right panel) (50,000 cpm each) in intact RRL were used for binding to lipid vesicles (Ves) containing 2 μg of WT TOM40 protein for 20 min at 30 °C. Liposomes were recovered by layering on 0.4 m sucrose followed by centrifugation at 125,000 × g for 1 h and washed twice with 20 mm Tris-HCl (pH 7.4) and 10 mm NaCl; equal portions were analyzed on two companion polyacrylamide gels. One gel was subjected to fluorography (upper panels), and the second gel was subjected to immunoblot analysis using TOM40 antibody (lower panels). D, effect of Hsp90 and Hsp70 on binding of CYP+33/1A1 to TOM40 vesicles. CYP+33/1A1 protein was translated in Hsp90- and Hsp70-depleted RRL, and 50,000 cpm each were incubated with liposomes containing WT TOM40 in the presence or absence of various amounts of purified Hsp90 and Hsp70 as indicated. E, Hsp90 and CYP+33/1A1 proteins were co-translated in Hsp90-depleted RRL, and 50,000 cpm each were incubated with liposomes containing WT and Δ143 TOM40 proteins (2 μg of protein each) as described in C. Liposomes were recovered and sequentially washed with Tris-HCl buffer containing 10, 40, and 80 mm NaCl. The washes and also residual proteins (RES) in the vesicle were analyzed by SDS-PAGE and fluorography. F and G, quantification of Hsp90 (F) and +33/1A1 (G) protein bands from autoradiogram in E. The input lanes in represent 20% of total counts used for binding. NS, nonspecific.
The proper orientation of reconstituted TOM40 and Δ143 TOM40 proteins and the presence of TOM40 pore were tested using [14C]sucrose and [3H]dextran (∼70 kDa) as permeable and impermeable substrates. As shown in Table 1, reconstituted vesicles with CYP1A1 showed nearly complete retention of both 14C and 3H radioactivity suggesting no pore activity. Vesicles containing increasing amounts of intact TOM40 as well as Δ143 TOM40 proteins showed increasing release of [14C]sucrose but complete retention of [3H]dextran, suggesting efficient pore activity.
TABLE 1.
Pore function of reconstituted TOM40 and Δ143 TOM40 proteins
Reconstitution of lipid vesicles with different proteins, introduction of radioactive sucrose and dextran into vesicles, and purification of vesicles were carried out as described under “Experimental Procedures.” S/D indicates ratio between [14C]sucrose and [3H]dextran retained in the vesicles.
| Vesicle type | Input radioactivity (cpm × 103) |
cpm × 103 retained in vesicles |
Ratio S/D | ||
|---|---|---|---|---|---|
| [3H]Dextran | [14C]Sucrose | [3H]Dextran | [14C]Sucrose | ||
| CYP1A1 (30 μg)a | 102 | 109 | 33 | 31 | 0.92b |
| TOM40 (4 μg) | 100 | 98 | 26 | 19 | 0.74 |
| TOM40 (30 μg) | 105 | 106 | 25 | 15 | 0.57 |
| ΔTOM40 (4 μg) | 109 | 102 | 28 | 25 | 0.86 |
| ΔTOM40 (30 μg) | 97 | 98 | 29 | 15 | 0.51 |
a Amounts of proteins were reconstituted in 1 mg of lipid vesicles.
b Values represent averages of assays run in duplicate.
In Fig. 8C, we tested the efficiencies of two types of chimeric signal-containing proteins, CYP+5/1A1 and CYP+33/1A1, for binding to reconstituted vesicles containing WT TOM40 protein. 35S-Labeled client proteins translated in intact RRL were incubated with lipid vesicles at 30 °C for 20 min, and the vesicles were recovered by centrifugation at 125,000 × g for 1 h, washed twice with buffer containing 10 mm NaCl, and used for analysis. The autoradiogram in Fig. 8C (left panel) shows that DHFR, a cytosolic protein, and CYP+5/1A1 did not bind significantly to TOM40 vesicles. The inability of CYP+5/1A1 protein to bind to TOM40 may be due to lack of TOM70 in the vesicle. CcOVb subunit (Fig. 8C, left panel) and also CYP+33/1A1 (right panel) bound efficiently to TOM40 in the absence of other TOMs. These results are consistent with our in vitro and in vivo results that only CYP+33/1A1 but not CYP+5/1A1 can bypass all the peripheral TOMs. In Fig. 8D we ascertained the roles of Hsp70 and Hsp90 for the CYP+33/1A1 protein binding to reconstituted TOM40 vesicles using the Hsp70- and Hsp90-depleted RRL system. 35S-Labeled CYP+33/1A1 protein was bound to TOM40 vesicles, and the vesicles were reisolated, washed twice, and analyzed by SDS-gel electrophoresis. Autoradiogram in Fig. 8D shows that CYP+33/1A1 did not bind to TOM40 in the absence of Hsp70 or Hsp90. Also, both Hsp90 and Hsp70 induced the binding in a concentration-dependent manner. These results further confirm the need for both the Hsp70 and Hsp90 for supporting the bypass pathway.
In Fig. 8E we tested whether Hsp90 could physically associate with the reconstituted TOM40 vesicles for delivering the CYP+33/1A1 protein and whether the binding requires the cytosol-exposed region of TOM40. In this experiment, Hsp90-depleted RRL was used for translating CYP+33/1A1 and Hsp90 proteins together in the presence of [35S]Met and used for binding to vesicles containing TOM40 and Δ143 TOM40 proteins. The vesicles were recovered and washed sequentially with buffers containing 10, 40, and 80 mm NaCl, and both the eluted proteins and that remaining with the vesicles (residual) were analyzed for assessing the type of association of Hsp90 and CYP+33/1A1 with TOM40. Fig. 8E shows that both the CYP+33/1A1 and Hsp90 bind to WT TOM40, and most of the bound Hsp90 protein was eluted with 40 mm NaCl (Fig. 8, E and F), whereas elution of CYP+33/1A1 required 80 mm NaCl suggesting a more stable interaction of the latter with TOM40 (Fig. 8, E and G). Additionally, Hsp90 and CYP+33/1A1 proteins bind at markedly reduced levels with Δ143 TOM40 (Fig. 8, E–G) suggesting that the binding requires intact cytosol-exposed Pro-rich domain of TOM40. These results provide insight into the mechanism of the bypass pathway that involves direct binding of Hsp90 for delivering the client protein to TOM40.
DISCUSSION
Mitochondrial targeting signals are heterogeneous, ranging from α-helical, β-sheet, and unstructured (13). The most common type of mitochondrial signals are pre-sequences, which consist of 15–40 NH2-terminal amino acids that form amphiphilic helices with positively charged residues lining one side of the helix. These signals are commonly observed in mitochondrial matrix- and inner membrane-targeted proteins and cleaved by matrix metalloprotease(s) after the protein is translocated into the mitochondrial compartment (13). The pre-sequences are critically important for the binding of precursor proteins to mitochondrial import receptors, TOMs, and translocases of inner mitochondrial membrane (51). The second category of mitochondrial targeting signals is internal; these reside deep inside the protein (52). A third type of signal is a variation of the internal signal, where a cryptic mitochondrial signal follows immediately downstream of an NH2-terminal transmembrane domain. This type of targeting signal is known to occur in a number of membrane-anchored proteins, such as Bcs1, Tim14, Mdj2, D-AKAP1, and others (13, 52–57). We have described a fourth type of mitochondrial targeting signal, designated as a chimeric signal, based on its ability to direct the same primary translation product to two different subcellular compartments, namely microsomes and mitochondria or cytosol and mitochondria (16, 18, 21, 22). The chimeric signals we defined are positionally similar to the Bcs1-type internal signal and follow immediately downstream of an NH2-terminal transmembrane domain. However, in contrast to the transmembrane localization of proteins containing internal signals (category 3), proteins containing chimeric signals are targeted to the mitochondrial matrix (15). The reason for this difference is probably because the stop transfer signal of CYPs, which enables the proteins to associate with the ER membranes through a single transmembrane anchor (55, 56), are incapable of eliciting a similar stop transfer event in the mitochondrial outer or inner membranes. Similar to the Bcs1-type signals, chimeric NH2-terminal or COOH-terminal signals are not cleaved by matrix proteases (17, 53).
Our studies with xenobiotic-inducible CYPs, glutathione S-transferases (GST), and APP led to the discovery of a new family of mitochondrial targeting signals that are referred to as chimeric signals (14–18, 21, 22). In the case of CYPs and APP, chimeric signals are located at the NH2 terminus, whereas in GSTs, these signals are located at the COOH terminus (14,16–18, 21–23). The characteristic features of these signals are that the first half (20–30 amino acids) of the signal domain hydrophobic and the second half (10–12 amino acids) of the signal domain consists of sequences rich in positively charged amino acids followed by a Pro-rich domain. Studies also showed that chimeric signals are not substrates for mitochondrial matrix proteases (14). Unlike the canonical signals that target proteins exclusively to mitochondria, the chimeric signals are dynamic in the sense that they direct the targeting of the protein to mitochondria as well as nonmitochondrial compartments, depending on the physiological demand of the cell.
In this study, we tested four different chimeric signals, including two NH2-terminally truncated forms to mimic endoproteolytic cleavage in the cytosol as in the case of CYP1A1 (CYP+5/1A1 and CYP+33/1A1) and two that are targeted to mitochondria as intact uncleaved signals, namely CYP2B1 and CYP2E1 (15, 16, 18, 21, 24), for their ability to bind different TOM receptor proteins under both in vitro and in vivo conditions. Mitochondrial targeting of both CYP2B1 and CYP2E1 was enhanced markedly by PKA-mediated phosphorylation at the unique target sites, Ser-128 and Ser-129, respectively (16, 18). Results presented here show that the client protein binding to Hsp70 and Hsp90 is enhanced by internal phosphorylation, which may be the reason for increased mitochondrial import. Our results also suggest that the chimeric signals fall into at least two distinct classes. One class, i.e. CYP+33/1A1 and CYP2B1, binds to TOM40 directly in the absence of other TOM proteins, and the second class, i.e. CYP+5/1A1 and CYP2E1, requires other peripheral TOMs for binding to reconstituted TOM40 (Fig. 8). In this respect, CcO Vb, which also binds to TOM40 in the absence of other TOMs, resembles the first category of chimeric signals. Our results () also suggest that intact TOM40 is needed for nascent protein binding to mitochondria suggesting cooperativity between the peripheral TOMs and TOM40 in the recruitment of client proteins.
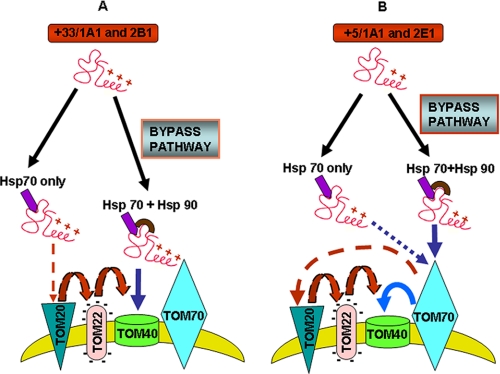
As shown in a model (Fig. 9), the chimeric signals of CYP2B1, CYP2E1, CYP+5/1A1, and CYP+33/1A1 are capable of targeting proteins to the channel-forming TOM40 protein in the absence of some or all peripheral TOM receptors (TOM70, TOM20, and TOM22 in the case of CYP2B1 and CYP+33/1A1, and TOM20 and TOM22 in the case of CYP2E1 and CYP+5/1A1). It was shown that mitochondria-destined proteins with pre-sequences first interact with TOM20 and then TOM22, whereas proteins with internal signals first interact with TOM70 followed by sequential interaction with TOM20 and TOM22 (58). Interestingly, the chimeric signals of CYP2B1 and CYP+33/1A1 first interact with TOM20 and TOM22 in the presence of Hsp70 (see Fig. 9A), thus mimicking the characteristic feature of canonical pre-sequences. In the presence of Hsp70 and Hsp90, CYP2B1 and CYP+33/1A1 are delivered directly to TOM40, thus bypassing all peripheral TOMs. The signals of CYP2E1 and CYP+5/1A1, on the other hand, mimic the features of internal-targeting signals by first interacting with TOM70 (Fig. 9B). Here again, in the presence of Hsp70 alone, these proteins require all three peripheral TOMs, whereas in the presence of Hsp70 and Hsp90, they can bypass TOM20 and TOM22 but require TOM70. These results clearly show the divergent nature of chimeric signals in addition to the functional evolution of the TOM complex, which is capable of interacting with diverse types of mitochondrial targeting signals.
FIGURE 9.
A proposed TOM bypass mechanism for the mitochondrial targeting of chimeric signal-containing proteins. A, in the presence of Hsp70 alone, CYP+33/1A1, and CYP2B1 proteins associate with TOM20 and TOM22 on their way to TOM40 without requiring TOM70. In the presence of Hsp70 and Hsp90, CYP2B1 and CYP+33/1A1 bypass all peripheral TOMs and associate directly with TOM40. B, in the presence of only Hsp70, CYP+5/1A1 and CYP2E1 proteins associate with TOM70, TOM20, and TOM22 and are delivered to TOM40. In the presence of Hsp70 and Hsp90, these proteins require only TOM70 and bypass TOM20 and TOM22 for association with TOM40.
Hsp family cytoplasmic chaperones are known to facilitate mitochondrial import of client proteins partly by preventing misfolding or protein aggregation (21, 59–62). More recent studies by Hartl and co-workers (49, 61) provided evidence for the direct physical and functional interactions between cytoplasmic chaperones, Hsp70 and Hsp90, and mitochondrial import machinery. Hsp70 and Hsp90 dock onto the TOM complex through the specialized tetratricopeptide domain of TOM70 and deliver client proteins to the TOM complex for import (61). In yeast, Hsp70 alone appears to be sufficient for pre-protein import, whereas in mammalian cells, both Hsp70 and Hsp90 are involved in pre-protein delivery and ATP-dependent translocation (63, 64). Using a combinatorial approach, we show that all four CYP proteins with chimeric signals interact with both Hsp70 and Hsp90 and, more importantly, that client protein binding to Hsp90 is dependent on the presence of Hsp70. Notably, chimeric signal-containing client proteins also facilitate the binding of Hsp90 to form the ternary complex. Furthermore, our results show that Hsp90 physically binds to the cytosol-exposed Pro-rich domain of TOM40 for client protein delivery through the bypass pathway. These results suggest a degree of cooperativity between the two chaperones. The only exception was CYP27A1, which did not bind to Hsp90 either in the presence or absence of Hsp70. Similarly, mitochondrial import of CYP27A1 did not require Hsp90.
Our observations on distinctly different requirements for TOM receptors and Hsp family chaperones by client proteins containing canonical and chimeric signals likely point to different phases of evolution of mitochondrial import machinery. Based on the fact that Hsp70 protein alone can support yeast mitochondrial protein import, it was suggested that the pathway requiring Hsp90 in mammalian cells might represent a later step of evolution of the import pathway as it exists in the mammalian cells (65–67). It is likely that the pathway for the mitochondrial import of chimeric signal-containing proteins represents a more recent evolutionary event, consistent with the increased metabolic and physiologic roles of mitochondria in mammalian cell function. Proteomic studies suggest that more than 50% of the nearly 2000 proteins associated with mammalian mitochondria lack canonical mitochondrial targeting signals (56, 67–69). This study therefore provides new insights into the mitochondrial transport/association of noncanonical signal-containing proteins. Our results showing the efficient mitochondrial import of CYP27A1 in the absence of Hsp90 is indeed intriguing. Nevertheless, because CYP27A1 function in steroid hormone biosynthesis marks some of the earliest stages of mammalian evolution, the transport pathway of this protein requiring only Hsp70 may represent a transition point from lower eukaryotes to mammals. In summary, we provide evidence for varying requirements for mitochondrial protein import based on signal type. Our results also define mechanisms by which noncanonical chimeric signal-containing proteins are imported into the mitochondria.
Supplementary Material
Acknowledgments
We are thankful to Dr. Nicolaus Pfanner for generously providing the yeast tomΔ and ρ° control strains used in this study and to Drs. Frank Luca, Kasinath Kuravi, and Gopa Biswas for helping with yeast cell expression experiments. We are also thankful to members of the Avadhani laboratory for their valuable suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-34883.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
- ER
- endoplasmic reticulum
- TOM
- translocase of outer mitochondrial membrane
- CYP
- cytochrome P450
- RRL
- rabbit reticulocyte lysate
- Hsp
- heat shock protein
- MOPS
- 4-morpholinepropanesulfonic acid
- GST
- glutathione S-transferase
- APP
- amyloid precursor protein
- WT
- wild type
- PKA
- cAMP-dependent protein kinase.
REFERENCES
- 1.Schatz G., Dobberstein B. ( 1996) Science 271, 1519– 1526 [DOI] [PubMed] [Google Scholar]
- 2.Wickner W., Schekman R. ( 2005) Science 310, 1452– 1456 [DOI] [PubMed] [Google Scholar]
- 3.Dolezal P., Likic V., Tachezy J., Lithgow T. ( 2006) Science 313, 314– 318 [DOI] [PubMed] [Google Scholar]
- 4.Subramani S. ( 1998) Physiol. Rev. 78, 171– 188 [DOI] [PubMed] [Google Scholar]
- 5.von Heijne G. ( 1985) J. Mol. Biol. 184, 99– 105 [DOI] [PubMed] [Google Scholar]
- 6.Nothwehr S. F., Gordon J. I. ( 1990) BioEssays 12, 479– 484 [DOI] [PubMed] [Google Scholar]
- 7.Van Ael E., Fransen M. ( 2006) Biochim. Biophys. Acta 1763, 1629– 1638 [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson B. M., Regnacq M., Stirling C. J. ( 1997) J. Membr. Biol. 155, 189– 197 [DOI] [PubMed] [Google Scholar]
- 9.Gilmore R., Walter P., Blobel G. ( 1982) J. Cell Biol. 95, 470– 477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wickner W. T., Lodish H. F. ( 1985) Science 230, 400– 407 [DOI] [PubMed] [Google Scholar]
- 11.Isenmann S., Khew-Goodall Y., Gamble J., Vadas M., Wattenberg B. W. ( 1998) Mol. Biol. Cell 9, 1649– 1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaussi R. ( 1995) Eur. J. Biochem. 228, 551– 561 [DOI] [PubMed] [Google Scholar]
- 13.Neupert W., Herrmann J. M. ( 2007) Ann. Rev. Biochem. 76, 723– 749 [DOI] [PubMed] [Google Scholar]
- 14.Addya S., Anandatheerthavarada H. K., Biswas G., Bhagwat S. V., Mullick J., Avadhani N. G. ( 1997) J. Cell Biol. 139, 589– 599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anandatheerthavarada H. K., Addya S., Dwivedi R. S., Biswas G., Mullick J., Avadhani N. G. ( 1997) Arch. Biochem. Biophys. 339, 136– 150 [DOI] [PubMed] [Google Scholar]
- 16.Anandatheerthavarada H. K., Biswas G., Mullick J., Sepuri N. B., Otvos L., Pain D., Avadhani N. G. ( 1999) EMBO J. 18, 5494– 5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhagwat S. V., Biswas G., Anandatheerthavarada H. K., Addya S., Pandak W., Avadhani N. G. ( 1999) J. Biol. Chem. 274, 24014– 24022 [DOI] [PubMed] [Google Scholar]
- 18.Robin M. A., Anandatheerthavarada H. K., Biswas G., Sepuri N. B., Gordon D. M., Pain D., Avadhani N. G. ( 2002) J. Biol. Chem. 277, 40583– 40593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karniely S., Pines O. ( 2005) EMBO Rep. 6, 420– 425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knox C., Sass E., Neupert W., Pines O. ( 1998) J. Biol. Chem. 273, 25587– 25593 [DOI] [PubMed] [Google Scholar]
- 21.Robin M. A., Prabu S. K., Raza H., Anandatheerthavarada H. K., Avadhani N. G. ( 2003) J. Biol. Chem. 278, 18960– 18970 [DOI] [PubMed] [Google Scholar]
- 22.Anandatheerthavarada H. K., Biswas G., Robin M. A., Avadhani N. G. ( 2003) J. Cell Biol. 161, 41– 54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boopathi E., Srinivasan S., Fang J. K., Avadhani N. G. ( 2008) Mol. Cell 32, 32– 42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sepuri N. B., Yadav S., Anandatheerthavarada H. K., Avadhani N. G. ( 2007) FEBS J. 274, 4615– 4630 [DOI] [PubMed] [Google Scholar]
- 25.Suzuki H., Kadowaki T., Maeda M., Sasaki H., Nabekura J., Sakaguchi M., Mihara K. ( 2004) J. Biol. Chem. 279, 50619– 50629 [DOI] [PubMed] [Google Scholar]
- 26.Pfanner N., Craig E. A., Meijer M. ( 1994) Trends Biochem. Sci. 19, 368– 372 [DOI] [PubMed] [Google Scholar]
- 27.Ryan K. R., Jensen R. E. ( 1995) Cell 83, 517– 519 [DOI] [PubMed] [Google Scholar]
- 28.Schneider H. C., Berthold J., Bauer M. F., Dietmeier K., Guiard B., Brunner M., Neupert W. ( 1994) Nature 371, 768– 774 [DOI] [PubMed] [Google Scholar]
- 29.Brix J., Dietmeier K., Pfanner N. ( 1997) J. Biol. Chem. 272, 20730– 20735 [DOI] [PubMed] [Google Scholar]
- 30.Brix J., Rüdiger S., Bukau B., Schneider-Mergener J., Pfanner N. ( 1999) J. Biol. Chem. 274, 16522– 16530 [DOI] [PubMed] [Google Scholar]
- 31.Meisinger C., Ryan M. T., Hill K., Model K., Lim J. H., Sickmann A., Müller H., Meyer H. E., Wagner R., Pfanner N. ( 2001) Mol. Cell. Biol. 21, 2337– 2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Wilpe S., Ryan M. T., Hill K., Maarse A. C., Meisinger C., Brix J., Dekker P. J., Moczko M., Wagner R., Meijer M., Guiard B., Hönlinger A., Pfanner N. ( 1999) Nature 401, 485– 489 [DOI] [PubMed] [Google Scholar]
- 33.Pfaller R., Pfanner N., Neupert W. ( 1989) J. Biol. Chem. 264, 34– 39 [PubMed] [Google Scholar]
- 34.Lithgow T., Junne T., Wachter C., Schatz G. ( 1994) J. Biol. Chem. 269, 15325– 15330 [PubMed] [Google Scholar]
- 35.Kurz M., Martin H., Rassow J., Pfanner N., Ryan M. T. ( 1999) Mol. Biol. Cell 10, 2461– 2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diekert K., de Kroon A. I., Ahting U., Niggemeyer B., Neupert W., de Kruijff B., Lill R. ( 2001) EMBO J. 20, 5626– 5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose M. D. ( 1987) Methods Enzymol. 152, 481– 504 [DOI] [PubMed] [Google Scholar]
- 38.Mumberg D., Müller R., Funk M. ( 1994) Nucleic Acids Res. 22, 5767– 5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niranjan B. G., Wilson N. M., Jefcoate C. R., Avadhani N. G. ( 1984) J. Biol. Chem. 259, 12495– 12501 [PubMed] [Google Scholar]
- 40.Hill K., Model K., Ryan M. T., Dietmeier K., Martin F., Wagner R., Pfanner N. ( 1998) Nature 395, 516– 521 [DOI] [PubMed] [Google Scholar]
- 41.Zalman L. S., Nikaido H., Kagawa Y. ( 1980) J. Biol. Chem. 255, 1771– 1774 [PubMed] [Google Scholar]
- 42.Sepuri N. B., Schülke N., Pain D. ( 1998) J. Biol. Chem. 273, 1420– 1424 [DOI] [PubMed] [Google Scholar]
- 43.Laemmli U. K. ( 1970) Nature 227, 680– 685 [DOI] [PubMed] [Google Scholar]
- 44.Su P., Rennert H., Shayiq R. M., Yamamoto R., Zheng Y. M., Addya S., Strauss J. F., 3rd, Avadhani N. G. ( 1990) DNA Cell Biol. 9, 657– 667 [DOI] [PubMed] [Google Scholar]
- 45.Kaufmann T., Schlipf S., Sanz J., Neubert K., Stein R., Borner C. ( 2003) J. Cell Biol. 160, 53– 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stan T., Brix J., Schneider-Mergener J., Pfanner N., Neupert W., Rapaport D. ( 2003) Mol. Cell. Biol. 23, 2239– 2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otera H., Taira Y., Horie C., Suzuki Y., Suzuki H., Setoguchi K., Kato H., Oka T., Mihara K. ( 2007) J. Cell Biol. 179, 1355– 1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamano K., Yatsukawa Y., Esaki M., Hobbs A. E., Jensen R. E., Endo T. ( 2008) J. Biol. Chem. 283, 3799– 3807 [DOI] [PubMed] [Google Scholar]
- 49.Young J. C., Hoogenraad N. J., Hartl F. U. ( 2003) Cell 112, 41– 50 [DOI] [PubMed] [Google Scholar]
- 50.Fan A. C., Bhangoo M. K., Young J. C. ( 2006) J. Biol. Chem. 281, 33313– 33324 [DOI] [PubMed] [Google Scholar]
- 51.Brunner M., Schneider H. C., Lill R., Neupert W. ( 1995) Cold Spring Harbor Symp. Quant. Biol. 60, 619– 627 [DOI] [PubMed] [Google Scholar]
- 52.Steger H. F., Söllner T., Kiebler M., Dietmeier K. A., Pfaller R., Trülzsch K. S., Tropschug M., Neupert W., Pfanner N. ( 1990) J. Cell Biol. 111, 2353– 2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fölsch H., Guiard B., Neupert W., Stuart R. A. ( 1996) EMBO J. 15, 479– 487 [PMC free article] [PubMed] [Google Scholar]
- 54.Mokranjac D., Sichting M., Neupert W., Hell K. ( 2003) EMBO J. 22, 4945– 4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bar-Nun S., Kreibich G., Adesnik M., Alterman L., Negishi M., Sabatini D. D. ( 1980) Proc. Natl. Acad. Sci. U. S. A. 77, 965– 969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szczesna-Skorupa E., Ahn K., Chen C. D., Doray B., Kemper B. ( 1995) J. Biol. Chem. 270, 24327– 24333 [DOI] [PubMed] [Google Scholar]
- 57.Ma Y., Taylor S. S. ( 2008) J. Biol. Chem. 283, 11743– 11751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chacinska A., Pfanner N., Meisinger C. ( 2002) Trends Cell Biol. 12, 299– 303 [DOI] [PubMed] [Google Scholar]
- 59.Beddoe T., Lithgow T. ( 2002) Biochim. Biophys. Acta 1592, 35– 39 [DOI] [PubMed] [Google Scholar]
- 60.Deshaies R. J., Koch B. D., Schekman R. ( 1988) Trends Biochem. Sci. 13, 384– 388 [DOI] [PubMed] [Google Scholar]
- 61.Hartl F. U., Hayer-Hartl M. ( 2002) Science 295, 1852– 1858 [DOI] [PubMed] [Google Scholar]
- 62.Murakami H., Pain D., Blobel G. ( 1988) J. Cell Biol. 107, 2051– 2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ren J., Bharti A., Raina D., Chen W., Ahmad R., Kufe D. ( 2006) Oncogene 25, 20– 31 [DOI] [PubMed] [Google Scholar]
- 64.Ellis R. J. ( 2003) Nature 421, 801– 802 [DOI] [PubMed] [Google Scholar]
- 65.Voos W., Röttgers K. ( 2002) Biochim. Biophys. Acta 1592, 51– 62 [DOI] [PubMed] [Google Scholar]
- 66.Voos W. ( 2003) Mol. Cell 11, 1– 3 [DOI] [PubMed] [Google Scholar]
- 67.Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. ( 2003) Cell 115, 629– 640 [DOI] [PubMed] [Google Scholar]
- 68.Gabaldón T., Huynen M. A. ( 2004) Biochim. Biophys. Acta 1659, 212– 220 [DOI] [PubMed] [Google Scholar]
- 69.Taylor S. W., Fahy E., Zhang B., Glenn G. M., Warnock D. E., Wiley S., Murphy A. N., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. ( 2003) Nat. Biotechnol. 21, 281– 286 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.