Abstract
Herpes simplex virus entry into cells requires a multipartite fusion apparatus made of glycoprotein D (gD), gB, and heterodimer gH/gL. gD serves as a receptor-binding glycoprotein and trigger of fusion; its ectodomain is organized in an N-terminal domain carrying the receptor-binding sites and a C-terminal domain carrying the profusion domain, required for fusion but not receptor binding. gB and gH/gL execute fusion. To understand how the four glycoproteins cross-talk to each other, we searched for biochemical defined complexes in infected and transfected cells and in virions. Previously, interactions were detected in transfected whole cells by split green fluorescent protein complementation (Atanasiu, D., Whitbeck, J. C., Cairns, T. M., Reilly, B., Cohen, G. H., and Eisenberg, R. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 18718–18723; Avitabile, E., Forghieri, C., and Campadelli-Fiume, G. (2007) J. Virol. 81, 11532–11537); it was not determined whether they led to biochemical complexes. Infected cells harbor a gD-gH complex (Perez-Romero, P., Perez, A., Capul, A., Montgomery, R., and Fuller, A. O. (2005) J. Virol. 79, 4540–4544). We report that gD formed complexes with gB in the absence of gH/gL and with gH/gL in the absence of gB. Complexes with similar composition were formed in infected and transfected cells. They were also present in virions prior to entry and did not increase at virus entry into the cell. A panel of gD mutants enabled the preliminary location of part of the binding site in gD to gB to the amino acids 240–260 portion and downstream with Thr304-Pro305 as critical residues and of the binding site to gH/gL at the amino acids 260–310 portion with Pro291-Pro292 as critical residues. The results indicate that gD carries composite-independent binding sites for gB and gH/gL, both of which are partly located in the profusion domain.
Introduction
Herpes simplex virus (HSV)2 entry into the cell occurs by fusion, and requires a multipartite apparatus made of a glycoprotein quartet: gD, gB, and the heterodimer gH/gL; for reviews, see Refs. 1–3). When ectopically expressed, the four glycoproteins mediate cell-cell fusion (4). gD serves as the receptor-binding glycoprotein and interacts with three alternative receptors, nectin1, herpesvirus entry mediator (HVEM) and modified heparan sulfate (5–8). It also represents the key player in the triggering of fusion, i.e. in inducing fusion execution by gB and gH/gL (9–12). These are among the most highly conserved proteins across the Herpesviridae family and constitute the fusion core apparatus. Their respective roles in fusion execution are unclear at present. Thus, gH carries elements typical of fusion glycoproteins, i.e. hydrophobic regions able to interact with target cells or artificial membranes and two heptad repeats potentially able to form a coiled coil (13–18). Peptides mimicking the hydrophobic regions of gH promote fusion of artificial membranes (19, 20). With respect to gB, the crystal structure shows a trimer with a central coiled-coil and an overall structure similar to that of the fusion glycoprotein G of vesicular stomatitis virus in its postfusion conformation (21, 22). Recently, a receptor for gB was described (23). Of note, although for virus-to-cell entry and for cell-cell fusion, fusion requires the simultaneous presence of gB and gH/gL, fusion of perinuclear virions with the outer nuclear membranes appears to necessitate either gH/gL or gB (24). Furthermore, it has been reported that HSV fusion may be preceded by a hemifusion intermediate mediated by gD and gH/gL (25).
A major focus of current research is definition of protein-protein interactions. With respect to HSV entry/fusion, to understand how the four glycoproteins cross-talk to each other, it is pivotal to detect which complexes are formed by the glycoproteins in their prefusion and fusion-active conformations, and, ultimately, the chain of interactions that signal the gD encounter with its cellular receptor and culminate in activation of gB and gH/gL. The proposed model of gD-activated entry/fusion (2, 9–12, 26, 27) envisions that (i) gD ectodomain is organized in two functionally and topologically distinct regions, the N-terminal one, spanning amino acids (aa) 1 to ∼240/260, carrying the receptor binding sites, and the C-terminal one (aa 240/260–310), carrying the profusion domain required for fusion but not for receptor binding; (ii) the unliganded gD adopts an auto-inhibited conformation, whereby the C-terminal domain folds around the N-terminal one and occupies or hinders the receptor-binding sites; (iii) gD undergoes a closed-to-open switch in conformation, whereby the C-terminal domain is dislodged from its binding site and exposes the profusion domain; and (iv) the active form of gD ultimately leads to the activation of gH/gL and gB. It was hypothesized that gB and gH/gL activation occurs through their recruitment to activated gD and that the C-terminal profusion domain carries the actual binding sites for gB and gH/gL (9, 12). An alternative possibility is that the C-terminal profusion domain simply enables the conformational changes in gD but does not carry the actual binding sites for gB and gH/gL.
Efforts to validate the model prompted the search of interactions among the glycoprotein quartet. So far, it was found that HSV-infected cells harbor a co-immunoprecipitable complex made of HVEM, gD, and gH (28). A number of interactions were detected in transfected whole cells by split green fluorescent protein (or variations thereof) complementation assay (29, 30). The interactions were gD-gH/gL, gD-gB, and gB-gH/gL. The former two were found in cells transfected with two or three glycoproteins and are believed to mirror interactions that take place before gD activation. In contrast, the gB-gH/gL interaction was detected at fusion. Whether such interactions occur in infected cells or are seen only in transfected cells under conditions of high overexpression and whether they lead to biochemically defined complexes have not been investigated so far. In addition, inasmuch as the split green fluorescent protein complementation irreversibly stabilizes weak and transients interactions, it tends to overemphasize interactions. For other herpesviruses, evidence is accumulating of complex formation among the glycoproteins involved in virus entry. Thus, Epstein-Barr virus gH and gL form a complex with the receptor-binding glycoprotein gp42; human cytomegalovirus and murine γ-herpesvirus 68 gH and gL form a complex with gB (31–33).
The objective of this work was to provide biochemical evidence for complex formation among the glycoprotein quartet, to verify whether complexes are present in infected cells and in virions and whether they are formed at virus entry into the cell. We analyzed the composition of the complexes by two approaches, co-immunoprecipitation and a pulldown assay that exploits the ability of One-strep-tagged proteins to be specifically retained by the Strep-Tactin resin. Complexes with undistinguishable composition were detected in infected and transfected cells and in virions prior to entry into the cell. A panel of mutants enabled the preliminary location of part of the gD regions critical to gB- and gH/gL-binding sites at the profusion domain.
EXPERIMENTAL PROCEDURES
Cells and Viruses
The cells were grown in Dulbecco's modified minimum essential medium containing 5–20% fetal calf serum. HSV-1(F) was described (34). The ΔgD F-gDβ (35), ΔgB-KΔT (36), ΔgH SCgHZ (37), and ΔgL (38) HSV mutants were grown and titrated in the respective complementing cells.
Antibodies
R8 polyclonal antibody (pAb) to gD and BD80 monoclonal antibody (mAb) to aa 264–275 epitope of mature gD were generously provided by Dr. G. H. Cohen and Dr. R. Eisenberg; mAbs HD1, HC1, and H233 were a gift of Dr. L. Pereira. pAbs to gH and gL were a gift from Dr. H. Browne (Cambridge, UK) and D. Johnson (Portland). mAb H170 (reactive to aa 1–23 epitope), H1817, and H633 were purchased from Goodwin Institute. mAbs 52S, 53S, 30, and 5E1 were described (39–41). mAb 52S reacts to a conformational-dependent epitope. mAb 53S reacts to a conformation-dependent and gL-dependent epitope. V5 mAb was from Invitrogen. pAb to gM was described (42). A pAb to gH/gL was derived to a soluble form of gH truncated at aa 789/gL produced in insect cells.3
Virus Yield Assay
Vero cells grown in 12-well plates were infected with the indicated viruses at 0.1 PFU/cell for 90 min at 37 °C. The inoculum was removed, and unpenetrated virions were inactivated by means of an acidic wash (40 mm citrate acid, 10 mm KCl, 135 mm NaCl, pH 3). Replicate cultures were frozen at indicated times, i.e. 3, 24, and 48 h after infection. The progeny virus was titrated in Vero cells.
Plasmids
The mammalian expression plasmids encoding gH, gL, and gB in MTS vector, and gD in pcDNA3.1, all under the cytomegalovirus promoter, were described (43). Plasmids encoding HVEM (pBEC) (5), HER2 (human epidermal growth factor receptor 2) (44), gDΔPFD (herein renamed gDΣ260–310), gDTP, and gDPP were described (9, 10).
Tagging of gH, gL, and gC
To enable detection or retention to resin, gH, gL, and gC were tagged with heterologous epitopes, as follows. The 5E1 epitope consists of a 27-aa-long sequence recognized by mAb 5E1, initially derived to human herpesvirus 7 (39). The 5E1 epitope was inserted in gH (gH5E1), and the thrombin plus 5E1 and His (polyhistidine) epitopes were inserted in gL (gL5E1.His), just upstream of the stop codon. To generate gH5E1, a SphI restriction site was inserted in the cytoplasmic tail of gH, in place of the stop codon, by site-directed mutagenesis with oligonucleotides 5′-CCG TTT TTT TGG AGA CGC ATG CAA AGT GGG CGT GAA TTC GGC CGT TTC TCC GCC C-3′ and 5′-GGG CGG AGA AAC GGC CGA ATT CAC GCC CAC TTT GCA TGC GTC TCC AAA AAA ACG G-3′. The oligonucleotides contained EcoRI restriction site for screening and introduced a single mutation, E838M. Two annealing oligonucleotides, encoding the 5E1 epitope 5′-ACA TGC ATG CAT GTT TCC AGA CCA GGA AGC ACT ACA CCC TCT GGG AAC TCT GCA AGA TAT GGG-3′ and 5′-GGA AGA TCT GGT ACC TTA CGG AGT TAT ACT TCT AGG TGT GTT ATT CCC ATA TCT TGC AGA GTT CCC-3′ were ligated to SphI/BglII-digested gH. The oligonucleotides contained the Asp718 restriction site for easiness of screening. The aa sequence of gH5E1 cytoplasmic tail was modified to ILKVLRTSVPFFWRRMHVSRPGSTTPSGNSARYGNNTPRSITP. To generate gHst.V5, indicated below also as gHst, we followed a similar strategy; the annealing oligonucleotides encoding the Factor Xa protease cleavage site, followed by V5 and One-strep-tag epitopes, were 5′-GGA GAC GCA TGC TAA TCG AAG GGC GAG GTA AGC CTA TCC CTA ACC CTC TCC TAG GCC TCG ATT CTA CGA GCG CTT GGA GCC ACC CGC AGT TCG AGA AAG G-3′ and 5′-GGT AGT AGA TCT CAT TTT TCG AAC TGC GGG TGG CTC CAC GAT CCA CCT CCC GAT CCA CCT CCG GAA CCT CCA CCT TTC TCG AAC TGC GGG TGG CTC CAA G-3′. They were ligated to the SphI/BglII-digested gH. The aa sequence of gHst.V5 cytoplasmic tail became ILKVLRTSVPFFWRRMLIEGRGLPIPNPLLGLDSTSAWSHPQFELGGGSGGGSGGGSWSHPQFEL. To generate gL5E1.His, we followed essentially a similar strategy and inserted the NheI restriction site by site-directed mutagenesis, in place of the stop codon, by means of oligonucleotides 5′-CCC CAC TCC CGG CGC CTG CTA GCA TGA ATT CAC GGA AAC CCG TCC GGG TTC GGG-3′ and 5′-CCC GAA CCC GGA CGG GTT TCC GTG AAT TCA TGC TAG CAG GCG CCG GGA GTG GGG-3′. The annealing oligonucleotides encoding the trombin-5E1-His epitopes were 5′-CTA TAG CTA GCC CTG GTT CCG CGT GGA TCC TCC AGA CCA GGA AGC ACT ACA CCC TCT GGG AAC TCT GCA AGA TAT GGG AA-3′ and 5′-GGA AGA TCT TCA ATG GTG ATG GTG ATG ATG CGG AGT TAT ACT TCT AGG TGT GTT ATT CCC ATA TCT TGC AGA GTT CCC-3′ and were ligated to NheI/BglII digested gL. The aa sequence of the gL C terminus was changed from SRRL to SRRLLALVPRGSSRPGSTTPSGNSARYGNNTPRSITPHHHHHH. To generate gLV5.His, the annealing oligonucleotides encoding the V5 and His epitopes were 5′-ATG CTC GCT AGC TGG TAA GCC TAT CCC TAA CCC TCT CCT CGG TCT CGA TTC TAC GC-3′ and 5′-TTA GCG AGA TCT CAA TGG TGA TGA TGG TGA TGA TGA ACG GTA CGC GTA GAA TCG AGA C-3′ and were ligated to NheI/BglII-digested gL. The aa sequence of gL C terminus was changed from SRRL to SRRLLAGLPIPNPLLGLDSTRTVHHHHHHH. gCV5 was generated as follows. The gC ORF was PCR-amplified from HSV-1(F) DNA with primers encoding the V5 epitope 5′-AGA TCT AGG CCT ATG GCC CCG GGG CGG GTG GGC CTT GCC G TG GTC CTG TGG AGC CTG-3′ and 5′-GCA CGG GGC GGC CGC TTA CGT AGA ATC GAG ACC GAG GAG AGG GTT AGG GAT AGG GAT AGG CTT ACC GGC TAG CCG CCG ATG ACG CTG CCG CGA CTG TGA TGT GCG G-3′. The StuI-NotI-digested gC amplimer was ligated to MTS vector. For all plasmids, the ORF was sequenced.
Genetic Engineering of HSV1(BAC)-gDst
HSV1(BAC)-gDst was generated by bacterial artificial chromosome (BAC) “galK recombineering,” employing the Escherichia coli strain SW102 and galK (galactokinase) positive/negative selectable marker, kindly provided by Dr. N. G. Copeland) (45). pYEbac102 HSV-BAC was provided by Y. Kawaguchi and carries the BAC sequences inserted between UL3 and UL4 genes. The galK cassette was recombined in HSV-1(BAC) to replace the gD stop codon. To this end, the cassette was PCR-amplified from pGalK plasmid with primers that annealed in their 5′ end to sequences upstream and downstream of gD stop codon. The oligonucleotides used for amplification were 5′-CCC ACA TCC GGG AAG ACG ACC AGC CGT CCT CGC ACC AGC CCT TGT TTT ACC CTG GTG ACA ATT AAT CAT CGGCA-3′ and 5′-CAT CCC AAC CCC GCA GAC CTG ACC CCC CCG CAC CCA TTA AGG GGG GGT ATT CAG CAC TGT CCT GCT CCTT-3′. The amplimer was recombined into the HSV-1 BAC, and the recombinant bacteria were selected on minimal media containing galactose as the only carbon source. Next, the galK cassette was substituted with sequences encoding the One-strep-tag by means of a double-stranded oligonuocletide that carried flanking sequences homologous to gD sequences upstream and downstream of the stop codon. The double-stranded oligonucleotide was generated by annealing-extension of two synthetic oligonucleotides 5′-CCC ACA TCC GGG AAG ACG ACC AGC CGT CCT CGC ACC AGC CCT TGT TTT ACA GCG CTT GGA GCC ACC CGC AGT TCG AGA AAG GTG GAG GTT CCG GAG GTG GAT CCG GAG GT-3′ and 5′-CAT CCC AAC CCC GCA GAC CTG ACC CCC CCG CAC CCA TTA AGG GGG GGT ATT CAT TTT TCG AAC TGC GGG TGG CTC CAC GAT CCA CCT CCG GAT CCA CCT CCG GAA CC-3′. Its homologous recombination into the HSV-BAC was achieved by selecting against the galK cassette, i.e. by resistance to 2-deoxy-galactose on minimal plates containing glycerol as carbon source. Chloramphenicol selection was kept throughout, to maintain the HSV-BAC sequences. The recombinant HSV1(BAC)-gDst DNA was extracted from bacteria and transfected into 293 T cells to reconstitute the virus. The 3′ of gD gene was sequenced for accuracy in DNA extracted from bacteria and in virus stock.
gD Constructs
The linear map of the gD constructs employed in this study is shown in Fig. 6. To generate gDΣ240–260, the starting plasmid contained an Asp718 restriction site at aa 260 (46). An additional Asp718 site was inserted at aa 240 by site-directed mutagenesis by means of oligonucleotides 5′-CAG CTT GAA GAT CGC GGT ACC GAA GCT TCC CAA GGC CCC ATA CAC GAG CAC CC-3′ and 5′-GGG TGC TCG TGT ATG GGG CCT TGG GAA GCT TCG GTA CCG CGA TCT TCA AGC TG-3′. Two synthetic annealing oligonucleotides encoding a 18-aa-long Ser-Gly linker 5′-PHO-GTA CCC AGT AGT GGC GGT GGC TCT GGA TCC GGC TCG AGC GGA GGC GGT AGC GGG-3′ and 5′-PHO-GGT CAT CAC CGC CAC CGA GAC CTA GGC CGA GCT CGC CTC CTC CAT CGC CCC ATG-3′ were ligated into Asp718-digested gD. Orientation was screened by colony PCR and confirmed by sequence. To generate gDΣ218–240, we followed essentially a similar strategy. Two Asp718 restrictions sites were introduced in gD plasmid at aa 218 and 240 by simultaneous double site-directed mutagenesis. The Asp718 site in aa 218 was inserted by means of oligonucleotides 5′-TGA CGG TGG ACA GCA TGG TAC CGC TGC CCC GCT TCA TCC-3′ and 5′-GGA TGA AGC GGG GCA GCG GTA CCA TGC TGT CCA CCG TCA-3′; the Asp718 site at aa 240 was inserted by means of the oligonucleotides described above for gDΣ240–260. The DNA encoding the 18-aa-long Ser-Gly linker was generate as above and ligated with Asp718-digested gD. Orientation was screened by colony PCR and confirmed by sequence.
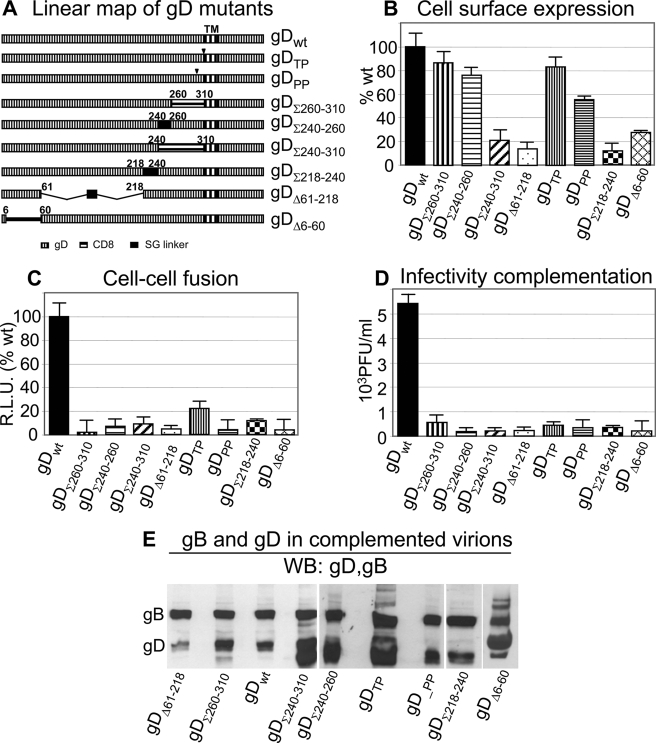
FIGURE 6.
Characterization of a panel of gD mutants. A, linear map of the gD mutants. gDTP and gDPP carry the gDP291L-P292A or gDT304-P305L substitutions (9). In gDΣ260–310 (previously named gDΔPFD) (10) and gDΣ240–310, the indicated sequences were substituted with the CD8 sequences corresponding to the pretransmembrane region. gDΣ240–260 carries a 18-aa-long Ser-Gly linker in place of the endogenous 240–260-aa sequence. In gDΣ218–240, the 218–240-aa segment, carrying α-helix3, was substituted with a 18-aa-long Ser-Gly linker. In gDΔ61–218, the deleted sequences were replaced with a 18-mer Ser-Gly linker. gDΔ6–60 carries the indicated deletion. B, gD cell surface expression measured in transfected 293T cells by CELISA with mAb H170 (or mAb BD80 for gDΔ6–60). The extent of expression is relative to cells expressing wt gD (100%). C, cell-cell fusion in 293T cells transfected with wt or mutant gD, plus gB, gH, and gL. All other details as in legend to Fig. 1C. D, infectivity complementation of gD mutants. 293T cells in T75 flasks were transfected with one of the gD mutants and 4 h later were infected with ΔgD-HSV (FgDβ). In this assay, progeny virus harvested at 18 h after infection is complemented by the transiently expressed transgenic gD; it was titrated in R6 cells (which express wt gD and allow plaque formation); titer was expressed as PFU/ml. It can be seen that all of the gD mutants were dramatically hampered in infection, relative to wt gD. In all histograms, each column represents the average of triplicate samples. The bars denote ± S.E. E, extracellular virions from the experiment shown in D ultracentrifuged and analyzed by WB for the presence of gD and for comparison of gB. It can be seen that, except gDΔ61–218, all mutant forms of gD were incorporated in virions.
The starting plasmid for gDΔ61–218 was generated by Dr. L. Menotti in the course of an independent study.4 Briefly, the gD ORF, engineered in a vector designed to enable homologous recombination of mutant gD into HSV-BAC, was mutagenized by insertion of NdeI restriction sites at aa 60 and 218. Two synthetic annealing oligonucleotides encoding a 18-aa Ser-Gly linker (5′-PHO-TAG TAG TGG CGG TGG CTC TGG ATC CGG CTC GAG CGG AGG CGG TAG CGG GAG TGG-3′ and 5′-pho-TACC CAC TCC CGC TAC CGC CTC CGC TCG AGC CGG ATC CAG AGC CAC CGC CAC TAC-3′) were ligated into the NdeI-digested gD. Finally, the SacII/HindIII fragment containing the partially deleted ORF was subcloned in pcDNA containing gD ORF. Orientation was screened by colony PCR and confirmed by sequence. To generate gDΣ240–310, Asp718 restriction sites were engineered at aa 240 and 310 of wt gD, respectively, by site-directed mutagenesis by means of oligonucleotides 5′-CAG CTT GAA GAT CGC GGT ACC GAA GCT TCC CAA GGC CCC ATA CAC GAG CAC CC 3′ and 5′-CAT CCC CCG GCG GTA CCG AAC AAC ATG GGC CTG 3′. A CD8 amplimer obtained with oligonucleotides 5′-CGG GGT ACC CAG TGG CGG TAG TTC GGC CCT GAG CAA CTC CATC3′ and 5′GCGC CCA GAT GTA GAT AGG TAC CGC GAA GTC CAG CCC CCT CGT GTGC 3′ and digested with Asp718 was then ligated into Asp718-digested gD. To generate gDΔ6–60, two EcoRI restrictions sites were introduced in pcDNA-gD plasmid by simultaneous double site-directed mutagenesis with prior elimination of the EcoRI site present in the vector. The EcoRI site at aa 6 was introduced by means of oligonucleotides 5′-CAA ATA TGC CTT GGC GGA GAA TTC TCT CAA GAT GGC CG-3′ and 5′-CGG CCA TCT TGA GAG AAT TCT CCG CCA AGG CAT ATT TG-3′; the EcoRI site at aa 59 was introduced by means of oligonucleotides 5′-CAC GGT TTA CTA CGC GAA TTC GGA GCG CGC CTG CCG-3′ and 5′-CGGC AG GC GCG CTC CGA ATT CGC GTA GTA AAC CGTG-3′. The EcoRI-digested gD was relegated, thus collapsing the EcoRI fragment. For all of the plasmids, the ORF was sequenced.
Co-immunoprecipitation and Pulldown Experiments
Co-immunoprecipitation was carried out from infected or transfected cells. Infected 293 T cells received 5 PFU/cell of one of the following viruses, HSV-1(F), ΔgD, ΔgB, ΔgH, or ΔgL HSV, as titrated in Vero or their respective complementing cells. To enable detection of gH and gL, the cells were transfected with gH5E1 and gL5E1.His (5 and 3 μg of DNA/flask, respectively) 6 h prior to infection with HSV-1(F), ΔgD, and ΔgB viruses. Infected cells were harvested 18 h after infection. U251, human embryonic lung, and human foreskin fibroblast cells were infected with HSV-1(F) or ΔgD virus (5 PFU/cell). We used 3-fold higher amount of cells than in the experiments with infected 293T cells. To enable detection of gH and gL, the cells were transfected with gH5E1 and gL5E1.His (15 and 9 μg of DNA/T75 flask, respectively) 6 h prior to infection. Infected cells were harvested 18 h after infection. Transfected 293T cells in T25 flasks received the appropriate plasmid mixture together with Arrest-in (Celbio, Milano), in the amount of 1.5 μg of DNA for each plasmid (1× amount). When indicated, pBEC was co-transfected at 3 μg/T25 flask (2X amount). Transfection mixtures in which one or more plasmids were omitted contained the HER2-encoding plasmid in place of the omitted plasmid, such that the total amount of DNA transfected in each flask did not vary. The cells were harvested 18 h after infection or transfection, prior to the appearance of syncytia, without freezing. For co-immunoprecipitation, in a typical experiment cells from two replicate T25 flasks (or one T75 flask) were solubilized in 200 (or 300) μl of EA1 buffer (50 mm HEPES, 250 mm NaCl, 0.1% Igepal) containing the protease inhibitors Nα-p-tosyl-l-lysine chloromethyl ketone hydrochloride and Nα-p-tosyl-l-phenylalanine chloromethyl ketone (final concentration, 0.3 mm each), as described (28), at 4 °C for 20 min. The lysates were centrifuged at 14,000 rpm for 30 min. The supernatants were cleared by incubation for 1 h at 4 °C with a preimmune rabbit serum (2 μl of serum/200 μl of supernatant) followed by absorption to protein A-coupled Sepharose (10 mg) (Sigma-Fluka, Milano). The cleared unbound fraction was then incubated with pAb R8 to gD (2 μl/200 μl) overnight at 4 °C, and thereafter with 10 mg of protein A-coupled Sepharose for 1 h at 4 °C. The beads were washed three times with EA1 buffer containing the protease inhibitors and once with 50 mm Tris-HCl, pH 7.5, and 15 mm NaCl.
gHst pulldown experiments were carried out from transfected 293T cells (2 T25 flasks/sample), and gDst pulldown experiments were carried out from HSV1(BAC)-gDst virions alone or absorbed to cells. For pulldown experiments from cells, two replicate T25 flasks (or virions in the indicated amounts) were solubilized with 200 μl of EA1plus buffer (50 mm HEPES, 250 mm NaCl, 0.5% Igepal, pH 8) containing 0.3 mm each Nα-p-tosyl-l-lysine chloromethyl ketone hydrochloride and Nα-p-tosyl-l-phenylalanine chloromethyl ketone at 4 °C for 20 min. The lysates were centrifuged at 14,000 rpm for 60 min. The supernatants were cleared by preabsorption for 1 h at 4 °C to protein A-coupled Sepharose (10 mg) (Sigma-Fluka). The unbound fraction was then incubated with Strep-Tactin Sepharose (IBA, GmbH, Gottingen, Germany) (30 μl/200 μl of lysate) for 1 h at 4 °C and washed five times with the resin washing buffer (100 mm Tris-HCl, pH 8, 150 mm NaCl, and 1 mm EDTA), according to the manufacturer.
For both co-immunoprecipitation and pulldown experiments, the material deriving from two flasks was solubilized with 120 μl of sample buffer (2% sodium dodecyl sulfate, 5% β-mercaptoethanol, 50 mm Tris-HCl, pH 7, and 2.5% sucrose), boiled, and loaded in two different gels: one for Western blot (WB) detection of gD and gB and one for detection of gH, gL, and gC.
Western Blot
Proteins separated by 8.5% PAGE were transferred to Hybond ECL nitocellulose membrane from GE Healthcare (Milano) and developed by Amersham Biosciences ECL Advance Western blotting detection kit (GE Healthcare).
Cell Surface Expression
Cell enzyme-linked immunosorbent assay (CELISA) was performed as described (48, 49). Briefly, 293T cells in 48 wells were transfected with plasmids encoding wt or mutant gD (125 or 375 ng/well, corresponding to 1× or 3× amounts, as specified). Sixteen h later they were reacted with mAb H170, or BD80 and fixed with paraformaldehyde, followed by anti-mouse peroxidase. To measure gH cell surface expression, 293 T cells were transfected with mixture of plasmids encoding wt gH/wt gL, gH5E1/gL5E1.His, or gHst/gLV5.His. Sixteen h later they were reacted with mAb 53S, fixed with paraformaldehyde, followed by anti-mouse peroxidase. All of the samples were run in triplicate.
Cell-Cell Fusion Assay
The luciferase-based cell-cell fusion assay was performed as detailed (49, 50) by means of a luciferase assay system from Promega (Florence, Italy) in 293T cells. The total amount of transfected plasmid DNA was made equal by the addition of human epidermal growth factor receptor 2 plasmid DNA. All of the samples were run in triplicate.
Infectivity Complementation
The assay was performed as detailed elsewhere (46). Briefly, 293T cells in T75 flasks were transfected with the appropriate gD plasmid. Four h later, the cells were infected with a gD−/+ stock of FgDβ (3 PFU/cell). Unpenetrated virions were removed by two phosphate-buffered saline rinses and inactivated by means of 40 mm citric acid, 10 mm KCl, 135 mm NaCl, pH 3, for 1 min. The monolayers were then rinsed twice with phosphate-buffered saline and overlaid with medium containing 1% fetal calf serum. The cells were incubated overnight at 37 °C. The extracellular progeny virions were ultracentrifuged, analyzed by WB, and titrated in the gD-complementing R6 cells.
RESULTS
Tagging of HSV-1 Glycoproteins
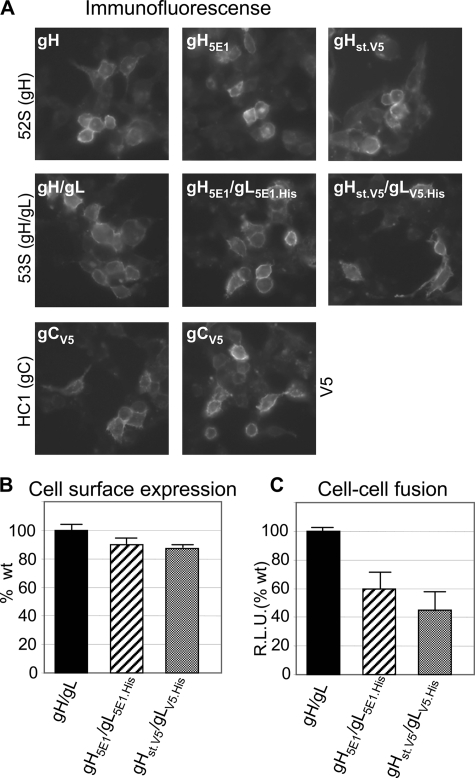
Biochemical evidence of complex formation among the HSV glycoprotein quartet by means of co-immunoprecipitation or pulldown experiments relied on WB-positive antibodies, able to detect the complexed glycoproteins. The most limiting factor throughout this study was the detection of gH and gL, which greatly affected the sensitivity of the assays. To improve their detection, gH and gL were tagged with the following epitopes: 5E1 (51) plus or minus polyhistidine, V5-polyhistidine, and One-strep-tag. All of the epitopes were engineered just upstream of the stop codon. gC was similarly tagged with the V5 epitope. As shown in Fig. 1, the tagged glycoproteins, named gH5E1, gL5E1.His, gHst.V5, and gLV5.His maintained immunofluorescence reactivity to conformation-dependent mAbs 52S, 53S, and HC1, reactive to gH/gL, and gC, respectively. They were transported to the cell surface, as measured in the CELISA test (Fig. 1B). When cotransfected with gD and gB, the tagged gH and gL maintained the ability to induce cell-cell fusion, even though efficiency was reduced, as compared with the respective untagged version (Fig. 1C).
FIGURE 1.
Characterization of tagged forms of gH, gL and gC, gH5E1, gHst.V5, gL5E1.His, gLV5.His, gCV5. A, immunofluorescence of 293T cells transfected with the indicated tagged versions of gH, gL, or gC, reacted with conformation-dependent mAbs 52S to gH, 53S to gH/gL, HC1 to gC, or V5. B, cell surface expression of the indicate gH/gL combinations, as measured by CELISA in transfected 293T cells, expressed as percentages relative to cells transfected with untagged gH/gL. C, cell-cell fusion in 293T cells transfected with the indicated combinations of gH/gL, plus gD and gB, measured by means of a luciferase based assay. R.L.U., relative luciferase units expressed as percentages relative to cells transfected with untagged gH/gL. In B and C, each column represents the mean of triplicate samples. The bars denote ± S.E.
The Glycoprotein Quartet Forms Complexes Co-immunoprecipitated with gD in Cells Infected with wt or Deletion HSV Mutants
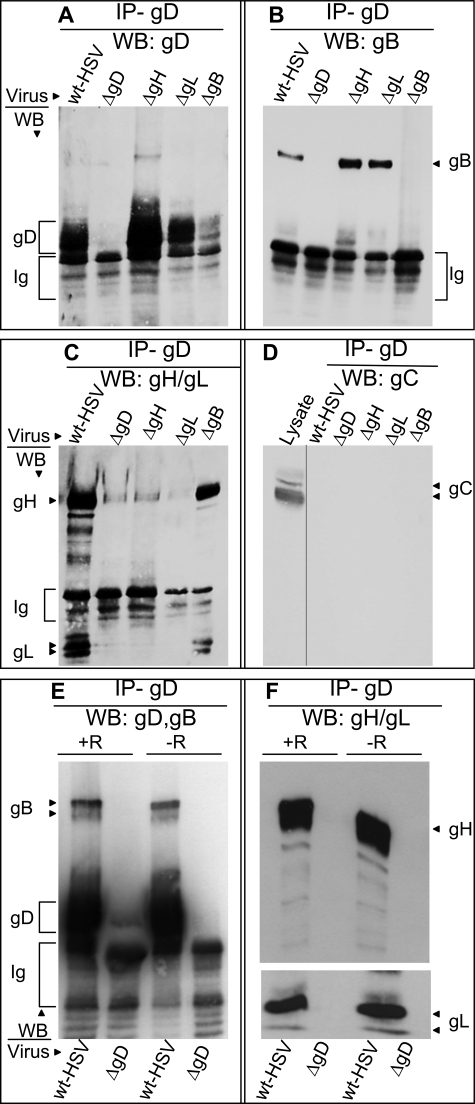
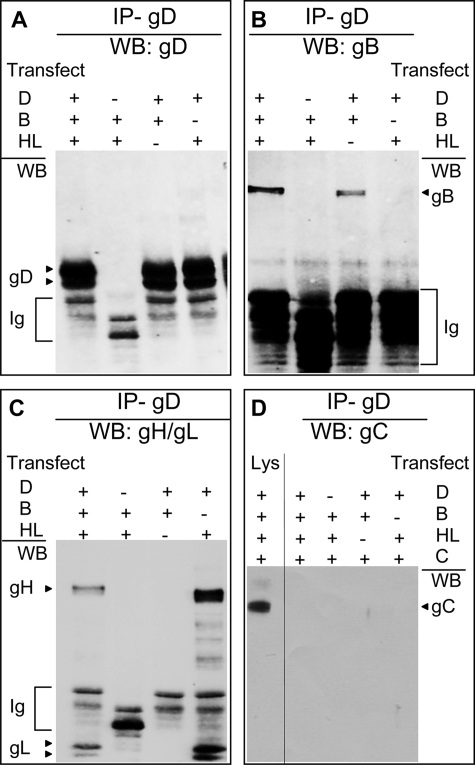
The aim of the this series of experiments was to detect the complexes formed by the glycoprotein quartet in infected cells by gD co-immunoprecipitation. Prior to infection, 293T cells were transfected with gH5E1 and gL5E1.His to facilitate their detection and with HVEM to potentially increase complexes dependent on receptor-bound gD. Six h later, they were infected with HSV-1(F) or HSV deletion mutants in the glycoprotein genes and lysed 18 h after infection. gD was immunoprecipitated with pAb R8; the proteins, separated by denaturing PAGE, were identified by WB. The results in Fig. 2 show that, from wt HSV-infected cells, gD co-immunoprecipitated not only gH (Fig. 2C), as expected (28) but also gB (Fig. 2B) and gL (Fig. 2C). The co-immunoprecipitation of gB, gH, and gL was specific by two criteria. First, gB, gH, and gL failed to be detected when cells were infected with ΔgD-HSV, instead of wt virus. Second, gC, which is present in the same subcellular compartments as the other glycoproteins but is not expected to form any complex with them, was indeed absent from the co-immunoprecipitate (Fig. 2D); the only gC-positive lane in Fig. 2D is that of an aliquot of the lysate, not subjected to immunoprecipitation.
FIGURE 2.
gD co-immunoprecipitates gB and gH/gL, and not gC, from infected cells. A–D, 293T cells were infected with wt HSV-1(F) or deletion ΔgD, ΔgB, ΔgH, or ΔgL mutant viruses (5 PFU/cell). To enable gH and gL detection, 6 h prior to infection, the cells to be infected with wt HSV, ΔgD, or ΔgB viruses were transfected with gH5E1 and gL5E1.His. E and F, 293 T cells were transfected with HVEM plasmid (+R) or no HVEM (−R) at the time of gH5E1 and gL5E1.His transfection. All of the cells were harvested 18 h after infection and immediately lysed with EA1 buffer. gD was immunoprecipitated with pAb R8 (IP-gD) and harvested with protein A-Sepharose beads. The beads from one sample were split into two portions and run in two separate gels: one for detection of gB and gD and one for detection of gH and gL; gC was detected in the same blot as gH and gL, after stripping. The gD co-immunoprecipitated proteins were detected by WB with mAb H170 to gD (A and E), mAb H1817 to gB (B and E), mAb5E1 to tagged gH and gL (C and F), and mAb H633 to gC (D). In D, the only positive lane is the one loaded with an aliquot of the infected cells lysate. The secondary antibody used for WB detected the pAb R8 Ig, whose electrophoretic mobility is faster than that of gD.
To determine the requirements for complex formation, the cells were infected with ΔgB, ΔgH, or ΔgL HSVs. gD was immunoprecipitated with pAb R8, and the co-immunoprecipitated proteins were identified as above. Fig. 2 shows that gD co-immunoprecipitated gB from cells infected with ΔgH or ΔgL viruses (Fig. 2B), i.e. in the absence of gH and gL. Conversely, gD co-immunoprecipitated gH and gL from cells infected with ΔgB virus (Fig. 2C), i.e. in the absence of gB.
In the experiments reported in Fig. 2 (A–D), HVEM was hyperexpressed by transfection. To assess whether HVEM hyperexpression was necessary or at least augmented detection of the glycoprotein complexes, 293T cells were infected with wt HSV-1(F) or ΔgD-HSV, with or without prior transfection of HVEM. All of the cells were transfected with gH5E1 and gL5E1.His. gD was co-immunoprecipitated with pAb R8, and the co-immunoprecipitated proteins were detected by WB, as above. The results in Fig. 2 (E and F) show that HVEM hyperexpression did not modify the ability of gD to co-immunoprecipitate gB, gH, and gL and affected the quantities of co-immunoprecipitated glycoproteins to a very minor extent.
Complex Formation among the HSV Glycoprotein Quartet Occurs in Cell Lines Relevant to HSV Infection
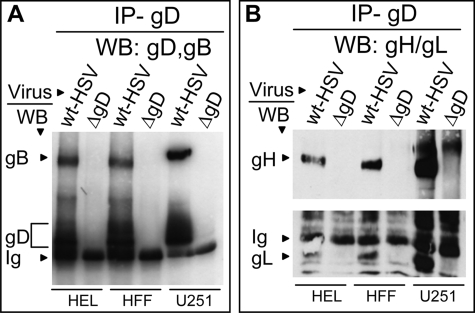
HSV exhibits a broad tropism in vivo and in cultured cells. In humans, it generally infects the skin and the central nervous system; under certain conditions, including immunosuppression, it infects a variety of organs, testifying for the wide tropism. Here, we asked whether complexes were detected in human cells relevant to HSV infection. We analyzed neural glioblastoma U251 cells and human fibroblasts. Glioblastoma is the target of HSV oncolytic virotherapy (52). Fig. 3 shows that gD co-immunoprecipitated gB and gH/gL in all cell lines.
FIGURE 3.
gD co-immunoprecipitates gB and gH/gL from infected human embryonic lung (HEL), human foreskin fibroblast (HFF) and U251 cells. The indicated cells were infected with wt HSV-1(F) or deletion mutant HSV-ΔgD. Experimental details as in the legend to Fig. 2. WB for gH/gL was carried out with pAb to a truncated form of gHt/gL.
The Glycoprotein Quartet Forms gD Co-immunoprecipitated Complexes in Transfected Cells
Next, we verified whether the complexes formed in infected cells were formed also in transfected cells. 293T cells were co-transfected with plasmid mixtures encoding the glycoprotein quartet plus HVEM (to increase the amounts of receptor-bound gD) and gCV5, as negative control. Where indicated, the transfection mixtures lacked gD, gB, or gH/gL. gD was immunoprecipitated with pAb R8; co-immunoprecipitated proteins were identified by WB. The results in Fig. 4 show that gB (Fig. 4B), and gH/gL (Fig. 4C) were co-immunoprecipitated from lysates of cells transfected with the quartet. From cells transfected with a mixture lacking gB, gD could still co-immunoprecipitate gH/gL (Fig. 4C). From cells transfected with mixtures lacking gH/gL, gD could still co-immunoprecipitate gB (Fig. 4B). The specificity of co-immunoprecipitation was assessed by the absence of gC from the co-immunoprecipitated proteins (Fig. 4D). The results indicate that complexes formed in transfected cells closely mirror those formed in the infected cells and imply that no viral protein other than those that are recruited to the complexes is required for complex formation. Because the complexes formed in transfected cells did not differ from those in infected cells, most of the subsequent experiments were carried out in transfected cells.
FIGURE 4.
gD co-immunoprecipitates gB and gH/gL, and not gC, from transfected cells. 293T cells were co-transfected with plasmid mixtures encoding gD (D), gB (B), gH5E1/gL5E1.His (HL), gCV5 (C), plus HVEM or with mixtures lacking the indicated glycoproteins (−). The cells were harvested 18 h after transfection. All other details are as in the legend to Fig. 2. Lys, lysate of transfected cells prior to immunoprecipitation.
Genetic Engineering of HSV-1 Carrying One-strep-tagged gD (HSV1(BAC)-gDst)
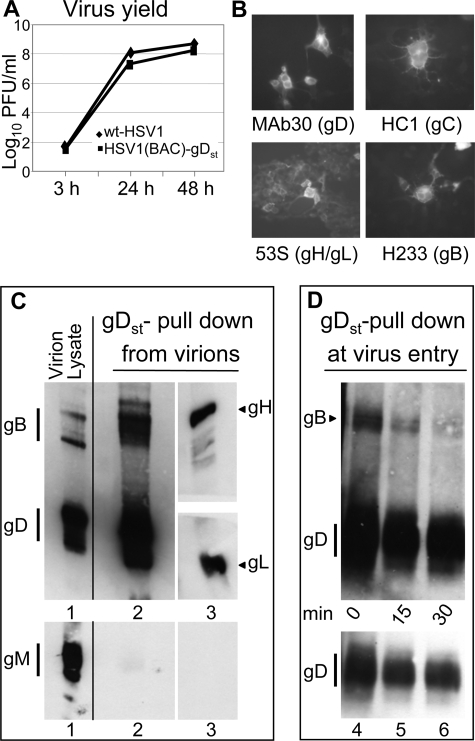
The HSV glycoprotein complexes identified in this study and elsewhere (29, 30) were detected in infected or transfected cells. As mentioned above, a constant major limit in our assays was detection of gH and gL. To investigate glycoprotein complexes in virions and to augment complex capture, we engineered an HSV carrying One-strep-tagged form of gD (gDst). One-strep-tagged proteins, herein named strep-tagged proteins, specifically absorb to Strep-Tactin resin (IBA GmbH, Goettingen, Germany) and retain any protein complexed to them. The virus was engineered through the BAC technology, as detailed under “Experimental Procedures,” and named HSV1(BAC)-gDst. The strep-tag was well tolerated by gD. In a virus yield experiment HSV1(BAC)-gDst replicated to very similar extent as wt HSV1(F) (Fig. 5A). gD, as well as gB, gH/gL, and gC, maintained reactivity to conformation-dependent antibodies (Fig. 5B).
FIGURE 5.
Characterization of HSV1(BAC)-gDst and analysis of glycoprotein complexes in virions and during virus entry. A, kinetics of HSV1(BAC)-gDst replication. Vero cells were infected with HSV1(BAC)-gDst or HSV-1(F), 0.1 PFU/cell. Replicated samples of infected cells were frozen at the indicated times after infection, and progeny virus titrated in Vero cells. No major difference was observed in the yield of HSV1(BAC)-gDst relative to wt HSV-1. B, replicate samples of HSV1(BAC)-gDst-infected 293T cells (0.1 PFU/cell) characterized for immunofluorescence reactivity. Infected cells were incubate with the indicated mAbs, reactive to conformation-dependent epitopes of the protein indicated in parentheses. C, detection of glycoprotein complexes in HSV1(BAC)-gDst virions. Partially purified extracellular HSV1(BAC)-gDst virions were lysed; a small aliquot of the lysate (corresponding to 0.2 × 108 PFU) was loaded in lane 1. The remaining lysate was absorbed onto strep-Tactin resin; gDst and complexed glycoproteins were detected by WB to gD (mAb H170), gB (mAb H1817), gH (pAb to gH), and gL (pAb to gL). The gD-gB lane (lane 2) was loaded with material, retained by the step-Tactin resin, derived from 2.8 × 108 PFU virions. The gH/gL lane (lane 3) was loaded with material, retained by the step-Tactin resin, derived from 1.4 × 109 PFU. The upper blot containing lanes 1–3 was stripped and reacted again with pAb to gM (lower panel); it can be seen that only the lane containing the virion lysate showed reactivity, indicating that gM was not present in the Strep-Tactin-retained fraction. D, detection of glycoprotein complexes at virus entry. Partially purified extracellular HSV1(BAC)-gDst virions were absorbed to 293T cells in T25 flask (4 × 108 PFU/T25 flask, ∼ 140 PFU/cell) at 4 °C for 2 h. The unabsorbed virus was removed and titrated and accounted for 75% of the input virus. The cells were rinsed twice with ice-cold medium, overlaid with warm medium, immediately shifted to 37 °C for 0, 15, and 30 min (lanes 4–6, respectively) and lysed. The lysate was absorbed to Strep-Tactin resin. The lane of each time point contained material derived from one T25 flask. gDst and complexed gB were detected by WB. The bottom panel shows a lighter exposure of the gD, to highlight the decrease in the cell-associated virion gDst during the 30-min time interval.
Glycoprotein Complexes Are Present in Virions
HSV1(BAC)-gDst was employed to ask whether glycoproteins already interact with each other in virions independent of entry into the cells and whether complexes formed at virus entry into the cell. Partially purified extracellular HSV1(BAC)-gDst virions were lysed, and gDst was absorbed to Strep-Tactin resin; complexed glycoproteins were analyzed by WB. The results in Fig. 5C show the WB reactivity of virion lysate (lane 1) and furthermore that small amounts of gB, gH, and gL were complexed to gDst in resting HSV(BAC)-gDst virions (lanes 2 and 3). The amounts of pulled down gB and gH/gL was small, and to visualize them the gB-gD and gH/gL lanes (Fig. 5C, lanes 2 and 3, respectively) were loaded with material pulled down from 2.8 × 108 PFU (lane 2) and 1.4 × 109 PFU (lane 3), respectively; for comparison, Fig. 5C, lane 1 contains lysates from 0.2 × 108 PFUs. To ensure that gB and gH/gL detected in the pulled down fraction were indeed pulled down by gDst and not simply present in the fraction because of incomplete lysis of virions, we checked for the presence of an unrelated membrane protein, gM, in lysed virions (Fig. 5C, lane 1) and in the pulled down fraction (Fig. 5C, lane 2). gM is a polytopic protein, expected to be solubilized from virions with difficulty (42). Fig. 5C shows that gM was present in virion lysate but not in the pulled down fraction, providing evidence that virions were effectively lysed.
To analyze the destiny of the pre-existing complexes and to ask whether new complexes were formed during virus fusion with cell, HSV(BAC)-gDst virions were allowed to attach to 293T cells for 2 h at 4 °C (∼140 PFU/cell), unabsorbed virus was rinsed, and the cells were shifted to 37 °C for 0, 15, or 30 min and lysed. The lysate was absorbed to the Strep-Tactin resin. Retained proteins were identified by WB. Fig. 5D shows that in the 30-min interval, the amount of virion gDst associated to cells decreased; gB decreased in parallel. We did not detect any gH/gL, likely because of limits of detection. The results indicate that virion gDst, and the complexed gB, tended to decrease rather than increase during virus fusion to 293T cells.
Characterization of gD Mutants Carrying Substitutions, Deletions, or Mutations across the Ectodomain
The objective of the next series of experiments was to preliminarily define the gD regions involved in complex assembly with gB and gH/gL. We employed a panel of gD mutants carrying substitutions, deletions, or mutations across the entire gD ectodomain (linear maps shown in Fig. 6A). Some were previously described, and some were generated for the purpose of this study. As mentioned in the Introduction, gD ectodomain can be schematically subdivided into two regions; the N-terminal region up to aa 240/260 includes the receptor-binding sites; the downstream region, spanning from aa 240/260 to 310 carries the profusion domain. In gDΣ260–310 (previously named gDΔPFD) (10) and in gDΣ240–310, the indicated sequences were substituted with the CD8 sequences corresponding to the pretransmembrane region. CD8 was initially chosen as donator of heterologous sequence because it is a transmembrane protein, totally unrelated to HSV attachment entry (9, 53). gDΣ240–260 carries an 18-aa-long Ser-Gly linker in place of the endogenous 240–260 aa sequence. The gDP291L-P292A and gDT304A-P305L mutants, herein and elsewhere referred as gDPP and gDTP, respectively, carry the indicated substitutions in the profusion domain. Both are partially impaired in infection (9). In gDΣ218–240, the 218–240-aa segment, carrying α-helix3, was substituted with an 18-mer Ser-Gly linker. gDΔ61–218 was generated in this study following the discovery that, in a very peculiar form of gD split into two fragments, one of which carried the Kringle domain from urokinase plasminogen activator, the 61–218-aa region was dispensable (54). Studies from our laboratory confirm that this sequence can be substituted with heterologous sequences.4 gDΔ6–60 was designed during the course of the study; of note, it is known that the gD N terminus can be deleted at least up to aa 38 (55) and leave a functional gD that cannot interact with HVEM any longer.
The panel of gD mutants was preliminarily characterized with respect to ability to be transported to the cell surface and to mediate cell-cell fusion and virus infection. Cell surface expression was measured in transfected 293T cells and expressed as a percentage relative to wt gD-transfected cells. Fig. 6B shows that the gDΣ260–310, gDΣ240–260, and gDTP, gDTP mutants were expressed at the cell surface in amounts ranging from 50 to 80% of wt gD; the remaining mutants were severely impaired. Increasing the amount of transfected plasmid DNA by 3-fold did not result in substantially increased cell surface expression (data not shown). We next measured the ability of the mutants to induce cell-cell fusion when co-transfected with gB, gH, and gL. The results in Fig. 6C show that all of the mutants were severely hampered, irrespective of their cell surface expression. Lastly, the ability of gD mutants to promote virus infection was measured in a complementation assay, whereby the gD deletion virus FgDβ, which does not encode gD, is grown in cells transiently expressing transgenic wt or mutant-gD. The complemented progeny virions were titrated in R6 cells that express wt gD and allow plaque formation once the virus has entered the R6 cells by means of the complementing gD (Fig. 6D). We further ascertained that the complemented virions carried in their envelope the mutants gD, and, for comparison, gB. Aliquots of virions were pelletted by ultracentrifugation and analyzed for the presence of gD and of gB by WB. As shown in Fig. 6E, all forms of mutant gD were incorporated at substantial amounts, except gDΔ61–218. Cumulatively the results indicated that the gD mutants were severely hampered in infectivity complementation, even though, with one exception, they were incorporated in virion envelopes.
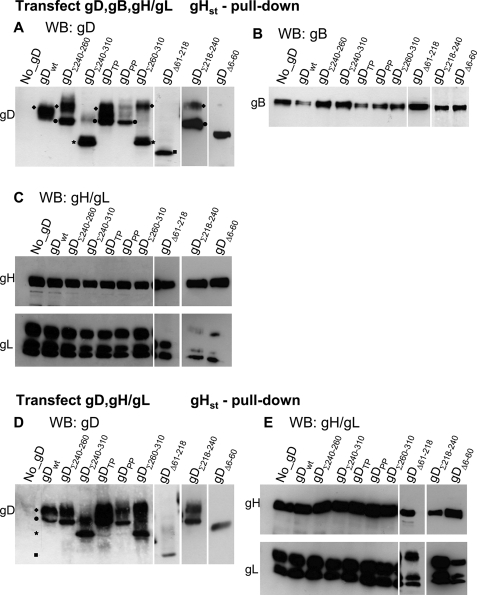
Ability of gD Mutants to Form Complexes with gH/gL Detected by gHst Pulldown
In the following two series of experiments, the panel of mutants was employed to define the gD regions critical for the interactions with gH/gL and with gB. To detect gD-gH complexes, we optimized a pulldown assay based on gHst. 293T cells were transfected with gHst, gLV5.His (both described in Fig. 1), wt, or mutant gD, plus or minus gB. The cleared lysates were adsorbed to Strep-Tactin resin. The retained proteins were analyzed by WB. Fig. 7A shows that gHst pulled down wt gD, gDΣ240–260, gDTP, gDΣ218–240, and gDΔ61–218, gDΔ6–60. It is worth noting that gDΣ240–310 and gDΣ260–310 accumulate as three bands (Fig. 8) with apparent molecular masses of ≥56, 50, and 38 kDa. The ≥56-kDa species (marked with diamonds in Figs. 7 and 8) represent the mature forms of the glycoprotein. The 50-kDa species (marked with circles) was observed in all forms of gD and represents the precursor form. The 38-kDa species (marked with stars) likely represents an N-terminal degradation product. Its reactivity to mAb H170, used for WB, which recognizes an epitope located at aa 1–23, indicates that this is an N-terminal peptide. Interestingly, with respect to gDΣ240–310 and gDΣ260–310, gHst pulled down the 38-kDa N-terminal species and only small amounts of the higher molecular mass species (Fig. 7A). A striking difference was observed also between gDPP and gDTP, which carry substitutions at Pro291-Pro292 or Thr304-Pro305, respectively. gDPP but not gDTP failed to be pulled down by gHst. The pattern of gD pulled down by gHst in cells transfected with gD, gH, and gL, in the absence of gB (Fig. 7D) was essentially similar to that in Fig. 7A. We interpret the results with gDΣ240–310 and gDΣ260–310 to mean that gD carries a composite contact surface to gH/gL. Part is made by N-terminal region and enables gHst to pull down the N-terminal 38-kDa degradation product. The other involves segments downstream of aa 240, is lost in gDΣ240–310 and gDΣ260–310 because of the substitutions, and accounts for inability of gHst to efficiently pull down the mature forms of these two chimeras. The Pro291-Pro292 doublet mutated in gDPP is critical to the latter interaction site.
FIGURE 7.
Ability of gHst to pulldown wt or gD mutants and gB. 293T cells were co-transfected with mixtures containing gHst, wt, or mutant forms of gD, gB, gLV5.His, and HVEM (A–C). In D and E, the transfection mixture contained gHst, wt, or mutant gD and no gB. The cells were harvested 18 h after transfection. Following cell lysis, gHst was allowed to react with Strep-Tactin resin. The proteins complexed to gHst were retained by the resin, separated by PAGE, and identified by WB. The lane marked no_gD contained all of the proteins except gD and served as control that the proteins under examination were specifically pulled down by gHst. In all panels, the lanes marked gDΔ61–218, gDΣ218–240, and gDΔ6–60 come from different blots. All other details are as described in the legend to Fig. 2. In A and D, it can be seen that different forms of gD migrate with different electrophoretic mobility and apparent molecular mass. Diamonds indicate 56-kDa mature forms. Circles indicate the 50-kDa precursor. Stars indicate the 38-kDa degradation product. Squares indicate the deleted low molecular mass gDΔ61–218.
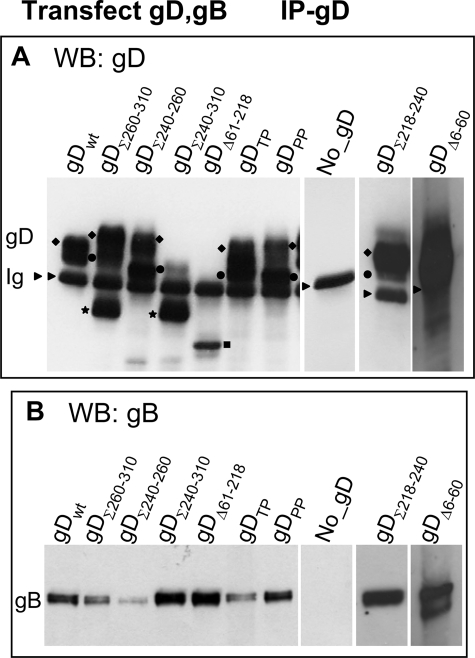
FIGURE 8.
Ability of gD mutants to co-immunoprecipitate gB. 293T cells were co-transfected with plasmid mixtures encoding wt or mutant forms of gD, plus gB, and HVEM. The lane marked No_gD indicates a transfection mixture lacking gD; it served as a control that the proteins under examination were specifically co-immunoprecipitated by gD. The cells were harvested 18 h after transfection. All other details are as in the legend to Figs. 2 and 7. Essentially, gD was immunoprecipitated with pAb R8 (IP-gD) (panel A); the co-immunoprecipitated proteins were identified by WB (panel B). In A it can be seen that different forms of gD migrate with different electrophoretic mobility and apparent molecular mass. The symbols are as explained in legend to Fig. 7. An arrowhead points to heavy IgG band.
Ability of gD Mutants to Form Complexes with gB
In this latter series of experiments 293T cells were transfected with wt or mutant gD plus gB. gD was immunoprecipitated with pAb R8. The results in Fig. 8 show that gDΣ240–260, and to a lesser extent gDΣ260–310, and gDTP co-immunoprecipitate gB in reduced amounts. The results suggest that a gD region critical for interaction with gB lies between aa 240 and 260 and likely extends downstream; critical residues are Thr304-Pro305.
DISCUSSION
An extensive literature has documented that the entry of HSV-1 into cells involves the fusion of the virion envelope with a cellular membrane and that a quartet of glycoproteins, gD gB, gH, and gL, are both necessary and sufficient for fusion (1, 3, 4). The discovery that gD serves as the trigger of fusion (9) implied that it cross-talks with at least one of the downstream glycoproteins gB and gH/gL and boosted a search on the interactions that take place among the glycoprotein quartet. Interactions were identified by co-immunoprecipitation between gD and gH/gL (28) and by split green fluorescent protein complementation assay between gD and gB and between gD and gH/gL (29, 30). In the latter assay weak and transient interactions are rendered irreversible, thus enhancing their detection. The studies presented here were designed to (i) provide biochemical evidence of the complexes formed by the four glycoproteins; (ii) to verify whether complexes form not only in transfected cells but also in infected cells and in virions; and (iii) to preliminarily identify the gD regions critical for complex formation with gB and gH/gL. We employed two independent assays, i.e. co-immunoprecipitation and a pulldown assay that made use of One-strep-tagged gD or gH. The salient features of the results were as follows.
The Four Glycoproteins Form Multiple Complexes in Infected Cells, and These Complexes Do Not Differ from Those Formed in Cells Transfected Solely with Plasmids Encoding the Four Glycoproteins
In cells infected with wt HSV or transfected with the four glycoproteins, we detected complexes that included gD, gB, gH, and gL. One limitation of the assays was that it was impossible to define the actual composition of the complexes, in particular whether (i) a same gD molecule interacted simultaneously with gB and gH/gL, (ii) some gD molecules interacted with the other (e.g. gB) in a bipartite complex and still other with gH/gL in a tripartite complex, or (iii) gD interacted with only one of the downstream glycoproteins (e.g. gH), which in turn recruited gB in a stepwise fashion. To shed light on the composition of the complexes, we infected cells with deletion mutant viruses in gB, gH, or gL or transfected cells with plasmids mixtures lacking gB or gH/gL. Immune precipitation and pulldown experiments from lysates of cells infected with mutant viruses lacking gB or from lysates of cells transfected with gD and gH/gL showed that gD forms a complex with gH/gL in the absence of gB. Similar experiments with cells infected with gH or gL deletion mutants, or with cells transfected with gD and gB, highlighted that gD forms a complex with gB in the absence of gH/gL. The same types of complexes were detected in physiologically relevant cells, including neural cells and human fibroblasts. The key conclusions are 2-fold. First, complexes are formed in infected cells; they do not differ in composition from those formed in transfected cells. This finding, which legitimates the use of transfected cells in studies of complex formation, implies that formation of the complexes requires no viral protein other than the aforementioned quartet. Moreover, the interactions detected previously (29, 30) are authentic and do not reflect artifacts caused by transient hyperexpression or irreversible stabilization of transient interactions. Second, gD can form more than one type of complex and carries distinct and independent binding sites for gB and for gH/gL. This property argues somehow against a stepwise recruitment of glycoproteins to gD, as predicted by hemifusion model (25).
gD Regions Critical for Enabling the Binding of gB and gH/gL
One aim of the current experiments was to identify the gD regions critical for complex formation with gB and with gH/gL. gD ectodomain can be divided into two functionally and topologically distinct regions. The N-terminal region up to aa 240/260 carries the binding sites for HVEM (aa 1–32) and for nectin1 (aa ∼30–240); it includes the Ig-folded core (aa 56–184). The C-terminal region carries the profusion activity, required for triggering of fusion but not for receptor binding. Here, we employed a panel of gD mutants carrying substitutions or deletions across the entire ectodomain. The key results were that (i) none of the mutants was completely hampered in ability to recruit gB or gH/gL, indicating that the binding sites in gD for gB and for gH/gL are each made of multiple contacts, or anyway by a composite contact surface. Consequently, alteration of anyone residue critical to the contact surface does not result in ablation of the overall binding, but in impaired interaction. (ii) The gD regions critical for recruitment of gH/gL and of gB were differently affected in different gD mutants; hence they are distinct one from the other. (iii) The gD region critical to gH/gL recruitment maps partly at the gD N terminus and partly at the gD C terminus; critical residues are Pro291-Pro292. (iv) The gD region critical to gB recruitment includes the aa 240–260 segment and extends downstream; the critical residues are Thr304-Pro305. Altogether, the current experiments provide the first evidence that the C terminus of gD ectodomain harbors regions critical for complex formation with gB and gH/gL.
Glycoprotein Complexes Are Present in Resting Virions, Albeit in Small Amounts
A key question is whether the glycoprotein complexes are present in resting virions or whether they form at the time of virus entry into the cells and of virion envelope fusion with cell membranes. To address this question we engineered a HSV carrying a strep-tagged form of gD, named HSV1(BAC)-gDst. gDst was found to be complexed with gB and with gH/gL in resting virions; the amount of complexed gB and gH/gL was indeed low. When HSV1(BAC)-gDst virions were allowed to enter cells for a 30-min time interval, we did not obtain evidence of de novo gD-gB complex formation; rather, the amount of gD-gB complex decreased. This finding is in agreement with an earlier report showing that gE from input virions first associate with and then disappear from cell surfaces (56).
The Significance of the Glycoprotein Complexes and Interactions Identified in These and Preceding Studies
A key question raised by the results presented in this report and elsewhere (29, 30) relates to the significance of the diverse complexes involving members of the glycoprotein quartet. There are two fundamental possibilities. Foremost, current evidence indicates that resting virions harbor small amounts of preformed complexes; these complexes did not increase during virus entry; rather, they tended to decrease. In principle, the process of HSV-mediated fusion must entail activation of gB and/or gH/gL from fusion-inactive to fusion-active conformations. A possibility compatible with the current results is that conformational changes in gD may be signaled from gD to the precomplexed gB or gH/gL and in this way induce conformational changes to the fusion executors. A nonexclusive possibility is that complexes made by the glycoproteins, or one of the complexes, either is disassembled or is triggered to become fusogenic, once the fusion executors encounter cellular proteins (e.g. receptors), without any appreciable biochemical modification to the glycoproteins themselves.
Second, the possibility exists that members of the glycoprotein quartet, like most HSV proteins analyzed to date, perform multiple functions. In support of this view is the observation that gB is targeted to the multivesicular body compartment and plays a role in HSV envelopment and exit (57), gD and gB traffic to inner nuclear membrane to enable primary envelopment (58), and gD encodes an anti-apoptotic function (47). Conceivably, the formation of these complexes requires the involvement (e.g. direct binding, posttranslational modifications, etc) of cellular proteins that enable the execution of these functions. A corollary of this notion is that a fusogenic complex, if it exists, is but one of the complexes formed by members of the quartet. The resolution of function of the various complexes will ultimately require cell-free assembly as well as identification and characterization of the cellular partners with which the glycoproteins interact.
Acknowledgments
We thank Dr. L. Menotti for carrying out the genetic engineering of HSV1(BAC)-gDst and for sharing with us unpublished plasmids and results, Dr. V. Gatta for invaluable help in the genetic engineering of HSV1(BAC)-gDst, Drs. G. Cohen and R. Eisenberg (University of Pennsylvania, Philadelphia, PA), L. Pereira (University of California, San Francisco), D. Johnson (Oregon University, Portland, OR), H. Browne (Cambridge University, Cambridge, UK) for gift of antibodies, Y. Kawaguchi (Tokyo, Japan) for pYEbac102 HSV-BAC, Drs. Copeland and U. Koszinowski for galK recombineering plasmid, and Elisabetta Romagnoli for invaluable help with cell cultures.
This work was supported by TargetHerpes European Union Grant LSHG-CT-2006-037517 (we specifically acknowledge the contribution of IBA GmbH) and by our department through Fondi Pallotti, by Progetti di Ricerca di Interesse Nazionale, Ministero dell'Istruzione Università e Ricerca, and by a University of Bologna Ricerca Fondamentale Orientata grant.
3 T. Gianni and G. Campadelli-Fiume, manuscript in preparation.
4 Menotti, L., Nicoletti, G., Gatta, V., Croci, S., Landuzzi, L., De Giovanni, C., Nanni, P., Lollini, P. L., and Campadelli-Fiume, G. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, in press.
- HSV
- herpes simplex virus
- aa
- amino acid
- HVEM
- herpesvirus entry mediator
- WB
- Western blot
- pAb
- polyclonal antibody
- mAb
- monoclonal antibody
- PFU
- plaque-forming unit(s)
- ORF
- open reading frame
- BAC
- bacterial artificial chromosome
- wt
- wild type
- CELISA
- cell enzyme-linked immunosorbent assay.
REFERENCES
- 1.Campadelli-Fiume G., Amasio M., Avitabile E., Cerretani A., Forghieri C., Gianni T., Menotti L. ( 2007) Rev. Med. Virol. 17, 313– 326 [DOI] [PubMed] [Google Scholar]
- 2.Rey F. A. ( 2006) EMBO Rep. 7, 1000– 1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spear P. G. ( 2004) Cell Microbiol. 6, 401– 410 [DOI] [PubMed] [Google Scholar]
- 4.Turner A., Bruun B., Minson T., Browne H. ( 1998) J. Virol. 72, 873– 875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montgomery R. I., Warner M. S., Lum B. J., Spear P. G. ( 1996) Cell 87, 427– 436 [DOI] [PubMed] [Google Scholar]
- 6.Geraghty R. J., Krummenacher C., Cohen G. H., Eisenberg R. J., Spear P. G. ( 1998) Science 280, 1618– 1620 [DOI] [PubMed] [Google Scholar]
- 7.Cocchi F., Menotti L., Mirandola P., Lopez M., Campadelli-Fiume G. ( 1998) J. Virol. 72, 9992– 10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shukla D., Liu J., Blaiklock P., Shworak N. W., Bai X., Esko J. D., Cohen G. H., Eisenberg R. J., Rosenberg R. D., Spear P. G. ( 1999) Cell 99, 13– 22 [DOI] [PubMed] [Google Scholar]
- 9.Cocchi F., Fusco D., Menotti L., Gianni T., Eisenberg R. J., Cohen G. H., Campadelli-Fiume G. ( 2004) Proc. Natl. Acad. Sci. U. S. A. 101, 7445– 7450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fusco D., Forghieri C., Campadelli-Fiume G. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 9323– 9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carfi A., Willis S. H., Whitbeck J. C., Krummenacher C., Cohen G. H., Eisenberg R. J., Wiley D. C. ( 2001) Mol. Cell 8, 169– 179 [DOI] [PubMed] [Google Scholar]
- 12.Krummenacher C., Supekar V. M., Whitbeck J. C., Lazear E., Connolly S. A., Eisenberg R. J., Cohen G. H., Wiley D. C., Carfí A. ( 2005) EMBO J. 24, 4144– 4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gianni T., Martelli P. L., Casadio R., Campadelli-Fiume G. ( 2005) J. Virol. 79, 2931– 2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gianni T., Menotti L., Campadelli-Fiume G. ( 2005) J. Virol. 79, 7042– 7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gianni T., Piccoli A., Bertucci C., Campadelli-Fiume G. ( 2006) J. Virol. 80, 2216– 2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galdiero S., Falanga A., Vitiello M., Browne H., Pedone C., Galdiero M. ( 2005) J. Biol. Chem. 280, 28632– 28643 [DOI] [PubMed] [Google Scholar]
- 17.Galdiero S., Vitiello M., D'Isanto M., Falanga A., Collins C., Raieta K., Pedone C., Browne H., Galdiero M. ( 2006) J. Gen. Virol. 87, 1085– 1097 [DOI] [PubMed] [Google Scholar]
- 18.Galdiero S., Falanga A., Vitiello M., Raiola L., Fattorusso R., Browne H., Pedone C., Isernia C., Galdiero M. ( 2008) J. Biol. Chem. 283, 29993– 30009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gianni T., Fato R., Bergamini C., Lenaz G., Campadelli-Fiume G. ( 2006) J. Virol. 80, 8190– 8198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galdiero S., Falanga A., Vitiello M., D'Isanto M., Cantisani M., Kampanaraki A., Benedetti E., Browne H., Galdiero M. ( 2008) Peptides 29, 1461– 1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heldwein E. E., Lou H., Bender F. C., Cohen G. H., Eisenberg R. J., Harrison S. C. ( 2006) Science 313, 217– 220 [DOI] [PubMed] [Google Scholar]
- 22.Roche S., Bressanelli S., Rey F. A., Gaudin Y. ( 2006) Science 313, 187– 191 [DOI] [PubMed] [Google Scholar]
- 23.Satoh T., Arii J., Suenaga T., Wang J., Kogure A., Uehori J., Arase N., Shiratori I., Tanaka S., Kawaguchi Y., Spear P. G., Lanier L. L., Arase H. ( 2008) Cell 132, 935– 944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farnsworth A., Wisner T. W., Webb M., Roller R., Cohen G., Eisenberg R., Johnson D. C. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 10187– 10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian R. P., Geraghty R. J. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 2903– 2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campadelli-Fiume G., Menotti L. ( 2007) Human Herpesviruses Biology, Terapy, and Immunoprophylaxis ( Arivin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., Yamanishi K. eds), Vol. 1, pp. 93– 111, Cambridge Press, Cambridge, UK: [PubMed] [Google Scholar]
- 27.Lazear E., Carfi A., Whitbeck J. C., Cairns T. M., Krummenacher C., Cohen G. H., Eisenberg R. J. ( 2008) J. Virol. 82, 700– 709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Romero P., Perez A., Capul A., Montgomery R., Fuller A. O. ( 2005) J. Virol. 79, 4540– 4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atanasiu D., Whitbeck J. C., Cairns T. M., Reilly B., Cohen G. H., Eisenberg R. J. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 18718– 18723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avitabile E., Forghieri C., Campadelli-Fiume G. ( 2007) J. Virol. 81, 11532– 11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillet L., Stevenson P. G. ( 2007) J. Virol. 81, 13082– 13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirschner A. N., Lowrey A. S., Longnecker R., Jardetzky T. S. ( 2007) J. Virol. 81, 9216– 9229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patrone M., Secchi M., Bonaparte E., Milanesi G., Gallina A. ( 2007) J. Virol. 81, 11479– 11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ejercito P. M., Kieff E. D., Roizman B. ( 1968) J. Gen. Virol. 2, 357– 364 [DOI] [PubMed] [Google Scholar]
- 35.Ligas M. W., Johnson D. C. ( 1988) J. Virol. 62, 1486– 1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai W. Z., Person S., Warner S. C., Zhou J. H., DeLuca N. A. ( 1987) J. Virol. 61, 714– 721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forrester A., Farrell H., Wilkinson G., Kaye J., Davis-Poynter N., Minson T. ( 1992) J. Virol. 66, 341– 348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roop C., Hutchinson L., Johnson D. C. ( 1993) J. Virol. 67, 2285– 2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stefan A., Secchiero P., Baechi T., Kempf W., Campadelli-Fiume G. ( 1997) J. Virol. 71, 5758– 5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Showalter S. D., Zweig M., Hampar B. ( 1981) Infect. Immun. 34, 684– 692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brandimarti R., Huang T., Roizman B., Campadelli-Fiume G. ( 1994) Proc. Natl. Acad. Sci. U. S. A. 91, 5406– 5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baines J. D., Roizman B. ( 1993) J. Virol. 67, 1441– 1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avitabile E., Lombardi G., Campadelli-Fiume G. ( 2003) J. Virol. 77, 6836– 6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rovero S., Amici A., Carlo E. D., Bei R., Nanni P., Quaglino E., Porcedda P., Boggio K., Smorlesi A., Lollini P. L., Landuzzi L., Colombo M. P., Giovarelli M., Musiani P., Forni G. ( 2000) J. Immunol. 165, 5133– 5142 [DOI] [PubMed] [Google Scholar]
- 45.Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G. ( 2005) Nucleic Acids Res. 33, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cocchi F., Menotti L., Di Ninni V., Lopez M., Campadelli-Fiume G. ( 2004) J. Virol. 78, 4720– 4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou G., Galvan V., Campadelli-Fiume G., Roizman B. ( 2000) J. Virol. 74, 11782– 11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menotti L., Cocchi F., Campadelli-Fiume G. ( 2002) Virology 301, 6– 12 [DOI] [PubMed] [Google Scholar]
- 49.Pertel P. E., Fridberg A., Parish M. L., Spear P. G. ( 2001) Virology 279, 313– 324 [DOI] [PubMed] [Google Scholar]
- 50.Milne R. S., Hanna S. L., Rux A. H., Willis S. H., Cohen G. H., Eisenberg R. J. ( 2003) J. Virol. 77, 8962– 8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stefan A., De Lillo M., Frascaroli G., Secchiero P., Neipel F., Campadelli-Fiume G. ( 1999) J. Clin. Microbiol. 37, 3980– 3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markert J. M., Gillespie G. Y., Weichselbaum R. R., Roizman B., Whitley R. J. ( 2000) Rev. Med. Virol. 10, 17– 30 [DOI] [PubMed] [Google Scholar]
- 53.Browne H., Bruun B., Whiteley A., Minson T. ( 2003) J. Gen. Virol. 84, 1085– 1089 [DOI] [PubMed] [Google Scholar]
- 54.Zhou G., Roizman B. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 4142– 4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menotti L., Cerretani A., Hengel H., Campadelli-Fiume G. ( 2008) J. Virol. 20, 10153– 10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Para M. F., Baucke R. B., Spear P. G. ( 1980) J. Virol. 34, 512– 520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calistri A., Sette P., Salata C., Cancellotti E., Forghieri C., Comin A., Göttlinger H., Campadelli-Fiume G., Palù G., Parolin C. ( 2007) J. Virol. 81, 11468– 11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torrisi M. R., Di Lazzaro C., Pavan A., Pereira L., Campadelli-Fiume G. ( 1992) J. Virol. 66, 554– 561 [DOI] [PMC free article] [PubMed] [Google Scholar]