Abstract
Premature intracellular activation of the digestive enzyme trypsinogen is considered to be the initiating event in pancreatitis. However, the direct consequences of intracellular trypsin activity have not previously been examined. In the current study, a mutant trypsinogen (paired basic amino acid cleaving enzyme (PACE)-trypsinogen), which is activated intracellularly by the endogenous protease PACE, was developed. This new construct allowed for the first time direct examination of the effects of intracellular trypsin on pancreatic acinar cells. We found that PACE-trypsinogen was expressed in the secretory pathway and was activated within acinar cells. Expression of PACE-trypsinogen induced apoptosis of HEK293 cells and pancreatic acinar cells, as indicated by histology, DNA laddering, PARP cleavage, and caspase-3 activation. Cell death was blocked by the trypsin inhibitor Pefabloc but not by the pancaspase inhibitor benzyloxycarbonyl-VAD, indicating that caspase-independent pathways were also involved. However, intracellular trypsin had no significant effect on the activity of the proinflammatory transcription factor NF-κB. In contrast, extracellular trypsin caused cell damage and dramatically increased NF-κB activity. These data indicate that localization of active trypsin determines its effects on pancreatic acinar cells. This new model will greatly improve our understanding of the role of active trypsin in pancreatitis and its associated inflammatory response.
Introduction
“Pancreatitis” is the general term for inflammatory diseases of the pancreas and includes both acute and chronic forms. Pancreatitis results in significant morbidity and mortality, and chronic pancreatitis is associated with an increased risk of pancreatic cancer (1). Unfortunately the mechanisms responsible for the initiation of pancreatitis are not fully understood, which probably explains the persistent lack of causal therapies. More than 100 years ago, Chiari (2) proposed that acute pancreatitis was an autodigestive process, and since that time, the major focus of research on this disease has centered on the role of digestive enzymes (3). Activated digestive enzymes capable of injuring the gland have been detected within the pancreas, pancreatic juice, and plasma in clinical as well as experimental forms of pancreatitis, and the morphological changes that characterize severe pancreatitis indicate that a digestive injury to pancreatic tissue has occurred (4). Furthermore, pretreatment with gabexate mesilate, a serine protease inhibitor, reduces endoscopic retrograde cholangiopancreatography (ERCP)-induced pancreatitis (5). These observations support the suggestion that pancreatitis results from the intrapancreatic activation of digestive enzyme zymogens.
Two decades ago, it was observed that serine proteases were activated intracellularly in experimental models of acute pancreatitis (6, 7). These results were taken as a confirmation of the existing paradigm and served to narrow the focus in the field of pancreatitis research to the hypothesis that prematurely activated trypsinogen within the pancreatic acinar cell is the principal initiator of pancreatitis, although the treatments utilized to induce pancreatitis do not solely activate trypsinogen (8–10). More direct support for this concept came from the observation that a missense mutation in human cationic trypsinogen, R122H, is the cause of hereditary pancreatitis, a rare form of chronic pancreatitis (11). However, this mutation has pleiotropic effects rather than being exclusively an activating mutation, and there is no direct evidence for increased intracellular trypsin activity in patients with the disease. Furthermore, patients with this mutation do not suffer continuous pancreatitis but rather experience episodic attacks of the disease. Therefore, despite being the central focus of many years of study in the field of pancreatitis, the role of intracellular trypsin activity in pancreatitis remains unclear.
Recently, studies have shown that the transcription factor NF-κB is activated early and largely responsible for the inflammatory response occurring during the development of pancreatitis. NF-κB regulates gene targets that influence inflammation, immunity, and cell death (12). Under normal conditions, NF-κB is present in the cell cytoplasm bound to an inhibitory subunit, IκB (inhibitory κB). Upon perturbation of the cell by a variety of stimuli, IκB becomes phosphorylated on specific residues, leading to its degradation and the release of NF-κB. Once freed from the inhibitory subunit, NF-κB translocates to the nucleus, where it interacts with other transcription factors and leads to the expression of a variety of genes, including many cytokines, chemokines, enzymes, and adhesion molecules. We and several other laboratories have firmly established that NF-κB is activated early in the course of several models of acute pancreatitis (13–15). Using adenovirus to express the NF-κB active subunit p65, we have also shown that direct activation of NF-κB causes pancreatic and systemic inflammation (16), and this was recently confirmed using transgenic models (17). Thus, NF-κB activation is a key mechanism involved in the initiation of inflammation associated with acute pancreatitis.
The relationship between trypsin and NF-κB activation, two early events in pancreatitis, are unclear. Mimicking the activation of NF-κB by expression of NF-κB subunit p65, it has been shown that NF-κB does not directly activate trypsin (18, 19). However, it remains unknown whether intracellular trypsin activates NF-κB. This important issue can only be addressed by specific activation of trypsinogen intracellularly.
Several animal models of acute pancreatitis have been widely utilized to study the pathophysiology of this disease, including secretagogue hyperstimulation, retrograde bile perfusion, and ischemia reperfusion (20, 21). These models have greatly advanced our understanding of the early cellular events that underlie the development of acute pancreatitis. However, these treatments cause generalized cell damage, rather than specific activation of trypsinogen, and tend to generate widespread nonspecific effects which do not fully mimic conditions of clinical pancreatitis. Thus, current models are not suitable to investigate the role of intracellular trypsinogen activation in pancreatitis.
To specifically examine the effects of intracellular trypsinogen activation on pancreatic acinar cells, we developed a mutant trypsinogen that can be delivered by adenoviral gene transfer and activated by endogenous cellular mechanisms. Through directly manipulating intracellular trypsin activity, we are able to characterize for the first time the direct effects of this enzyme on acinar cell function. We observed that spontaneous intracellular trypsinogen activation rapidly led to acinar cell apoptosis through caspase-dependent and -independent pathways. However, intracellular trypsin activity was unable to activate NF-κB and generate inflammatory mediators. In contrast, we found that extracellular trypsin caused a rapid and dramatic increase of NF-κB activity and its target gene expression. Therefore, the site of active trypsin determines its effects on pancreatic acinar cells. These observations made on the direct effects of trypsin on acinar cells have important implications for understanding the development of pancreatitis.
EXPERIMENTAL PROCEDURES
Construction of a PACE3-activable Trypsinogen
To construct a PACE-activable trypsinogen, the original enteropeptidase cleavage site, -DDDDK-, was replaced by a PACE cleavage site, -RTKR-, using site-specific mutagenesis of the wild type rat trypsinogen cDNA. To facilitate detection of the PACE-trypsinogen, a peptide sequence, -RDPMYPYDVPDYAFPVD-, containing an influenza hemagglutinin tag (HA tag) was added between the signal peptide and the PACE cleavage site (PACE-trypsinogen). As a control, a catalytically inactive “dead” trypsin was also produced by mutating the first amino acid, isoleucine, of active trypsin (-IVGG-, p.I24) to -LVGG- (Dead-trypsinogen, p.I24L). These trypsin mutants were first cloned into pcDNA3.1, tested for functionality, and then used for generation of adenoviral vectors according to the method of He et al. (22). Briefly, the PACE-trypsinogen or Dead-trypsinogen was cloned to pAd-Track-CMV shuttle vector. This plasmid expresses the trypsin mutants and, in parallel, green fluorescent protein under the control of distinct CMV promoters. The constructs were linearized with PmeI and co-transfected together with the adenoviral backbone vector, pAd-Easy-1, into Escherichia coli strain BJ5183. Recombinants were selected with kanamycin and screened by BamHI and PacI digestion. The DNA sequence of positive clones was verified by sequencing, and recombinants were then cleaved with PacI and transfected (Lipofectamine; Invitrogen) into a packaging HEK293 cell line. Following stepwise amplification in HEK293 cells, recombinant high titer virus stocks were obtained by CsCl gradient purification. Adenoviruses bearing an NF-κB luciferase reporter (BD Biosciences, San Jose CA) or β-galactosidase were also constructed with the same strategy. The titers of the recombinant virus stocks were determined based on the counting of green fluorescent protein-expressing cells, as described previously (23).
PACE-trypsinogen Purification and PACE Cleavage
To obtain purified PACE-trypsinogen, pAd-Track-CMV-PACE-trypsinogen was transfected to HEK293 cells. Cells were incubated for 24 h, lysed, and subsequently immunoprecipitated with anti-HA high affinity matrix (Roche Applied Science). Purified PACE-trypsinogen was then incubated with recombinant PACE (New England Biolabs, Ipswich, MA) at 30 °C in 100 mm HEPES (pH 7.5) with 0.5% Triton X-100, 1 mm CaCl2, 1 mm 2-mercaptoethanol for 2 h. Cleavage of PACE-trypsinogen was verified by Western blotting, and functionality of the enzyme was detected by trypsin activity assays.
Trypsin Activity Assay
Trypsin activity was measured fluorometrically by using Boc-Gln-Ala-Arg-MCA (Bachem, Torrance, CA) as the substrate according to the method of Kawabata et al. (24). Briefly, cells were washed twice with phosphate-buffered saline and then homogenized in ice-cold MOPS buffer containing 250 mm sucrose, 5 mm MOPS, and 1 mm MgSO4 (pH 6.5) with a motorized glass-Teflon homogenizer. After centrifugation (5,000 × g for 5 min), supernatant was collected for measurement. A 100-μl sample was added to a cuvette containing assay buffer (50 mm Tris, 150 mm NaCl, 1 mm CaCl2, and 0.1% bovine serum albumin, pH 8.0). The reaction was initiated by adding substrate, and the fluorescence emitted at 440 nm after excitation at 380 nm was monitored on a LS55 Luminescence Spectrophotometer (PerkinElmer Life Sciences). Results were expressed as slope of fluorescence increase over time and matched to an internal purified trypsin standard (Sigma). Activity values were corrected for their protein concentration and expressed as -fold change of control.
Preparation of Acini and Adenovirus-mediated Gene Delivery
All experiments were conducted with the consent of the Ethics Committee for the Use of Experimental Animals at the University of Texas. Acini were prepared by methods described previously (23). In brief, the pancreas was excised from freely fed adult male C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME). Obtained tissue was enzymatically digested with purified collagenase (Worthington), followed by mechanical shearing. The resulting suspension of acini was then filtered through 150-μm Nitex mesh, purified by sedimentation through 4% bovine serum albumin in Dulbecco's modified Eagle's medium (Invitrogen). The acini were infected with corresponding virus preparations and incubated in Dulbecco's modified Eagle's medium containing 1% bovine serum albumin, 0.01 mg/ml soybean trypsin inhibitor (Sigma). All viruses were used at equal titer, and amounts were optimized to obtain complete delivery efficiency. Infected acini were then incubated at 37 °C in a humidified 5% CO2 atmosphere. In some experiments, the acini were treated with a trypsin inhibitor Pefabloc (Roche Applied Science), a pancaspase inhibitor Z-VAD (Bachem, Torrance, CA), cholecystokinin, TNF-α, or trypsin (Sigma) at the indicated concentrations.
Immunofluorescence
Pancreatic acini were infected with virus encoding PACE-trypsinogen for 8 h and fixed for 20 min in freshly prepared 4% paraformaldehyde. Infected acini were then embedded in 20% sucrose (water) and O.C.T. embedding medium (2:1 ratio). Frozen sections were cut (10 μm) and placed on Superfrost glass slides. The sections were postfixed for 10 min with acetone at −20 °C, air-dried, and then blocked with 5% bovine serum albumin in phosphate-buffered saline containing 0.02% Triton X-100. For antibody labeling of HA and amylase, sections were incubated overnight with rabbit polyclonal antibody (1:250) against HA (Rockland Immunochemicals, Gilbertsville, PA) and rabbit anti-amylase (1:1000; Sigma). Secondary antibodies were Cy3-labeled anti-rabbit and Cy5-labeled anti-rat IgG (Vector Laboratories, Burlingame, CA) The nuclei were counterstained with 4,6-diamidino-2-phenylindole blue fluorescent dye. Fluorescent imaging was performed on a Zeiss LSM510 confocal scanning microscope (Zeiss, Thornwood, NY).
Flow Cytometry
HEK293 cells were transfected with PACE-trypsinogen plasmid or an equal amount of β-galactosidase control plasmid. After 48 h, they were trypsinized and collected by centrifugation. The cell pellets were then suspended in 500 μl of propidium iodide-hypotonic lysis buffer containing 0.1% sodium citrate, 0.1% Triton X-100, 100 μg/ml RNase, and 50 μg/ml propidium iodide (BD Biosciences). The cell suspension was protected against light, incubated for 1 h on ice, and then analyzed with a FACSCalibur Cytometer (BD Biosciences).
Immunoblot Analysis
Whole cell extracts were prepared in radioimmune precipitation buffer (10 mm Tris (pH 7.5), 150 mm NaCl, 10 mm EDTA, 1% Nonidet P-40, 0.1% SDS, 1% sodium sarcosyl, 0.2 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 1 mm dithiothreitol). Samples (40 μg of protein each) were separated by electrophoresis in polyacrylamide-SDS gels and transferred to nitrocellulose membrane. Membranes were incubated in blocking buffer (5% nonfat dry milk in Tris-buffered saline, 0.1% Tween 20) for 1 h at room temperature and probed with primary antibodies (anti-HA (1:10,000), Rockland (Gilbertsville, PA); anti-caspase-3 and anti-PARP (1:1000), Cell Signaling (Danvers, MA)) overnight at 4 °C, followed by hybridization with a horseradish peroxidase-conjugated secondary antibody mouse IgG (1:4000) (Amersham Biosciences). Signals were detected by chemiluminescence using the ECL detection system (Amersham Biosciences).
DNA Isolation and Fragmentation Assay
Equal numbers of mouse acinar cells were infected with virus encoding PACE-trypsinogen, Dead-trypsinogen, or β-galactosidase control virus for 16 h. Cells were collected by centrifugation, and DNA was extracted using an apoptotic DNA ladder kit (Roche Applied Science) according to the manufacturer's instructions. Equal amounts of DNA from each sample were analyzed by electrophoresis on a 1.5% agarose gel containing ethidium bromide.
LDH Release Assay
Equal numbers of mouse acinar cells were infected with virus encoding PACE-trypsinogen or β-galactosidase control virus. Supernatant was analyzed at either 16 or 24 h for LDH activity according to manufacturer's instructions (LDH cytotoxicity assay kit, Cayman Chemical Co., Ann Arbor, MI).
NF-κB Luciferase Assay
Mouse pancreatic acinar cells were co-infected with control or PACE-trypsinogen virus together with the adenovirus-bearing NF-κB luciferase reporter gene (100:1) and incubated for 16 h. As positive control, isolated acini were treated with TNF-α (10 ng/ml) starting from 6 h after viral infection. Luciferase activity was measured using a luciferase assay kit according to the manufacturer's instructions (Promega, Madison, WI).
Real Time RT-PCR
Quantitative real time RT-PCR was conducted by previously published methods with modifications (25). Total RNA was prepared from pancreatic acini using TRIzol reagent (Invitrogen). RNA was purified by digestion for 15 min with DNase, and recovery of RNA was obtained using the RNeasy kit (Qiagen, Valencia, CA). Reverse transcription was conducted for 45 min at 48 °C from 70 ng of purified total RNA in a 25-μl volume of Access RT-PCR reaction mixture (Promega, Madison, WI) followed by 40 cycles of PCR (10-s denaturation at 94 °C, 1-min combined annealing and extension at 60 °C). SYBR Green I was used to monitor the PCR products on the I-Cycler thermal cycler and IQ real time PCR detection system (Bio-Rad). Primers designed for mob-1 (GenBankTM accession NM_001565) were as follows: forward, 5′-TGA AAG CAG TTA GCA AGG AAA TGT-3′; reverse, 5′-ATA CTC CAT GTA GGG AAG TGA TGG-3′. 18 S rRNA was used as an internal control (forward, 5′-GAG CGG TCG GCG TCC CCC AAC TTC-3′; reverse, 5′-GCG CGT GCA GCC CCG GAC ATC TAA-3′).
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay was performed for measuring NF-κB activity, as described previously (16). Mouse pancreatic acinar cells were treated with various concentrations of trypsin in the absence of trypsin inhibitor in the medium for 2 h. Nuclear extracts were prepared with a nuclear extraction kit (Pierce). Aliquots of nuclear extract with equal amounts of protein (10 μg) were used in 20-μl reactions in a buffer containing 10 mmol/liter HEPES (pH 7.9), 10% glycerol (v/v), 1 mmol/liter dithiothreitol, 1 μg of poly(dI-dC), and 5 μg of nuclease-free bovine serum albumin. The binding reaction was started by adding 100,000 cpm of the 22-base pair oligonucleotide 5′-AGT TGA GGG GAC TTT CCC AGG C-3′ containing the NF-κB consensus sequence (Promega, Madison, WI) labeled with [32P]adenosine triphosphate (10 mCi/mmol) by T4 polynucleotide kinase. To determine the specificity of NF-κB binding, unlabeled “cold” NF-κB oligonucleotide was added 10 min before adding the 32P-labeled NF-κB oligonucleotide. All reaction mixtures were subjected to polyacrylamide gel electrophoresis on a 4.5% gel in 0.5× Tris borate-EDTA buffer (44.5 mmol/liter Tris base, 44.5 mmol/liter boric acid, and 1 mmol/liter disodium EDTA, pH 8.3) at 100 V for 3 h. Gels were dried and directly exposed to a B-1 phosphorimaging screen and visualized with the use of a GS-505 Molecular Imaging System (Bio-Rad).
Electron Microscopy
For these studies, mouse pancreatic acini were harvested 10 h after infection with corresponding virus preparations. Samples were fixed with a solution containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1 m cacodylate buffer, pH 7.3, for 1 h. After fixation, the samples were washed and treated with 0.1% Millipore-filtered cacodylate-buffered tannic acid, postfixed with 1% buffered osmium tetroxide for 1 h, and stained en bloc with 1% Millipore-filtered uranyl acetate. The samples were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in Spurr's low viscosity medium. The samples were polymerized in an oven at 70 °C for 2 days. Ultrathin sections were cut in a Leica Ultracut microtome (Leica, Deerfield, IL), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL, Peabody, MA) at an accelerating voltage of 80 kV. Digital images were obtained using the AMT Imaging System (Advanced Microscopy Techniques Corp., Danvers, MA).
Statistical Analysis
If not otherwise stated, experiments were repeated three times, and descriptive analysis with mean ± S.E. was performed. To test whether results were significantly different from each other, data were analyzed using the unpaired t test.
RESULTS
A PACE-activable Trypsinogen Was Developed
To directly address the effects of intracellular trypsinogen activation, we developed a mutant trypsinogen (PACE-trypsinogen) that can be activated by PACE. The wild-type trypsinogen possesses internal sequences that direct it to the secretory pathway (signal peptide (SP)) and that maintain it in an inactive state (trypsinogen activation peptide (TAP)) (Fig. 1a). PACE-trypsinogen was developed by inserting a PACE recognition sequence -Arg-Thr-Lys-Arg-(-RTKR-) immediately before the critical Ile-24 (p.I24) site of trypsinogen (Fig. 1b). This chimeric trypsinogen is expected to be recognized by intracellular PACE and cleaved at the second arginine residue, leaving an active trypsin within the cell. To facilitate the immunodetection of the PACE-activable trypsinogen, an HA tag was added before the dibasic residues (Fig. 1b). The wild-type SP and TAP were kept in PACE-trypsinogen to ensure correct compartmentalization. As a control, we also produced a catalytically inactive trypsinogen (Dead-trypsinogen) by replacing the Ile24 with Leu24 (p.I24L) (Fig. 1c).
FIGURE 1.
Structure of wild type trypsinogen and recombinant trypsinogen mutants. a, wild type trypsinogen contains an SP for direction into the secretory pathway as well as the TAP sequence, which inhibits enzymatic activity. Within the duodenum, enteropeptidase cleaves off the TAP sequence and subsequently activates trypsinogen to trypsin by exposing the critical amino-terminal isoleucine. b, a mutant trypsinogen (PACE-trypsinogen) contains a PACE cleavage site (-RTKR-), which replaces the enteropeptidase cleavage site. Upon synthesis, trans-Golgi-localized PACE cleaves at its recognition site on PACE-trypsinogen and leaves activated trypsin within the secretory compartment. An HA tag was added to facilitate the detection of PACE-trypsinogen expression. c, by replacing the first critical amino acid of trypsin from Ile to Leu, an enzymatic inactive trypsinogen (Dead-trypsinogen, p.I24L) was developed.
PACE-trypsinogen Was Activated by PACE and Correctly Entered the Secretory Pathway
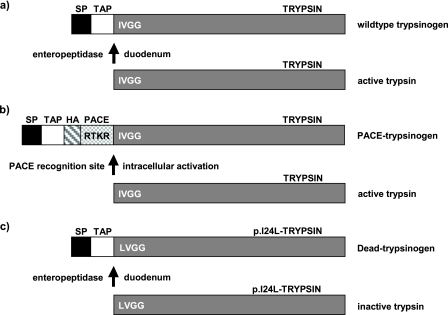
The PACE-trypsinogen was cloned to pAdTrack-CMV plasmid and transfected to HEK293 cells. 24 h later, cells were harvested, and lysates were used for Western blotting. Immunodetection with an anti-HA antibody showed successful expression (Fig. 2a). To test the functionality of the PACE-trypsinogen construct, the ability of exogenous PACE to activate this molecule was evaluated. PACE-trypsinogen was expressed in HEK293 cells and purified from cell lysates using an anti-HA high affinity antibody matrix. Incubation of purified PACE-trypsinogen with recombinant PACE led to the disappearance of the HA-tagged band in Western blots, indicating that PACE-trypsinogen was cleaved as predicted (Fig. 2b). Enzymatic activity of trypsin was detected by a fluorometric assay in PACE-trypsinogen samples treated with recombinant PACE (Fig. 2c). Untreated and therefore intact PACE-trypsinogen did not lead to any detectable enzymatic activity. These data indicate that PACE was able to cleave and activate PACE-trypsinogen as predicted in purified preparations.
FIGURE 2.
Expression, cleavage, and enzymatic functionality of PACE-trypsinogen. a, HEK 293 cells were transfected with pAd-Track-CMV-PACE-trypsinogen plasmid or empty pAd-Track-CMV control plasmid for 24 h, and cell lysates were subjected to Western blot with an anti-HA antibody. HA-tagged PACE-trypsinogen was detected in HEK293 cells transfected with pAd-Track-CMV-PACE-trypsinogen plasmid but not in empty vector control cells. b, PACE-trypsinogen was purified from HEK293 cell lysates, which were transfected with pAd-Track-CMV-PACE-trypsinogen plasmid using an anti-HA high affinity antibody matrix. Incubation of purified PACE-trypsinogen with recombinant PACE for 2 h led to the disappearance of the HA band in Western blots, indicating that the PACE-trypsinogen was completely cleaved by PACE. c, affinity-purified PACE-trypsinogen cleaved by recombinant PACE, but not uncleaved PACE-trypsinogen, showed a prominent increase in enzymatic activity, as detected by a fluorometric assay. In this assay, trypsin activates a specific fluorescent substrate, and the slope of increasing fluorescence intensity reflects trypsin activity. The arrow indicates when samples were added. Shown is an experiment representative of three similar responses.
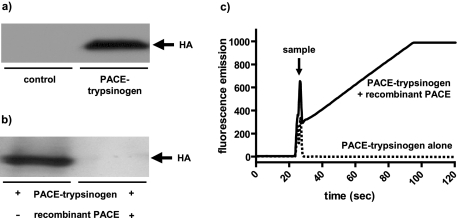
Next, we wished to determine whether the PACE-trypsinogen mutant would enter the correct, physiologically relevant compartment, which is the secretory pathway. As designed, PACE-trypsinogen retained the wild type trypsinogen SP sequence (Fig. 1b) that should direct the expressed molecule to the secretory compartment. To test this assumption, PACE-trypsinogen was expressed in mouse pancreatic acini using adenovirus-mediated gene transfer and was localized using immunofluorescence. Infection of isolated mouse acini with PACE-trypsinogen-bearing virus led to the expression of the HA-tagged protein as early as 5 h postinfection (Fig. 3a). Confocal imaging of HA antibody directed immunofluorescence in pancreatic acini 8 h postinfection revealed that PACE-trypsinogen was discretely colocalized with amylase to the Golgi region and the early secretory compartment, thus indicating that PACE-trypsinogen had correctly entered the secretory pathway (Fig. 3b). Punctate instead of diffuse localization of PACE-trypsinogen indicated the packaging of the enzyme into early granules. These granules were found mostly in proximity of the Golgi region, and HA-staining appeared to be lost in mature granules. This supports the hypothesis that the SP-TAP-HA-PACE recognition portion of PACE-trypsinogen is removed and that the protein is therefore activated in the early post-Golgi compartment.
FIGURE 3.
Expression and subcellular localization of PACE-trypsinogen in isolated pancreatic acini. a, isolated mouse acini were infected with PACE-trypsinogen-bearing viruses, and cell lysates were subjected to Western blot with an anti-HA antibody at the indicated times. The expression of the HA-tagged protein was detected as early as 5 h postinfection. b, confocal imaging of HA-directed immunofluorescence of isolated acini infected with PACE-trypsinogen virus and incubated for 8 h revealed that PACE-trypsinogen (red) was colocalized with amylase (green) to the early secretory pathway (yellow, colocalization; blue, nucleus).
PACE-trypsinogen Was Activated Intracellularly and Induced Apoptosis in HEK293 Cells
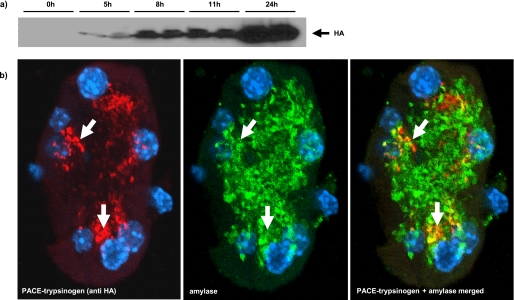
To determine whether PACE-trypsinogen could be activated intracellularly, we initially investigated the effects of expression in HEK293 cells. Within 24–96 h of transfection, the expression of PACE-trypsinogen caused a dramatic reduction in the number of transfected cells attached to the culture dish (Fig. 4a). The PACE-trypsinogen-transfected cells had a shrunken appearance compared with control cells, suggesting that apoptosis might be occurring (Fig. 4a). To evaluate the level of apoptosis arising within 48 h, flow cytometry was utilized to quantify the number of cells with a fractionated sub-G1 content of DNA. We found that there was a 2.5-fold increase in the number of cells in this population when HEK293 cells were transfected with PACE-trypsinogen compared with empty vector control (Fig. 4b). Additionally, expression of the enzymatic inactive mutant Dead-trypsinogen did not affect cell viability within 24–96 h (data not shown). As early as by 24 h, PACE-trypsinogen transfection also dramatically increased the cleavage of PARP, a 116-kDa nuclear poly(ADP-ribose) polymerase that is one of the main cleavage targets of caspase-3 during the initiation of apoptosis (Fig. 4c). These data indicate that expression of PACE-trypsinogen in HEK293 cells led to intracellular trypsin activity and initiated cellular apoptosis through caspase-dependent pathways.
FIGURE 4.
Intracellular expression of PACE-trypsinogen induced apoptosis in HEK293 cells. a, transfection of pAd-Track-CMV-PACE-trypsinogen, which expresses both green fluorescent protein and PACE-trypsinogen, dramatically eliminated transfected HEK293 cells after 4 days compared with a vector expressing Dead-trypsinogen. Also, most PACE-trypsinogen-transfected cells appeared shrunken compared with control cells, indicating apoptosis. b, cells with sub-G1 DNA were quantified by flow cytometry 48 h after transfection with PACE-trypsinogen or empty control plasmid in HEK293 cells. There was a significant increase of the cell population with sub-G1 DNA in the PACE-trypsinogen-transfected group. Data shown are means ± S.E. for three separate experiments (*, p < 0.01). c, PACE-trypsinogen transfection increased PARP cleavage in HEK293 cells within 24 h. Shown are three separate samples from cells transfected with either PACE-trypsinogen or empty control plasmid.
PACE-trypsinogen Was Activated in Pancreatic Acinar Cells and Induced Apoptosis
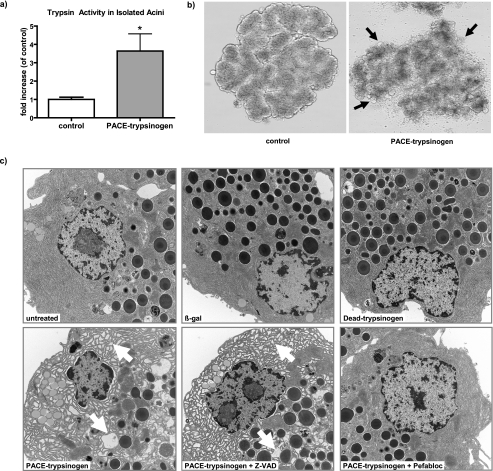
Pancreatic acinar cells are specialized to produce digestive enzymes and have additional protective mechanisms that are not found in other cell types, such as HEK293 cells. Therefore, we next determined the effects of intracellular trypsinogen activation on the native cell type, pancreatic acinar cells. To this end, isolated mouse acinar cells were infected with adenoviruses expressing PACE-trypsinogen or β-galactosidase as a control. Abundant trypsin activity was detected in PACE-trypsinogen virus-infected acini but not cells infected with control virus (Fig. 5a). These data indicated that the PACE-trypsinogen was indeed activated in pancreatic acinar cells.
FIGURE 5.
Expression of PACE-trypsinogen caused increased intracellular trypsin activity that disrupted pancreatic acinar cell morphology. a, elevated trypsin activity was detected in lysates from mouse pancreatic acinar cells that had been infected with the PACE-trypsinogen virus (24 h) but not acini infected with control virus (β-galactosidase; β-gal). Data shown are means ± S.E. for three experiments. (*, p < 0.05). b, infection with PACE-trypsinogen for 24 h caused profound acinar cell blebbing (arrows). Blebbing was not prominent with control virus expressing only β-galactosidase. c, on the level of electron microscopy, PACE-trypsinogen expression caused profound changes in pancreatic acinar cell morphology, including ER swelling and vacuolization (arrows). These changes were blocked by a trypsin inhibitor Pefabloc (100 μm) but not a pancaspase inhibitor Z-VAD (50 μm). Neither β-galactosidase nor Dead-trypsinogen-expressing viruses caused any changes in acinar cell morphology compared with untreated control. Pancreatic acini were infected with the indicated viruses, incubated for 10 h, fixed, and subjected to electron microscopy (magnification ×10,000).
Expression of PACE-trypsinogen in pancreatic acinar cells caused dramatic changes in cell architecture despite the multifaceted protective mechanisms known to exist in these cells. PACE-trypsinogen caused profound acinar cell blebbing at the light microscopy level (Fig. 5b, arrows). Closer examination of the cells using electron microscopy indicated that the expression of intracellular active trypsin led to severe ER swelling, formation of vacuoles (Fig. 5c, arrows), and apoptosis. The morphological changes were inhibited by the trypsin inhibitor Pefabloc but not by a pancaspase inhibitor, Z-VAD, suggesting that these effects were mediated by caspase-independent pathways (Fig. 5c). The absence of ER swelling in acini infected with catalytically inactive Dead-trypsinogen and β-galactosidase virus controls further confirmed that the damage was related to trypsin activity rather than nonspecific overexpression of foreign proteins.
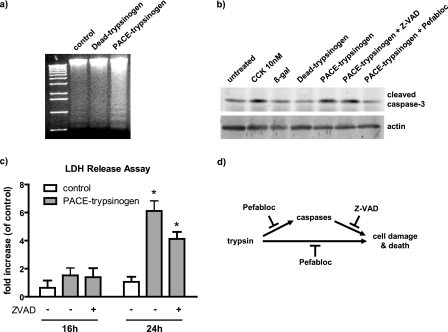
Next, we examined the effects of intracellular active trypsin on acinar cell apoptosis. An increased rate of acinar cell apoptosis after infection with the PACE-trypsinogen virus was indicated, as shown by analysis of DNA fragmentation (DNA laddering) (Fig. 6a). Similarly, caspase-3 cleavage was increased when pancreatic acini were infected with PACE-trypsinogen virus, indicating that intracellular trypsin activity also caused activation of caspase-3 (Fig. 6b). Notably, the cleavage of caspase-3 was not inhibited by a pancaspase inhibitor, suggesting that the cleavage might be mediated directly by trypsin (Fig. 6b). In this case, we hypothesized that inhibition of trypsin should inhibit the cleavage. Indeed, caspase-3 cleavage was blocked by treatments with the trypsin inhibitor Pefabloc (Fig. 6b).
FIGURE 6.
Expression of PACE-trypsinogen caused acinar cell death. a, DNA fragmentation was elevated in PACE-trypsinogen virus-infected acinar cells compared with Dead-trypsinogen and β-galactosidase control virus-infected cells at 16 h postinfection. b, intracellular trypsin increased the level of cleaved caspase-3. Cells were infected with viruses bearing β-galactosidase, Dead-trypsinogen, or PACE-trypsinogen. After 16 h, cells were harvested, and proteins were blotted with an antibody specific for active caspase-3. PACE-trypsinogen-expressing acini showed increased caspase-3 cleavage, which could be blocked by Pefabloc (100 μm) but not by pancaspase inhibitor Z-VAD (50 μm). Acini treated with supraphysiologic cholecystokinin (10 nm) served as a positive control, and β-galactosidase- or Dead-trypsinogen-expressing virus-infected acini were used as negative controls. c, as a measure of more general cell damage, LDH release was evaluated in acini infected with PACE-trypsinogen virus at different times in the presence and absence of Z-VAD (50 μm). LDH release was not affected by 16 h after infection of acini. However, increased release of LDH was noted after 24 h, indicating advanced cell damage. This increase was not prevented by the presence of Z-VAD. Data presented show LDH -fold increase compared with untreated acini. Green fluorescent protein adenovirus-infected cells served as control. Results presented are means ± S.E. for three experiments. (*, p < 0.05 versus control). d, intracellular active trypsin caused cell death through caspase-dependent and -independent pathways. Inhibition of active trypsin with Pefabloc blocks both caspase activation and cell damage and death. However, inhibition of caspase activity with Z-VAD did not prevent cell death initiated by intracellular trypsin.
In contrast to the rapid effects of intracellular trypsin on acinar cell apoptosis, LDH release, a marker of plasma membrane damage, was not altered by expression of PACE-trypsinogen at either 8 (data not shown) or 16 h (Fig. 6c). LDH release was significantly increased in the PACE-trypsinogen virus-infected acini compared with the control virus-treated group after 24 h. The release of LDH was not significantly inhibited by the pancaspase inhibitor Z-VAD (Fig. 6c). These data suggest that pancreatic acini expressing intracellular active trypsin may undergo further cell damage at this late stage and that intracellular trypsin caused cell damage independent of its ability to activate caspase-3. Taken together, these data provide direct evidence that intracellular activation of trypsinogen leads to pancreatic acinar cell death through caspase-dependent and -independent pathways (Fig. 6d).
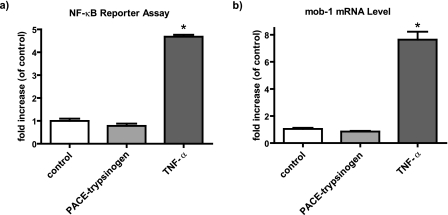
NF-κB Was Not Activated by Intracellular Trypsin
NF-κB is a central mediator of the inflammatory response observed in acute pancreatitis. Therefore, we analyzed whether intracellular trypsinogen activation could trigger NF-κB activation. The effects of PACE-trypsinogen expression on NF-κB were examined in HEK293 (data not shown) and isolated mouse pancreatic acinar cells using an NF-κB luciferase reporter and by measuring mRNA levels of a known NF-κB target gene, mob-1 (Fig. 7). Expression of PACE-trypsinogen did not increase expression of an NF-κB luciferase reporter (Fig. 7a). Intracellular trypsin activity also did not stimulate expression of NF-κB target gene mob-1 in either pancreatic acinar cells (Fig. 7b) or HEK293 cells (data not shown). In contrast, TNF-α, a known activator of NF-κB, caused a large increase in luciferase activity and mob-1 expression on isolated pancreatic acinar cells (Fig. 7, a and b).
FIGURE 7.
Intracellular trypsin activity did not activate NF-κB in pancreatic acinar cells. a, activity of NF-κB luciferase reporter was not altered by infection with PACE-trypsinogen as compared with β-galactosidase control-bearing adenoviruses at 16 h postinfection. In contrast, treatment with TNF-α (10 ng/ml) caused a large increase in luciferase signal. b, expression of PACE-trypsinogen (or β-galactosidase control) for 16 h did not increase expression of the NF-κB target gene, mob-1, mRNA as measured by quantitative real time RT-PCR. However, mob-1 expression was strongly induced by TNF-α (10 ng/ml), used as a positive control. Data shown are means ± S.E. for three experiments (*, p < 0.05).
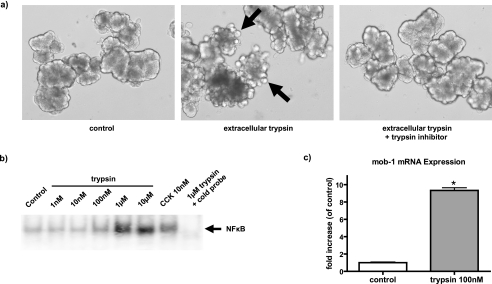
Extracellular Trypsin Caused Cell Damage and Activated NF-κB
We hypothesized that the activation of NF-κB and the inflammatory response observed after trypsinogen activation in pancreatitis could be caused by extracellular trypsin that may leak from dying cells. To test this hypothesis, we treated isolated mouse pancreatic acini in primary culture with active trypsin. Extracellular trypsin rapidly caused pancreatic acinar cells to bleb, and this effect could be totally blocked by a specific trypsin inhibitor (soybean trypsin inhibitor) (Fig. 8a). Active trypsin also caused a rapid increase in NF-κB DNA binding activity, as determined by EMSA (Fig. 8b), and NF-κB target gene mRNA expression, as measured by quantitative RT-PCR (Fig. 8c). These effects of extracellular trypsin were dose-dependent, with significant effects noted at 100 nm and maximal effects at 1 μm trypsin (Fig. 8b). These data support the hypothesis that extracellular but not intracellular trypsin activity leads to NF-κB activation in pancreatic acinar cells.
FIGURE 8.
Extracellular trypsin disrupted acinar cell morphology and strongly activated NF-κB in pancreatic acinar cells. a, extracellular trypsin (1 μm) caused blebbing within 2 h when it was added to the incubation medium. This effect was totally blocked by a specific trypsin inhibitor (soybean trypsin inhibitor; 0.01%). b, extracellular trypsin dose-dependently increased NF-κB binding activity as determined by EMSA. Cholecystokinin (10 nm)-treated acinar cells served as a positive control. The binding can be competed by unlabeled cold probe, indicating that the binding is NF-κB-specific. Data shown are representative of two separate experiments. c, NF-κB target gene mob-1 mRNA expression was up-regulated within 2 h by extracellular trypsin, as measured by quantitative real time RT-PCR. Data shown are means ± S.E. of three experiments (*, p < 0.05).
DISCUSSION
In the current study, we developed a new model in which trypsinogen is specifically targeted for activation within the acinar cell. Using this model, we observed for the first time that intracellular trypsin activity leads to acinar cell apoptosis without activation of the proinflammatory transcription factor NF-κB. In contrast, extracellular trypsin activity was a powerful inducer of the inflammatory signaling cascade. These observations have important implications for the mechanisms involved in the initiation of acute pancreatitis.
The central paradigm of pancreatitis research has long been that premature activation of trypsinogen within the acinar cell is the initiator of pancreatitis (16, 18, 26–28). However, the mechanisms underlying the trypsin-induced injury remain unclear. Our understanding of the role of trypsinogen activation in pancreatitis has been hampered by the complexity of the animal models utilized in its study. Currently, there are no models that allow direct control of trypsin activity in the pancreas. Instead, models of pancreatitis involve administration of high concentrations of secretagogues, retrograde injection of bile salts into the pancreatic duct, or other harsh procedures that caused generalized cell damage and cell signaling pathway activation. For example, caerulein hyperstimulation, which is the most common animal model of acute pancreatitis, causes both intracellular trypsinogen activation and NF-κB activation (29). Thus, these models are not suitable to investigate specifically the role of intracellular trypsinogen activation and the relationship between trypsin activity and NF-κB activation. To overcome this obstacle, we were prompted to develop a unique mutant trypsinogen that is activated intracellularly.
Trypsin is a member of the large and diverse serine peptidase family. Trypsinogen is normally synthesized within the pancreatic acinar cells as a zymogen. A signal peptide ensures correct localization to the secretory pathway and storage in zymogen granules. Upon secretion, trypsinogen is flushed out through the pancreatic duct to the duodenum where the brush border enzyme, enteropeptidase, converts it to active trypsin by removal of an inhibitory peptide, TAP, from the amino terminus. Upon enteropeptidase cleavage, the nascent amino-terminal isoleucine (p.I24) fits into a hydrophobic pocket in the protein, thus directing the amino terminus to form a salt bridge to Asp-194, which appears to be a critical site to obtain enzymatic activity of trypsin (30). Active trypsin subsequently activates other zymogens, including trypsinogen, by removing inhibitory peptides in a similar manner.
The key to the current study was the development of a mutant form of trypsinogen that is activated by an endogenous mechanism. This was achieved by replacing the endogenous enteropeptidase cleavage site with a sequence that could be recognized and cleaved by PACE. PACE (also called furin) is a mammalian propeptide-processing endoprotease that is present in the trans-Golgi network of virtually all nonendocrine cells. PACE proteolytically activates a large number of proprotein substrates, such as TGF-β, neural growth factor, insulin-like growth factor, and natriuretic peptide, as they enter the secretory pathway (31). PACE has previously been utilized to process a modified insulin precursor to mature insulin in the pancreas (32). Thus, expression of PACE-activable trypsinogen is predicted to be cleaved intracellularly by PACE and allows the evaluation of the effects of trypsinogen activation in the absence of the confounding factors present in conventional models. Indeed, we found that purified mutant trypsinogen was cleaved and activated by recombinant PACE.
The location of the pathological activation of trypsinogen within human acinar cells during pancreatitis is uncertain. In the caerulein-induced pancreatitis model, it is suggested that zymogen activation is mediated by lysosomal hydrolases, such as cathepsin B, and made possible by perturbations in intracellular trafficking of proteins, which cause lysosomal hydrolases and digestive enzyme zymogens to become co-localized within intracellular membrane-bounded organelles (33–35). This co-localization phenomenon is thought to lead to trypsinogen activation, its release to the cytosol, and subsequent cell injury. For the mutant trypsinogen utilized in the current study, the wild-type signal peptide was unaltered, ensuring that it was directed into its normal physiological compartment, the secretory pathway. Based on the known localization of PACE, we predicted that the mutant trypsinogen would be activated in the trans-Golgi network of the secretory pathway. This prediction was supported by the observation that intact PACE-trypsinogen was primarily localized in proximity to the Golgi region. Furthermore, punctate localization indicated packaging of PACE-trypsinogen into granules. Loss of staining in apical parts of the acinar cells, where mature zymogen granules are located, suggested that PACE-trypsinogen was cleaved, resulting in the loss of the HA tag, and was therefore activated within the early secretory pathway. However, due to its ability to cause cell apoptosis, it seems likely that the PACE-activated protease may have gained access to the cytosol. The determination of the precise intracellular sites of action of the active trypsin in this model will require further studies.
We found that the presence of intracellular active trypsin rapidly induced prominent DNA laddering, indicating that apoptosis occurred at early times. This apoptosis may partially be executed through a caspase-dependent pathway, because PARP and caspase-3 were observed to be cleaved. However, inhibition of caspase activity with a pancaspase inhibitor did not prevent trypsin-induced cell morphological changes at early times (Fig. 5c) or cell death at later times (Fig. 6c), suggesting that intracellular trypsin causes cell damage through caspase-independent pathways as well. At later times, there was evidence of advanced cell damage, as indicated by increased release of LDH in the culture medium. However, this late severe response may have been influenced by prolonged culture of the acini in addition to excessive trypsin activity generated by adenoviral expression. There are also important differences in the sensitivity of acinar cells from different species to apoptotic insults (36). Therefore, it remains unclear which form of cell death may predominate when trypsin is activated intracellularly in human pancreatic acinar cells. Nonetheless, the current data support apoptosis rather than necrosis as the primary response to intracellular trypsin activity.
Another concern in the current study is the possibility that excessive expression of a foreign protein may induce a nonspecific activation of ER stress mechanism, and ER stress itself may lead to pancreatitis (37). Moreover, one of the prominent features of this model was swollen ER, a sign of excessive ER stress. To address this issue, we compared the effects of the active trypsin construct with controls that overexpressed either β-galactosidase or a catalytically inactive mutant trypsinogen (Dead-trypsinogen). Both constructs lacking trypsin activity caused no obvious changes in HEK293 cells or pancreatic acinar cells, indicating that the effects observed were not caused by nonspecific effects of gene expression on the ER. This was further confirmed by the fact that an intracellular trypsin inhibitor, Pefabloc, blocked the deleterious biological effects of active trypsin expression.
NF-κB activation has been found to be central to the inflammatory response which occurs early during the initiation of acute pancreatitis. Therefore, it seems reasonable that NF-κB activation may be important in transforming the signal from intracellular trypsin activity to an inflammatory response. However, the relationship between intracellular trypsin activity and NF-κB activation has largely been unclear. Previously, it has been shown that active NF-κB does not induce trypsinogen activation (18, 19). In the current study, we determined for the first time that intracellular trypsin activity does not activate NF-κB. In contrast, we found that extracellular trypsin caused dramatic effects on acinar cells, including extensive cell blebbing and activation of NF-κB, even at very low concentrations. These effects may be mediated by proteinase-activated receptors, since trypsin is a strong activator of PAR-2 (proteinase-activated receptor-2) and PAR-4 and also a weak activator of PAR-1 and PAR-3 (38). It has previously been demonstrated that PAR-2 and PAR-4 exist on pancreatic acinar cells (39–41). Thus, it is likely that trypsin could activate proteinase-activated receptors on acinar cells and subsequently cause activation of NF-κB. Further research will be necessary to investigate the details of this response.
Taken together, this study provides the first direct test of the biological effects of intracellular trypsin activity on pancreatic acinar cell function. The results indicate that intracellular trypsin induces acinar cell apoptosis without activation of NF-κB. In contrast, extracellular trypsin is a powerful inducer of NF-κB. This may prove to be an important new finding, since NF-κB is a critical component of the inflammatory response seen during pancreatitis. Therefore, the localization of enzymatically active trypsin seems to be the key to determining its biological effects within the pancreas.
Acknowledgments
We thank Diane Simeone, Yan Bi, and John A. Williams for helpful discussions and insights and also Brad Nelson for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK52067-05 (to C. D. L.), R21 DK068414-02 (to B. J.), and 2P30 DK034933-20 (to B. J.). Resources from the combined Morphology and Image Analysis Core of the Michigan Gastrointestinal Peptide Center (National Institutes of Health Grant P30 DK-34933) and the Michigan Diabetes and Research Training Center (National Institutes of Health Grant P60 DK-20572) further contributed to this research.
- PACE
- paired basic amino acid cleaving enzyme
- HA
- hemagglutinin
- CMV
- cytomegalovirus
- MOPS
- 4-morpholinepropanesulfonic acid
- Z
- benzyloxycarbonyl
- BOC
- butyloxycarbonyl
- MCA
- methylcoumarinamide
- TNF
- tumor necrosis factor
- RT
- reverse transcription
- TAP
- trypsinogen activation peptide
- SP
- signal peptide
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Mitchell R. M., Byrne M. F., Baillie J. ( 2003) Lancet 361, 1447– 1455 [DOI] [PubMed] [Google Scholar]
- 2.Chiari H. ( 1896) Z. Heilk. 17, 69– 96 [Google Scholar]
- 3.Pandol S. J. ( 2005) Curr. Opin. Gastroenterol. 21, 538– 543 [DOI] [PubMed] [Google Scholar]
- 4.Geokas M. C., Rinderknecht H. ( 1974) Am. J. Dig. Dis. 19, 591– 598 [DOI] [PubMed] [Google Scholar]
- 5.Cavallini G., Tittobello A., Frulloni L., Masci E., Mariana A., Di Francesco V. ( 1996) N. Engl. J. Med. 335, 919– 923 [DOI] [PubMed] [Google Scholar]
- 6.Gilliland L., Steer M. L. ( 1980) Am. J. Physiol. 239, G418– 426 [DOI] [PubMed] [Google Scholar]
- 7.Koike H., Steer M. L., Meldolesi J. ( 1982) Am. J. Physiol. 242, G297– 307 [DOI] [PubMed] [Google Scholar]
- 8.Niederau C., Grendell J. H. ( 1988) J. Clin. Invest. 81, 229– 236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leach S. D., Modlin I. M., Scheele G. A., Gorelick F. S. ( 1991) J. Clin. Invest. 87, 362– 366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bialek R., Willemer S., Arnold R., Adler G. ( 1991) Scand. J. Gastroenterol. 26, 190– 196 [DOI] [PubMed] [Google Scholar]
- 11.Whitcomb D. C., Gorry M. C., Preston R. A., Furey W., Sossenheimer M. J., Ulrich C. D., Martin S. P., Gates L. K., Jr., Amann S. T., Toskes P. P., Liddle R., McGrath K., Uomo G., Post J. C., Ehrlich G. D. ( 1996) Nat. Genet. 14, 141– 145 [DOI] [PubMed] [Google Scholar]
- 12.Senftleben U., Karin M. ( 2002) Crit Care Med. 30, Suppl. 1, 18– 26 [PubMed] [Google Scholar]
- 13.Grady T., Liang P., Ernst S. A., Logsdon C. D. ( 1997) Gastroenterology 113, 1966– 1975 [DOI] [PubMed] [Google Scholar]
- 14.Gukovsky I., Gukovskaya A. S., Blinman T. A., Zaninovic V., Pandol S. J. ( 1998) Am. J. Physiol. 275, G1402– 1414 [DOI] [PubMed] [Google Scholar]
- 15.Steinle A. U., Weidenbach H., Wagner M., Adler G., Schmid R. M. ( 1999) Gastroenterology 116, 420– 430 [DOI] [PubMed] [Google Scholar]
- 16.Chen X., Ji B., Han B., Ernst S. A., Simeone D., Logsdon C. D. ( 2002) Gastroenterology 122, 448– 457 [DOI] [PubMed] [Google Scholar]
- 17.Baumann B., Wagner M., Aleksic T., von Wichert G., Weber C. K., Adler G., Wirth T. ( 2007) J. Clin. Invest. 117, 1502– 1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han B., Ji B., Logsdon C. D. ( 2001) Am. J. Physiol. Cell Physiol. 280, C465– 472 [DOI] [PubMed] [Google Scholar]
- 19.Hietaranta A. J., Saluja A. K., Bhagat L., Singh V. P., Song A. M., Steer M. L. ( 2001) Biochem. Biophys. Res. Commun. 280, 388– 395 [DOI] [PubMed] [Google Scholar]
- 20.Banerjee A. K., Galloway S. W., Kingsnorth A. N. ( 1994) Br. J. Surg. 81, 1096– 1103 [DOI] [PubMed] [Google Scholar]
- 21.Kaiser A. M., Saluja A. K., Sengupta A., Saluja M., Steer M. L. ( 1995) Am. J. Physiol. 269, C1295– 1304 [DOI] [PubMed] [Google Scholar]
- 22.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 2509– 2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji B., Kopin A. S., Logsdon C. D. ( 2000) J. Biol. Chem. 275, 19115– 19120 [DOI] [PubMed] [Google Scholar]
- 24.Kawabata S., Miura T., Morita T., Kato H., Fujikawa K., Iwanaga S., Takada K., Kimura T., Sakakibara S. ( 1988) Eur. J. Biochem. 172, 17– 25 [DOI] [PubMed] [Google Scholar]
- 25.Ji B., Bi Y., Simeone D., Mortensen R. M., Logsdon C. D. ( 2001) Gastroenterology 121, 1380– 1390 [DOI] [PubMed] [Google Scholar]
- 26.Grady T., Mah'Moud M., Otani T., Rhee S., Lerch M. M., Gorelick F. S. ( 1998) Am. J. Physiol. Gastrointest. Liver Physiol. 275, G1010– 1017 [DOI] [PubMed] [Google Scholar]
- 27.Steer M. L., Meldolesi J. ( 1988) Annu. Rev. Med. 39, 95– 105 [DOI] [PubMed] [Google Scholar]
- 28.Gukovskaya A. S., Vaquero E., Zaninovic V., Gorelick F. S., Lusis A. J., Brennan M. L., Holland S., Pandol S. J. ( 2002) Gastroenterology 122, 974– 984 [DOI] [PubMed] [Google Scholar]
- 29.Gukovskaya A. S., Gukovsky I., Jung Y., Mouria M., Pandol S. J. ( 2002) J. Biol. Chem. 277, 22595– 22604 [DOI] [PubMed] [Google Scholar]
- 30.Gráf L., Hegyi G., Likó I., Hepp J., Medzihradszky K., Craik C. S., Rutter W. J. ( 1988) Int. J. Pept. Protein Res. 32, 512– 518 [DOI] [PubMed] [Google Scholar]
- 31.Thomas G. ( 2002) Nat. Rev. Mol. Cell Biol. 3, 753– 766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanagita M., Hoshino H., Nakayama K., Takeuchi T. ( 1993) Endocrinology 133, 639– 644 [DOI] [PubMed] [Google Scholar]
- 33.Berndt W., Müller-Wieland K. ( 1970) Pol. Arch. Med. Wewn 4, 535– 537 [PubMed] [Google Scholar]
- 34.Saluja A. K., Donovan E. A., Yamanaka K., Yamaguchi Y., Hofbauer B., Steer M. L. ( 1997) Gastroenterology 113, 304– 310 [DOI] [PubMed] [Google Scholar]
- 35.Lerch M. M., Halangk W., Krüger B. ( 2000) Adv. Exp. Med. Biol. 477, 403– 411 [DOI] [PubMed] [Google Scholar]
- 36.Mareninova O. A., Sung K. F., Hong P., Lugea A., Pandol S. J., Gukovsky I., Gukovskaya A. S. ( 2006) J. Biol. Chem. 281, 3370– 3381 [DOI] [PubMed] [Google Scholar]
- 37.Kubisch C. H., Logsdon C. D. ( 2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1804– 1812 [DOI] [PubMed] [Google Scholar]
- 38.Nguyen T. D., Moody M. W., Steinhoff M., Okolo C., Koh D. S., Bunnett N. W. ( 1999) J. Clin. Invest. 103, 261– 269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawabata A., Nishikawa H., Kuroda R., Kawai K., Hollenberg M. D. ( 2000) Br. J. Pharmacol. 129, 1808– 1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bohm S. K., Kong W., Bromme D., Smeekens S. P., Anderson D. C., Connolly A., Kahn M., Nelken N. A., Coughlin S. R., Payan D. G., Bunnett N. W. ( 1996) Biochem. J. 314, 1009– 1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu W. F., Andersen H., Whitmore T. E., Presnell S. R., Yee D. P., Ching A., Gilbert T., Davie E. W., Foster D. C. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 6642– 6646 [DOI] [PMC free article] [PubMed] [Google Scholar]