Abstract
The RIG-I like receptor (RLR) comprises three homologues: RIG-I (retinoic acid-inducible gene I), MDA5 (melanoma differentiation-associated gene 5), and LGP2 (laboratory of genetics and physiology 2). Each RLR senses different viral infections by recognizing replicating viral RNA in the cytoplasm. The RLR contains a conserved C-terminal domain (CTD), which is responsible for the binding specificity to the viral RNAs, including double-stranded RNA (dsRNA) and 5′-triphosphated single-stranded RNA (5′ppp-ssRNA). Here, the solution structures of the MDA5 and LGP2 CTD domains were solved by NMR and compared with those of RIG-I CTD. The CTD domains each have a similar fold and a similar basic surface but there is the distinct structural feature of a RNA binding loop; The LGP2 and RIG-I CTD domains have a large basic surface, one bank of which is formed by the RNA binding loop. MDA5 also has a large basic surface that is extensively flat due to open conformation of the RNA binding loop. The NMR chemical shift perturbation study showed that dsRNA and 5′ppp-ssRNA are bound to the basic surface of LGP2 CTD, whereas dsRNA is bound to the basic surface of MDA5 CTD but much more weakly, indicating that the conformation of the RNA binding loop is responsible for the sensitivity to dsRNA and 5′ppp-ssRNA. Mutation study of the basic surface and the RNA binding loop supports the conclusion from the structure studies. Thus, the CTD is responsible for the binding affinity to the viral RNAs.
Introduction
A variety of pathogen-associated molecular patterns, including microbial peptidoglycan, lipopolysaccharide, β-1,3-glucan, and viral DNA or RNA are recognized by pattern recognition receptors that evoke the innate immune responses of host cells. In viral infections, double-stranded RNA (dsRNA)2 is recognized by Toll-like receptor-3 in the early endosome and by RIG-I like receptors (RLRs) in the cytoplasm. These two receptors initiate the innate immune responses including the production of cytokines and type-I interferon, which are critical for the subsequent adaptive immune response (1).
The RLR comprises three homologs: RIG-I (retinoic acid-inducible gene I), MDA5 (melanoma differentiation-associated gene 5), and LGP2 (laboratory of genetics and physiology 2) (see Fig. 1A) (2), and they sense a viral infection by recognizing replicating viral RNA in the cytoplasm. The RIG-I and MDA5 consist of three functional domains: tandem-CARDs (caspase activation and recruitment domain), a DEAD box helicase-like domain, and a well conserved C-terminal domain (CTD), whereas LGP2 has only the DEAD box helicase like domain and well conserved CTD. The three RLRs are considered to play different roles in the recognition of pathogen-associated molecular patterns and to be activated by different viruses and different viral RNAs. RIG-I is activated by a variety of viruses, including paramyxovirus, rhabdovirus, and orthomyxovirus, recognizing not only dsRNA but also 5′-triphosphated single-stranded RNA (5′ppp-ssRNA) (3, 4), and MDA5 is mainly activated by picornavirus (5, 6), whereas LGP2 lacking the tandem CARDs was originally identified as a negative regulator, but gene disruption study showed that it may function as a positive regulator (2, 7).
FIGURE 1.
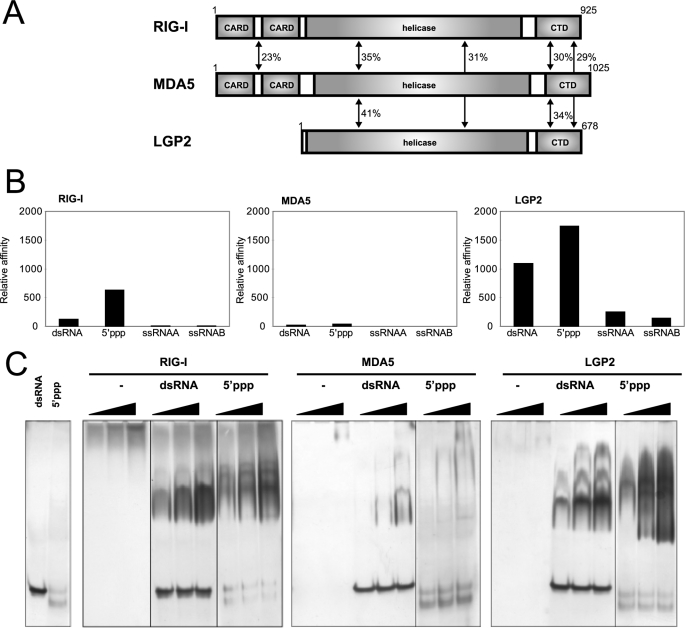
Functional analysis of RLR CTDs. A, domain structures of RIG-I, MDA5, and LGP2. Sequence identities of each domain among the RLRs are indicated. B, RNA-binding activity of RLR CTDs determined by SPR. GST-RIG-I CTD, GST-MDA5 CTD, and GST-LGP2 CTD were captured by the anti-GST anti-body immobilized onto the sensor chip, then dsRNA, 5′ppp-ssRNA, and two ssRNAs (ssRNAA and ssRNAB) were injected. Each resonance unit of RNA bound to a GST fused protein are standardized by molecular weight of the RNAs, then normalized by the resonance unit and molecular weight of the captured GST-fused protein. Normalized data are summarized and shown as bar graphs. RNAs bound to RIG-I, MDA5, and LGP2 are indicated from left to right in the panels. C, RNA-binding activity of RLR CTDs determined by EMSA. CTDs without the GST tag were prepared and subjected to EMSA. Increasing amounts of CTD (10, 20, and 40 pm) were reacted with the indicated probe and analyzed by native PAGE. The gels were silver-stained to visualize protein and RNA probe. −, no RNA; dsRNA, 25/25c probe; 5′ppp: 5′pppGG25 probe.
Our previous study has shown that the three domains of RIG-I cooperatively take part in pathogen-associated molecular pattern recognition and signal transduction (8). Further, the tandem CARDs are essential to transduce the signal via CARD-CARD interaction with the downstream CARD containing signal element, IPS-1/MAVS/VISA/Cardif (9–12). The CTD plays a critical role in the specific recognition of dsRNA and 5′ppp-ssRNA, while the DEAD box helicase cooperatively enhances the affinity to dsRNA through a conformational change. In the resting state, RIG-I is believed to be in closed conformation via interaction with these three domains (13). Upon viral infection, CTD specifically recognizes viral RNA and induces a domain rearrangement that allows RIG-I to form a stable complex with viral RNA and simultaneously exposes the tandem CARDs so that RIG-I is assumed to become an open conformation to interact with IPS-1/MAVS/VISA/Cardif (8, 14).
In the previous report, we have solved the solution structure of RIG-I CTD and shown the presence of a large positively charged surface that is responsible for viral RNA binding. In this report, we compare RNA-binding activity of the three CTDs (RIG-I, MDA5, and LGP2) and determine the solution structures of MDA5 and LGP2 CTDs. The RNA-binding activity is correlated to a conserved RNA binding loop, including a critical phenylalanine residue of RIG-I and LGP2. This residue is not conserved in MDA5, reflecting the distinct specificity of this sensor. Our results provide new insights into the recognition of viral RNA in cytoplasm.
EXPERIMENTAL PROCEDURES
Recombinant Proteins
The C-terminal domains of human MDA5 (896–1025) and LGP2 (546–678) were amplified by PCR and inserted into a pGEX-6P-1 vector to produce GST fusion protein (Amersham Biosciences). Expression vectors were introduced into Escherichia coli BL21(DE3) cells and cultured in M9 medium. The MDA5 CTD expression was induced by addition of 0.01 mm isopropyl 1-thio-β-d-galactopyranoside when the absorbance at 600 nm was ∼0.4. The cells were then grown at 16 °C for 20 h; LGP2 CTD expression was induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside then incubated at 25 °C for 20 h. The cells were suspended in PBS containing 1 mm 4-(2-aminoethyl)benzenesulfonylfluoride hydrochloride, lysed via sonication, and centrifuged. The supernatant was mixed with glutathione-Sepharose 4B (Amersham Biosciences) for 12 h, and the protein bound to glutathione-Sepharose 4B was washed first with PBS then with PBS containing 500 mm NaCl to remove the nucleic acid bound to the protein. The proteins were eluted with 50 mm Tris, pH 8, 150 mm NaCl, 20 mm reduced glutathione. The eluted protein was digested by PreScission protease (Amersham Biosciences). The proteins were further purified with size-exclusion Superdex 75 columns (Amersham Biosciences) with PBS containing 500 mm NaCl.
Site-directed Mutagenesis and Preparation of Mutant Proteins Using 293T Cells
Mutations of the RLR were introduced by the overlap extension PCR (15). The mutagenized cDNA, which was designed to possess an N-terminal FLAG tag, was subcloned into pEFBOS. The expression vectors were transiently transfected to 293T cells and purified by anti FLAG beads (8). The mutations did not alter the stability of the RLR proteins drastically as the production levels, and the recoveries from the transfected mammalian cells are comparable (Figs. 5–7).
FIGURE 5.
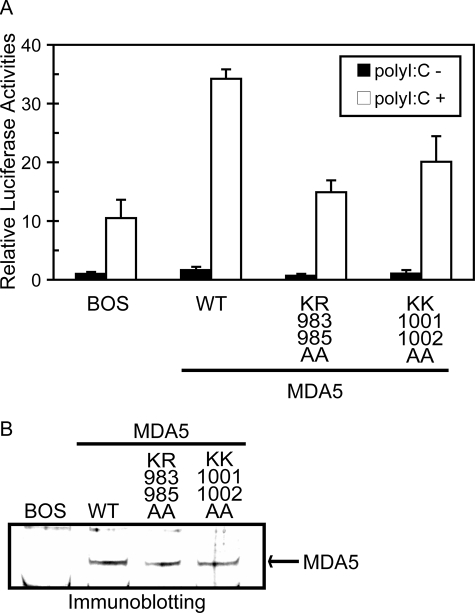
Functional analysis of basic residue mutations of MDA5 on the basic surface. A, MDA5−/− mouse embryo fibroblasts were transfected with the reporter gene, p-125Luc, and pRL-tk, together with the expression vector for MDA5 and mutants. Cells were stimulated by transfection with poly(I:C) and subjected to a dual-luciferase assay. The values are the means ± S.D. from triplicate experiments. The relative luciferase activity was calculated by considering the luciferase activity from cells transfected with empty vector (BOS) as 1.0. The cell lysates were analyzed for expression of MDA5 and mutants by Immunoblotting (B).
FIGURE 6.
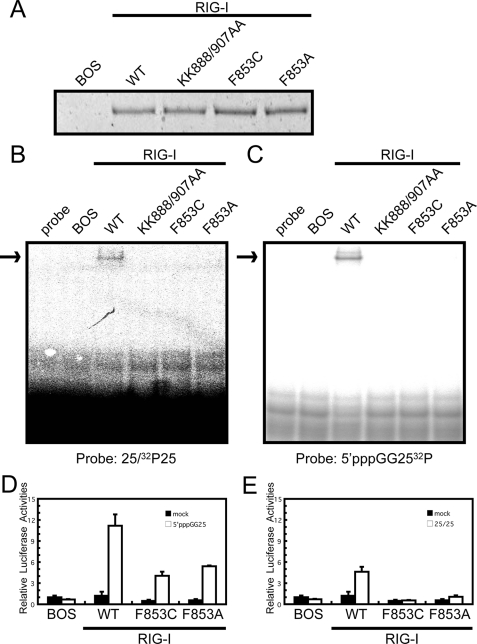
Functional analysis of the basic surface and the RNA binding loop mutants of RIG-I. Full-length wild type RIG-I and mutants were produced in 293T cells and purified by anti FLAG antibody. A, silver staining of the purified recombinant RIG-I. The recombinant RIG-I was subjected to EMSA using 32P-labeled dsRNA (B; 25/25c) or 5′ppp-ssRNA (C; 5′pppGG25) as probe. D and E, interferon-β promoter activation by wt RIG-I and mutants. RIG-I−/− mouse embryo fibroblasts were transfected with the reporter gene, p-125Luc, and pRL-tk, together with the expression vector for RIG-I and mutants. Cells were stimulated by transfection with 25/25c (D) or 5′pppGG25 (E) and subjected to a dual-luciferase assay. The values are the means ± S.D. from triplicate experiments. The relative luciferase activity was calculated by considering the luciferase activity from cells transfected with empty vector (BOS) as 1.0.
FIGURE 7.
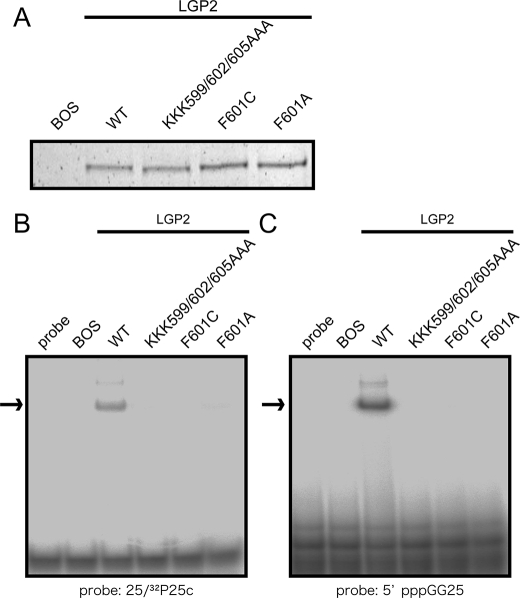
Effect of the mutations of LGP2 on RNA recognition. Full-length wild-type LGP2 and mutants on the RNA binding loop were produced in 293T cells and purified by anti FLAG antibody. A, silver staining of the purified recombinant LGP2. The recombinant LGP2 were subjected to EMSA using (B) 32P-labeled dsRNA (25/25c) or (C) 5′ppp-ssRNA (C; 5′pppGG25) as probe. The arrows indicate the RNA-LGP2 complex.
Preparation of Uniformly Labeled Proteins
Uniformly Labeled MDA5 and LGP2 CTDs were expressed as above except here using medium containing 15N-ammonium chloride and d-glucose (or [d-13C]glucose), and purified similarly. The NMR samples for the structure determination were prepared at 0.94 mm MDA5 CTD in 20 mm Bis-Tris, pH 7.0, 250 mm NaCl, 10 mm DTT, and 1.00 mm LGP2 CTD in 50 mm Tris, pH 7, 250 mm NaCl, and 1 mm DTT.
NMR Measurements
The NMR spectra were acquired at 25 °C on Varian Unity Inova 500, 600, and 800 spectrometers. The data were processed using NMRPipe (16) and analyzed using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). For assignments of the protein backbone and side-chain 1H, 15N, and 13C resonances, two- and three-dimensional (2D and 3D) spectra were obtained. Backbone resonances were assigned using the 2D 1H-15N HSQC, 3D HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HNCO, (HCA)CO(CA)NH, HNCAHA, and HBHA(CO)NH spectra. The aliphatic atoms of the side chains were assigned using 2D 1H-13C HSQC, 3D C(CO)NH, H(CCO)NH, CCH-TOCSY, and HCCH-TOCSY spectra, while aromatic side-chain atoms were assigned using the 2D 1H-13C HSQC, (Hb)Cb(CgCd)Hd, (Hb)Cb(CgCdCe)He, and 3D HCCH-TOCSY spectra.
Structural Determination
The 3D 15N-edited NOESY and 13C-edited NOESY spectra (tmix = 80 ms) were measured to obtain NOE distance constraints. Backbone ϕ and ψ dihedral angle constraints were generated using the TALOS program (17). The structures of MDA5 CTD and LGP2 CTD were determined using CANDID/CYANA 2.1 (18, 19). Structural statistics for the best 20 structures of MDA5 CTD and LGP2 CTD are shown in Tables 1 and 2. The 20 lowest energy structures of both MDA5 CTD and LGP2 CTD were deposited at the Protein Data Bank (PDB codes 2RQB and 2RQA, respectively).
TABLE 1.
Structural statistics for 20 MDA5 CTD
| NOE distance constraints | |
| Total | 2970 |
| Short range |i-j|< = 1 | 1662 |
| Medium range 1<|i-j|<5 | 372 |
| Long range |i-j|> = 5 | 982 |
| Dihedral angle constraints | |
| ϕ | 76 |
| ψ | 73 |
| Residual NOE violations | |
| Number > 0.3 Å | 0 |
| Residual angle violations | |
| Number > 5.0° | 0 |
| Ramachandran statistics (%) | |
| Residues in most favored region | 74.0 |
| Residues in additionally allowed region | 24.1 |
| Residues in generally allowed region | 1.9 |
| Residues in disallowed region | 0 |
| Structural coordinates root mean square deviation (Å) (residues 897–1017) | |
| Backbone atoms for the final ensemble | 0.40 |
| All heavy atoms for the final ensemble | 0.76 |
TABLE 2.
Structural statistics for 20 LGP2 CTD
| NOE distance constraints | |
| Total | 2593 |
| Short range |i-j|< = 1 | 1517 |
| Medium range 1<|i-j|<5 | 322 |
| Long range |i-j|> = 5 | 754 |
| Dihedral angle constraints | |
| ϕ | 58 |
| ψ | 62 |
| Residual NOE violations | |
| Number > 0.3 Å | 2 |
| Residual angle violations | |
| Number > 5.0° | 0 |
| Ramachandran statistics (%) | |
| Residues in most favored region | 71.0 |
| Residues in additionally allowed region | 27.4 |
| Residues in generally allowed region | 1.7 |
| Residues in disallowed region | 0 |
| Structural coordinates root mean square deviation (Å) (residues 549–657) | |
| Backbone atoms for the final ensemble | 0.49 |
| All heavy atoms for the final ensemble | 0.84 |
NMR Titration Study of LGP2
The chemical shift perturbation study of the amide nitrogen and proton signals of LGP2 CTD in 1H-15N HSQC spectra was performed upon addition of the following RNAs: GG25/2 + 25c (dsRNA), 5′-triphosphated GG25 (5′ppp-ssRNA), two single-stranded RNAs GG25 (ssRNAA), and 2 + 25C (ssRNAB) were also titrated as controls. The GG25/2 + 25c was prepared by mixing equal molar GG25 and 2 + 25c, and then annealed in 1×M buffer (10 mm Tris, pH 7.5, 10 mm MgCl2, 1 mm DTT). The final concentration of dsRNA was 0.71 mm. To equalize the condition of the titration experiments, GG25, 2 + 25C, and 5′-triphosphated GG25 was prepared at 0.71 mm in 1×M buffer. The 15N-labeled LGP2 CTD was prepared at 92.4 μm in 250 μl of 50 mm Tris, pH 7, 250 mm NaCl, 1 mm DTT, and 10% D2O. The volume of RNA at the addition of 1.0 equivalent molar ratio was 32.5 μl. 1×M buffer alone was also titrated, and it was confirmed that there was no effect on the NMR spectrum by the buffer. GG25, 5′-GGAAACUAAAAGGGAGAAGUGAAAGUG-3′; 2 + 25C, 5′-AUCACUUUCACUUCUCCCUUUCAGUUU-3′.
NMR Titration Study of MDA5
The experiment was performed in the same manner as in the study of LGP2 except for slightly different conditions of RNA and protein concentrations. Final concentration of RNAs were 0.644 mm, and the MDA5 CTD was prepared at 105 μm in 250 μl of 50 mm Tris, pH 7, 250 mm NaCl, 1.5 mm DTT, and 10% D2O. The volume of RNA at the addition of 1.0 equivalent molar ratio of RNA was 40.8 μl.
Surface Plasmon Resonance Analysis
A Biacore X (Amersham Biosciences) was used for the SPR study. First, GST-fused RIG-I, MDA5, and LGP2 CTD were trapped by anti-GST antibody immobilized on the surface of a sensor CM4 chip, then RNAs were injected to detect the interaction. The standard running buffer used in the analysis was 50 mm Tris, pH 7.5, 150 mm NaCl, and the proteins and RNAs were prepared in the same buffer. All experiments were performed at 25 °C. Immobilization of anti-GST antibody and regeneration of the surface was carried out using a GST Capture Kit and an Amine Coupling Kit (Amersham Biosciences) according to the instruction manuals. The GST-RLR CTDs were prepared at 20 μg/ml and injected for 300 s at the flow rate of 20 μl/min confirming the amount of CTDs captured to the anti-GST antibody with the Resonance Unit (RU) to be ∼800. As a control, GST was captured in an anti-GST antibody immobilized reference cell. The RNAs (dsRNA, 5′ppp-ssRNA, ssRNAA, and ssRNAB) were prepared at 1 μm and injected for 120 s at the flow rate of 20 μl/min and data were acquired as the difference in RU between GST-RLR CTDs captured cells and reference cells.
EMSA
EMSA was performed essentially as described previously (8). The native gel was silver-stained (for unlabeled probe and protein), or subjected to radioactivity detection (for a 32P-labeled probe). The 5′ppp-ssRNA was labeled by using 5′-32P-pCp (PerkinElmer Life Sciences) and T4 RNA ligase (Takara, Ohtsu, Siga, Japan) following the manufacturer's instructions.
Preparation of dsRNA Docked with RIG-I and LGP2 CTDs
The crystal structure of RIG-I CTD (20) and short dsRNA derived from the crystal structure (PDB ID: 1YYW) were docked with the Molecular Docking Algorithm Patchdock (21). Ten residues from the basic surface of RIG-I CTD were applied as potential binding sites for RIG-I CTD, whereas no sites were applied for dsRNA. Then ten highest scoring structures were checked to establish whether they satisfy the NMR titration data. 6 of the 10 structures satisfied the NMR data, and orientations of dsRNA in those structures were similar. A dsRNA-docked model with LGP2 CTD was similarly calculated and 5 of 10 structures showed similar orientations of the dsRNA.
RESULTS
Characterization of RLR CTDs for Specific Binding to RNAs by SPR and EMSA
To investigate the RNA binding specificity of LGP2 CTD (546–678) and MDA5 CTD (896–1025), both CTDs were expressed as GST fusion proteins and were applied to the SPR analysis together with RIG-I CTD (792–925) (Fig. 1B and supplemental Fig. 1). The GST fusion proteins were captured on the sensor chip, and their affinities to 5′ppp-ssRNA, dsRNA, and the two complementary ssRNAs were tested. In the SPR experiment, RIG-I CTD was bound to dsRNA and 5′ppp-ssRNA specifically, but was not bound to the two ssRNAs as has also previously been observed by EMSA (8). The LGP2 CTD was strongly bound to dsRNA and 5′ppp-ssRNA, but it exhibited lower binding affinity to ssRNA. The MDA5 CTD did not bind to any of the RNAs tested under these conditions.
The RIG-I CTD has been reported to specifically bind to both dsRNA and 5′ppp-ssRNA by EMSA (8). We produced CTDs without the GST tag and compared the RNA-binding activity by EMSA (Fig. 1C). The CTD of RIG-I and LGP2 specifically bound to dsRNA and 5′ppp-ssRNA, but not to ssRNAs (data not shown). This result is consistent with that of the SPR analysis. The EMSA data shows the appearance of slower mobility complexes, most clearly observed with LGP2 CTD and dsRNA. These complexes are likely corresponding to dsRNA bound to multiple CTD molecules (see “Discussion”). The MDA5 CTD exhibited low binding affinity to dsRNA, and its biding affinity to 5′ppp-ssRNA was very low. This is reminiscent of the previous observation that full-length MDA5 bound much more weakly to poly(I:C)-Sepharose than to RIG-I (22) (supplemental Fig. 3). Summarizing, these results suggest that RLR CTDs play a critical role in the RNA recognition by RLR.
Solution Structures of MDA5 and LGP2 CTDs
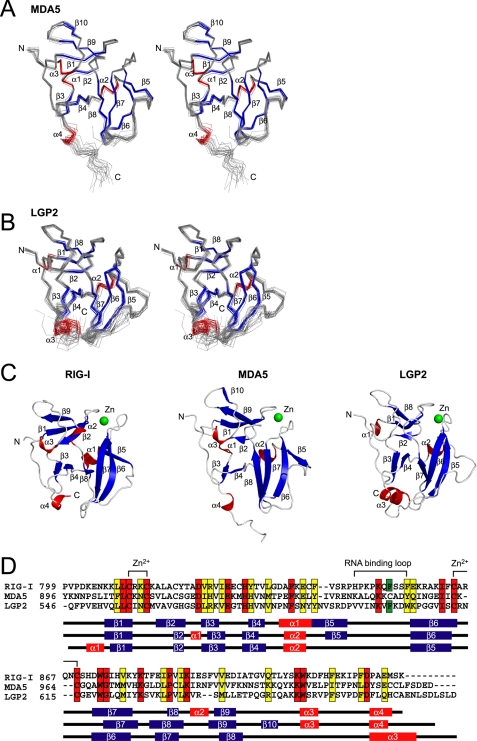
The solution structures of MDA5 and LGP2 CTDs were determined by NMR (Fig. 2, A and B), and both MDA5 and LGP2 CTDs have a fold similar to that of RIG-I CTD and contain a single conserved Zn2+ binding site (Fig. 2, C and D). We confirmed that both CTDs contain a single Zn2+ ion, by atomic absorption spectroscopy, similar to RIG-I CTD (supplemental Table 1). The core of the structure is composed of a central anti-parallel β sheet (Fig. 2, C and D) (β3–β8 in RIG-I and MDA5, β3–β7 in LGP2). A further anti-parallel β sheet (β1, -2, and -9 in RIG-I; β1, -2, -9, and -10 in MDA5; β1, -2, and -8 in LGP2) is located on top of the central β sheet, and there are several short helices (α1–3 in RIG-I and MDA5, α1 and -2 in LGP2) attached to the two β sheets that help stabilize the structure of the CTDs. The C-terminal region of RIG-I, MDA5, and LGP2 CTDs has a long loop that is surrounding one edge of the central β sheet and the C-terminal helix (α4 in RIG-I and MDA5, α3 in LGP2) is located at the bottom of the central β sheet. The long loop between β5 and β6 that was implicated in RNA recognition in RIG-I, is also present in all RLR CTDs (termed “RNA Binding Loop” below). Recently, the crystal structure of LGP2 CTD was reported. Although the RNA binding loop was not observed in the crystal structure, the author implied that the loop is important for RNA specificity (23).
FIGURE 2.
Solution structure of MDA5 CTD and LGP2 CTD. A and B, best fit superposition of the backbone atoms of 20 NMR-derived MDA5 CTD (A) and LGP2 CTD (B). Structures are shown in stereo. β-Strands and α-helices are shown in blue and red, respectively. C, ribbon diagrams of the structure of RIG-I CTD, MDA5 CTD, and LGP2 CTD (left to right). Secondary structure elements are labeled. The figure was prepared using PyMOL. D, sequence alignment of human RIG-I, MDA5, and LGP2 CTDs. ClustalX was used to align the sequences. The secondary structure elements of each CTD are indicated below the alignment. The amino acids in red and yellow indicate conserved (red) and type-conserved (yellow) residues with the Zn2+ binding Cys-X-X-Cys motifs and RNA binding loop. The Phe residues conserved in RIG-I and LGP2 in the RNA binding loop are colored green.
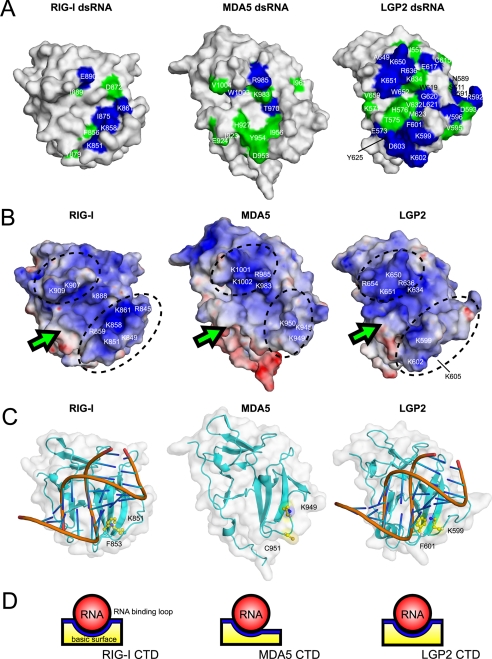
Although the RLR CTDs have a similar global fold with a large basic surface on the central β-sheet (see Fig. 4B), a close inspection of the RLR CTD structures shows small but appreciable differences (Fig. 2C). Similar to RIG-I, the C-terminal helices of LGP2 and MDA5 are located at the bottom of the central β-sheet and interacts with the loop between β6 and β7 (β7 and β8 in MDA5), whereas the C-terminal helix of MDA5 is projected outward. However, the regions following the C-terminal helix are flexible, because these regions exhibit small steady-state NOEs (<0.3 in supplemental Fig. 2). There is also an appreciable difference in the RNA binding loop. The N-terminal part of the RNA binding loop in MDA5 CTD is clipped onto the surface of β6 so that the conformation of the rest of the RNA binding loop is restricted and gives rise to an open and flat conformation (Fig. 2, A and C, middle). Thus, the basic surface becomes a more open structure (Figs. 2C, 4B, and 4D (middle)). Although the C-terminal RNA binding loop in LGP2 is restrained but stands upright on β6 and encloses the central β-sheet together with the central part of the loop between β8 and α3, a loop N terminus to β8, forming a basic groove (Figs. 2C, right, and 4B, right). A similar basic groove was identified as the RNA binding surface in RIG-I (Figs. 2C and 4B, left). Both RIG-I and LGP2 but not MDA5 share the common structural feature for the RNA binding loop. It is to be noted that the RNA binding loops in MDA5 and LGP2 are relatively restricted, based on the steady-state NOE measurements (>0.5 in supplemental Fig. 2).
FIGURE 4.
Structural comparisons of RLR CTDs. A, surface representation of the dsRNA affected surface of RLR CTDs (left to right, RIG-I CTD, MDA5 CTD, and LGP2 CTD). The residues that disappeared in NMR titration experiments upon addition of 0.25 and 0.5 equivalents of dsRNA are colored in blue and green, respectively. The residue numbers are also shown. The data of the dsRNA affected surface of RIG-I CTD was derived from our previous study (8). B, electrostatic surface potentials of the RLR CTDs (left to right, RIG-I CTD, MDA5 CTD, and LGP2 CTD). Dotted circles indicate the banks surrounding the basic surface. C, left, dsRNA-bound model of RIG-I CTD. Right, dsRNA-bound model of LGP2 CTD. The Lys and Phe residues in the RNA binding loop conserved in RIG-I and LGP2 are shown in ball-and-stick models and labeled. Middle, structure of MDA5 CTD, the residues corresponding to the Lys-851 and Phe-853 in RIG-I are shown. All figures are shown in the same structural orientation as in Fig. 2A. D, schematic diagrams of dsRNA bound to the basic surface of RIG-I CTD (left), MDA5 CTD (mid), and LGP2 CTD (right) viewed from the arrows indicated in Fig. 4B.
NMR Titration of MDA5 and LGP2 CTDs
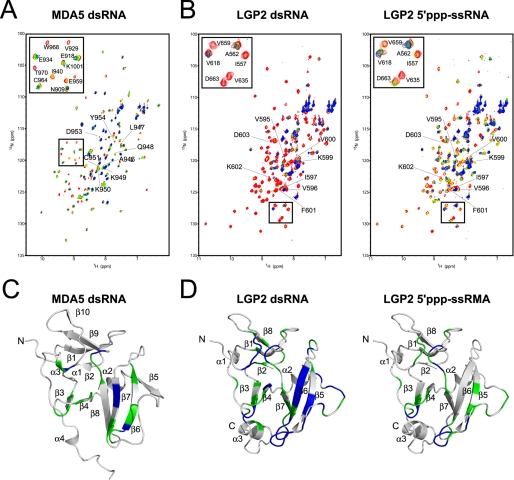
To establish whether the basic surfaces of MDA5 and LGP2 CTDs are responsible for the binding to viral RNAs, we performed NMR titration with the RNAs (dsRNA, 5′ppp-ssRNA, and ssRNAs), used in the SPR and EMSA assays reported above, against 15N-labeled MDA5 and LGP2 CTDs (Fig. 3, A and B).
FIGURE 3.
NMR titration results for MDA5 CTD and LGP2 CTD. A, NMR titration of MDA5 CTD with dsRNA. An overlay of 1H-15N HSQC spectra are shown in red, yellow, green, and blue at 0, 0.25, 0.5, and 1.0 equivalents (molar ratio of dsRNA to proteins), respectively, where the residues in the RNA binding loop are labeled. The inset shows the excerpt of the enclosed region of the spectrum with assignment. B, NMR titration of LGP2 CTD with dsRNA (left panel) and 5′ppp-ssRNA (right panel). The figures are prepared in the same manner as in A. C, mapping of the residues of MDA5 CTD affected by the addition of dsRNA on the ribbon diagram. The residues whose peaks disappeared on addition of 0.25 and 0.5 equivalent molar ratios of dsRNA to CTD are colored blue and green, respectively. D, mapping of the residues of LGP2 CTD affected by the addition of dsRNA and 5′ppp-ssRNA (left and right panels) on the ribbon diagram, colors are the same as in C. The orientations are the same as in Fig. 2, A and B.
In MDA5 CTD, there were negligibly small chemical shift changes in the 1H-15N HSQC spectra even with addition of 2.0 equivalent molar ratios of 5′ppp-ssRNA and two ssRNAs, indicating an absence of interaction between MDA5 CTD and these RNAs (data not shown). However, upon addition of 0.5 equivalent molar ratio of dsRNA, an appreciable number of peaks disappeared possibly due to intermediate exchange processes (Fig. 3, A and C). The disappeared peaks were assigned to the residues on β3, β4 and their connecting loop, β6, β7, the loop C terminus to β8, and α3. However, most of the residues on the RNA binding loop other than Asp-953 and Tyr-954 did not disappear (Fig. 3C). This strongly suggests that the MDA5 CTD uses the basic surface to interact with dsRNA similar to RIG-I, but the RNA binding loop does not appear to be involved in RNA binding. This result is consistent with EMSA (Fig. 1C) but not with the SPR analysis (Fig. 1B), where there was no interaction between MDA5 and dsRNA. This inconsistency may be due to the higher concentration of MDA5 CTD and dsRNA used in the EMSA and NMR titration studies than in the SPR studies.
For LGP2, even with addition of 2.0 equivalent molar ratios of ssRNAs, 1H-15N HSQC of LGP2 CTD spectra showed neither chemical shift changes nor any loss of signal intensities (data not shown), indicating that LGP2 CTD does not recognize ssRNAs. However, the addition of a 0.25 equivalent molar ratio of dsRNA and 0.5 equivalent molar ratio of 5′ppp-ssRNA caused the disappearance of a large number of peaks (Fig. 3B). When the residues corresponding to the peaks, which disappeared by dsRNA titration, were mapped on the three-dimensional structure of LGP2 CTD, these were located on β3, β4 and the connecting loop, the RNA binding loop, β5, β6 and the loop C terminus to β6, β7 and the loop between β7 and β8, and the C terminus to β8 (α3 in RIG-I) (Fig. 3D, left). The binding surface for 5′ppp-ssRNA is similar to that of dsRNA but is less extensive (Fig. 3D, right). It is to be noted that unlike MDA5, a large number of the peaks assigned on the RNA binding loop (Fig. 3B) disappeared in both cases, indicating that the RNA binding loop plays a critical role in the recognition of RNA ligands in LPG2. Residues of CTD involved in the interaction with dsRNA along with surface charges are mapped for comparison among RLRs (8) (Fig. 4, see “Discussion”).
Analysis of the Structure-Function Relationship of RLR by Mutagenesis
We generated MDA5 mutants considering the results of the chemical shift perturbation study and the electrostatic surface potential (Fig. 4, A and B, middle). The KR983/985AA and KK1001/1002AA mutants were expressed in MDA5−/− mouse embryo fibroblasts, and next the ability to mediate the virus-responsive reporter gene activation was examined. To stimulate MDA5, the cells were transfected with poly(I:C), a known chemical ligand for MDA5 (Fig. 5A). Irrespective of the absence of MDA5, vector-transfected cells exhibited reporter activation upon stimulus by poly(I:C), presumably due to the activation of RIG-I by short poly(I:C) present in this batch of poly(I:C) (300–3000 bp, data not shown). Expression of wt MDA5 strongly enhanced this response, suggesting this increase is mediated by ectopically expressed MDA5. However both of the mutants exhibited only slight activation. Comparable levels of wt and mutant MDA5 were expressed in these cells (Fig. 5B), suggesting that mutated basic amino acid residues perform critical functions.
The solution structures and chemical shift perturbation studies of RIG-I and LGP2 may highlight the presence of the RNA binding loop. Because the specificity of RIG-I and LGP2 CTDs to viral RNAs are similar, we focused on a Phe residue that are conserved in the RNA binding loop of RIG-I and LGP2 but is not conserved in MDA5. First, we produced and purified full-length wt RIG-I and mutants with F853A and F853C that mimic MDA5 (Fig. 2D) and KK888/907AA (8) in 293T cells (Fig. 6A). Comparable levels of wt and mutant proteins were produced and recovered, suggesting that these mutations did not alter the stability of RIG-I (Fig. 6A). The purified proteins were subjected to EMSA using dsRNA and 5′ppp-RNA as probes (Fig. 6, B and C). The results clearly show that, along with the previously identified basic residues on the basic surface, Phe-853 in the RNA binding loop is critical for binding with dsRNA and 5′ppp-ssRNA. Further, the mutation of Phe-853 reduced the signaling activity of RIG-I when stimulated with these RNA species, particularly with dsRNA (Fig. 6, D and E). Next the involvement of the RNA binding loop in RNA recognition by LGP2 was examined. Three basic residues in the RNA binding loop or the conserved Phe were substituted (KKK599/602/605AAA, F601A, and F601C). These LGP2 mutant proteins along with the wt protein were produced and recovered at similar levels suggesting that these mutations did not affect protein stability dramatically (Fig. 7A). The purified LGP2 proteins were examined for RNA binding. The results in Fig. 7 (B and C) clearly show that the residues on the RNA binding loop are critical for LGP2 to recognize dsRNA and 5′ppp-ssRNA.
To directly compare RNA-binding activity of full-length RIG-I, MDA5, and LGP2 and their mutants, EMSA was performed under the same conditions (supplemental Fig. 3). The result shows that the binding properties of full-length RLRs are closely reflecting those of CTD (Fig. 1C) and that mutations at critical residues prevented the RNA binding.
DISCUSSION
The experiments reported here compared the RNA-binding activity of CTDs of RLRs. The results of SPR and EMSA assays suggested that LGP2 CTD bound to dsRNA and 5′ppp-ssRNA with higher affinity than RIG-I, but that MDA5 CTD exhibited weaker binding.
The solution structure of the CTD revealed several critical features shared among or unique to each RLR. There is an RNA binding loop in all the CTDs, and the NMR structure and titration studies as well as the functional studies showed that the RNA binding loop in RIG-I and LGP2 are critical for RNA recognition but that it seems to be less important in MDA5. In silico docking with RIG-I (Fig. 4C) suggests that dsRNA lies on the basic groove of the RIG-I CTD, and that multiple interactions between the basic residues and phosphate backbone of the RNA maintains the complex. In the structure of the dsRNA-bound model of RIG-I CTD, the aromatic moiety of Phe-853 is stacked into the groove of the dsRNA via the hydrophobic interaction with a ribose moiety of the dsRNA. This interaction would anchor the position of both dsRNA and the RNA binding loop and allow the side chain of the conserved Lys-851 to form a stable electrostatic interaction with dsRNA. Whereas, in MDA5, the aromatic residue in this position is replaced by Cys, which would impair the hydrophobic interaction. Moreover, the flat surface formed by the RNA binding loop is not favorable for the binding between MDA5 and dsRNAs. On the other hand, the Phe-601 of LGP2 is located in a similar position and may similarly interact with the sugar moiety as is suggested in RIG-I (Fig. 4C). Taken together, both RIG-I CTD and LGP2 CTD bind to dsRNA and 5′ppp-ssRNA with high affinity. It is to be noted that slower migrating complexes are observed at higher protein concentrations, particularly with LGP2 (Fig. 1C). These complexes are likely multiple LGP2 molecules bound to single dsRNA molecules. Our model (Fig. 4C) allows binding of multiple CTDs with single dsRNA. Formation of RLR multimers on dsRNA or 5′ppp-ssRNA may facilitate signaling through CARD oligomerization (14).
Further, despite a low binding affinity, MDA5 CTD also recognizes dsRNA, and the residues present in the basic surface are critical for the signaling activity of MDA5 (Figs. 4 and 5), suggesting that MDA5 CTD participates in dsRNA recognition. However it is also known that full-length MDA5 exhibits lower binding affinity compared with LGP2 and RIG-I (22), suggesting that MDA5 may utilize some other RNA recognition protein. Related to this, LGP2−/− mice exhibit hypersensitivity to encephalomyocarditis virus (EMCV) infection (7), detection of which is mediated by MDA5 (5). Further, our preliminary result suggests that LGP2 and MDA5 bind to poly(I:C) in a cooperative manner (not shown).
Both RIG-I and LGP2 recognize dsRNA and 5′ppp-ssRNA, the two distinct non-self RNA patterns. In the case of 5′ppp-ssRNA recognition, 5′-end triphosphate is an apparent determinant of a non-self RNA. Although the model here provides a recognition mechanism of dsRNA by the CTD of RIG-I and LGP2, the recognition of 5′ppp-ssRNA by RIG-I and LGP2 need to be further substantiated. Interestingly, mutagenesis studies indicate that critical residues for dsRNA recognition are also indispensable for 5′ppp-ssRNA recognition (8) (Figs. 5–7), suggesting common structural features of these patterns. In summary, our functional and structural analyses have uncovered the mechanism underlying the different functions of each RLR molecule. During revision of the manuscript a report describing crystal structure of LGP2 CTD and dsRNA was published (24). The report essentially describes recognition of dsRNA ends by LGP2. However, the EMSA result shows that the end structure of dsRNA does not affect recognition by LGP2 (supplemental Fig. 4). Apparently, further analyses are required to elucidate the mechanism of dsRNA recognition by RLR.
Supplementary Material
This work was supported by grants-in-aid (Tokutei-ryoiki (Matrix of Infection Phenomena), Kiban (S)) and National Projects on Targeted Proteins Research Program from the Japanese Ministry of Education, Culture, Sports, Science, and Technology, by a Research Grant of the Uehara Memorial Foundation, and by Nippon Boehringer Ingelheim Co., Ltd.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and supplemental Table 1.
The atomic coordinates and structure factors (codes 2RQB and 2RQA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- dsRNA
- double-stranded RNA
- ssRNA
- single-stranded RNA
- RLR
- RIG-I-like receptor
- CARD
- caspase recruitment domain
- IPS-1
- interferon promoter stimulator-1
- RIG-I
- retinoic acid-inducible gene I
- MDA5
- melanoma differentiation-associated gene 5
- LGP2
- laboratory of genetics and physiology 2
- 5′ppp-ssRNA
- 5′-triphosphated single-stranded RNA
- GST
- glutathione S-transferase
- CTD
- C-terminal domain
- PBS
- phosphate-buffered saline
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DTT
- dithiothreitol
- NOESY
- nuclear Overhauser effect spectroscopy
- wt
- wild type
- EMSA
- electrophoretic mobility shift assay
- SPR
- surface plasmon resonance.
REFERENCES
- 1.Akira S., Uematsu S., Takeuchi O. ( 2006) Cell 124, 783– 801 [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. ( 2004) Nat. Immunol. 5, 730– 737 [DOI] [PubMed] [Google Scholar]
- 3.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. ( 2006) Science 314, 994– 997 [DOI] [PubMed] [Google Scholar]
- 4.Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis E., Sousa C. ( 2006) Science 314, 997– 1001 [DOI] [PubMed] [Google Scholar]
- 5.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. ( 2006) Nature 441, 101– 105 [DOI] [PubMed] [Google Scholar]
- 6.Loo Y. M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M. A., García-Sastre A., Katze M. G., Gale M., Jr. ( 2008) J. Virol. 82, 335– 345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkataraman T., Valdes M., Elsby R., Kakuta S., Caceres G., Saijo S., Iwakura Y., Barber G. N. ( 2007) J. Immunol. 178, 6444– 6455 [DOI] [PubMed] [Google Scholar]
- 8.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. ( 2008) Mol. Cell 29, 428– 440 [DOI] [PubMed] [Google Scholar]
- 9.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. ( 2005) Nat. Immunol. 6, 981– 988 [DOI] [PubMed] [Google Scholar]
- 10.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. ( 2005) Nature 437, 1167– 1172 [DOI] [PubMed] [Google Scholar]
- 11.Seth R. B., Sun L., Ea C. K., Chen Z. J. ( 2005) Cell 122, 669– 682 [DOI] [PubMed] [Google Scholar]
- 12.Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. ( 2005) Mol. Cell 19, 727– 740 [DOI] [PubMed] [Google Scholar]
- 13.Saito T., Hirai R., Loo Y. M., Owen D., Johnson C. L., Sinha S. C., Akira S., Fujita T., Gale M., Jr. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 582– 587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneyama M., Fujita T. ( 2008) Immunity 29, 178– 181 [DOI] [PubMed] [Google Scholar]
- 15.Higuchi R., Krummel B., Saiki R. K. ( 1988) Nucleic Acids Res. 16, 7351– 7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. ( 1995) J. Biomol. NMR 6, 277– 293 [DOI] [PubMed] [Google Scholar]
- 17.Cornilescu G., Delaglio F., Bax A. ( 1999) J. Biomol. NMR 13, 289– 302 [DOI] [PubMed] [Google Scholar]
- 18.Güntert P., Mumenthaler C., Wüthrich K. ( 1997) J. Mol. Biol. 273, 283– 298 [DOI] [PubMed] [Google Scholar]
- 19.Herrmann T., Güntert P., Wüthrich K. ( 2002) J. Mol. Biol. 319, 209– 227 [DOI] [PubMed] [Google Scholar]
- 20.Cui S., Eisenächer K., Kirchhofer A., Brzózka K., Lammens A., Lammens K., Fujita T., Conzelmann K. K., Krug A., Hopfner K. P. ( 2008) Mol. Cell 29, 169– 179 [DOI] [PubMed] [Google Scholar]
- 21.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. ( 2005) Nucleic Acids Res. 33, W363– 367, web server issue [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr., Akira S., Yonehara S., Kato A., Fujita T. ( 2005) J. Immunol. 175, 2851– 2858 [DOI] [PubMed] [Google Scholar]
- 23.Pippig D. A., Hellmuth J. C., Cui S., Kirchhofer A., Lammens K., Lammens A., Schmidt A., Rothenfusser S., Hopfner K. P. ( 2009) Nucleic Acids Res. 37, 2014– 2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Ranjith-Kumar C. T., Brooks M. T., Bharmaiah S., Herr A. B., Kao C., Li P. ( 2009) J. Biol. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.