Abstract
Cardiolipin (CL) is an anionic membrane lipid present in bacteria, plants, and animals, but absent from archaea. It is generally thought that bacteria use an enzyme belonging to the phospholipase D superfamily as cardiolipin synthase (Cls) catalyzing a reversible phosphatidyl group transfer from one phosphatidylglycerol (PG) molecule to another PG to form CL and glycerol. In contrast, in eukaryotes a Cls of the CDP-alcohol phosphatidyltransferase superfamily uses cytidine diphosphate-diacylglycerol (CDP-DAG) as the donor of the phosphatidyl group, which is transferred to a molecule of PG to form CL. Searching the genome of the actinomycete Streptomyces coelicolor A3(2) we identified a gene coding for a putative Cls of the CDP-alcohol phosphatidyltransferase superfamily (Sco1389). Here we show that expression of Sco1389 in a CL-deficient Rhizobium etli mutant restores CL formation. In an in vitro assay Sco1389 condenses CDP-DAG with PG to form CL and therefore catalyzes the same reaction as eukaryotic cardiolipin synthases. This is the first time that a CDP-alcohol phosphatidyltransferase from bacteria is shown to be responsible for CL formation. The broad occurrence of putative orthologues of Sco1389 among the actinobacteria suggests that CL synthesis involving a eukaryotic type Cls is common in actinobacteria.
Introduction
Cardiolipin (CL)3 is an anionic phospholipid almost ubiquitous in bacteria and throughout the eukaryotic kingdom, which was first isolated from beef heart (1). The presence of cardiolipin seems to be limited to specific energy-transducing membranes, such as the bacterial cytoplasmic membrane, mitochondrial membranes, and the membranes of hydrogenosomes (2). CL presents a number of interesting biophysical properties; for example, in the presence of certain divalent cations CL has the potential to form non-bilayer structures (3). This polymorphic phase behavior could introduce discontinuity into the bilayer structures and enable dynamic membrane functions, such as membrane fusion during cell division, formation of adhesion sites between the inner and outer membrane, integration of proteins into the membrane, and their stabilization within the membrane (4), among others. Another interesting characteristic of CL is that, due to the free hydroxyl group of the central glycerol moiety, the second pK of the headgroup is drastically increased, thereby creating an acid-anion domain. This means that CL might function as a proton trap, playing a role in proton conductance across energy-transducing membranes (5, 6).
In bacteria, CL is thought to be synthesized via a reversible phosphatidyl group transfer from one phosphatidylglycerol (PG) molecule to another PG, yielding glycerol in addition to CL (7) (see Fig. 1). This bacteria-specific transesterification is catalyzed by the enzyme cardiolipin synthase, i.e. Cls-I, which belongs to the phospholipase D (PLD) superfamily that includes type I phosphatidylserine synthases, poxvirus envelope proteins, phospholipases D, and nucleases (8). In contrast, eukaryotes synthesize CL from cytidine diphosphate-diacylglycerol (CDP-DAG) and PG thereby forming CL and cytidine monophosphate (see Fig. 1). Cls of the so-called “eukaryotic type,” i.e. Cls-II, belongs to the CDP-alcohol phosphatidyltransferase superfamily (7). The latter also includes type II phosphatidylserine synthases (9), phosphatidylcholine synthases (10), phosphatidylglycerolphosphate synthases (11), phosphatidylinositol synthases (12), CDP-choline:1,2-diacylglycerol cholinephosphotransferases (13), and CDP-ethanolamine:1,2-diacylglycerol ethanolaminephosphotransferases (13).
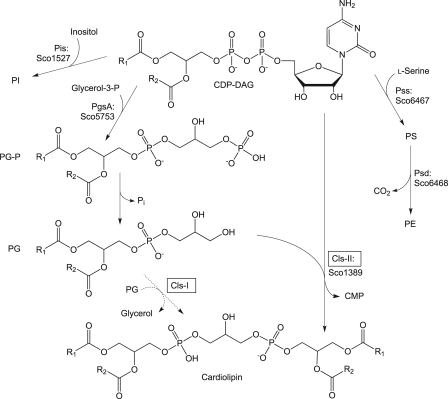
FIGURE 1.
In silico reconstruction of phospholipid biosynthesis in S. coelicolor. The gene products whose functions were assigned or predicted in the present work are indicated. CDP-DAG, cytidine diphosphate-diacylglycerol; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PG, phosphatidylglycerol; PS, phosphatidylserine; PG-P, phosphatidylglycerol 3-phosphate; Pss, phosphatidylserine synthase; Psd, phosphatidylserine decarboxylase; PgsA, phosphatidylglycerol-3-phosphate synthase; Pis, phosphatidylinositol synthase; Cls-I, bacterial type CL synthase; and Cls-II, eukaryotic type CL synthase.
Streptomycetes are high (G+C) Gram-positive soil-dwelling bacteria belonging to the class actinobacteria, which includes several other genera of medical or industrial relevance, such as Mycobacterium and Corynebacterium. Streptomyces species show a complex life-cycle involving growth through a mycelium of branching-hyphal filaments and the formation of reproductive spores. The latter morphological differentiation is often linked to the synthesis of a wide variety of secondary metabolites (14). Thus, species from the genus Streptomyces are of great interest as producers of antibiotics and molecules with other medical and biotechnological applications (15).
Streptomyces coelicolor A3(2) was the first streptomycete with a completely sequenced and annotated genome (16). This information, together with a cumulative body of knowledge on the physiology of Streptomyces (17, 18), allowed a genome-scale metabolic reconstruction for this strain (19). The value and quality of the S. coelicolor genome-scale metabolic reconstruction, which includes 700 genes, 500 metabolites, and 700 reactions, is supported by a computational and experimental network evaluation, a level of sophistication that not many model bacteria have reached (20). Thus, the S. coelicolor metabolic network accounts for all the biosynthetic pathways that are required for the synthesis of the components needed for growth and biomass production (e.g. membranes), albeit the fact that certain network gaps had to be closed (“missing genes”), even in the absence of direct genetic, biochemical, or physiological data. One such example is the synthesis of CL (19), which in Streptomyces hygroscopicus (21) and Mycobacterium tuberculosis (22) accounts for ∼30% of the total phospholipid content.
In this work we aimed to identify a gene responsible for Cls activity in S. coelicolor. By using phylogenomic analyses, Sco1389 belonging to the CDP-alcohol phosphatidyltransferase superfamily was predicted to encode a Cls. We show that Sco1389 from S. coelicolor condenses CDP-DAG with PG yielding CL and therefore catalyzes the same reaction as eukaryotic Cls. This is the first time that a CDP-alcohol phosphatidyltransferase from bacteria is shown to be responsible for CL formation. The broad occurrence of putative orthologues of Sco1389 among the actinobacteria indicates that CL synthesis involving a eukaryotic type Cls seems to be common in actinobacteria (Fig. 1).
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
S. coelicolor A3(2) (16) was grown in YEME liquid medium (3 g of yeast extract, 5 g of Bacto-Peptone, 3 g of Oxoid malt extract, 10 g of glucose, 340 g (34% final) of sucrose, and up to 1000 ml of distilled water) (23) at 30 °C. Rhizobium etli CE3 wild type (24) and mutant R. etli CFNX185 (25) were grown at 30 °C in peptone/yeast extract (PY) medium with CaCl2 in a final concentration of 4.5 mm (26). CFNX185 lacks the gene coding for the predicted Cls due to a large deletion in its plasmid E and does not form CL. Escherichia coli was grown at 37 °C in LB medium. Antibiotics were added to the medium when required to obtain the following final concentrations (mg/liter): nalidixic acid (30), tetracycline (10 and 20 for R. etli and E. coli, respectively), and carbenicillin (100).
Cloning and Expression of Sco1389 in Rhizobium etli
Recombinant DNA techniques were performed according to standard protocols. Sco1389 was amplified from cosmid St1A8A (27). PCR was performed using XL polymerase (Applied Biosystems) and primers incorporating NdeI and BamHI restriction sites (underlined), respectively (5′-GCGCAGCCATATGAGCATCATTGGCTCGTTTC-3′ and 5′-GCGGATCCATGTCGGAAGCGTCCTCCTCTG-3′). After digestion with NdeI and BamHI, the amplified DNA fragment was cloned into the expression vector pET15b (Novagen) to obtain plasmid pMSC01. The sequence of the cloned insert was confirmed by sequencing. Plasmids pET15b and pMSC01 were linearized with BamHI and cloned into the BamHI restriction site of the broad host-range plasmid pRK404 (28) to obtain plasmids pMSC03 and pMSC04, respectively. Both broad host-range plasmids were mobilized into the R. etli mutant strain CFNX185 by triparental mating using the mobilizing plasmid pRK2013 as described previously (29).
Determination of the Membrane Lipid Compositions of S. coelicolor and R. etli
Cultures of S. coelicolor and R. etli (1 ml) were inoculated from fresh overnight cultures to an A620 of 0.1. After addition of 1 μCi of [1-14C]acetate (Amersham Biosciences, 58 mCi/mmol), the cultures were labeled for 24 h. The cells were harvested by centrifugation and resuspended in 100 μl of water. Lipids were extracted according to Bligh and Dyer (30). The chloroform phase was used for lipid analysis on TLC plates (high-performance TLC aluminum sheets, Silica Gel 60, Merck). For one-dimensional TLC, the solvent system used was chloroform-methanol-glacial acetic acid (13:5:2, v/v). For two-dimensional TLC analysis, a chloroform-methanol-water (14:6:1, v/v) mixture was used in the first dimension, and chloroform-methanol-glacial acetic acid (13:5:2, v/v) in the second dimension. The TLCs were exposed to autoradiography film (Kodak) or to a PhosphorImager screen (Amersham Biosciences). The individual lipids were quantified by using a Storm 820 PhosphorImager and ImageQuant software (Amersham Biosciences).
Lipid Extraction and Purification
Cultures (1 liter) of R. etli CE3, CFNX185.pMSC03, and CFNX185.pMSC04 were inoculated from overnight cultures and grown to an A620 of 0.8. Lipids were extracted according to Bligh and Dyer (30), and half of the resulting samples were used to purify the CL-containing fractions using an 8-ml DEAE-cellulose (Whatman DE-52) column with minor modifications to the protocol previously described (31). The column was washed with 16 ml of each of the following: chloroform/methanol/water (2:3:1, v/v), chloroform/methanol/30 mm ammonium acetate in water (2:3:1, v/v), chloroform/methanol/60 mm ammonium acetate in water (2:3:1, v/v), chloroform/methanol/90 mm ammonium acetate in water (2:3:1, v/v), chloroform/methanol/120 mm ammonium acetate in water (2:3:1, v/v), chloroform/methanol/240 mm ammonium acetate in water (2:3:1, v/v), and chloroform/methanol/480 mm ammonium acetate in water (2:3:1, v/v). Fractions of 2–4 ml were recovered, and the CL-containing fraction was identified by iodine staining after separation with one-dimensional TLC. Total lipid extracts and the purified CL were then analyzed by liquid chromatography (LC)-electrospray ionization (ESI)/mass spectrometry.
Mass Spectrometry Analysis
Normal phase LC-ESI/MS of lipids was performed using an Agilent 1200 Quaternary LC system coupled to a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems, Foster City, CA). An Ascentis® Si HPLC column (5 μm, 25 cm × 2.1 mm) was used. Mobile phase A consisted of chloroform/methanol/aqueous ammonium hydroxide (800:195:5, v/v). Mobile phase B consisted of chloroform/methanol/water/aqueous ammonium hydroxide (600:340:50:5, v/v). Mobile phase C consisted of chloroform/methanol/water/aqueous ammonium hydroxide (450:450:95:5, v/v). The elution program consisted of the following: 100% mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 11 min. The LC gradient was then changed to 100% mobile phase C over 3 min and held at 100% C for 3 min, and then returned to 100% A over 0.5 min and held at 100% A for 5 min. The total LC flow rate was 300 μl/min. The post-column splitter diverted ∼10% of the LC flow to the ESI source of the Q-Star XL mass spectrometer, with MS settings as follows: IS = −4500 V, CUR = 20 p.s.i., GS1 = 20 p.s.i., DP = −55 V, and FP = −150 V. Nitrogen was used as the collision gas for MS/MS experiments. Data acquisition and analysis were performed using the instrument's Analyst QS software.
Preparation of R. etli Membrane Extracts
Cell-free extracts were prepared with some modifications to the method described previously (32). Cultures (1 liter) of mutant or wild-type strains were inoculated from an overnight 20-ml culture, grown to an A620 of 0.8, and harvested by centrifugation for 20 min at 3200 × g. The pellets were washed with 100 ml of suspension buffer (0.1 m Tris/HCl, pH 8) and then resuspended in an 8-ml total volume of the same buffer. The cell suspension was passed three times through a French pressure cell press at 20,000 lb/in2. Cell debris and unbroken cells were removed by centrifugation at 2,000 × g for 10 min. The supernatant was recovered and centrifuged for 90 min at 100,000 × g to spin down the membranes. The pellet obtained was resuspended in 2 ml of suspension buffer, aliquoted, and stored at −20 °C. The protein concentration was determined by the method of Dulley and Grieve (33).
Determination of Cls Activity
[32P]CDP-1,2-diacyl-sn-glycerol was synthesized as described previously (34) using a two-step enzymatic conversion. First, diacylglycerol kinase (Calbiochem, 12 units/mg of protein) was used to prepare 32P-labeled phosphatidic acid from 1,2-dipalmitoyl-sn-glycerol and [γ-32P]ATP (Amersham Biosciences, 10 mCi/ml, ∼3000 Ci/mmol). Then, 32P-labeled phosphatidic acid was converted to [32P]CDP-diacylglycerol using CTP and the CDP-diglyceride synthase reaction. The enzymatic assays were performed with TLC-purified samples using chloroform/methanol/acetic acid/water (32:4:5:1, v/v) as the solvent system. The [32P]CDP-DAG had a specific radioactivity of 5.04 × 106 cpm/nmol. The in vitro conversion of [32P]CDP-DAG and PG into CL was assayed using cell-free membrane extracts from R. etli CE3, R. etli CFNX185 mutant carrying the pMSC03 vector, and R. etli CFNX185 harboring the plasmid pMSC04. The standard reaction mixture contained, in a total volume of 50 μl in Eppendorf tubes, 50 μg of protein, 50 mm Tris/HCl, pH 7.5, 10 mm CoSO4, 50 μm [32P]CDP-DAG (6.3 × 104 cpm/nmol), 50 μm PG, and 0.05% (w/v) Triton X-100. In some assays CoSO4 was replaced with 10 mm MgSO4 or 10 mm MnSO4. The reaction mixtures were incubated for 60 min in a water bath at 30 °C, and reactions were stopped by adding 425 μl of methanol/chloroform/water (2:1:0.4, v/v). Phase separation was achieved by addition of 125 μl of chloroform and 125 μl of water, and after washing the chloroform phase once with 200 μl of water, the lipids in the chloroform phase were separated by one-dimensional TLC using chloroform-methanol-glacial acetic acid (13:5:2, v/v) as solvent system. Incorporation of 32P into CL was measured by quantifying the intensity of the CL spot using a Storm PhosphorImager and ImageQuant, and the intensity units were later converted to Bq by interpolation of a [γ-32P]ATP standard curve.
Phylogenetic Analyses
The sequences of putative genes encoding for CDP-alcohol phosphatidyltransferases were retrieved by PSI-BLAST (35) using Sco1389 as query and confirmed to have this domain using the PROSITE (36) and Pfam (37) databases. The sequence alignment was performed with the multiple alignment software MUSCLE (38) using the default parameters and then edited with the Jalview alignment editor (39). The phylogenetic tree was constructed via the PHYML web server (40), using the LG amino acid substitution model, four substitution rate categories, and estimating the topology, branch lengths, rate parameters, the proportion of invariable sites, and the Gamma distribution parameter. To support the bipartitions, 100 bootstrap replicas were performed.
RESULTS
S. coelicolor Membranes Contain Cardiolipin
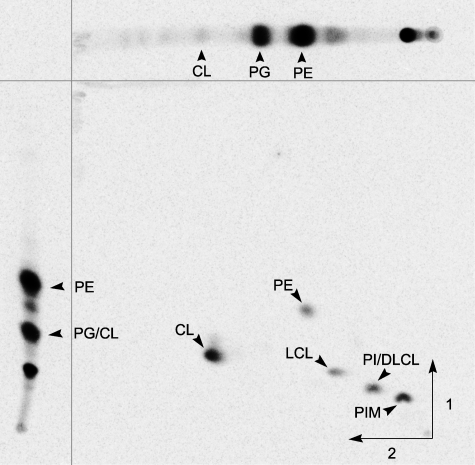
The presence of CL had been described earlier in S. hygroscopicus (21). To verify that CL is also present in S. coelicolor total lipids were labeled with [14C]acetate, extracted, and then separated by two-dimensional TLC (Fig. 2). A major spot that migrates similar to cardiolipin was identified by comparison with the known lipid profile of Sinorhizobium meliloti (10, 32) and with commercial standards. A quantification of the lipids present in the membrane of S. coelicolor shows that CL accumulates to ∼30% of total lipids (data not shown), which is similar to what has been described for M. tuberculosis (22) and S. hygroscopicus (21). The putative identity of the remaining lipids of S. coelicolor was assigned by comparison with the published migration pattern of lipids from S. hygroscopicus (21) (Fig. 2). Interestingly, in agreement with the lipid composition previously determined for the above-mentioned actinomycetes (21, 22), S. coelicolor does not accumulate PG.
FIGURE 2.
S. coelicolor lipid profile: separation of [14C]acetate-labeled lipids from S. coelicolor by two-dimensional TLC. As a reference to facilitate the identification of CL, labeled lipids from S. meliloti 1021 were run in one dimension only in each of the two solvent systems. The CL spot was identified as the intersection of the corresponding spots in S. meliloti. Lysocardiolipin (LCL), dilysocardiolipin (DLCL), phosphatidylinositol (PI), and phosphatidylinositol mannoside (PIM) were inferred from the data published by Hoischen et al. (21) for S. hygroscopicus.
In Silico Functional Genomics of cls Genes in S. coelicolor
After having confirmed the presence of CL in S. coelicolor, the genome of this organism was searched for genes coding for putative cls genes. Because bacteria are expected to have only Cls enzymes of the PLD type, a PLD-like Cls sequence was first used as query. Standard sequence searches using the cls gene from E. coli and Bacillus subtilis failed to recognize a “clear-cut” homologue (>25% sequence identity) in S. coelicolor. Earlier reports had speculated about the presence of a eukaryote-like Cls belonging to the CDP-alcohol phosphatidyltransferase family in M. tuberculosis and M. smegmatis (19, 22, 41). Eight genes encoding for putative CDP-alcohol phosphatidyltransferases (Pfam accession: PF01066) (37) were identified in the S. coelicolor genome using PSI-BLAST searches (35): Sco1389, Sco1527, Sco3457, Sco5753, Sco6467, Sco6647, Sco6752, and Sco6755. Seven of these candidates encode proteins contain the motif DGX2ARX8GX3DX3D (PROSITE accession: PS00379) (36), characteristic for this protein superfamily, whereas one protein, i.e. Sco6755, presents a motif that differs in one amino acid.
Four of these genes, i.e. Sco3457, Sco6647, Sco6752, and Sco6755, are poorly conserved among other actinobacteria beyond the genus Streptomyces (data not shown), which makes it improbable that they are involved in cardiolipin biosynthesis; Sco1527 is very similar in sequence (38% identity) and gene context to pgsA1, the phosphatidylinositol synthase gene from M. tuberculosis (22); Sco5753 appears to be a homologue of pgsA3 (43% identity), which has been proposed to encode for a phosphatidylglycerol phosphate synthase (PgsA) in the same organism (22); PssA from B. subtilis (42) is the closest homologue of Sco6467 with an experimentally verified annotation, and in the S. coelicolor genome it is located downstream of a gene encoding a putative phosphatidylserine decarboxylase (i.e. Sco6468), suggesting that the product of Sco6467 is a type II phosphatidylserine synthase involved in the biosynthesis of phosphatidylserine. Therefore, it was hypothesized that the remaining gene, i.e. Sco1389, might code for a Cls-II in S. coelicolor, implying that in this organism PG and CDP-DAG are used as substrates for CL biosynthesis.
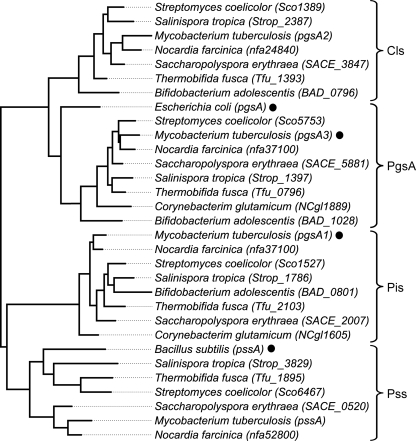
Phylogenetic analyses involving homologues of these genes are consistent with this functional annotation (Fig. 3). The sequences compared cluster in four well supported clades represented by phosphatidylglycerol phosphate synthases (i.e. Sco5753), phosphatidylserine synthases (i.e. Sco6467), phosphatidylinositol synthases (i.e. Sco1527), and the so-called “eukaryote-like” cardiolipin synthases, or Cls-II (i.e. Sco1389).
FIGURE 3.
Phylogenetic tree of proteins from the CDP-alcohol phosphatidyltransferase family present in actinobacteria. Sequences with experimental evidence for their activities are indicated (●): PgsA, phosphatidylglycerol-3-phosphate synthases; Pis, phosphatidylinositol synthases; Pss, phosphatidylserine synthases; and Cls, cardiolipin synthases.
Sco1389 Encodes a Cardiolipin Synthase
CL is formed in low amounts in E. coli cells during logarithmic growth and is accumulating to higher concentrations during stationary growth phase. We wanted to find out if expression of the putative Cls Sco1389 in E. coli would cause an increase in CL formation in E. coli. Sco1389 was amplified by PCR and cloned into the expression vector pET15b to obtain pMSC01. Plasmids pET15b and pMSC01 were used to transform E. coli BL21(DE3) (43), and transformants were grown in complex medium in the presence of [14C]acetate. Total lipids were extracted as mentioned before and subsequently separated by TLC. The amount of CL almost doubled from 4.6% in the wild type to 8.1% in the strain expressing the S. coelicolor gene, whereas the percentage of the rest of the lipids remained almost unaffected (data not shown). To get further support that Sco1389 encodes a cardiolipin synthase activity, Sco1389 was cloned into the broad host range vector pRK404 (28). This construct and a vector control were mobilized by conjugation into the CL-deficient Rhizobium etli mutant CFNX185 (25). The latter strain lacks the only cls homologue (gi: 3895503) identified in this organism due to a deletion of a part of its p42E plasmid.
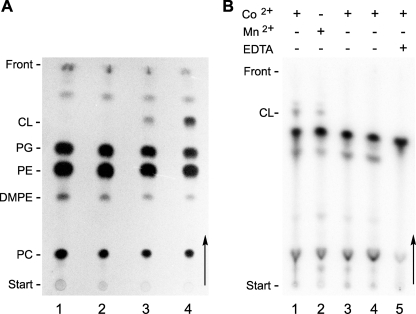
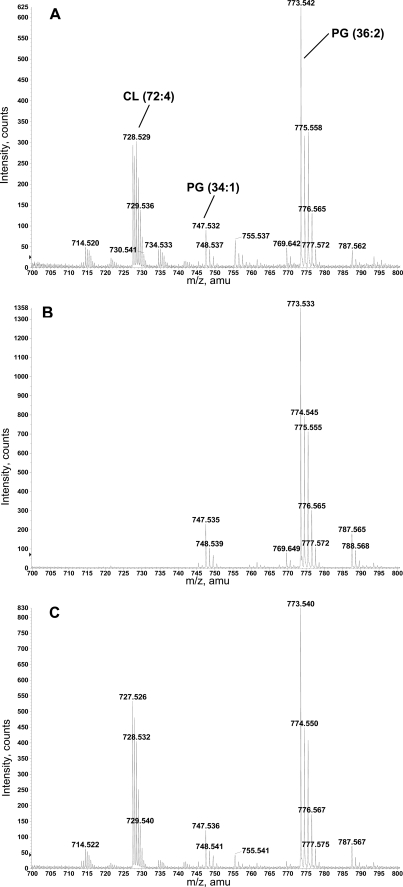
The membrane lipid composition of R. etli CE3 wild type, CFNX185, and CFNX185 derivatives harboring different plasmids was analyzed. Whereas no CL was detected in the mutant CFNX185 and in CFNX185 harboring the empty plasmid, a lipid migrating as CL was detected in the wild-type CE3, and more importantly, in the mutant CFNX185 expressing Sco1389 (Fig. 4A). The presence of CL in the lipid extracts was confirmed by LC/MS analysis of the total lipid extracts of CE3 and CFNX185.pMSC04 (Fig. 5, A and B). The major CL species, detected as [M-2H]2− at m/z 727.5 in the negative ion mode, contains in its fatty acid portion a total of 72 carbons and 4 double bonds, which probably translates into 4 lactobacillic acid residues. In comparison, no CL was detected in total lipids extracted from cultures of CFNX185.pMSC03 grown and extracted under identical conditions (Fig. 5C). CL was also purified from the total lipid extract of CE3 and CFNX185.pMSC04 by ion exchange chromatography and detected using LC/MS. No CL was detected in the corresponding lipid fractions isolated from CFNX185.pMSC03 (data not shown). These results demonstrate unambiguously that Sco1389 encodes for cardiolipin synthase activity in vivo.
FIGURE 4.
Sco1389 synthesizes CL using CDP-DAG as a substrate. A, expression of the Cls Sco1389 in the CL-deficient R. etli mutant CFNX185. Lipids of R. etli strains were radiolabeled with [14C]acetate during growth in complex medium, separated by one-dimensional TLC, and visualized by autoradiography. The following strains were analyzed: CL-deficient R. etli mutant CFNX185 (lane 1), CFNX185.pMSC03 (empty vector, lane 2), CFNX185.pMSC04 (Sco1389-expressing, lane 3), R. etli wild-type CE3 (lane 4). B, in vitro assays for the transfer of [32P]phosphatidate from [32P]CDP-DAG into CL. Autoradiography of a TLC of lipid products obtained from Cls enzymatic assays using membrane extracts of different R. etli strains. Cls activity assays were performed with membrane extracts of the strains CFNX185.pMSC04 (lanes 1, 2, and 5), CFNX185 (lane 3), and R. etli wild-type CE3 (lane 4) were incubated with 50 μm [32P]CDP-DAG, 50 μm PG, 0.05% Triton X-100 and 10 mm CoSO4 or MnSO4, 20 mm EDTA were added when indicated. CL was identified with commercial standards. CL, cardiolipin; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; MMPE, monomethyl phosphatidylethanolamine; DMPE, dimethyl phosphatidylethanolamine; and PC, phosphatidylcholine.
FIGURE 5.
Total lipid extracts from R. etli CE3 wild type (A), CFNX185.pMSC03 (CL-deficient mutant with empty plasmid) (B), and CFNX185.pMSC04 (CL-deficient mutant expressing Sco1389) (C) were analyzed by normal phase LC/ESI-MS for the presence of CL. The major CL species (with 72 carbons and 4 double bonds) in A and C are detected as [M-2H]2− ions at m/z 727.5 in the negative ion mode. Major PG species detected in all three samples are PG (34:1) and PG (36:2), and the corresponding peaks are labeled in A. The mass spectra were averaged from the spectra acquired during 15–16 min during the LC/MS.
Sco1389 Uses CDP-DAG and PG as Substrates
The fact that Sco1389 is a member of the CDP-alcohol phosphatidyltransferase family implies that this enzyme uses PG and CDP-DAG as substrates. Because the putative protein product of Sco1389 is predicted to contain four transmembranal helices, membrane extracts of CFNX185.pMSC03 and CFNX185.pMSC04 were assayed in the presence of the substrates PG and [32P]CDP-DAG. As expected, no CL formation was observed using an extract obtained from CFNX185.pMSC03, whereas the membrane extract obtained from CFNX185.pMSC04 was able to incorporate [32P]phosphatidate, from [32P]CDP-DAG, into CL (Fig. 4B). In agreement with this, a membrane protein extract from the wild-type strain CE3, which has a Cls-I enzyme but lacks a Cls-II enzyme, could not synthesize CL from these substrates.
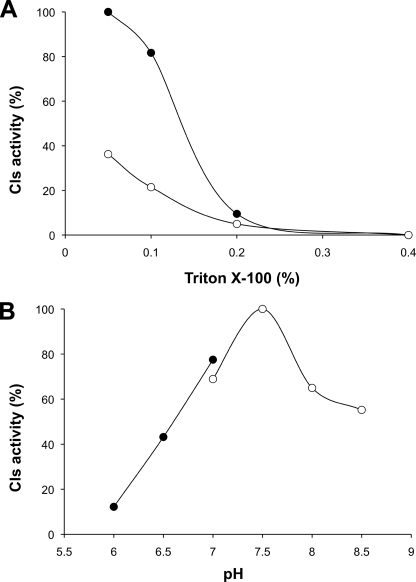
Because the membrane extracts already contain substantial amounts of PG, cardiolipin synthase activity was detected even without addition of exogenous PG. However, incorporation of [32P]phosphatidate from [32P]CDP-DAG into CL was clearly increased by addition of 50 μm PG (Fig. 6A). Triton X-100 had to be added to all reactions to solubilize the lipid mixture and make it accessible to the membrane-bound Cls. High concentrations of Triton X-100 have proven to decrease the formation of product by other enzymes of this family (34), probably due to substrate dilution. Therefore, the effect of the Triton X-100 concentration on the standard Sco1389 assay was studied. Under our assay conditions there was a strong Triton X-100 dependence of the Cls activity with a maximal activity at 0.05% (w/v) and at concentrations higher than 0.2%, Triton X-100 clearly interfered with Sco1389-dependent CL formation (Fig. 6A).
FIGURE 6.
Effects of pH and Triton X-100 on Sco1389 activity. A, cardiolipin synthase activity was assayed at the indicated Triton X-100 concentrations with 50 μm PG (●) or without exogenous PG (○). B, cardiolipin synthase activity was assayed at the indicated pH values with 50 mm BisTris/HCl (●) or Tris/HCl (○). Otherwise, conditions of the standard assay were used, and activity was measured as described in the text.
Ion Dependence and pH Optimum of Sco1389 Cls Activity
Because the activities of Cls of eukaryotic origin that have been studied previously depend on the presence of bivalent cations (44–46), the influence of different bivalent cations on CL formation catalyzed by Sco1389 was tested. The largest activity was found with Co2+, whereas Mn2+ supported a lower activity; there was no detectable cardiolipin synthase activity when using Ni2+, Ca2+, or Mg2+ (Table 1). The addition of 20 mm EDTA to the reaction mix caused the loss of Cls activity, confirming the metal dependence of Sco1389 (Fig. 4B and Table 1). Furthermore, Cls activity was measured at pH 6.0–7.0 (BisTris/HCl buffer) and at pH 7.0–9.0 (Tris/HCl buffer). The largest activity was observed using Tris/HCl at pH 7.5, and the activity decreased as the pH was raised or lowered (Fig. 6B).
TABLE 1.
Cation dependency of the S. coelicolor Cls in vitro
The effect of selected bivalent cations on [32P]phosphatidate transfer from [32P]CDP-DAG into CL was tested with membrane extracts from CFNX185.pMSC04. The reaction mixture contained CDP-DAG, PG, Triton X-100, Tris/HCl buffer (pH 7.5), the indicated ion at 10 mm, and, when added, EDTA at 20 mm.
| Added cation | CLS activity |
|---|---|
| % | |
| Co2+ | 100 |
| Mn2+ | 38 |
| Mg2+ | 7.2 |
| Ni2+ | 2.4 |
| Ca2+ | 0.6 |
| Co2+, EDTA | 1.8 |
| – | 2.1 |
DISCUSSION
Cardiolipin is a membrane lipid present in organisms from bacteria to plants and animals, but is absent from archaea. The precursor of cardiolipin is phosphatidylglycerol, which is universally synthesized from CDP-DAG and glycerolphosphate with phosphatidylglycerolphosphate as the intermediate. However, eukaryotes and prokaryotes are thought to employ different reactions to convert PG into CL. In bacteria, a phosphatidyl group is transferred from one PG to another PG to form CL, whereas in eukaryotes, CDP-DAG is the donor of the phosphatidyl group which is transferred to a molecule of PG to form CL. It is generally thought that prokaryotes use a protein belonging to the PLD superfamily as Cls, whereas in eukaryotic organisms a Cls of the CDP-alcohol phosphatidyltransferase superfamily is present.
Characterization of several PLD enzymes isolated from cultures of Streptomyces species has been previously reported (47, 48). All of these enzymes appear to be bona fide PLD, exhibiting specificity for a broad range of substrates, and an inherent catalytic promiscuity, which includes the ability to synthesize cardiolipin in vitro. The genome of S. coelicolor contains a gene belonging to the PLD superfamily, i.e. Sco7081. However, the lack of conservation of this gene across the actinomycetes suggests that Sco7081 is unlikely to encode a real Cls. In contrast, eight genes encoding for putative CDP-alcohol phosphatidyltransferases were identified in the genome of S. coelicolor. The function of four of these eight candidate genes could be tentatively assigned after phylogenomic analyses (Fig. 3), including phosphatidylinositol synthase (i.e. Sco1527), type II phosphatidylserine synthase (i.e. Sco6467), phosphatidylglycerolphosphate synthase (i.e. Sco5753), and cardiolipin synthase (i.e. Sco1389), the implication being that in S. coelicolor CL is synthesized from PG and CDP-DAG (Fig. 1). Interestingly, the clade in which Sco1389 is present contains genes from most actinobacteria, implying a broad occurrence of a eukaryote-like Cls in this family of organisms. An exception to this rule is Corynebacterium glutamicum, which has been shown to have a Cls from the PLD family (49) and to lack an orthologue of Sco1389 (data not shown). From an evolutionary standpoint, moreover, the occurrence of a eukaryote-like Cls in bacteria is in agreement with the endosymbiotic origin of mitochondria from independent prokaryotic cells, as it has been previously suggested for hydrogenosomes and mitochondria (7). Under the light of our results it might be worthwhile to search bacterial genome sequences for genes encoding orphan CDP-alcohol phosphatidyltransferases with unknown function to find out if the eukaryote-like pathway for CL biosynthesis is restricted to actinobacteria or is more common in bacteria.
This is the first time that a CDP-alcohol phosphatidyltransferase from bacteria is shown to be responsible for CL formation. In the in vitro enzyme assays that were established (Fig. 4) no CL formation could be observed when the native Cls-I from R. etli CE3 was present; in contrast, CL formation was clearly catalyzed by Sco1389 expressed from R. etli from PG and CDP-DAG. Furthermore, our initial characterization of the Cls encoded by Sco1389 shows similarity in its enzymatic properties to other proteins from the same family (34) and to previously studied eukaryotic cardiolipin synthases (44, 45, 50), such as dependence of the enzyme activity on the presence of specific bivalent cations and substrate dilution kinetics at increasing concentrations of Triton X-100. All Cls characterized so far require either Mg2+, Mn2+, or Co2+ as cofactor; for example, in the case of mammalian Cls-II Co2+ works best (46), whereas the Cls from Saccharomyces cerevisiae prefers Mg2+ over Co2+ (44). In the case of Sco1389 the enzyme works with Co2+ and Mn2+, but no activity was observed with Mg2+.
An obvious experiment to show the function of Sco1389 in vivo is to construct a S. coelicolor mutant deficient in Sco1389 and to compare CL formation in mutant and wild type. Unfortunately, a S. coelicolor mutant deficient in Sco1389 could not be obtained following well established Streptomyces mutagenesis protocols, suggesting an essential role for this gene (data not shown).
Genes responsible for CL biosynthesis have been described from several bacteria such as B. subtilis (51) and E. coli (7), and mutants deficient in Cls have been described in some of these organisms. In most of these mutants, trace amounts of CL were still detectable, indicating that a second enzyme activity, responsible for the formation of CL, is still present. It has been speculated that, in these organisms the enzyme phosphatidylserine synthase is responsible for the residual Cls activity observed. Surprisingly, in the R. etli mutant CFNX185 lacking its predicted cls gene due to a deletion, no CL could be detected (Fig. 5). Because the mutant CFNX185 still carries a functional Pss, a role for Pss in CL biosynthesis can be excluded in this case.
The reason why actinobacteria use CDP-DAG and PG as substrates for CL synthesis, instead of the simpler PG transesterification used by other bacteria, has yet to be addressed. The transesterification reaction used by bacteria is a near-equilibrium reaction that is mainly controlled by substrate availability, whereas CL synthesis in eukaryotes is performed by an irreversible reaction that involves the cleavage of a high energy anhydride bond from CDP-DAG (52).
Apparently, most bacteria studied so far synthesize CL using a Cls-I and tend to accumulate large amounts of PG in addition to smaller amounts of CL. In contrast, looking at the membrane lipid composition of the studied actinobacteria (21, 22, 41), which we predicted to have Cls-II, CL seems to be the major membrane lipid in all cases, and it is obvious that in any of the cases significant amounts of PG are present. From a thermodynamic viewpoint a membrane lipid composition such as present in actinobacteria cannot be achieved using the transesterification reaction that is predominantly present in bacteria. To shift the equilibrium of the reaction almost completely toward the product CL as in actinobacteria, the reaction has to proceed with a release of significant amounts of Gibbs free energy in the form of the cleavage of the acid anhydride bond of CDP-DAG. It is not clear if the accumulation of CL and the lack of PG are simply a result of the distinct CL biosynthesis pathway used in actinobacteria or if this apparently unusual membrane composition reflects some physiological need of the latter.
Thus, the use of CDP-DAG and PG for CL biosynthesis by actinobacteria may implicate a tighter control of CL synthesis. It is therefore anticipated that study of CL biosynthesis in actinobacteria may eventually lead to a better understanding of the role of CL in bacterial and mitochondrial membrane dynamics.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-069338 (a LIPID MAPS glue grant to Z. G. and the mass spectrometry facility in the Department of Biochemistry, Duke University Medical Center). This work was also supported by the Consejo Nacional de Ciencia y Tecnologia, Mexico (Grants 46020-N, 82614, and 82319) and Dirección General de Asuntos del Personal Académico/Universidad Nacional Autónoma de México (Grant IN217907).
- CL
- cardiolipin
- PG
- phosphatidylglycerol
- CDP-DAG
- cytidine diphosphate-diacylglycerol
- LCL
- lysocardiolipin
- DLCL
- dilysocardiolipin
- PI
- phosphatidylinositol
- PIM
- phosphatidylinositol mannoside
- Cls
- cardiolipin synthase
- TLC
- thin-layer chromatography
- MS
- mass spectrometry
- PLD
- phospholipase D
- LC
- liquid chromatography
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Pangborn M. C. ( 1942) J. Biol. Chem. 143, 247– 256 [Google Scholar]
- 2.de Andrade Rosa I., Einicker-Lamas M., Roney Bernardo R., Previatto L. M., Mohana-Borges R., Morgado-Díaz J. A., Benchimol M. ( 2006) Eukaryot. Cell 5, 784– 787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasilenko I., De Kruijff B., Verkleij A. J. ( 1982) Biochim. Biophys. Acta 684, 282– 286 [DOI] [PubMed] [Google Scholar]
- 4.Dowhan W. ( 1997) Annu. Rev. Biochem. 66, 199– 232 [DOI] [PubMed] [Google Scholar]
- 5.Kates M., Syz J. Y., Gosser D., Haines T. H. ( 1993) Lipids 28, 877– 882 [DOI] [PubMed] [Google Scholar]
- 6.Haines T. H., Dencher N. A. ( 2002) FEBS Lett. 528, 35– 39 [DOI] [PubMed] [Google Scholar]
- 7.Nishijima S., Asami Y., Uetake N., Yamagoe S., Ohta A., Shibuya I. ( 1988) J. Bacteriol. 170, 775– 780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting C. P., Kerr I. D. ( 1996) Protein Sci. 5, 914– 922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sohlenkamp C., de Rudder K. E., Geiger O. ( 2004) J. Bacteriol. 186, 1667– 1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohlenkamp C., de Rudder K. E., Rohrs V., Lopez-Lara I. M., Geiger O. ( 2000) J. Biol. Chem. 275, 18919– 18925 [DOI] [PubMed] [Google Scholar]
- 11.Gopalakrishnan A. S., Chen Y. C., Temkin M., Dowhan W. ( 1986) J. Biol. Chem. 261, 1329– 1338 [PubMed] [Google Scholar]
- 12.Nikawa J., Kodaki T., Yamashita S. ( 1987) J. Biol. Chem. 262, 4876– 4881 [PubMed] [Google Scholar]
- 13.Hjelmstad R. H., Bell R. M. ( 1991) J. Biol. Chem. 266, 5094– 5103 [PubMed] [Google Scholar]
- 14.Chater K. F. ( 2006) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 361, 761– 768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demain A. L., Adrio J. L. ( 2008) Mol. Biotechnol. 38, 41– 55 [DOI] [PubMed] [Google Scholar]
- 16.Bentley S. D., Chater K. F., Cerdeño-Tárraga A. M., Challis G. L., Thomson N. R., James K. D., Harris D. E., Quail M. A., Kieser H., Harper D., Bateman A., Brown S., Chandra G., Chen C. W., Collins M., Cronin A., Fraser A., Goble A., Hidalgo J., Hornsby T., Howarth S., Huang C. H., Kieser T., Larke L., Murphy L., Oliver K., O'Neil S., Rabbinowitsch E., Rajandream M. A., Rutherford K., Rutter S., Seeger K., Saunders D., Sharp S., Squares R., Squares S., Taylor K., Warren T., Wietzorrek A., Woodward J., Barrell B. G., Parkhill J., Hopwood D. A. ( 2002) Nature 417, 141– 147 [DOI] [PubMed] [Google Scholar]
- 17.Barona-Gómez F., Hodgson D. A. ( 2003) EMBO Rep. 4, 296– 300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgson D. A. ( 2000) Adv. Microb. Physiol. 42, 47– 238 [DOI] [PubMed] [Google Scholar]
- 19.Borodina I., Krabben P., Nielsen J. ( 2005) Genome Res. 15, 820– 829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed J. L., Famili I., Thiele I., Palsson B. O. ( 2006) Nat. Rev. Genet. 7, 130– 141 [DOI] [PubMed] [Google Scholar]
- 21.Hoischen C., Gura K., Luge C., Gumpert J. ( 1997) J. Bacteriol. 179, 3430– 33436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson M., Crick D. C., Brennan P. J. ( 2000) J. Biol. Chem. 275, 30092– 30099 [DOI] [PubMed] [Google Scholar]
- 23.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. ( 2000) Practical Streptomyces Genetics, p. 412, The John Innes Foundation, Norwich, UK [Google Scholar]
- 24.Noel K. D., Sanchez A., Fernandez L., Leemans J., Cevallos M. A. ( 1984) J. Bacteriol. 158, 148– 155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brom S., García de los Santos A., Stepkowsky T., Flores M., Dávila G., Romero D., Palacios R. ( 1992) J. Bacteriol. 174, 5183– 5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beringer J. E. ( 1974) J. Gen. Microbiol. 84, 188– 198 [DOI] [PubMed] [Google Scholar]
- 27.Redenbach M., Kieser H. M., Denapaite D., Eichner A., Cullum J., Kinashi H., Hopwood D. A. ( 1996) Mol. Microbiol. 21, 77– 96 [DOI] [PubMed] [Google Scholar]
- 28.Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. ( 1985) Plasmid 13, 149– 153 [DOI] [PubMed] [Google Scholar]
- 29.Ruvkun G. B., Ausubel F. M. ( 1981) Nature 289, 85– 88 [DOI] [PubMed] [Google Scholar]
- 30.Bligh E. G., Dyer W. J. ( 1959) Can. J. Biochem. Physiol. 37, 911– 917 [DOI] [PubMed] [Google Scholar]
- 31.Raetz C. R., Purcell S., Meyer M. V., Qureshi N., Takayama K. ( 1985) J. Biol. Chem. 260, 16080– 16088 [PubMed] [Google Scholar]
- 32.de Rudder K. E., Thomas-Oates J. E., Geiger O. ( 1997) J. Bacteriol. 179, 6921– 6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dulley J. R., Grieve P. A. ( 1975) Anal. Biochem. 64, 136– 141 [DOI] [PubMed] [Google Scholar]
- 34.de Rudder K. E., Sohlenkamp C., Geiger O. ( 1999) J. Biol. Chem. 274, 20011– 20016 [DOI] [PubMed] [Google Scholar]
- 35.Schäffer A. A., Aravind L., Madden T. L., Shavirin S., Spouge J. L., Wolf Y. I., Koonin E. V., Altschul S. F. ( 2001) Nucleic Acids Res. 29, 2994– 3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hulo N., Bairoch A., Bulliard V., Cerutti L., Cuche B. A., de Castro E., Lachaize C., Langendijk-Genevaux P. S., Sigrist C. J. ( 2008) Nucleic Acids Res. 36, D245– D249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. ( 2008) Nucleic Acids Res. 36, D281– D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar R. C. ( 2004) Nucleic Acids Res. 32, 1792– 1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. ( 2009) Bioinformatics 25, 1189– 1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guindon S., Gascuel O. ( 2003) Syst. Biol. 52, 696– 704 [DOI] [PubMed] [Google Scholar]
- 41.Mathur A. K., Murthy P. S., Saharia G. S., Venkitasubramanian T. A. ( 1975) Can. J. Microbiol. 22, 354– 358 [DOI] [PubMed] [Google Scholar]
- 42.Okada M., Matsuzaki H., Shibuya I., Matsumoto K. ( 1994) J. Bacteriol. 176, 7456– 7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. ( 1990) Methods Enzymol. 185, 60– 89 [DOI] [PubMed] [Google Scholar]
- 44.Tamai K. T., Greenberg M. L. ( 1990) Biochim. Biophys. Acta 1046, 214– 222 [DOI] [PubMed] [Google Scholar]
- 45.Nowicki M., Müller F., Frentzen M. ( 2005) FEBS Lett. 579, 2161– 2165 [DOI] [PubMed] [Google Scholar]
- 46.Hostetler K. Y., Galesloot J. M., Boer P., Van Den Bosch H. ( 1975) Biochim. Biophys. Acta 380, 382– 389 [DOI] [PubMed] [Google Scholar]
- 47.‘D’Arrigo P., De Ferra L., Pedrocchi-Fantoni G., Scarcelli D., Servi S., Strini A. ( 1996) J. Chem. Soc., Perkin Trans. I, 2657– 2660 [Google Scholar]
- 48.Piazza G. J., Marmer W. N. ( 2007) J. Am. Oil Chem. Soc. 84, 645– 651 [Google Scholar]
- 49.Nampoothiri K. M., Hoischen C., Bathe B., Möckel B., Pfefferle W., Krumbach K., Sahm H., Eggeling L. ( 2002) Appl. Microbiol. Biotechnol. 58, 89– 96 [DOI] [PubMed] [Google Scholar]
- 50.Schlame M., Hostetler K. Y. ( 1991) J. Biol. Chem. 266, 22398– 22403 [PubMed] [Google Scholar]
- 51.Kawai F., Shoda M., Harashima R., Sadaie Y., Hara H., Matsumoto K. ( 2004) J. Bacteriol. 186, 1475– 1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlame M. ( 2008) J. Lipid Res. 49, 1607– 1620 [DOI] [PMC free article] [PubMed] [Google Scholar]