Abstract
DNA ligases seal 5′-PO4 and 3′-OH polynucleotide ends via three nucleotidyl transfer steps involving ligase-adenylate and DNA-adenylate intermediates. DNA ligases are essential guardians of genomic integrity, and ligase dysfunction underlies human genetic disease syndromes. Crystal structures of DNA ligases bound to nucleotide and nucleic acid substrates have illuminated how ligase reaction chemistry is catalyzed, how ligases recognize damaged DNA ends, and how protein domain movements and active-site remodeling are used to choreograph the end-joining pathway. Although a shared feature of DNA ligases is their envelopment of the nicked duplex as a C-shaped protein clamp, they accomplish this feat by using remarkably different accessory structural modules and domain topologies. As structural, biochemical, and phylogenetic insights coalesce, we can expect advances on several fronts, including (i) pharmacological targeting of ligases for antibacterial and anticancer therapies and (ii) the discovery and design of new strand-sealing enzymes with unique substrate specificities.
DNA Ligase
The discovery of DNA ligases in 1967 by the Gellert, Lehman, Richardson, and Hurwitz laboratories was a watershed event in molecular biology (reviewed in Ref. 1). By joining 3′-OH and 5′-PO4 termini to form a phosphodiester, DNA ligases are the sine qua non of genome integrity. They are essential for DNA replication and repair in all organisms. Ligases were critical reagents in the development of molecular cloning and many subsequent ramifications of DNA biotechnology, including molecular diagnostics and SOLiD sequencing methods. Ligases are elegant and versatile enzymes and are enjoying a research renaissance in light of discoveries that most organisms have multiple ligases that either function in DNA replication (by joining Okazaki fragments) or are dedicated to particular DNA repair pathways, such as nucleotide excision repair, base excision repair, single-strand break repair, or the repair of double-strand breaks via nonhomologous end joining (2–4). Genetic deficiencies in human DNA ligases have been associated with clinical syndromes marked by immunodeficiency, radiation sensitivity, and developmental abnormalities (3). The physiology and division of labor among cellular DNA ligases are the subjects of recent reviews (2–4) and will not be covered here. This minireview will focus on new insights to ligase mechanism and evolution from ligase structure.
DNA ligation entails three sequential nucleotidyl transfer steps (Fig. 1). In the first step, nucleophilic attack on the α-phosphorus of ATP or NAD+ by ligase results in release of PPi or NMN and formation of a covalent ligase-adenylate intermediate in which AMP is linked via a P–N bond to N-ζ of a lysine. In the second step, the AMP is transferred to the 5′-end of the 5′-phosphate-terminated DNA strand to form DNA- adenylate. In this reaction, the 5′-phosphate oxygen of the DNA strand attacks the phosphorus of ligase-adenylate, and the active-site lysine is the leaving group. In the third step, ligase catalyzes attack by the 3′-OH of the nick on DNA-adenylate to join the polynucleotides and liberate AMP. The pathway entails a series of bond transformations: from phosphoanhydride (ATP) to phosphoramidate (ligase-adenylate) to phosphoanhydride (DNA-adenylate) to phosphodiester (sealed DNA). All three chemical steps depend on a divalent cation cofactor.
FIGURE 1.
Three-step pathway of nick sealing by DNA ligase.
ATP-dependent DNA Ligases
DNA ligases are grouped into two families, ATP-dependent ligases and NAD+-dependent ligases, according to the substrate required for ligase-adenylate formation (supplemental Fig. S1). All known eukaryal cellular DNA ligases are ATP-dependent (3). The complexity of the ligase “menu” varies among eukaryal taxa. For example, humans have four ATP-dependent DNA ligases, whereas fungi have two. ATP-dependent ligases are also found in all known archaea, consistent with a common ancestry for the archaeal/eukaryal DNA replication machinery.
The essential elements of the ATP-dependent ligase clade are exemplified by Chlorella virus DNA ligase (ChVLig),2 the smallest eukaryal ligase known (5). ChVLig consists of an N-terminal nucleotidyltransferase (NTase) domain and a C-terminal OB domain. Within the NTase domain is an adenylate-binding pocket composed of the six peptide motifs that define the covalent NTase enzyme superfamily of polynucleotide ligases and RNA-capping enzymes (supplemental Fig. S2) (6). Motif I (KxDGxR) contains the lysine to which AMP becomes covalently linked in the first step of the ligase reaction. Amino acids in motifs I, Ia, III, IIIa, IV, and V contact AMP and play essential roles in one or more steps of the ligation pathway (supplemental Fig. S2). The OB domain consists of a five-stranded antiparallel β-barrel plus an α-helix. Although ChVLig lacks the large N- or C-terminal flanking domains found in eukaryal cellular DNA ligases, it can sustain mitotic growth, DNA repair, and nonhomologous end joining in budding yeast when it is the only source of ligase in the cell.
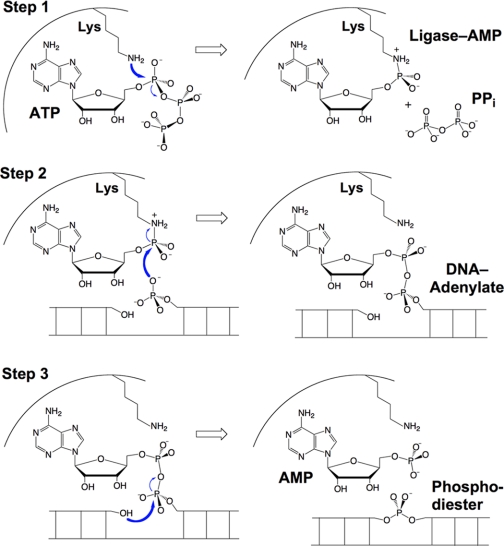
It is postulated that ChVLig represents a stripped-down pluripotent ligase due to its intrinsic nick-sensing function, the basis of which was illuminated by the crystal structure of ChVLig-AMP bound to a nicked duplex DNA (7). ChVLig encircles the DNA as a C-shaped protein clamp (Fig. 2A). The NTase domain binds to the broken and intact DNA strands in the major groove flanking the nick and also in the minor groove on the 3′-OH side of the nick (supplemental Fig. S3). The OB domain binds across the minor groove on the face of the duplex behind the nick. A “latch” module (consisting of a β-hairpin loop that emanates from the OB domain) occupies the major groove and completes the circumferential clamp via contacts between the tip of the loop and the surface of the NTase domain. The clamp-closing contacts are positioned at “six o'clock” on the DNA circumference (denoted by the red arrow in Fig. 2A). The latch is critical for clamp closure and is a key determinant of nick sensing.
FIGURE 2.
DNA ligases engage the nicked duplex as C-shaped protein clamps. A and B, ribbon diagrams are shown of the DNA-bound structures of ChVLig (Protein Data Bank code 2Q2T) and HuLig1 (code 1X9N). The proteins were superimposed with respect to their NTase (cyan) and OB (beige) domains. The interdomain contacts that close the ligase clamps are indicated by red arrows. In ChVLig, the latch and NTase domains make kissing contacts. By contrast, HuLig1 closes its clamp via contacts between the N-terminal DBD and the C-terminal OB domain. The peptide linker between the DBD and NTase domain of HuLig1 is indicated by the black arrow. C, the structures of the free ChVLig-AMP intermediate (left; Protein Data Bank code 1FVI) and ligase-AMP bound at a nick (right; with DNA omitted) were superimposed with respect to their NTase domains and then offset laterally to reveal the large movement of the OB domain triggered by nick recognition. The movement entails a rigid body rotation about the hinge segment connecting the NTase and OB domains (indicated by the arrow in free ligase). The surface peptide loop destined to become the latch in the DNA-bound ligase is disordered in the free ligase (dotted line). A sulfate ion is bound on the surface of the free ligase-AMP in a position that mimics the 5′-PO4 of the nick.
Comparison of the crystal structures of the free and nick-bound ChVLig-AMP reveals a large domain rearrangement accompanying nick recognition (Fig. 2C). In the free ChVLig-AMP, the OB domain is reflected away from the NTase domain to fully expose the DNA-binding surface above the AMP-binding pocket. The peptide segment that is destined to become the latch is disordered in the free ligase and sensitive to proteolysis, but this segment is protected from proteolysis when ChVLig binds to nicked DNA. DNA binding entails a nearly 180° rotation of the OB domain around a swivel (denoted by the black arrow in Fig. 2C) so that the concave surface of the OB β-barrel fits into the DNA minor groove. This transition elicits a 63-Å movement of the OB domain and places the latch deep in the DNA major groove.
Ellenberger and co-workers (8) had intuited exactly such a movement of the OB domain when they solved the crystal structure of the much larger human DNA ligase I (HuLig1; 919 amino acids) bound to an adenylylated nick. Their landmark structure was the first to document the circumferential envelopment of the DNA duplex by a ligase clamp and the resulting bending and distortion of the duplex that force the base pairs on the 3′-OH side of the nick into an RNA-like A helical conformation (8). Similar distortions were observed in the ChVLig·DNA complex (7).
A side-by-side comparison of the DNA-bound ChVLig and HuLig1 structures highlights the radical differences in the topology and connectivity of their respective protein clamps (Fig. 2, A and B). Whereas the NTase and OB domains adopt similar conformations and angular positions when docked on the nicked duplex, HuLig1 has a large N-terminal DNA-binding domain (DBD; with an all-α-helical tertiary structure) that occupies roughly the same angular position in the clamp toroid as the β-hairpin latch of ChVLig. The peptide covalently linking the DBD and NTase domain of HuLig1 is situated in the same six o'clock position on the DNA circumference as the noncovalent kissing contacts that close the clamp in ChVLig. HuLig1 closes its clamp by contacts between the N-terminal DBD and the C-terminal OB module, located at “one o'clock” on the DNA circumference (red arrow in Fig. 2B), which is the same angular position as the covalent connections between the ChVLig OB domain and the strands of the latch. Thus, Nature has found very different structural solutions to the problem of DNA envelopment by ATP-dependent ligases. I suspect there are additional solutions out there waiting to be discovered. For example, the bacteriophage T7 DNA ligase, another minimized ATP-dependent enzyme consisting of only NTase and OB domains (9), is a good candidate to engage the DNA duplex in a novel fashion, via a T7-specific peptide loop inserted within its NTase domain.
NAD+-dependent DNA Ligases
NAD+-dependent DNA ligases (referred to as LigA) are a distinctive and structurally homogeneous clade of enzymes found in all bacteria. Escherichia coli LigA (671 amino acids) is the prototype of this family. LigA has a modular architecture (Fig. 3A) built around a central ligase core composed of an NTase domain and an OB domain. The core is flanked by an N-terminal “Ia” domain and three C-terminal modules: a tetracysteine zinc finger, a helix-hairpin-helix (HhH) domain, and a BRCT domain. Each step of the ligation pathway depends upon a different subset of the LigA domains, with only the NTase domain being required for all steps. Domain Ia is unique to NAD+-dependent ligases, is responsible for binding the NMN moiety of NAD+, and is required for the reaction with NAD+ to form the ligase-AMP intermediate (10, 11).
FIGURE 3.
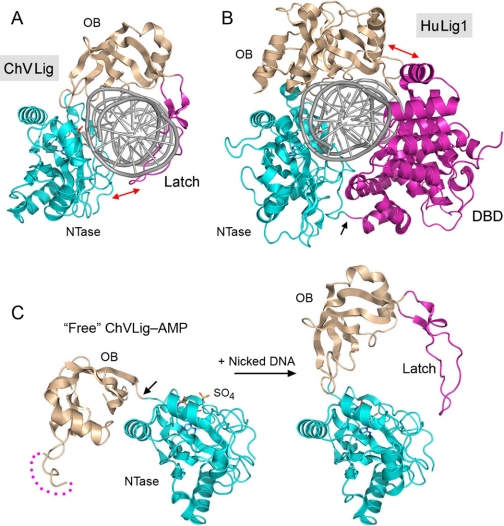
Structural images of LigA bound to nicked DNA, NAD+, or a small molecule inhibitor. A, space-filling model of the E. coli LigA protein clamp (Protein Data Bank code 2OWO) encircling nicked duplex DNA, which is rendered as a schematic trace. The modular domain structure of the LigA polypeptide is depicted below the structure. The LigA clamp is closed by kissing contacts between the NTase (cyan) and HhH (beige) domains. B, shown are surface views of the E. faecalis LigA·NAD+ complex (upper panel; Protein Data Bank code 1TAE) and the LigA·inhibitor complex (lower panel; code 3BA9) looking into the hydrophobic tunnel of the NTase domain that leads from the surface to the back end of the adenosine-binding pocket. The C-2 atom of adenine is pointing into the tunnel, which is empty in the NAD+ complex but filled in the inhibitor complex. C, shown are superimposed structures of NAD+ and the pyridopyrimidine inhibitor in their respective LigA complexes. The pyrimidine ring of the inhibitor overlies the adenine base. The bulky pyrido substituent of the inhibitor that fills the tunnel is an offshoot of the equivalent of the adenine C-2 (denoted by the black arrows) and N-3 edge of the ring.
The crystal structure of E. coli LigA bound to the nicked DNA-adenylate intermediate (12) revealed that LigA also encircles the DNA helix as a C-shaped protein clamp (Fig. 3A). The protein-DNA interface entails extensive DNA contacts by the NTase, OB, and HhH domains over a 19-bp segment of duplex DNA centered about the nick (supplemental Fig. S3). The NTase domain binds to the broken DNA strands at and flanking the nick; the OB domain contacts the continuous template strand surrounding the nick; and the HhH domain binds both strands across the minor groove at the periphery of the footprint. The zinc-finger module plays a structural role in bridging the OB and HhH domains. Domain Ia makes no contacts with the DNA duplex, consistent with its dispensability for catalysis of strand closure on an AppDNA substrate. (No electron density was observed for the C-terminal BRCT domain.)
The LigA NTase and OB domains are positioned similarly on the DNA circumference to the NTase and OB domains of the ATP-dependent DNA ligases, and they “footprint” similar segments of the DNA strands (supplemental Fig. S3), yet the topology of the LigA clamp is starkly different from that of the clamps formed by ChVLig and HuLig1. The kissing contacts that close the LigA clamp are sui generis, involving the NTase and HhH domains. Based on available structural data, it is clear that DNA ligases have evolved at least three different means of encircling DNA.
Comparisons of the E. coli LigA·AppDNA complex with structures of other bacterial ligases captured as the binary LigA·NAD+ complex (step 1 substrate), binary LigA·NMN complex (the post-step 1 leaving group), and covalent ligase-AMP intermediate (step 1 product after leaving group dissociation) highlight massive protein domain rearrangements (on the order of 50–90 Å) that occur in sync with substrate binding and catalysis (11, 12). DNA binding and clamp formation by LigA entail a nearly 180° rotation of the OB domain so that the concave surface of the OB β-barrel fits into the minor groove, similar to what is seen or inferred for ChVLig and HuLig1. The four-point binding of the HhH domain at the periphery of the LigA·DNA footprint stabilizes a DNA bend centered at the nick. The LigA-DNA interactions immediately flanking the nick induce a local DNA distortion, resulting in adoption of an RNA-like A-form helix, again echoing the findings for the HuLig1·DNA cocrystal.
DNA Ligases as Drug Targets
LigA is essential for the viability of E. coli and all other bacteria tested. Although NAD+-dependent DNA ligases have been discovered in sporadic cellular or viral niches outside the bacterial domain of life (supplemental Fig. S1), there is no instance in which a NAD+-dependent ligase is present in a eukaryal organism. The narrow phylogenetic distribution, unique substrate specificity, and distinctive domain structure of LigA compared with ATP-dependent human DNA ligases recommend the NAD+ ligases as targets for the development of new antibacterial drugs. Blocking the reaction of LigA with NAD+ is the obvious goal. Indeed, there has been a series of reports describing small molecule inhibitors of bacterial LigA, acting competitively with NAD+, discovered either by experimental high-throughput screening (13–15) or by “virtual” screening by computational docking of ligands into the LigA crystal structures (16, 17).
Inspection of the E. coli LigA·AppDNA structure and comparison with the Enterococcus faecalis LigA·NAD+ complex highlighted an eminently “druggable” active site, with a through-and-through tunnel from the exterior surface of the NTase domain to the adenosine-binding pocket that exposes the N-1, C-2, and N-3 edge of the adenine base (Fig. 3B, upper panel) (12). In particular, the adenine C-2 is pointed directly into the tunnel, which is formed by a cage of hydrophobic amino acids. This tunnel is present in all LigA NTase domains that have been crystallized, with similar exposure of the C-2 edge of the adenine base. By contrast, there is no such tunnel emanating from the adenosine-binding pockets of HuLig1, ChVLig, and other ATP-dependent DNA ligases. This situation invites the design of C-2-substituted analogs of adenosine nucleotides and NAD+ as selective inhibitors of LigA. Indeed, it was reported recently that 2-methylthio-ATP is a potent inhibitor of the adenylylation reaction of E. coli LigA (IC50 = 0.5 μm), being 200-fold more active than unmodified ATP (14).
Pyridochromanone exemplifies a specific and potent non-nucleotide LigA inhibitor in vitro, for which there is convincing genetic evidence that LigA is the immediate target of its antibacterial activity in vivo (13). Christopher Pinko and colleagues have deposited in the Protein Data Bank a suite of crystal structures of Enterococcus LigA bound to four different small molecule inhibitors, including pyridochromanone (code 3BA8) and three pyridopyrimidine compounds (codes 3BA9, 3BAA, and 3BAB). In each case, the inhibitor occupies the adenosine-binding pocket of the NTase domain in a fashion whereby an aromatic ring of the inhibitor overlaps the adenine base of NAD+ and a bulky aromatic/aliphatic moiety emanates from the ring system (at the sites equivalent to the adenine C-2 and N-3 atoms) and penetrates into the “drug tunnel” (Fig. 3, B and C). This mechanism of active-site occlusion is easily appreciated for the pyridopyrimidine inhibitor shown in Fig. 3C, where the phenoxybenzamide group fills the portion of the tunnel closest to the protein surface (Fig. 3B).
Human DNA ligases are also candidate drug targets for adjuvant cancer chemotherapy, predicated on potentiating the effects of DNA-damaging antineoplastics by transiently impeding DNA repair in tumor cells. Such a maneuver might permit lower dosing with DNA-damaging drugs, thereby avoiding off-target effects or idiosyncratic toxicities. Chen et al. (18) have conducted a virtual screen for small molecules that could dock into the N-terminal DBD of HuLig1, a domain conserved in all mammalian DNA ligases. They thereby discovered compounds that (i) inhibited nick sealing by human ligases in vitro at a step other than ligase adenylylation and (ii) sensitized tumor cells to DNA damage (18).
Is There an “Undifferentiated” Ligase with Respect to the Substrate for Enzyme Adenylylation?
The weight of structural evidence favors the idea that an ATP-dependent ligase is the ancestral state from which NAD+-dependent ligases evolved, by acquisition of a specialized structural domain (Ia) that binds the NMN leaving group. All known NAD+-dependent ligases have domain Ia, and they cannot use ATP as a substrate for ligase adenylylation, presumably because LigA enzymes lack “motif IV” of the OB domain, the component of ATP-dependent DNA ligases that is thought to interact with the PPi leaving group of ATP (5, 19). This “either/or” scenario of DNA ligase substrate specificity has been roiled by several seemingly conflicting reports concerning the nucleotide specificity of archaeal DNA ligases. All archaeal proteomes include an ATP-dependent DNA ligase that resembles in all structural respects (amino acid sequence; conservation of NTase motifs, including motif VI; and tertiary structure composed of the N-terminal DBD and the central NTase and C-terminal OB domains) (20, 21) the canonical ATP-dependent DNA ligases of eukaryal cells. Biochemical characterization of several archaeal DNA ligases verified their strict dependence on ATP and failure to utilize NAD+ (summarized in Ref. 22). By contrast, other archaeal ligases were reported to have “dual specificity,” variously using ATP and NAD+ or ATP and ADP (supplemental Fig. S4). Sulfophobococcus zilligii DNA ligase can apparently use any of three substrates: ATP, ADP, and GTP (23).
The undifferentiated nucleotide specificities of certain archaeal ligases have tantalizing implications for enzyme evolution and the possible use of ADP as a substrate by ancestors of modern DNA ligases. The ability of some archaeal ligases to utilize ATP and NAD+ or ATP and ADP as substrates might be explained if they recognized only the ADP component common to all three nucleotides. However, there are several missing pieces of the puzzle with respect to biochemistry and structure. The case for utilization of ADP as the substrate for ligase adenylylation would be fortified by more direct evidence entailing (i) demonstration of label transfer from [α-32P]ADP to the ligase to form the covalent intermediate and (ii) proof that 32Pi is released from [β-32P]ADP in 1:1 stoichiometry with ligase-adenylate formation. Moreover, Chen et al. (24) have recently raised a caveat to the assertion that ADP is an archaeal DNA ligase substrate by suggesting that contamination of the recombinant archaeal ligase protein preparations with the apparently thermostable E. coli adenylate kinase can result in conversion of ADP into ATP and AMP and thereby confound interpretation of the ADP/ATP specificity experiments. Solving the conundrum of how some dual-specificity archaeal ligases might utilize ATP and NAD+ will depend on capturing crystal structures of these enzymes with each nucleotide bound. Armed with this information, it would be most informative to trace how archaeal enzymes from closely related taxa that have extremely similar primary structures came to differ in their nucleotide specificities, culminating by introducing amino acids changes that elicit a gain of function, converting an ATP-only DNA ligase into a dual-specificity derivative.
Prospects for Designer Ligases
Can structural insights to ligase mechanism and substrate specificity be exploited to create novel catalysts, ones that will aid in comprehending the genetics and cell biology of DNA repair and in the practice of enzymatic joining and synthesis of nucleic acids? There is good reason to think so. Nature has already confounded the partitioning of ligases into rigid DNA-specific and RNA-specific clades. We now appreciate that many DNA ligases are adept at sealing substrates containing an all-RNA 3′-OH strand (8, 25, 26) and that some RNA ligases readily seal molecules with as few as two or three ribonucleotides at the 3′-OH terminus in an otherwise all-DNA context (27, 28). In a novel twist on this theme, some bacterial ATP-dependent DNA ligases actually require a single ribonucleotide at the reactive 3′-OH end for optimal sealing activity (29). T4 RNA ligase 1 (Rnl1), although profoundly useful as a reagent for joining RNA single strands, actually has an intrinsic specificity for tRNA tertiary structure that was recognized only recently (30). By contrast, another Rnl1-type RNA ligase encoded by a thermophilic bacteriophage is quite adept at sealing DNA single strands, the first example of such a broad specificity (31). The structural bases for these distinctive nucleic acid specificities remain obscure.
In light of what we do know, I can imagine several types of “designer ligase” projects worthy of pursuit, including (i) creation of “analog-sensitive” ligases by mutational remodeling of the adenosine-binding pocket and (ii) creation of step 3 arrest ligases that can efficiently generate DNA-adenylate but not synthesize a phosphodiester. The engineering of analog-sensitive protein kinases exemplifies the power of chemical genetics to illuminate biological signaling pathways (32). Ideally, one would want to engineer a DNA ligase mutant capable of binding an adenosine analog that is normally excluded from the active site of the wild-type ligase. Replacing the wild-type ligase with the analog-sensitive ligase in an organism that has multiple ligase isozymes could allow selective and rapid inhibition of strand sealing, without depleting the ligase protein and without affecting the numerous other proteins that interact with DNA ligases in mammalian cells (2, 3).
An engineered step 3 arrest mutant of DNA ligase would, if widely active, pepper the genome with unresected 5′-adenylates at sites of DNA repair or joining of Okazaki fragments. This would be catastrophic if the level of DNA 5′-adenylylation exceeds the cell's capacity to remove the lesions (33). Such a scenario is analogous to “poisoning” of topoisomerase IB by camptothecin drugs or drug-mimetic enzyme mutations that selectively impede the DNA ligation step of the topoisomerase pathway. Transient expression of a step 3 arrest ligase could thereby (i) illuminate cellular responses to an abortive repair intermediate, (ii) discriminate the relative impact of different ligase isozymes on the overall lesion burden, and (iii) aid in mapping genomic sites of ligase activity by recovering and sequencing the pool of 5′-adenylylated DNA strands.
Supplementary Material
This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and additional references.
- ChVLig
- Chlorella virus DNA ligase
- NTase
- nucleotidyltransferase
- HuLig1
- human DNA ligase I
- DBD
- DNA-binding domain
- HhH
- helix-hairpin-helix.
REFERENCES
- 1.Lehman I. R. ( 1974) Science 186, 790– 797 [DOI] [PubMed] [Google Scholar]
- 2.Tomkinson A. E., Vijayakumar S., Pascal J. M., Ellenberger T. ( 2006) Chem. Rev. 106, 687– 699 [DOI] [PubMed] [Google Scholar]
- 3.Ellenberger T., Tomkinson A. E. ( 2008) Annu. Rev. Biochem. 77, 313– 338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuman S., Glickman M. S. ( 2007) Nat. Rev. Microbiol. 5, 852– 861 [DOI] [PubMed] [Google Scholar]
- 5.Odell M., Sriskanda V., Shuman S., Nikolov D. ( 2000) Mol. Cell 6, 1183– 1193 [DOI] [PubMed] [Google Scholar]
- 6.Shuman S., Lima C. D. ( 2004) Curr. Opin. Struct. Biol. 14, 757– 764 [DOI] [PubMed] [Google Scholar]
- 7.Nair P. A., Nandakumar J., Smith P., Odell M., Lima C. D., Shuman S. ( 2007) Nat. Struct. Mol. Biol. 14, 770– 778 [DOI] [PubMed] [Google Scholar]
- 8.Pascal J. M., O'Brien P. J., Tomkinson A. E., Ellenberger T. ( 2004) Nature 432, 473– 478 [DOI] [PubMed] [Google Scholar]
- 9.Subramanya H. S., Doherty A. J., Ashford S. R., Wigley D. B. ( 1996) Cell 85, 607– 615 [DOI] [PubMed] [Google Scholar]
- 10.Sriskanda V., Shuman S. ( 2002) J. Biol. Chem. 277, 9685– 9700 [DOI] [PubMed] [Google Scholar]
- 11.Gajiwala K., Pinko C. ( 2004) Structure 12, 1449– 1459 [DOI] [PubMed] [Google Scholar]
- 12.Nandakumar J., Nair P. A., Shuman S. ( 2007) Mol. Cell 26, 257– 271 [DOI] [PubMed] [Google Scholar]
- 13.Brötz-Oesterhelt H., Knezevic I., Bartel S., Lampe T., Warnecke-Eberz U., Ziegelbauer K., Häbich D., Labischinski H. ( 2003) J. Biol. Chem. 278, 39435– 39442 [DOI] [PubMed] [Google Scholar]
- 14.Miesel L., Kravec C., Xin A. T., McMonagle P., Ma S., Pichardo J., Feld B., Barrabee E., Palermo R. ( 2007) Anal. Biochem. 366, 9– 17 [DOI] [PubMed] [Google Scholar]
- 15.Meier T. I., Yan D., Peery R. B., McAllister K. A., Zook C., Peng S. B., Zhoa G. ( 2008) FEBS J. 275, 5258– 5271 [DOI] [PubMed] [Google Scholar]
- 16.Srivastava S. K., Dube D., Tewari N., Dwivedi N., Tripathi R. P., Ramachandran R. ( 2005) Nucleic Acids Res. 33, 7090– 7101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava S. K., Dube D., Kukshal V., Jha A. K., Hajela K., Ramachandran R. ( 2007) Proteins 69, 97– 111 [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Zhong S., Zhu X., Dziegielewska B., Ellenberger T., Wilson G. M., MacKerrell A. D., Tomkinson A. E. ( 2008) Cancer Res. 68, 3169– 3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Håkansson K., Doherty A. J., Shuman S., Wigley D. B. ( 1997) Cell 89, 545– 553 [DOI] [PubMed] [Google Scholar]
- 20.Pascal J. M., Tsodlikov O. V., Hura G. L., Song W., Cotner E. A., Classen S., Tomkinson A. E., Tainer J. A., Ellenberger T. ( 2006) Mol. Cell 24, 279– 291 [DOI] [PubMed] [Google Scholar]
- 21.Nishida H., Kiyonari S., Ishino Y., Morikawa K. ( 2006) J. Mol. Biol. 360, 956– 967 [DOI] [PubMed] [Google Scholar]
- 22.Keppetipola N., Shuman S. ( 2005) J. Bacteriol. 187, 6902– 6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y., Seo M. S., Kim J. H., Kim Y. J., Kim G. A., Lee J. I., Lee J. H., Kwon S. T. ( 2008) Environ. Microbiol. 10, 3212– 3224 [DOI] [PubMed] [Google Scholar]
- 24.Chen B., Sysoeva T. A., Chowhury S., Guo L., Nixon B. T. ( 2008) FEBS J. 276, 807– 815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekiguchi J., Shuman S. ( 1997) Biochemistry 36, 9073– 9079 [DOI] [PubMed] [Google Scholar]
- 26.Bullard D. R., Bowater R. P. ( 2006) Biochem. J. 398, 135– 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nandakumar J., Shuman S. ( 2004) Mol. Cell 16, 211– 221 [DOI] [PubMed] [Google Scholar]
- 28.Martins A., Shuman S. ( 2004) J. Biol. Chem. 279, 50654– 50661 [DOI] [PubMed] [Google Scholar]
- 29.Zhu H., Shuman S. ( 2008) J. Biol. Chem. 283, 8331– 8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L. K., Nandakumar J., Schwer B., Shuman S. ( 2007) RNA 13, 1235– 1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blondal T., Thorisdottir A., Unnsteinsdottir U., Hjorleifsdottir S., Aevarsson A., Ernstsson S., Fridjonsson O. H., Skirnisdottir S., Wheat J. O., Hermansdottir A. G., Sigurdsson A. T., Hreggvisson G. O., Smith A. V., Kristjansson J. K. ( 2005) Nucleic Acids Res. 33, 135– 142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight Z. A., Shokat K. M. ( 2007) Cell 128, 425– 430 [DOI] [PubMed] [Google Scholar]
- 33.Ahel I., Rass U., El-Khamisy S. F., Katyal S., Clements P. M., McKinnon P. J., Caldecott K. W., West S. C. ( 2006) Nature 443, 713– 716 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.