Abstract
The nematode cuticle is a protective collagenous extracellular matrix that is modified, cross-linked, and processed by a number of key enzymes. This Ecdysozoan-specific structure is synthesized repeatedly and allows growth and development in a linked degradative and biosynthetic process known as molting. A targeted RNA interference screen using a cuticle collagen marker has been employed to identify components of the cuticle biosynthetic pathway. We have characterized an essential peroxidase, MoLT-7 (MLT-7), that is responsible for proper cuticle molting and re-synthesis. MLT-7 is an active, inhibitable peroxidase that is expressed in the cuticle-synthesizing hypodermis coincident with each larval molt. mlt-7 mutants show a range of body morphology defects, most notably molt, dumpy, and early larval stage arrest phenotypes that can all be complemented with a wild type copy of mlt-7. The cuticles of these mutants lacks di-tyrosine cross-links, becomes permeable to dye and accessible to tyrosine iodination, and have aberrant collagen protein expression patterns. Overexpression of MLT-7 causes mutant phenotypes further supporting its proposed enzymatic role. In combination with BLI-3, an H2O2-generating NADPH dual oxidase, MLT-7 is essential for post-embryonic development. Disruption of mlt-7, and particularly bli-3, via RNA interference also causes dramatic changes to the in vivo cross-linking patterns of the cuticle collagens DPY-13 and COL-12. This points toward a functionally cooperative relationship for these two hypodermally expressed proteins that is essential for collagen cross-linking and proper extracellular matrix formation.
Introduction
Collagenous extracellular matrices (ECMs)3 serve numerous critical roles and are found throughout the animal kingdom. The cuticle is an ECM that makes up the most external surface of nematodes and its roles are diverse. This tough but flexible exoskeleton maintains body shape, provides a protective barrier to the environment, and permits motility via its attachments to muscles (1). There are two distinct sets of discernible cuticular structures on the surface of the nematode Caenorhabditis elegans, circumferential indentations termed annulae and longitudinal ridges termed alae. The latter are present only in the first and alternative third stage (dauer) larvae and in adult C. elegans (2). At the transition between consecutive larval stages, a new cuticle is synthesized from the underlying hypodermis. The term molting is used for the removal and re-synthesis of the cuticle. The cuticle is a complex and versatile tissue made up of multiple layers, each of which has distinct components and levels of structural integrity (3, 4). The principal components of the cuticle are the collagens. Over 180 cuticle collagens are encoded in the C. elegans genome, and their temporal expression is cyclical and corresponds to the larval molts (5). Each of these synthetic periods is further subdivided into distinct peaks as follows: 4 h prior to, 2 h prior to, and coincident with the molt. Collagens whose temporal expression profiles coincide are predicted to interact with each other and in turn form distinct cuticle substructures (4). The processing of collagens into a functional cuticle entails a complex sequence of modifications, triple helix formation, proteolysis, and cross-linking events that are described in a number of reviews (1, 6–8). Mutations in specific collagens and their processing enzymes result in aberrant cuticle formation that leads to distinctive body shape phenotypes such as Dpy (dumpy, short and fat), Rol (roller), or Bli (blistered).
Here we focus on a peroxidase enzyme and its role in the cross-linking steps of cuticle biogenesis during which the pre-formed collagen triple helices are joined by intermolecular disulfide (9), glutamine-derived (10, 11), and tyrosine-derived bonds (12, 13). Disulfide bonds are catalyzed by enzymes of the thioredoxin class and are soluble in detergent and reducing agents, while glutamate and tyrosine bonds, which are resistant to reducing conditions (2, 14), are catalyzed by transglutaminases and peroxidase enzymes, respectively (15). A proportion of the adult cuticle remains intact after treatment in reducing conditions and represents the components that are cross-linked by the latter two enzymes. Differing extents of cuticle solubility are observed in different larval stages, suggesting function-related, stage-specific cross-linking. For example, the extensively cross-linked dauer cuticle may facilitate the extreme resilience of this stage to adverse conditions (14). Mutants of potential cross-link catalyzing enzymes exhibit morphological aberrations that range from Dpy to lethality (Let), and this exemplifies the requirement of these bonds in cuticle biogenesis (1, 6).
Tyrosine linkages are di- and tri-tyrosines and, specifically in nematodes, isotri-tyrosine bonds, all of which are biphenyl linkages of tyrosine molecules (12). In nematodes, these bonds link both collagens and cuticulins (12, 16, 17). This is in contrast to the ECMs of vertebrates in which the occurrence of tyrosine-derived cross-links is rare; instead cross-links are predominantly derived from hydroxylated lysine residues (7). Lysyl hydroxylase-derived linkages are absent from the C. elegans cuticle (14, 18) but do form critical cross-links in the nematode basement membrane collagens (18).
Peroxidase enzymes form cross-linked bonds by a reaction involving the oxidation of halides. The extensive family of peroxidases are found in all animals, fungi, bacteria, and plants (19) and includes the myeloperoxidase, eosinophil peroxidase, lactoperoxidase, thyroid peroxidase, and yeast cytochrome c peroxidase (20). In higher animals, peroxidases protect against bacteria, parasites, reactive-oxygen damage, and are also biosynthetic in nature, for example in the generation of thyroid hormone. Specificity within these functions is dictated by the halide substrates that the different peroxidases can catalyze (21); for example, when tyrosine is a substrate, tyrosine cross-links are formed. All peroxidase reactions require H2O2, a substrate that is generated by the NADPH oxidases. As a consequence of this dependence, peroxidases are most often found to co-localize with their partner NADPH oxidases (22).
In a class of enzymes termed the dual oxidases (Duox), a unique relationship between a peroxidase and its H2O2 donor is found in which a C-terminal NADPH oxidase domain is directly coupled to an N-terminal peroxidase-like domain. The topology of these enzymes facilitates the efficient coupling of the H2O2-donating and H2O2-utilizing moieties (13). C. elegans possesses two Duox genes, bli-3 (23) and its gene duplication f53g12.3. Although the proteins share 94% similarity, f53g12.3 is partially truncated at its C terminus and is predicted to abolish its NADPH-oxidative activity (13). The putative peroxidase domains of BLI-3 and f53g12.3 lack many of the essential heme-binding residues (13), and the functional relevance of this domain is therefore in doubt. However, evidence supporting a role for Duox-derived cross-links in the C. elegans cuticle comes from the blistered cuticle phenotype (Bli) of bli-3 alleles, such as bli-3(e767), and the strong Bli phenotype of RNAi-treated nematodes (13). Biochemical analysis definitively demonstrated the corresponding loss of tyrosine-derived cross-links in the cuticles of bli-3 RNAi worms (13).
Following a focused reverse genetic screen with a GFP-tagged collagen matrix marker (24), we have identified a novel heme peroxidase called MoLT-7 (MLT-7) that in combination with BLI-3 plays an essential role in cuticle collagen cross-linking, ECM formation, and viability of the nematode. In this study, we describe its biochemical, cell biological, and genetic characterization.
EXPERIMENTAL PROCEDURES
Strain Maintenance, Mutant Generation, and Backcrossing Strategy
Worms were cultured and manipulated following standard C. elegans methods (25). N2 (wild type), JR667 unc-119(e2498::Tc1); wIs51, DR96 unc-76(e911), and CB767 bli-3(e767) nematodes were obtained from the Caenorhabditis Genetics Centre. The mlt-7 deletion allele tm1794 was obtained from Shohei Mitani of the National Bioresource genome deletion project (Japan). This was backcrossed four times to N2 worms, using a PCR strategy with the following primers: zk4308f1, 5′-gtgtgcctacttcatgcctg-3′; zk4308r1, 5′-cattgagaactgttacagtg-3′; and zk4308r2, 5′-ctaggtagacacgagaatg-3′. The backcrossed mlt-7 mutant strain, TP89A, was maintained as a heterozygote and segregates homozygous mutants that are molt/larval arrest (mlt-7 Lva). The strain TP89B was derived from the occasional homozygous viable progeny from TP89A and segregates predominantly viable DPY homozygotes (mlt-7 Dpy). TP89C represents the mlt-7 heterozygous strain that was derived from a further backcross of TP89B with N2 animals. All genotypes were confirmed by single worm multiplex PCR (26). The compound mutant strain TP112, mlt-7 (tm1794); bli-3 (e767), is homozygous for e767 and was maintained by selecting tm1794 heterozygotes.
Ty-tagged Collagen Transgenic Strains
The construction of the dpy-13::Ty epitope-tagged transgenic strain IA139, dpy-13(e458); unc-76(e911); ijEx33 [dpy-13::Ty, unc-76(+)], has been described previously (4). The col-12 gene was tagged, and the transgenic strain IA132, unc-76(e911); ijEx32[col-12::Ty, unc-76(+)], was made using the same strategy (contact authors for details of primers used). The col-12::Ty transgene was constructed from three separate PCR-amplified fragments as follows: a promoter fragment, an N-terminal coding fragment, and a C-terminal coding fragment plus 3′-untranslated fragment, engineered to create a HindIII site toward the N-terminal end of the coding sequence that was used for the subsequent insertion of the Ty tag sequence. The completed plasmid pJM01 was assembled within pBluescript II (Stratagene). The transgenic strain IA132 was generated by co-microinjection of the strain DR96 (unc-76 mutant) with pJM01 (2 μg/ml) and the unc-76 rescue plasmid p7616B (100 μg/ml). The COL-12::Ty protein is secreted and localized within the cuticle, and details of its immunofluorescent localization are to be published elsewhere. Transgenic animals can be selected by rescue of the Unc-76 phenotype.
COL-19::GFP Screen for Cuticle Defects
The marker strain TP12, kaIs12 [col-19::gfp], and the RNAi methods used to identify cuticle defects are described in detail elsewhere (8, 24). Using a selected library of RNAi clones (27) based on a proposed function in extracellular matrix formation, RNAi feeding experiments were carried out on our marker strain, and disruption of collagen localization in the cuticle was assessed microscopically. For the study of peroxidase enzymes, we used the Wormbase data base to select predicted heme peroxidases that displayed associated body morphology defects following genome-wide RNAi studies (27); these were f56c11.1 (bli-3), f53g12.3, zk430.8 (mlt-7), c16c8.2, and T06d8.10 (supplemental Fig. 1).
Recombinant Protein Expression
A 1772-bp PCR product encoding the full-length peroxidase domain of MLT-7 was amplified from N2 cDNA with the primers below and cloned into the pQE30 vector (Qiagen) to provide an N-terminal His tag to the encoded protein as follows: zk430pqefwd, 5′-gccctgcagtaatgtccatctcccg-3′, and zk430pqerev, 5′-gcgaagcttctagagagctctcc-3′. Protein production was auto-induced in M15 cells at 25 °C for 40 h, using published method and buffers (28). Briefly, transformed colonies were used to inoculate MDG medium with trace metals, 0.5% glucose, 0.25% aspartic acid (28) supplemented with 50 μg/ml ampicillin and 25 μg/ml chloramphenicol and cultured overnight at 200–250 rpm at 37 °C. This was used to inoculate ZYM-5052 medium (28) supplemented with 50 μg/ml ampicillin and 25 μg/ml chloramphenicol for 40 h at 250–300 rpm at 25 °C. Induced cells were centrifuged and resuspended in lysis buffer, pH 8.0 containing 1 mg/ml lysozyme and 10 mm imidazole. The lysed cells were sonicated, and soluble protein was collected by centrifugation and purified using nickel-nitrilotriacetic acid columns with ProBond resin (Invitrogen) following the manufacturer's protocol. Fractions were analyzed by SDS-PAGE and concentrated using Millipore Microcon centrifugal filters (YM-30), and Western blotting was performed with an anti-HisG antibody (Invitrogen).
Peroxidase Activity Assays
Functional assays (13) were performed using various concentrations of the MLT-7 and NAS-36 (negative control) recombinant proteins to determine peroxidase activity. 0.0125 μg/ml horseradish peroxidase was used as the positive control. Samples were run in triplicate in 96-well plates using the 3,3′,5,5′-tetramethylbenzidine liquid substrate system (Sigma). The reactions were incubated for 30–60 min at room temperature, in the dark, before measuring the absorbance at 630 nm. Peroxidase inhibition assays were similarly performed with 4-aminobenzoic hydrazide (Sigma), using 100 μg/ml recombinant MLT-7 and 0.0125 μg/ml horseradish peroxidase, following a 30–60-min preincubation at room temperature in the dark prior to addition of substrate.
RNAi Feeding and Injection
The mlt-7 RNAi feeding clone was made by PCR amplification from N2 cDNA using primers zk430.8 rnai forward, 5′-ttgtaagacttgtgacccgg-3′, and zk430.8 rnai reverse, 5′-acgatgtggtcgtttgacggc-3′. The resulting 802-bp product was cloned into pCRScript (Stratagene) and subcloned into the RNAi feeding vector (pPD129.36) by digestion with NotI and PstI. mlt-7 in vitro-transcribed RNA was made by linearizing the RNAi feeding plasmid with NotI and HindIII. Double-stranded RNA was made as described previously (29) and injected at 0.5 mg/ml into CB767. The library bli-3 RNAi feeding clone (23) was sequence-verified and applied to TP89A mutants.
Construction and Expression of mlt-7 Promoter Reporter Gene Fusion
The 2103-bp putative promoter region of the mlt-7 gene was amplified from N2 genomic DNA with the primers zk430.8 promoter forward, 5′-cgcgcatgcggcaggtgcaggtaaaatgaagg-3′, and zk430.8 promoter reverse, 5′-gcgtctagatggagtcttctcatggtgttcc-3′. This region encompasses position −2089 to +14 relative to the initiation ATG. This fragment was cloned into the multi-intron promoterless reporter vector pPD96.04, and semi-stable transgenic lines were obtained following microinjection with a combination of 10 μg/ml of the reporter plasmid and 100 μg/ml p7616B (unc-76 rescue) plasmid into strain DR96 (unc-76 mutants). Transgenic lines were stained for β-galactosidase activity following published methods (29).
Semi-quantitative Reverse Transcriptase-PCR
A set of staged cDNA samples representative of the mRNA population at every 2 h of post-embryonic development (5) was used to amplify ama-1 and mlt-7 using the following primers: ama1 forward, 5′-gcgctggaaatttgacagattgc-3′, and ama-1 reverse, 5′-gaatacaattgtccacaagagttgc-3′; hpx-1 forward, 5′-gtccatctcccgactaatcaacc-3′, and hpx-1 reverse, 5′-gctctgtgagcttctgagc-3′. The PCRs were performed following published protocols (5), and the ratio of mlt-7 to ama-1 was calculated from ethidium gel images using ImageQuant software (Amersham Biosciences).
Microscopy
Worms were mounted on sodium azide agar pads, viewed under differential interference contrast (DIC) or epifluorescence optics on a Zeiss Axioscope 2, and images captured on an Axiocam MRm camera using Axiovision software. Immunocytochemistry was carried out as described previously (11) using anti-DPY-7 (4) and anti-MH27 (30) monoclonal antibodies and Alexa Fluor 488 anti-mouse secondary antibody (Molecular Probes).
Transmission Electron Microscopy
Larval stage N2 and mlt-7 Lva mutant worms were selected, washed in 1× M9 buffer, and fixed at 4 °C in 3% glutaraldehyde/PIPES buffer (0.2 m PIPES, pH 7.0). Samples were incubated on ice for 1 h and then washed in PIPES buffer. In preparation for transmission electron microscopy, samples were fixed for 1 h in 1% osmium tetroxide and then embedded in 1.5% agarose prior to dehydration through increasing alcohol from 30 to 100%. The dehydrated worms were embedded in LRW resin, and 60–70 nm sections were cut using a Reichert-Jung Ultracut E Microtome. Sections were contrast-stained with 2% uranyl acetate and then with Reynolds lead citrate and viewed under a Leo 912AB Omega EFTEM microscope.
mlt-7 Rescue and Overexpression
Cosmid M02B10, which overlaps with cosmid ZK430 and contains more upstream mlt-7 sequence, was obtained from the Sanger Institute (Cambridge, UK) and injected at 10 μg/ml into mlt-7 heterozygotes. Rescued homozygotes were selected from transgenic lines, and segregating progeny was scored visually. A mlt-7 gene rescue plasmid was constructed by digesting two fragments from cosmid M02B10; these were then sequentially ligated into the plasmid pGEM-T Easy (Promega). Cloning of an ∼4.7-kb SacII/AatII fragment (containing ∼400 bp of 3′-untranslated) was followed by an ∼11.3-kb NdeI/SacII fragment (containing ∼3.8 kb of 5′ upstream sequence); insert sequences were joined at the SacII site. This mlt-7 gene rescue plasmid was injected at 5 μg/ml into mlt-7 heterozygotes and the viable homozygous strain (TP89B). Overexpression of mlt-7 was achieved by injection of the gene rescue plasmid into wild type worms at 5 and 50 μg/ml. All experiments were performed by co-injection with 5 μg/ml Pdpy-7::gfp, and circularized pTAg was added to make a final mix concentration of 115 μg/ml. Three independent transgenic lines were examined in each case.
Brugia malayi mlt-7 Cloning
The B. malayi sequence with TIGR locus identifier (12787.m00394) had the highest BLAST homology score to MLT-7. Amplification of the 5′ end of the B. malayi mlt-7 cDNA was performed using the Invitrogen 5′-RACE system with the following primers: Bm hpx-1 RT 5′-ccgtaaatcattgaaccatc-3′, Bm hpx-1 R1 5′-catgtcgtgactcag-3′, and Bm hpx-1 R2 5′-cgatatctgagatgatagc-3′. Primer Bm hpx-1 RT was used for the cDNA synthesis reaction, and primers Bm hpx-1 R1 and R2 were used in subsequent nested PCR amplifications with the primers AAP and AUAP (Invitrogen). This 5′-RACE sequence allowed amplification of the full-length coding sequence.
Iodination
Groups of 100 adult nematodes (N2 and mlt-7 Dpy mutants) were collected and washed extensively in cold phosphate-buffered saline, 0.1% SDS, pH 6.5, before being transferred to IODO-GEN (Pierce)-coated tubes. Worms were then labeled with 200 μCi of 125I on ice for 10 min with gentle agitation. Saturated tyrosine was added to terminate the reactions. The labeled nematodes were then washed extensively in phosphate-buffered saline with protease inhibitor mixture, snap-frozen, and then homogenized with a micro-pestle. Extracts and resultant pellets were prepared with 1% SDS with 5% 2-mercaptoethanol, and replicates were counted via trichloroacetic acid precipitation. All samples were analyzed by SDS-PAGE and visualized by autoradiography.
Detection of the Ty-tagged Collagens by Western Blotting
50 RNAi-treated animals were collected into M9 buffer, and an equal volume of 2× Laemmli sample buffer (4% SDS, 20% glycerol, 200 mm dithiothreitol, 120 mm Tris, pH 6.8, bromphenol blue) was added. The samples were heated to 80 °C for 10 min, separated by SDS-PAGE, and Western blotting was performed with anti-Ty antibody (31) and then anti-mouse IgG horseradish peroxidase-conjugated antibody (Promega) followed by ECL (GE Healthcare). Equivalent loading was confirmed by reprobing the membranes with a β-actin antibody (Sigma).
Amino Acid Analysis
Wild type, bli-3 (e767), and mlt-7 Dpy mutant nematodes were grown on NGM plates, washed in M9 buffer, and frozen at −80 °C prior to extraction. Nematodes were resuspended in 0.5 ml of sonication buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 1 mm phenylmethanesulfonyl fluoride) and sonicated four times for 20 s. Protein concentration was determined with the Bradford assay using bovine serum albumin as a standard. Whole worm extracts were lyophilized and then acid-hydrolyzed for 24 h at 110 °C. Hydrolyzed samples were analyzed by Alta Bioscience (University of Birmingham) in a high resolution lithium buffer using a di-tyrosine marker kindly provided by Dr. Yoji Kato (University of Hyogo, Japan).
Hoechst Staining
Nematodes were incubated in M9 buffer containing 1 mg/ml Hoechst 33258 (Sigma) at room temperature for 15 min with gentle agitation. Following several washes in M9 buffer, the worms were viewed under epifluorescence (32).
RESULTS
mlt-7 Encodes an Animal Heme Peroxidase
Reverse genetic screens were performed to identify important components of the cuticle collagen biosynthetic pathway (24). Extensive disruption of the GFP-labeled cuticle collagen COL-19 was observed following mlt-7 RNAi and found to be comparable with bli-3 (f56c11.1 and f53g12.3) disruption (supplemental Fig. 1) suggesting that the encoded protein plays a key role in cuticle formation. The remaining two heme peroxidase-encoding transcripts, c16c8.2 and t06d8.10, although showing body morphology defects following RNAi (27), failed to disrupt the native COL-19 expression pattern (data not shown). The mlt-7 gene prediction from Wormbase (zk430.8) was confirmed by RT-PCR (data not shown). Following cleavage of a 24-amino acid signal peptide, the 724-residue mature peptide (CE05084) includes a metridin ShK toxin (ShKT) domain and a 536-residue peroxidase domain. Comparison of the peroxidase domain with that of other animal heme peroxidases, including myeloperoxidase and thyroperoxidase, highlighted the conserved catalytic and Ca2+-binding residues, including the active site distal and proximal histidines, and the distal arginine (33), shown for MLT-7 in Fig. 1A. In addition, 13 conserved cysteine residues, which have been implicated in the formation of six inter-chain disulfide bonds as well as in dimerization of peroxidases (33), are all found in MLT-7. This C. elegans protein therefore has the potential to fold correctly and to exhibit peroxidase activity. The gene structure, the physical location of the deletion mutation, and the promoter construct are depicted in Fig. 1B.
FIGURE 1.
A, alignment of nematode peroxidase-like domains. MLT-7 from C. elegans (CE05084), C. briggsae (CAP24519), B. malayi (Bm1_03125), and H. contortus (contained in Sanger H. contortus supercontig 0058423) together with the C. elegans BLI-3 dual oxidase (CE28463). Sequences were aligned using ClustalW and Boxshade. Conserved cysteines are indicated above the line with a C, and catalytic domain residues are indicated with an asterisk. Percentage identity/similarity to the C. elegans MLT-7 peroxidase domain are shown. B, structure and characterization of the mlt-7 gene and its encoded protein. Gene structure, position of the promoter construct, and the location of the deletion mutant tm1794 are shown. ShK, metridin ShK toxin domain.
Homologs of MLT-7 are conserved in other nematode species, including Caenorhabditis briggsae (CbMLT-7), Hemonchus contortus (HcMLT-7), and B. malayi (BmMLT-7, Fig. 1A). The B. malayi homolog was originally identified from the TIGR data base (locus 12787.m00394), but 5′-RACE demonstrated that the encoded protein differs from the data base prediction, being 48 residues longer at the N terminus. Overall, MLT-7 shares 95 and 98% identity to B. malayi and C. briggsae homologs, respectively. The C. elegans peroxidase domain is 98, 82, and 50% identical to that of C. briggsae, H. contortus, and B. malayi, respectively, but is only 24% identical to C. elegans BLI-3 peroxidase domains (Fig. 1A).
Peroxidase Activity of Recombinant Protein
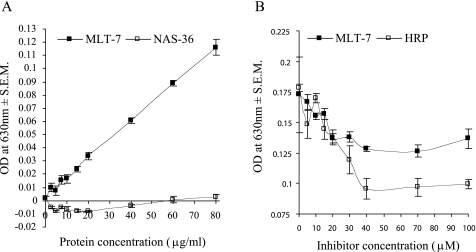
To demonstrate peroxidase activity for MLT-7, we tested the recombinantly expressed histidine-tagged peroxidase domain in an in vitro assay. A nickel-nitrilotriacetic acid-purified peptide of ∼65 kDa was highlighted following Western blots with an anti-His tag antibody (data not shown). In vitro peroxidase activity of this protein (0–80 μg/ml) was assayed using horseradish peroxidase enzyme as a positive control. Activity was found to correlate with increasing enzyme concentration confirming that MLT-7 possesses peroxidase activity (Fig. 2A). In contrast, an unrelated recombinant C. elegans enzyme, NAS-36, possessed no peroxidase activity (Fig. 2A). Activity of recombinant MLT-7 was inhibited, in a dose-responsive manner, following incubation with a known peroxidase inhibitor, 4-aminobenzoic hydrazide, further confirming the specificity of the peroxidase assay (Fig. 2B). Incubation of 100 μg/ml protein with varying concentrations of inhibitor (0–100 μm) for 30–60 min prior to the addition of substrate was sufficient to prevent the production of end product in these assays. This result was consistent with the inhibition of the positive control enzyme horseradish peroxidase (Fig. 2B). These results demonstrate that the peroxidase domain of MLT-7 is capable of activity that can be specifically inhibited.
FIGURE 2.
Peroxidase activity and inhibition of recombinant MLT-7. A, recombinant MLT-7 was expressed by auto-induction and assayed to determine the peroxidase activity of the protein. The control horseradish peroxidase (positive) and NAS-36 (negative) enzymes were also assessed. B, 100 μg/ml recombinant MLT-7 was assayed with increasing concentrations of the inhibitor 4-aminobenzoic hydrazide to establish the specificity of the peroxidase activity.
mlt-7 Expression Pattern Points to a Role in Cuticular Biogenesis
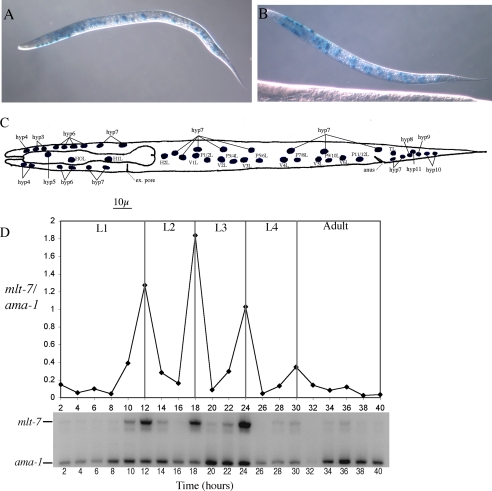
A distinctive clue as to whether a gene performs a function in cuticle biogenesis comes from its hypodermal and cyclical temporal expression pattern. Cuticle collagens and their associated biosynthetic enzymes are expressed in this tissue in distinct waves that correspond to the molting cycle (1). The spatial expression pattern of mlt-7 was characterized using a nuclearly localized lacZ reporter construct, containing the putative promoter region of mlt-7. Staining was observed in all larval and adult stages, localizing to the nuclei of the hypodermal cells, specifically the head (hyp3, hyp4, hyp6, and hyp7) and seam/body (hyp7, H, V, and P) hypodermal cells (Fig. 3, A and B). A semi-quantitative reverse transcriptase-PCR approach was used to examine the temporal expression pattern of mlt-7. Fragments of mlt-7, as well as a constitutively expressed RNA polymerase subunit, ama-1, which functions as a control (5), were amplified from a set of cDNAs that represent the mRNA population at every 2 h of development between hatching and 10 h after the L4-adult molt. These experiments showed that expression of mlt-7 is cyclical, peaking at 12, 18, 24, and 30 h post-hatch (Fig. 3, D and E), which is coincident with each of the larval molts. Such a cyclical hypodermal expression pattern is entirely consistent with mlt-7 playing a role in cuticle biogenesis.
FIGURE 3.
Post-embryonic spatial and temporal expression pattern of MLT-7. A and B represent typical head (hyp3, -4, -6, and -7) and seam/body hypodermal (hyp7, H, V, and P) nuclearly localized staining pattern noted in newly hatched L1 larva transformed by Pmlt-7::lacZ. C depicts a schematic representation of the head and body hypodermal cells of a newly hatched L1 larva (based on Ref. 29). D, relative transcript levels were assessed by comparing the abundance of mlt-7 compared with that of the constitutively expressed RNA polymerase gene ama-1. cDNA samples represent 2-h intervals after the initiation of feeding of starved L1 larvae. L1 to L4 depict larval stages. D depicts a representative plot and the corresponding gel.
mlt-7 Mutants Exhibit Gross Morphological Abnormalities
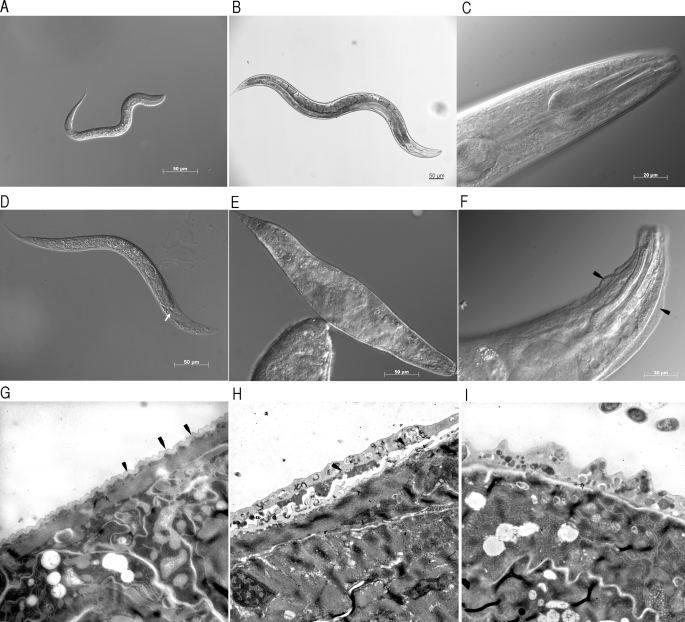
Cuticle-related mutants are characteristically associated with molting defects (Mlt), body morphology defects (Dpy, Rol, and Bli), and in some severe cases embryonic (Emb) or larval (Lva) arrest (1). To characterize the effects of ablating MLT-7 function, we obtained, from the National Bioresource Project (Japan), tm1794, a mutant allele of mlt-7 with a 524-bp deletion that removes all of exon II and regions of the flanking intron sequences, corresponding to positions 4710/4711–5234/5235 of cosmid zk430. The tm1794 deletion is predicted to remove part of the ShKT domain resulting in a frameshift that terminates translation (Fig. 1A). Following extensive backcrossing to remove unwanted deleterious mutations, we characterized the offspring of both heterozygous and homozygous mothers and found different results for each (Table 1). Offspring from heterozygote mothers show a low level of embryonic lethality such that 3.9% of embryos failed to hatch at 20 °C. At all temperatures tested, ∼22% of the remaining animals exhibit gross morphological abnormalities; these animals represent the homozygote population, and this mutant strain was termed TP89A. The predominant mutant phenotype of these animals is classed as larval arrest (Lva) with associated molt defects (Fig. 4D), whereas a significantly smaller proportion of animals exhibited a Dpy appearance (Table 1). Both phenotypic classes were confirmed to be homozygous following genotyping by single worm PCR (data not shown). Synchronizing development following feeding of starved first stage larvae (L1) revealed that the mlt-7 Lva phenotype occurred late in the L1 stage as all homozygous animals were retained in the L1 cuticle and did not develop beyond the early L2 stage.
TABLE 1.
Characterization of mlt-7 mutant strains
| Strain | Total progeny | Hatched | Unhatched (%) | Lva (%) | Dpy (%) |
|---|---|---|---|---|---|
| TP89A | 257 | 247 | 10 (3.9) | 57 (22.2) | 1 (0.4) |
| TP89B | 268 | 267 | 1 (0.4) | 89 (33.2) | 178 (66.4) |
| TP89C | 466 | 462 | 4 (0.9) | 108 (23.2) | 1 (0.2) |
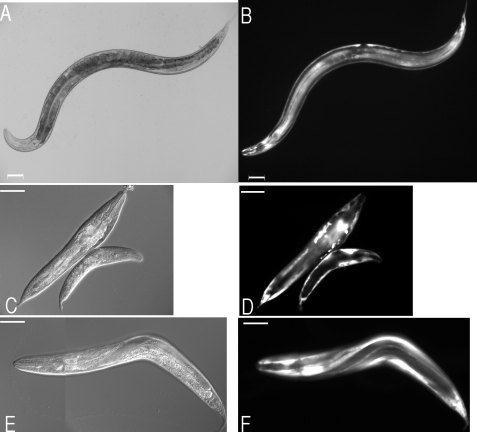
FIGURE 4.
Morphological characterization of mlt-7 mutants. A, typical wild type morphology of an L1 stage N2 strain. B, wild type morphology of adult stage N2 strain. C, typical slender wild type head morphology of adult N2 strain. D, typical larval arrest and associated molt defects in TP89A strain, depicting L2 larvae trapped in unshed L1 cuticle (arrow depicts L2 head). E, severe dumpy phenotype of viable adult TP89B mutant strain. F, associated blister phenotype (arrowhead) in severely dumpy viable adult. Scale bars represent 50 μm in A, B, D, and E and 20 μm in C and F. G, longitudinal transmission EM of N2 larval cuticle revealing regular appearance of the cuticle. Representative annuli are indicated by arrows. H, longitudinal TEM of mlt-7 Lva mutant, depicting unshed cuticle and cellular material within the cuticle matrix (arrow). I, cross-section TEM of mlt-7 Lva mutant cuticle depicting cellular infiltrate in the cuticle matrix (asterisk). TEMs G, H, and I were taken at 3000× magnification.
For the progeny of viable homozygous mothers (TP89B), embryonic lethality (Table 1) is negligible, being similar to that described for wild type animals. The progeny from this strain exhibits gross morphology defects, with Dpy phenotypes (Fig. 4E) predominating over the Lva and being in contrast to that described for TP89A. There are ∼2-fold more Dpy animals than Lva. It must be noted that animals described as Dpy displayed a range of phenotypes, ranging from slightly to extremely Dpy (Fig. 4E), and many additionally exhibit cuticular blisters that result from separation of the cuticle layers (Fig. 4F). It is significant to note that returning this allele to the heterozygous state by crossing TP89B with wild type animals results in reversion of this strain (TP89C) to the original segregation described for strain TP89A (Table 1).
Cuticle of mlt-7 Mutant Animals Is Aberrant
We performed transmission electron microscopy of cross-sections taken from wild type and mutant larvae to analyze the cuticle of the mlt-7 Lva mutants. Compared with wild type animals (Fig. 4G), the cuticles of mlt-7 Lva animals are thickened and contain a cellular infiltrate (Fig. 4, H and I). In Fig. 4H, the infiltrated outer-most layer corresponds to the unshed L1 larval cuticle. Such infiltrates are absent from the matrix of the wild type cuticle (Fig. 4G). Circumferential annular furrows are visible in wild type cuticles (Fig. 4G) but cannot be discerned in the cuticle of mutant animals (Fig. 4, H and I).
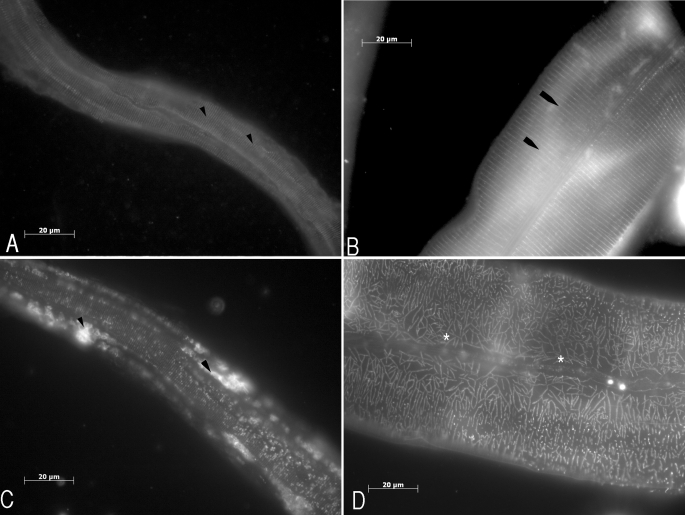
The effects of this mutation on the cuticle structure were further characterized by observing the localization of an integral cuticle collagen, DPY-7, via immunolocalization on freeze-cracked and fixed animals (Fig. 5). The DPY-7 anti-collagen antibody localizes to the annular furrows (4) that in wild type animals appear as regularly spaced circumferential rings (Fig. 5, A and B). The L1 DPY-7 localization in the mlt-7 mutants displaying the Lva phenotype is severely disrupted (Fig. 5C). The annular rings are incomplete, being present in focal spots. In addition, a proportion of the collagen failed to be secreted and was retained in a perinuclear location that corresponded to the lateral seam cells (Fig. 5C). The cuticle of mlt-7 mutants displaying the Dpy phenotype is also highly disrupted, with annular rings being severely fragmented and branched, having a shattered appearance in all regions of the cuticle (Fig. 5D).
FIGURE 5.
Cuticle collagen DPY-7 immunolocalization in wild type and mlt-7 mutant nematodes. A, epifluorescent image of N2 L1 cuticle, depicting the regular annular ring staining pattern of the DPY-7 cuticle collagen (arrowheads). B, epifluorescent image of N2 adult cuticle, depicting the wild type annular ring staining pattern of the DPY-7 cuticle collagen (arrowheads). C, epifluorescent image of mlt-7 Lva mutant cuticle depicting aberrant, irregular DPY-7 staining pattern; arrowheads denote focal spots of nonsecreted DPY-7. D, epifluorescent image of adult stage mlt-7 dumpy mutant cuticle highlighting the aberrant DPY-7 expression pattern; highly branched DPY-7 denoted by asterisk. Scale bars represent 20 μm.
The C. elegans cuticle is secreted from the hypodermis, a syncytium that includes the seam cells. These cells underlie, and secrete, the collagens that form the tri-laminate alae (1). Seam cell defects are often associated with aberrant cuticles, and therefore an antibody recognizing the seam cell boundaries, MH27 (30), and the seam cell nuclear marker strain, JR667, are often used to assess seam cell defects (11). In wild type animals, the multiple seam cells fuse in the adult stage to form a single continuous band of cells that stretches from the head to the tail. By applying these markers in the mlt-7 mutant animals, the seam cells are predominantly wild type in appearance, but some cells do not fuse, and these areas correlate with the dumpiest parts of the worm (see supplemental Fig. 2).
Mutation of mlt-7 Impairs Cuticle Function
The structural integrity, composition, and cuticle cross-linking of the mlt-7 mutant Dpy phenotype worms were investigated by performing iodination and dye permeability assays and by analyzing the amino acid content of protein extracts.
Radioactive iodine labels free and accessible tyrosine residues in proteins and has previously been used to analyze the cuticle collagens of nematodes (34). Equal numbers of live wild type, mlt-7 Dpy, and bli-3 adults were collected and labeled via surface iodination then extracted in detergent with reducing agent before being separated into soluble and insoluble fractions (Fig. 6A). The soluble fractions of the mutant worms incorporated higher trichloroacetic acid-precipitated counts than the wild type worms, and this was particularly significant for the mlt-7 mutants (Fig. 6, A and B). A comparable proportion of label was found in the insoluble pellet of wild type and mutant worms (Fig. 6B), but because of the insoluble nature of these proteins, they could not be separated by electrophoresis (results not shown). These experiments indicate that the cuticles of the mlt-7 Dpy and to a lesser extent the bli-3 mutant worms are either more accessible to label or contained fewer cross-linked tyrosine residues compared with the wild type animals. The latter hypothesis was supported following the amino acid analysis of whole worm extracts from mixed stage populations of the same strains. Using the appropriate standards, we were able to detect a trace of di-tyrosine. We found 0.0993 μg of dityrosine from 172 μg of total extract of wild type worms, 0.257 μg from 714 μg of total bli-3 mutant extract, but were unable to detect this modified amino acid in the mlt-7 mutant extract (from 140 μg total protein); this represents 0.058, 0.036, and 0%, respectively, of the total protein following hydrolysis. In comparison, hydroxyproline, an additional enzymatically modified amino acid found in the cuticle, was detected at comparable levels between strains, being found at 1, 0.68, and 0.84% in the wild type, bli-3, and mlt-7 total extracts, respectively.
FIGURE 6.
Iodination of wild type and mlt-7 mutant worms correlates to increased permeability of the cuticle. A, SDS-polyacrylamide gel separation of extrinsically labeled N2, mlt-7 Dpy mutants, and bli-3(e767) adults. Live worms were iodinated and then extracted in 1% SDS and 5% mercaptoethanol and boiled, and supernatants (equal nematode numbers) were loaded. B, values per worm, with standard error bars, of tricarboxylic acid-precipitable counts for each strain following the sequential extractions shown in gel A. Permeability of the cuticle was examined following the incubation of live nematodes with the fluorescent nuclear dye Hoechst 33258. C, DIC image of an N2 adult. D, animal in C viewed under epifluorescence shown to exclude the dye. E, DIC image of adult stage mlt-7 Dpy mutant. F, animal in E, viewed under epifluorescence showing nuclearly localized staining due to permeability of the cuticle. All scale bars represent 50 μm.
The integrity of the nematode cuticle was investigated by measuring the permeability of this exoskeleton to a nuclei marker, Hoechst 33258. In wild type animals, the cuticle provides a barrier to this dye, as can be seen from the lack of nuclei staining, as only gut auto-fluorescence is observed (Fig. 6D). Conversely, when mlt-7 Dpy animals are exposed to the dye, the nuclei are fluorescently labeled (Fig. 6D), confirming that the dye is able to permeate the cuticle of these mutant worms.
MLT-7 and BLI-3 Are Required for Normal Cuticle Cross-link Formation
To test if MLT-7 and BLI-3 are required for the formation of the normal pattern of cross-linking of cuticular collagens, Ty epitope-tagged versions of the cuticle collagens DPY-13 and COL-12 were used. DPY-13::Ty is fully functional as it efficiently rescues the phenotype of a dpy-13(e458) mutant (4). No mutants are available in col-12, so it was not possible to test the function of COL-12::Ty in this way. However, by immunofluorescent detection, both the COL-12::Ty and the DPY-13::Ty proteins are located within the C. elegans cuticle. The details of COL-12::Ty cuticular localization will be presented elsewhere. Significantly, it is probable that DPY-13 and COL-12 contribute to different cuticular sub-structures as their genes are expressed approximately 2 h apart during cuticle synthesis (5).
The monomeric forms of DPY-13 and COL-12 have a predicted molecular mass of ∼30 kDa; however, when extracted from whole animals where they are assembled into the extracellular matrix of the cuticle, DPY-13::Ty and COL-12::Ty are detected as ladders of high molecular weight species (Fig. 7, A–C), most probably as a result of their nonreducible cross-linking. For DPY-13::Ty, the most abundant species detected is 80 kDa; however, a ladder extending from 30 kDa up to well above 200 kDa exists (Fig. 7A). The pattern of these cross-linked species differs considerably between L4 larvae and adults (Fig. 7A), indicating that the cross-linking of DPY-13::Ty varies developmentally. For COL-12::Ty, two major species are detected, ∼140 and ∼200 kDa (their size is too large to estimate accurately) (Fig. 7C). There is significantly more COL-12::Ty present in adults than L4 larvae, but the cross-linked pattern appears similar with the larger of the two species being most abundant in both cases.
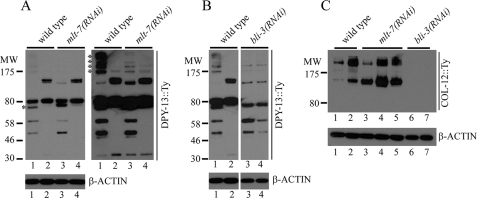
FIGURE 7.
RNAi of mlt-7 and bli-3 affects the cross-linking and assembly of the collagens DPY-13 and COL-12 in developmentally regulated ways. A, DPY-13::Ty cross-linked species vary developmentally and are affected by mlt-7(RNAi). The DPY-13::Ty protein species were detected by Western blotting of protein extracts from C. elegans strain IA139, from untreated (wild type) or with mlt-7(RNAi) treatment for one generation. Both a short (left side) and long (right side) exposure of the same blot are shown. Lanes 1 and 3, protein extracts from L4 larvae; lanes 2 and 4, protein extracts from adult animals. The DPY-13::Ty species marked with an asterisk in lane 1 of the short exposure is altered in molecular weight and relative abundance by mlt-7(RNAi) (lane 3). The multiple high molecular weight species (>175 kDa) of DPY-13::Ty, marked with an asterisk, and visualized with long exposure in lane 1 are much less abundant with mlt-7(RNAi) (lane 3). The blot was stripped and re-probed for β-actin as a loading control; data shown below. B, DPY-13::Ty cross-linked species are affected by bli-3(RNAi). The DPY-13::Ty protein species were detected by Western blotting of protein extracts from C. elegans strain IA139, from untreated (wild type), or with bli-3(RNAi) treatment for one generation. Lanes 1 and 3, protein extracts from L4 larvae; lanes 2 and 4, protein extracts from adult animals. The blot was stripped and re-probed for β-actin as a loading control; data shown below. C, COL-12::Ty cross-linked species vary developmentally and are affected by both mlt-7 and bli-3(RNAi). The COL-12::Ty protein species were detected by Western blotting of protein extracts from C. elegans strain IA132, from untreated (wild type), or with bli-3(RNAi) or mlt-7(RNAi) treatment for one generation. Lanes 1, 3, and 6, protein extracts from L4 larvae; lanes 2, 4, and 7, protein extracts from young adult animals; lane 5, protein extracts from old adult animals. The blot was stripped and re-probed for β-actin as a loading control; data shown below. The molecular mass (MW) of protein markers is indicated in kDa at the left-hand side of the blots.
mlt-7(RNAi) significantly reduces the abundance of a ladder of DPY-13::Ty species above 175 kDa in size in L4 larvae (Fig. 7A, long exposure), and it has a minor effect on the relative abundance and molecular mass of an ∼75-kDa species (Fig. 7A). There was no observable effect on adults. mlt-7(RNAi) causes a significant change in the relative abundance of the two species of COL-12::Ty in L4 larvae and in adults (Fig. 7C). In the L4 and adult controls, the larger species is substantially more abundant than the smaller, a finding that is reversed by mlt-7(RNAi) where the smaller species is substantially more abundant, consistent with a reduction in the level or pattern of nonreducible cross-linking.
bli-3(RNAi) has a more severe effect on both collagens. For DPY-13::Ty it results in the complete loss of the major 80-kDa species and of an additional larger species in adults (Fig. 7B). Also, much of the >175-kDa material is greatly reduced or absent. Noticeably, where the pattern of DPY-13::Ty is distinct between L4 larvae and adults in untreated animals, it is identical with bli-3(RNAi). We conclude that BLI-3 is essential for the normal cross-linking of DPY-13 and for the developmental difference in its pattern of cross-linking between L4 larvae and adults. The effect of bli-3(RNAi) on COL-12::Ty is dramatic causing the complete absence of any detectable species (Fig. 7C), including anything of a size consistent with monomer, in both L4 larvae and adults.
Rescue and Overexpression of mlt-7 Supports an Enzymatic Function for This Encoded Protein
The mlt-7 Lva mutant phenotype exhibited by the TP89A strain was fully rescued following transgenic injection of a wild type copy of the mlt-7 gene (Fig. 8, A and B), both as part of a cosmid (data not shown) and as a plasmid that contains only the mlt-7 gene. Significantly, we also successfully rescued the mlt-7 Dpy phenotype of the TP89B strain following complementation with the same rescue construct (data not shown). These results confirm that loss of function of mlt-7 results in the range of gross morphological phenotypes associated with both the predominantly heterozygous-derived Lva phenotype (TP89A) and the predominantly homozygous-derived Dpy phenotype (TP89B) and is consistent with MLT-7 playing a key role in cuticle biogenesis.
FIGURE 8.
mlt-7 gene rescue of mlt-7 (tm1794) and overexpression in a wild type background. Restoration of wild type body shape in mlt-7 mutant following transgenic rescue with a wild type copy of the mlt-7 gene is shown. A, DIC image of rescued mlt-7 Dpy mutant. B, correlation of wild type body shape with expression of GFP transgenic marker. C, wild type nematodes transformed with mlt-7 rescue construct at 50 μg/ml (C and D) or 5 μg/ml (E and F), together with transformation marker gene Pdpy-7::gfp (D and F). C, severe dumpy roller phenotype conferred by high concentration of mlt-7. E, less severe dumpy left roller phenotype, conferred by lower concentration of mlt-7. Scale bars represent 50 μm.
The mlt-7 rescue construct was also transgenically introduced to wild type worms at various concentrations to examine the effects of overexpressing the encoded peroxidase enzyme. At the high concentration of 50 μg/ml, the wild type copy of this enzyme resulted in a low-penetrance severe Dpy phenotype (Fig. 8, C and D), similar to the mlt-7 Dpy mutants (Fig. 4E) and a predominant Rol phenotype (Fig. 8, E and F). At the lower concentration of 5 μg/ml, both phenotypes were also observed with the Rol phenotype occurring at a lower incidence.
RNA Interference of mlt-7 Supports the Deletion Mutant Loss of Function Phenotype
To corroborate the deletion mutant analysis, we applied RNAi feeding as a means of knocking down mlt-7 function in a wild type background. The F1 generation of RNAi-treated animals was analyzed for embryonic/larval lethality, and gross morphological defects and observations were based on two separate experiments to account for variations in the RNAi feeding conditions. No embryonic lethality was observed, but early larval lethality, predominantly at 25 °C, was noted. The morphological phenotypes ranged from Lva, Bli, Mlt, and Dpy and reflected the range of phenotypes that we described for the Lva and Dpy classes of mlt-7 mutants. Identical mlt-7 RNAi feeding experiments carried out in the mlt-7 Dpy mutant strain did not result in an additional phenotype, confirming the fact that this strain represents a null mutant. The differences observed between the Lva and Dpy phenotypic classes of mlt-7 mutants were not because of a maternal component of this gene. This was determined by applying mlt-7 RNAi to mlt-7 heterozygous mothers, both by feeding and micro-injection. RNAi treatment using either delivery method caused no increase in incidence of viable homozygotes segregating from these animals compared with controls (results not shown).
mlt-7 and bli-3 Genes in Combination Are Essential for Post-embryonic Viability
BLI-3 represents a previously characterized protein that is involved in cuticle cross-linking. This enzyme is expressed in the hypodermis, and bli-3 RNAi results in a mutant cuticle with a reduced di-tyrosine content (13). BLI-3 differs from most peroxidases, however, in having an H2O2 donor domain, namely a homolog of gp91phox, coupled to its peroxidase domain. To address the hypothesis that the gp91phox domain of BLI-3 may function as a H2O2 donor for MLT-7, we performed crosses to combine the bli-3(e767) and mlt-7(tm1794) mutants (Table 2 and Fig. 9). Adult stages of bli-3 are morphologically Bli and have Mlt defects (Fig. 9A) in contrast to the Lva phenotype of the mlt-7 mutants (Fig. 9B). In combination, the double homozygous mutants display an earlier first larval stage arrest phenotype (Lva) with more severe body morphology defects (Fig. 9, C and D). These L1 larvae do hatch but subsequently fail to develop further and become grossly abnormal and begin to collapse (Fig. 9, C and D). This death occurs earlier than the aforementioned mlt-7 larval arrest, which happens at the end of the L1/start of the L2 stage. Therefore, the combination of both mutations does not allow survival beyond embryogenesis, and both enzymes in combination are essential for post-embryonic morphogenesis. This finding was further supported following either bli-3 RNAi feeding in the mlt-7 Lva mutant background (TP89A) or mlt-7 RNAi injection in a bli-3(e767) mutant background, both of which, although showing variable penetrance, did produce a more severe phenotype (data not shown).
TABLE 2.
Compound mutant analysis of TP112 mlt-7 (tm1794); bli-3 (e767)
| Strain | mlt-7 genotype of mothers | Total progeny | Hatched | Unhatched (%) |
|---|---|---|---|---|
| TP112 | Heterozygous | 631 | 507 | 124 (19.6) |
| TP112 | Wild type | 355 | 355 | 0 (0) |
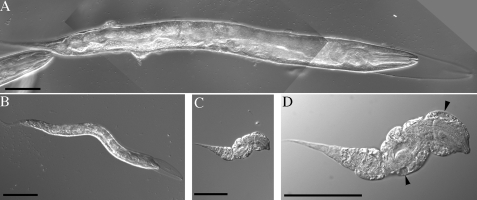
FIGURE 9.
Combined bli-3 (e767) and mlt-7 Lva mutants are embryonic lethal. A, bli-3(e767) mutant with associated blister and molt phenotypes. B, mlt-7 Lva mutant depicting L1 larval arrest and molt phenotype. C, combined homozygous bli-3(e767); mlt-7(tm1794) double mutant demonstrating the severely dumpy and early L1 larval lethal phenotypes. D, higher magnification image of double mutant depicted in C, denoting prominent blisters (arrowhead). Scale bars represent 50 μm.
DISCUSSION
In this study we have identified MLT-7 as an important novel enzyme involved in nematode cuticle biogenesis. Our results indicate that it is required for the formation of tyrosine cross-links in the cuticle collagens via its peroxidase activity. Previous studies have established that the integrity of the C. elegans cuticle requires formation of such chemical cross-links between collagen triple helices (17). Here, we have shown that mlt-7 encodes an animal heme peroxidase, whose activity has been demonstrated in vitro and whose cuticle function is supported by its temporal and spatial expression pattern and by its mutant phenotypes.
The MLT-7 protein includes a short ShkT domain followed by the peroxidase domain. The ShkT domain, which has also been termed SXC (35) on the basis of the presence of six regularly spaced, probably disulfide-bonded, conserved cysteine residues, is interesting as these domains are as follows: 1) implicated in extracellular matrix-specific protein-protein interactions; 2) associated with proteins containing tyrosinase, astacin, CUB, matrixin, peroxidase, zinc carboxypeptidase, and serine protease domains; and 3) found in known cuticle-associated proteins such as the mucins and some matrix metalloproteases. The C. elegans genome encodes 102 individual proteins that contain this domain. The vast majority of these proteins are predicted to encode a signal peptide to target them to the secretory pathway, 21 of which are associated with ECM formation (Wormbase WS195). Twelve out of the 40 matrix-associated nematode astacins possess an ShkT domain. To date, the best characterized ShkT protein is MAB-7 (36), which is essential for male tail development in C. elegans. The ShkT domain was shown via point mutant sequencing and protein deletion mutant analysis to perform a key role in ECM formation (36).
The MLT-7 protein possesses a functionally active type 3 animal heme peroxidase domain (Interpro:IPR002007). All the characteristic catalytic residues are conserved, including the catalytic distal and proximal histidines and the distal arginine, features that are not conserved in the peroxidase domain of BLI-3 (Fig. 1B). In addition, MLT-7 also encodes the 13 cysteines, which are highly conserved in the peroxidase family and which are believed to facilitate inter-chain interactions and dimerization (33). Thus mlt-7 encodes the residues necessary for the correct folding, dimerization, and peroxidase activity, and we have demonstrated that this activity is inhibitable in vitro.
The conservation of MLT-7 in other nematodes was also investigated. MLT-7 and its B. malayi and H. contortus homologs share 38 and 60.8% identity, respectively. The presence of this peroxidase in C. elegans, H. contortus, B. malayi, and C. briggsae suggests that it serves a conserved role in these, and perhaps other, nematode species. It is interesting to note that outside of the aligned nematode homolog (Fig. 1A), MLT-7 shares highest peroxidase domain homology with thyroid peroxidase (data not shown). Although the main function of most peroxidases is in the oxidation of halide ions, thyroid peroxidase differs because it predominantly uses protein-bound tyrosine as a co-substrate in the formation of thyroid hormone. We can therefore speculate that thyroid peroxidase and MLT-7 may perform similar functions, both allowing efficient access of protein bound-tyrosine into the active site, a feature that would be essential for MLT-7-mediated tyrosine cross-linking activity.
Our results support the contention that the peroxidase activity of MLT-7 is required for the biogenesis of the C. elegans cuticle. These include the gross morphological phenotypes (Dpy, Mlt, Bli, and Lva) of mlt-7 mutants and RNAi-treated animals that are all consistent with a defective cuticle. The observed cuticular defects at the higher order level are depicted in the EM analysis. The establishment that the cuticle barrier function was impaired in the dye exclusion assays together with the iodination and amino acid analysis all support a central role for the lack of tyrosine-derived cross-links to the mutant phenotypes observed in this structure. In addition, the hypodermal spatial and molting cycle temporal expression patterns are likewise consistent with MLT-7 playing a role in cuticle biogenesis.
It is significant to note that MLT-7 loss in the deletion mutant and its overexpression in wild type animals both produced cuticular defects. The observation that the overexpression effect was dose-dependent further supports an enzymatic role for MLT-7 and may reflect the presence of ectopic cross-links in the cuticle. Indeed, the range of overexpression phenotypes observed is reminiscent of the squat (Sqt) cuticle collagen mutants in C. elegans that range from Rol to severe Dpy depending on allelic variation or whether the mutation is homozygous or heterozygous (reviewed in Ref. 6).
An interesting observation from the mlt-7 mutant was that, depending on whether the progeny were derived from heterozygotic or homozygotic mothers, they exhibited different proportions of morphological phenotypes. The progeny of homozygotic mothers were predominantly Dpy, with a smaller population showing molting defects. On the other hand, the homozygous progeny from heterozygous mothers were prin-cipally late L1 Lva and Mlt. A significantly smaller proportion of homozygous progeny showed the viable Dpy phenotype. It is plausible that the Dpy phenotype may actually be the more severe manifestation in terms of disruption to the cuticle. However, although such Dpy animals are able to develop to adulthood, the predominant Lva phenotype of the heterozygous-derived animals ultimately leads to L1/L2 larval lethality as a consequence of the ensuing inability to molt, escape from the old cuticle, and feed. It would be easy to envisage that, during shedding, a more disrupted cuticle would be easier to degrade, explaining why no major Mlt defects are observed in Dpy animals. This scenario also supports a potential dual function for MLT-7, namely being involved in both cuticle cross-link degradation prior to molting in addition to tyrosine cross-link formation at cuticle synthesis, and would also be consistent with the overexpression phenotypes we have observed. We have not been able to test the relative levels of cuticle disruption between the different progeny, but it is reassuring to note that rescue of all phenotypic classes from all genetic backgrounds is obtained with the wild type copy of the mlt-7 gene. The phenotype differences of homozygotic mutants that are dependent on the maternal genotype may be explained if there was a maternal mlt-7 function present in the embryo. However, this possibility is unlikely because mlt-7 RNAi of the heterozygotic mothers did not cause an increase in the incidence of viable homozygotic progeny.
As demonstrated, MLT-7 is expressed in the hypodermis in a repeated cyclical manner, similar to cuticle collagens and their processing enzymes (6). Each of the four molts is subdivided into distinct periods of collagen expression that occur prior to or coincident with the shedding of the cuticle. Our results show that MLT-7 is expressed coincident with the molt. It is expressed at all larval stages, but the relative level of expression is reduced in the L4-adult molt compared with the other molts. It is also significant to note that BLI-3, the potential substrate-producing partner of MLT-7, has a comparable but lower abundance expression pattern (data not shown). It is known that for each of the four larval molts a specific assemblage of collagens and enzymes are secreted, enabling each larval cuticle to be distinct. It has previously been shown that interactions are formed between collagens that are expressed within the same expression period and, furthermore, that such similarly expressed collagens form distinct structures within the cuticle (4). It may therefore be envisaged that the collagens secreted prior to or concurrently with MLT-7 would be cross-linked by this enzyme.
Our mlt-7 and bli-3 RNAi results on the Ty-tagged collagen strains confirm that these enzymes do indeed play a role in cuticle collagen cross-linking patterns. The fact that DPY-13::Ty and COL-12::Ty display different patterns of ladders of detectable material indicates that they are not cross-linked to one another; however, the precise nature of the cross-linked species is currently unknown. The DPY-13 collagen has an obligate partner, DPY-5, without which it fails to assemble, so we would predict that DPY-5 might be included in the DPY-13 species, and it will be interesting to test this. The absence of COL-12::Ty with bli-3(RNAi) was unexpected, and currently we do not know its fate, i.e. if it is secreted then lost, degraded or reabsorbed, or if some other mechanism underpins this effect.
Of particular interest was the potential functional relationship between MLT-7 and the second class of animal heme peroxidases, the Duoxes (BLI-3 and f53g12.3). From the total of 25 C. elegans peroxidase-containing genes, five showed an RNAi-induced phenotype consistent with a role in cuticle biogenesis (27), and of these only three displayed collagen marker disruption patterns (supplemental Fig. 1). Although many peroxidases are encoded by the C. elegans genome, only one functional NADPH oxidase, namely BLI-3, is present. It was therefore pertinent to establish if the BLI-3 NADPH oxidase activity was required for MLT-7 function. The cuticular defects of mlt-7 and bli-3 mutants indicate that neither protein can fully compensate for the loss of the other. The increased phenotypic severity of mlt-7 mutants in comparison with bli-3 mutants may however suggest that the peroxidase activity of MLT-7 is of more importance in cuticle assembly than that of BLI-3. In addition, the combination of bli-3 and mlt-7 mutations leads to complete post-embryonic lethality. However, it remains to be conclusively proven that both enzymes are indeed acting in the same pathway because our results cannot discount the possibility that the synthetic lethality is arising from parallel pathways.
The bli-3(e767) mutant exhibits a mutagen-induced point mutation (246 Gly to Asp) in the BLI-3 peroxidase-like domain (23) and is therefore not considered to be a complete null, suggesting that the NADPH oxidase activity is potentially still functional. In contrast, treatment of animals with bli-3 RNAi ablates both the peroxidase and the NADPH oxidase activities. The fact that RNAi of bli-3 leads to more potent effects on gross morphology, compared with peroxidase domain mutants (13), is also consistent with the premise that BLI-3 is the H2O2 donor for MLT-7. However, bli-3 RNAi does not lead to the complete lethality that we describe for the bli-3/mlt-7 combined mutants. This may however be explained by either incomplete knockdown by RNAi or the presence of an alternative, as yet unidentified, source of H2O2.
It may therefore be proposed that MLT-7 is the key peroxidase responsible for tyrosine-derived cross-linking in the nematode cuticle and that BLI-3 is the main source of the H2O2 substrate, through its functional NADPH oxidase domain. The production and location of this substrate must be critically controlled, and the interaction of these two proteins through their peroxidase, MLT-7, and peroxidase-like, BLI-3, domains may provide a mechanism for this control.
Supplementary Material
Acknowledgments
We thank Dr. Shohei Mitani (National Bioresource Project, Tokyo Women's Medical University) and the Caenorhabditis Genetics Center (University of Minnesota) for providing strains. We also thank Dr. Yoji Kato (University of Hyogo) for supplying the dityrosine standards and Alta Bioscience (University of Birmingham) for carrying out the amino acid analysis.
This work was supported by the Medical Research and the Biotechnology and Biological Sciences Research Councils of the United Kingdom.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- ECM
- extracellular matrix
- RNAi
- RNA interference
- Duox
- dual oxidase
- RACE
- rapid amplification of cDNA ends
- GFP
- green fluorescent protein
- PIPES
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Page A. P., Johnstone I. L. ( 2007) WormBook ( The Caenorhabditis elegans Research Community, eds) WormBook 10.1895/wormbook.1.138.1 [DOI] [Google Scholar]
- 2.Cox G. N., Kusch M., Edgar R. S. ( 1981) J. Cell Biol. 90, 7– 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnstone I. L. ( 2000) Trends Genet. 16, 21– 27 [DOI] [PubMed] [Google Scholar]
- 4.McMahon L., Muriel J. M., Roberts B., Quinn M., Johnstone I. L. ( 2003) Mol. Biol. Cell 14, 1366– 1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnstone I. L., Barry J. D. ( 1996) EMBO J. 15, 3633– 3639 [PMC free article] [PubMed] [Google Scholar]
- 6.Page A. P., Winter A. D. ( 2003) Adv. Parasitol. 53, 85– 148 [DOI] [PubMed] [Google Scholar]
- 7.Myllyharju J., Kivirikko K. I. ( 2004) Trends Genet. 20, 33– 43 [DOI] [PubMed] [Google Scholar]
- 8.Page A. P., McCormack G., Birnie A. J. ( 2006) Int. J. Parasitol. 36, 681– 689 [DOI] [PubMed] [Google Scholar]
- 9.Schofield J. D., Uitto J., Prockop D. J. ( 1974) Biochemistry 13, 1801– 1806 [DOI] [PubMed] [Google Scholar]
- 10.Mehta K., Rao U. R., Vickery A. C., Fesus L. ( 1992) Mol. Biochem. Parasitol. 53, 1– 15 [DOI] [PubMed] [Google Scholar]
- 11.Eschenlauer S. C., Page A. P. ( 2003) J. Biol. Chem. 278, 4227– 4237 [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto D., Horiuchi K., Hirama M. ( 1981) Biochem. Biophys. Res. Commun. 99, 637– 643 [DOI] [PubMed] [Google Scholar]
- 13.Edens W. A., Sharling L., Cheng G., Shapira R., Kinkade J. M., Lee T., Edens H. A., Tang X., Sullards C., Flaherty D. B., Benian G. M., Lambeth J. D. ( 2001) J. Cell Biol. 154, 879– 891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox G. N., Staprans S., Edgar R. S. ( 1981) Dev. Biol. 86, 456– 470 [DOI] [PubMed] [Google Scholar]
- 15.Greenberg C. S., Birckbichler P. J., Rice R. H. ( 1991) FASEB J. 5, 3071– 3077 [DOI] [PubMed] [Google Scholar]
- 16.Sakura S., Fujimoto D. ( 1984) Photochem. Photobiol. 40, 731– 734 [DOI] [PubMed] [Google Scholar]
- 17.Fetterer R. H., Rhoads M. L., Urban J. F., Jr. ( 1993) J. Parasitol. 79, 160– 166 [PubMed] [Google Scholar]
- 18.Norman K. R., Moerman D. G. ( 2000) Dev. Biol. 227, 690– 705 [DOI] [PubMed] [Google Scholar]
- 19.Taurog A. ( 1999) Biochimie 81, 557– 562 [DOI] [PubMed] [Google Scholar]
- 20.Daiyasu H., Toh H. ( 2000) J. Mol. Evol. 51, 433– 445 [DOI] [PubMed] [Google Scholar]
- 21.Morrison M., Schonbaum G. R. ( 1976) Annu. Rev. Biochem. 45, 861– 888 [DOI] [PubMed] [Google Scholar]
- 22.Donkó A., Péterfi Z., Sum A., Leto T., Geiszt M. ( 2005) Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 2301– 2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmer F., Moorman C., van der Linden A. M., Kuijk E., van den Berghe P. V., Kamath R. S., Fraser A. G., Ahringer J., Plasterk R. H. ( 2003) PLoS Biol. 1, E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thein M. C., McCormack G., Winter A. D., Johnstone I. L., Shoemaker C. B., Page A. P. ( 2003) Dev. Dyn. 226, 523– 539 [DOI] [PubMed] [Google Scholar]
- 25.Sulston J., Hodgkin J. ( 1988) The Nematode Caenorhabditis elegans ( Wood W. B. the Community of Caenorhabditis elegans Researchers, eds), pp. 587– 606, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26.Winter A. D., McCormack G., Page A. P. ( 2007) Dev. Biol. 308, 449– 461 [DOI] [PubMed] [Google Scholar]
- 27.Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Welchman D. P., Zipperlen P., Ahringer J. ( 2003) Nature 421, 231– 237 [DOI] [PubMed] [Google Scholar]
- 28.Studier F. W. ( 2005) Protein Expr. Purif. 41, 207– 234 [DOI] [PubMed] [Google Scholar]
- 29.Winter A. D., Page A. P. ( 2000) Mol. Cell. Biol. 20, 4084– 4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francis G. R., Waterston R. H. ( 1985) J. Cell Biol. 101, 1532– 1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastin P., Bagherzadeh Z., Matthews K. R., Gull K. ( 1996) Mol. Biochem. Parasitol. 77, 235– 239 [DOI] [PubMed] [Google Scholar]
- 32.Moribe H., Yochem J., Yamada H., Tabuse Y., Fujimoto T., Mekada E. ( 2004) J. Cell Sci. 117, 5209– 5220 [DOI] [PubMed] [Google Scholar]
- 33.Nomura K., Hoshino K., Suzuki N. ( 1999) Arch. Biochem. Biophys. 367, 173– 184 [DOI] [PubMed] [Google Scholar]
- 34.Selkirk M. E., Nielsen L., Kelly C., Partono F., Sayers G., Maizels R. M. ( 1989) Mol. Biochem. Parasitol. 32, 229– 246 [DOI] [PubMed] [Google Scholar]
- 35.Blaxter M. L. ( 1998) Science 282, 2041– 2046 [DOI] [PubMed] [Google Scholar]
- 36.Tsang S. W., Nguyen C. Q., Hall D. H., Chow K. L. ( 2007) Dev. Biol. 312, 353– 366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.