Abstract
Coelomocytes, the heterogeneous population of sea urchin putative immune cells, were found to express a complex set of transcripts featuring scavenger receptor cysteine-rich (SRCR) repeats. SRCR domains define a metazoan superfamily of proteins, many of which are implicated in development and regulation of the immune system of vertebrates. Coelomocytes transcribe multiple SRCR genes from among a multigene family encoding an estimated number of 1,200 SRCR domains in specific patterns particular to each individual. Transcription levels for given SRCR genes may range from pronounced to undetectable, yet all tested animals harbor the genomic loci encoding these genes. Analysis of several SRCR genes revealed multiple loci corresponding to each type. In the case of one SRCR type, a cluster of at least three genes was detected within a 133-kb bacterial artificial chromosome insert, and conserved as well as unique regions were identified in sequences of three genomic clones derived from a single animal. Array hybridizations with repeated samples of coelomocyte messages revealed substantial alterations in levels of expression of many SRCR genes, with fluctuations of up to 10-fold in 1 week and up to 30-fold over a period of 3 months. This report is the first demonstration of genomic and transcriptional complexity in molecules expressed by invertebrate coelomocytes. The mechanisms controlling SRCR gene expression and the functional significance of this dynamic system await elucidation.
The immune system of the purple sea urchin (Strongylocentrotus purpuratus) is the most studied among invertebrate deuterostomes (1–7). Putative immune effector cells are coelomocytes, on average 7.5 × 106 cells per ml of coelomic fluid. Coelomocytes are free-wandering cells that populate the coelomic cavity. About two-thirds are phagocytic, and the rest are vibratile cells, or colorless and red spherule cells. Coelomocytes accumulate at sites of injury and form cellular clots, clear bacteria and other foreign substances from the coelomic cavity, and partake in allograft rejection (5, 8). Recently, two coelomocyte proteins of the complement system were reported, one homologous to factor B (3) and the other a homologue of vertebrate C3/C4/C5 complement proteins (1, 2). We have shown that coelomocytes display an extensive transcriptional response to challenge and injury (4). Introduction of live bacteria into the coelomic cavity sharply affected the levels of mRNAs encoding S. purpuratus transcription factors SpNFkB, SpRunt-1, and SpGATAc. Surprising aspects of coelomocytes' repertoire were the genes that were differentially transcribed among animals. These genes are members of the scavenger receptor cysteine-rich (SRCR) superfamily (4).
SRCR domains define a metazoan superfamily of proteins featuring one or more motifs of 110-aa residues displaying a conserved spacing of six to eight cysteines, which form intradomain disulfide bonds (9, 10). Sequences containing SRCR domains have been found among representatives of diverse animal phyla such as marine sponges (11, 12), nematodes (accession no. E330348), fruit flies (accession no. AC005732), echinoderms (4, 13), tunicates (accession no. BAA82522.1), sea lampreys (14), as well as various mammals (9, 15, 16). Two invertebrate members of the SRCR superfamily have been shown to function as cell surface receptors: the purported aggregation receptor of a marine sponge (12) and the sea urchin sperm activation receptor for the egg-jelly peptides (13).
Many of the vertebrate SRCR proteins are implicated in development of the immune system and in regulation of immune responses (9, 15, 16). Among the multidomain SRCR receptors of lymphoid cells are CD5 (17) and CD6 (18). CD5 is thought to be involved in development and regulation of T cell activation (19), whereas the precise biological role of CD6 is not well understood (20). Other functions of immune-related SRCR proteins include inhibition of apoptosis in a variety of cells (21), inhibition of B lymphocyte proliferation (22), regulation of monocyte and macrophage immune responses (23), modulation of the host response to endotoxin and binding and phagocytosis of bacteria (24–30), binding the lung microbial-opsonizing lectin (31), endocytosis (32), and regulation of the complement cascade (33). Of special interest is a multigene family of SRCR receptors expressed in T cells of cattle, pig, and sheep, collectively named T19 or WC1 antigens (16, 34). Multiple SRCR receptors are expressed selectively by discrete subsets of ungulate T cell populations, which are distinguished by amino acid polymorphism and by the presence or absence of complete protein domains (16, 35). This multigene family is characterized by many variable genomic versions, with more than 50 genes in the sheep genome (36). Evidence suggests a role for T19 receptors in signaling T cell growth arrest via down-regulation of mitogen-activated protein kinase cascades (37, 38).
Here we describe a highly heterogeneous and transcriptionally dynamic set of SRCR-containing molecules expressed specifically in sea urchin coelomocytes. Heterogeneity is demonstrated at genomic and transcriptional levels for multiple SRCR molecules.
Materials and Methods
Sea Urchins.
Live specimens of S. purpuratus were maintained at the California Institute of Technology Kerckhoff Marine Laboratory as described (39). Bacteria and fungi for pathogen challenges were isolated and cultured as described (4).
Probes.
SpSRCR1: Exon 1 PCR of 5′ untranslated region (UTR) and leader peptide, nucleotides 1–161 in SpSRCR1 (accession no. AF076513); SRCR1 PCR of one SRCR domain, nucleotides 2,009–2,246; extracellular matrix (ECM) PCR of nucleotides 807–1,340; von Willebrand factor (vWF) PCR of nucleotides 151–462; gSRCR1 type 1 SRCR 5′ flanking clone, 1,078-bp SalI insert (see below); gSRCR1–5′ PCR of nucleotides 147–952 in gSRCR1. SpSRCR7: EC an SRCR domain, ClaI–SacI fragment corresponding to nucleotides 1,972–2,587 in SpSRCR7.1; IC1 PCR of nucleotides 3,088–3,400 in SpSRCR7.1. An SpNFkB probe was generated by PCR across one exon (nucleotides 1,166–1,359 in accession no. AF064258).
Genomic and cDNA Clones.
An SRCR1 5′ flanking probe of 1,078 bp was PCR-cloned in pGEM-T (Promega) from a genomic library in λFIXII (40) by using gene-specific reverse primer SRCR.R1 (nucleotides 26–46 in SpSRCR1) and vector T3 primer. Arrayed libraries of coelomocyte cDNA and genomic bacterial artificial chromosomes (BAC) were screened for SRCR genes as described (4, 40). Genomic clones of type 1 SRCR genes were isolated with the gSRCR1 probe from 5 × 105 phage clones in λFIXII. Inserts from three positive clones were subcloned in pUC18 and partially sequenced: gSRCR1.1, 4,296 nucleotides of a 15-kb SalI insert; gSRCR1.3, 3,671 nucleotides of an 18-kb SalI insert; gSRCR1.5, 4,308 nucleotides of a 10-kb SalI insert.
DNA and RNA Blots.
Sperm DNA from individual sea urchins was isolated as described (40). BAC plasmid DNA was purified (41). Then 5 μg per lane was digested with NotI, EcoRI, or SalI; separated by pulsed-field gel electrophoresis in 1% agarose; and blotted. Hybridization and wash conditions are described below. Coelomocyte RNA was processed as described (4).
96-Spot Membrane Array.
The array consisted of 92 test spots and 4 plasmid spots (pUC18) to monitor background hybridization. SRCR fragments were cloned by PCR with degenerate primers (4). There were 87 test spots that represented unique sequence SRCR fragments about 230 bp long, cloned from sea urchin cDNA of coelomocytes and developmental stages and from genomic DNA. Inserts of test clones were PCR-amplified with flanking vector primers (T7 and SP6) and purified (QIAquick Spin Columns, Qiagen, Chatsworth, CA), and then 100 ng of DNA per spot was vacuum blotted (Bio-Dot Microfiltration Apparatus, Bio-Rad) onto a 7.5 × 11.5-cm Nylon membrane (Hybond Nfp, Amersham Pharmacia).
Probes for array hybridization were synthesized from 0.5 ml of coelomic fluid containing an average of 7 × 106 cells per ml (range 1.5–18), which was drawn into a syringe filled with 1 ml of filtered (0.22 μm, Schleicher & Schuell) calcium- and magnesium-free artificial sea water (per liter: 31 g of NaCl/0.8 g of KCl/0.25 g of NaHCO3/1.6 g of Na2SO4, pH 8) supplemented with 30 mM EDTA. Cells were pelleted, resuspended in 1 ml of RNA STAT-60 (Leedo Medical Laboratories, Houston), and stored at −70°C until use. After extraction, the RNA was precipitated with 20 μg of glycogen, and mRNA was selected with Dynabeads mRNA purification kit (Dynal, Great Neck, NY).
First-strand cDNA synthesis was primed with 2.5 μg of random hexamers anchored to T3 RNA polymerase promoter (RP-T3: 5′CGGAATTAACCCTCACTAAAGGGANNNNNN) according to the manufacturer's instructions (SuperScript First Strand cDNA Synthesis, GIBCO/BRL). Reactions were incubated for 10 min at 25°C, then ramped to 42°C at 0.1°C/sec and maintained for 30 min at 42°C and 15 min at 50°C (PTC200, MJ Research, Cambridge, MA). After RNase H treatment, second-strand cDNA was primed again with RP-T3 in the same tube. Samples in 21 μl were incubated for 1 min at 94°C and 1 min at 4°C. Then, 76 μl of PCR mix was added (12.8 μl of 10× PCR buffer/1.6 μl of 10 mM dNTP mix/61.6 μl of water), and the samples were incubated for 5 min at 25°C. Then, 10 units of Klenow exo− (New England Biolabs) and 5 units of AmpliTaq (Perkin–Elmer) were added. Reactions were incubated for 10 min at 25°C, ramped to 74°C at 0.1°C/sec, and maintained for 15 min at 74°C. The double-stranded cDNA was purified (NucleoTrap PCR Purification, CLONTECH), eluted in 45 μl of 10 mM Tris (pH 8.5), and PCR amplified in 100-μl reactions by using 20 pmol of T3 primer (5′CGGAATTAACCCTCACTAAAG) with the following parameters: 1 min at 94°C; then 15 cycles of 30 sec at 94°C, 1 min at 55°C, and 3 min at 74°C; and a final extension of 10 min at 74°C. Samples were purified (QIAquick Spin Column) and drop-dialyzed (VMWP02500, Millipore) for 1 h against water; then 40 pmol of the T3 primer was added, and the volume was reduced to 8.4 μl (SpeedVac, Savant). Radioactive labeling was with T3 RNA polymerase (MAXIscript, Ambion, Austin, TX). The cDNA was denatured for 3 min at 94°C and annealed to T3 primer while cooling to room temperature. Then, in vitro transcription mix was added; this mix contained 2.4 μl of 10× buffer; 1.2 μl each of 10 mM ATP, CTP, and GTP; 1 mM of UTP; 6 μl of [32P]UTP (20 mCi/ml, 800 Ci/mmol; 1 Ci = 37 GBq; Amersham Pharmacia); and 2.4 μl of T3 RNA polymerase. Samples were incubated for 3 h at 37°C; then 2 units of DNase I was added for 15 min. Labeled RNA was purified by G-50 chromatography (mini Quick Spin Columns, Roche, Indianapolis), yielding about 1 μg of RNA probe with specific activity 1.6 × 108 cpm/μg.
Duplicate arrayed membranes were hybridized for 21 h at 65°C in 10 ml of solution containing 5× standard saline phosphate/EDTA (20× SSPE per liter: 174 g of NaCl/27.6 g of NaH2PO4/7.4 g of Na2EDTA, pH 7.4), 5% SDS, 0.1% sodium pyrophosphate (Na4P2O70.10H2O), and 100 μg/ml total yeast RNA. Membranes were washed twice for 10 min at room temperature in 2× SSPE/0.1% SDS and twice for 15 min at 65°C in 1× SSPE/0.1% SDS. Hybridization intensities after 21 h of exposure were digitized by using a PhosphorImager (Storm 820, Molecular Dynamics). Values of hybridization intensity in arbitrary units were calculated as follows. For each sample, the average value of the four pUC18 spots was subtracted from values of the 92 test spots; then corresponding values from duplicate membranes were averaged. Pairwise correlation for sets of 15 SRCR markers that produced strong hybridization signals was calculated as the covariance of two data sets divided by the product of their standard deviation.
Results
Structure of Sea Urchin Coelomocyte SRCR Proteins.
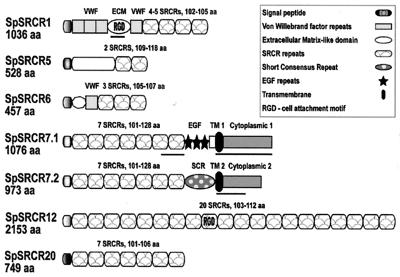
Four new types of coelomocyte SRCR molecules are described here. These are types 6, 7, 12, and 20, which are shown in Fig. 1 together with the previously reported SpSRCR1 and SpSRCR5 (4). These SRCR genes are expressed predominantly in coelomocytes, as evidenced by hybridization to RNA gel blots from embryonic and larval stages, as well as coelomocytes and adult tissues (ref. 4 and unpublished data). SpSRCR1 and SpSRCR6 contain vWF repeats, and a segment in SpSRCR1 is similar to an ECM domain of another sea urchin. Two completely different C-terminal variants were documented in SpSRCR7, immediately after the seventh SRCR. In SpSRCR7.1, there are three EGF-like repeats, whereas in SpSRCR7.2 there is one short consensus repeat of the complement control superfamily, and unique sequences encompass the putative transmembrane domains, the cytoplasmic segments, and the 3′ UTRs. These variants could represent either alternatively spliced gene products or the products of two genes with a conserved N terminus (11 amino acid substitutions in residues 1–795). The other five SRCR gene products contain predicted secretion peptides but no membrane anchor domains.
Figure 1.
Structure of six purple sea urchin coelomocyte SRCR proteins. Four of these SRCR molecules are mosaic, including vWF repeats in SpSRCR1 and SpSRCR6 (61–62 amino acids long) and another domain in SpSRCR1 (ECM, 347 amino acids), which is similar to an ECM protein of another sea urchin. SpSRCR7.1 features three epidermal growth factor (EGF) repeats (34–38 amino acids), whereas in SpSRCR7.2, there is one short consensus repeat (SCR) of the complement control superfamily (68 amino acids). In both SpSRCR7 variants, the N terminus is conserved (11-aa substitutions in residues 1–795), but immediately after the seventh SRCR, the predicted transmembrane domains, C terminus, and 3′ UTR regions are different. Other domains are not similar to any published sequences, such as the N-terminal domains in SpSRCR5 (amino acid 285) and SpSRCR6 (amino acid 53) and the domain that separates the EGF-like repeats from the membrane retention domain in SpSRCR7.1 (amino acid 52). Clones of SpSRCR1 differ in the number of SRCRs, displaying four or five domains within otherwise similar transcripts. Segments in SpSRCR1 and SpSRCR7 that were used as domain-specific probes are underlined: the ECM, the seventh SRCR, and cytoplasmic domains 1 and 2. Accession numbers for these sequences are SpSRCR1, AF076513; SpSRCR5, AF076514; SpSRCR6, AF228823; SpSRCR7.1, AF228824; SpSRCR7.2, AF228825; SpSRCR12, AF064259; and SpSRCR20, AF228826.
SRCR gene types 1 and 6 contain similar domains. The leader peptides are identical in 19 of 20 amino acids; the SRCR domains share 71–85% pairwise identities; and the vWF repeats are 68–75% identical. In contrast, the SRCRs in other types are highly divergent within and among genes. SpSRCR7 consists of 7 domains, each with a unique sequence (25–50% amino acid identities); SpSRCR12 consists of 20 domains with 33–83% identity; and the 7 SRCR domains in SpSRCR20 are 51–91% identical.
Diversity in Coelomocyte SRCR Messages.
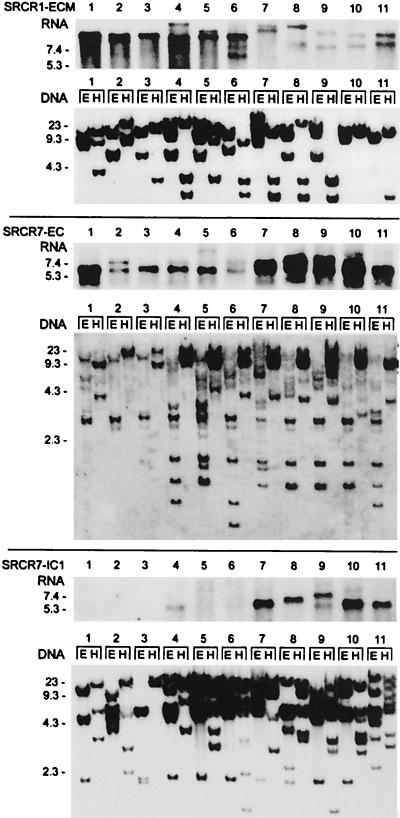
Sea urchin coelomocytes were shown to transcribe differentially type 1 SRCR genes (4). To study these differences, blots of coelomocyte RNA and the corresponding genome blots of 11 animals were compared after hybridization with specific SRCR1 and SRCR7.1 probes (Fig. 2). Among these animals, transcription of SpSRCR1 and SpSRCR7.1 genes range from pronounced to undetectable, but the genomic loci were detected in all. For instance, animals 7–10 express low levels of SpSRCR1; in animals 1–3, transcripts containing the C-terminal segment of SpSRCR7.1 are undetectable, but all genomes in the experiment harbor these segments. The genome of S. purpuratus reveals a high level of polymorphism, estimated at 4–5% differences in base pairs between any two haploid genomes (42). An example for polymorphism can be seen in the pattern of bands hybridizing with the SRCR1 ECM probe, which reveals extensive restriction site length polymorphism among the animals (Fig. 2 Top, DNA blot).
Figure 2.
Phenotypic variation in expression of sea urchin SRCR genes types 1 and 7. Blots of coelomocyte RNA and the corresponding genome blots of 11 animals were repeatedly hybridized with specific SpSRCR1 and SpSRCR7 probes. (Top) SpSRCR1: SRCR1-ECM, ECM domain probe; (Middle) SpSRCR7: SRCR7-EC, an SRCR domain probe; (Bottom) SpSRCR7.1: SRCR7-IC1, cytoplasmic 1 domain probe (the segments corresponding to these probes are underlined in Fig. 1). In the RNA blot, there are 10 μg of total RNA per lane, and in the DNA blot, there are 5 μg of sperm DNA per lane, digested with EcoRI (E) or HindIII (H). Numbers to the left of the blots indicate length (kb).
Genomic Arrangement of Coelomocyte SRCR Genes.
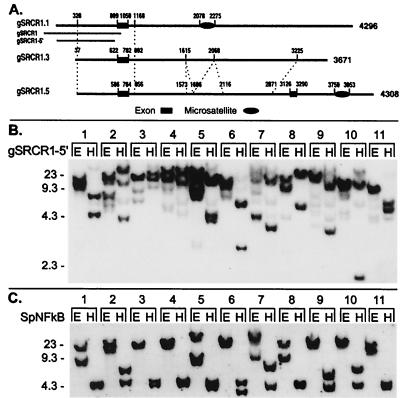
The complex genomic hybridization patterns detected with various SRCR probes may indicate the presence of multiple versions of these genes (Figs. 2 and 3 and unpublished data). To test this hypothesis, a genomic library constructed from the sperm DNA of a single sea urchin was screened with an SRCR1 5′ flanking probe, and three of the positive clones were analyzed (Fig. 3A). Alignment of the sequences revealed high conservation of the 5′ flanking regions, the first exon (5′ UTR and leader peptide in SpSRCR1), and about 110 nt at the beginning of the first intron (96% nucleotide identity). Further downstream, the sequence of gSRCR1.1 diverges. Composite dinucleotide CT microsatellites were identified in gSRCR1.1 and gSRCR1.5, but they are located at different sites in these clones. Clones gSRCR1.3 and gSRCR1.5 share further regions of similarity along the first intron except that one segment is duplicated in gSRCR1.3 (nucleotides 1,612–2,051 and nucleotides 2,079–2,482 share 89.5% identity in 440-nt overlap, with two gaps of 10 and 24 nt). However, the last 446 nucleotides in gSRCR1.3 are unlike the sequence of gSRCR1.5. A second exon is included in the sequence of gSRCR1.5, which is nearly identical to the N-terminal half of type 1 SRCR domain.
Figure 3.
Genomic diversity in type 1 SRCR genes. (A) Schematic presentation of similarity among three genomic clones that were detected with an SRCR type 1 5′ flanking probe. Domains displaying high similarity among the sequences are connected by dashed lines. Exon 1 corresponds to the 5′ UTR and leader peptide in SpSRCR1 (nucleotides 1–161), and exon 2 in gSRCR1.5 corresponds to the N-terminal half of the SRCR1 domain (nucleotides 1,939–2,103 in SpSRCR1). Composite dinucleotide CT microsatellites are denoted by an oval symbol. Corresponding regions of the SRCR1 5′ flanking probes are marked underneath gSRCR1.1: gSRCR1 (1,078-bp, 46-nt overlap with the first exon) and gSRCR1–5′ (805 bp, nucleotides 147–952 in gSRCR1). (B) Genome blot with the gSRCR1–5′ probe. (C) Genome blot probed with the single copy gene marker SpNFkB. The SpNFkB probe corresponds to a region located at the center of the Rel homology domain. Blot is the same as that shown in Fig. 2 (E; EcoRI; H; HindIII). Numbers to the left of the blots indicate length (kb).
Although two of these clones could represent allelic variants, the third indicates the presence of an additional SRCR type 1 gene. Further evidence for an additional gene is shown in a genome blot probed with an SRCR1 5′ flanking probe (Fig. 3B). A complex pattern was detected with the gSRCR1–5′ probe, consisting in several cases of at least three intense bands, for example in animals 2 and 5 (Fig. 3B, EcoRI and HindIII digests), whereas only in two animals did a single DNA band resolve (Fig. 3B, nos. 3 and 11, EcoRI digest). Because no recognition sites for EcoRI or HindIII exist in the probe gSRCR1–5′ or in the corresponding regions that were PCR amplified from the DNA of animals 2, 5, 7, 9, and 10 (data not shown), the pattern of three or more bands is most likely to represent a locus encoding linked genes. In contrast, the locus encoding SpNFkB revealed no more than two bands per lane and a single EcoRI band hybridizing in six animals (Fig. 3C), a pattern that is consistent with a single copy gene revealing diallelic restriction site polymorphism.
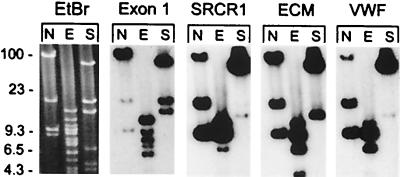
The number of SRCR-encoding loci was estimated by the hybridization of specific probes to an array of a genomic library constructed from the sperm DNA of a single sea urchin in BAC vector. This library contains inserts with an average length of 130 kb, and the complete library represents 13 haploid copies of the S. purpuratus genome (40, 43). There were 26 clones of the whole library that hybridized with the genomic SRCR1 5′ flanking probe, consistent with the presence of either two loci or a cluster of genes spanning well over the average size of a single BAC insert. Screening the BAC library with a type 1 SRCR repeat probe yielded an estimate of eight type 1 SRCR loci, only two of which colocalized with the SRCR1 5′ flanking probe. For SpSRCR7, a single BAC contained the SRCR repeats and the C terminus of SpSRCR7.2, whereas the C terminus of SpSRCR7.1 colocalized with the SRCR probe only in 25% of the cases, indicating that this segment resides apart from the rest of the gene. Although not shown here, two loci were estimated for SpSRCR5, and two were estimated for SpSRCR12, one of which neighbored one of the four loci detected with an Mo6 probe (Mo6 sequence is 5′ incomplete, consisting of nine SRCRs, two short census repeats, predicted transmembrane, and intracellular domains; accession no. AF228827), whereas another probe detected three additional loci (incomplete SRB18.13U; accession no. AF228861). Analysis of one BAC clone that was detected with the SRCR1 5′ flanking probe (Fig. 4) reveals a cluster of at least three closely related type 1 genes that were detected by four specific probes: exon 1, SRCR1 domain, the ECM domain, and the vWF domain.
Figure 4.
Clustered genes encoded in type 1 SRCR gene locus. An SRCR 1 clone of sea urchin genomic DNA in BAC plasmid vector (133-kb insert) was digested with NotI (N), EcoRI (E), or SalI (S), and the blot was probed repeatedly with specific segments of SpSRCR1: exon 1, the SRCR1 repeat element, the ECM domain and vWF domain. A cluster consisting of at least three closely related type 1 SRCR genes was detected by all four probes. Also shown is the ethidum bromide (EtBr) image of the pulsed-field gel corresponding to the blotted DNA; the BAC vector migrates at 8.9 kb in the N lane.
The calculated genome size of S. purpuratus is 8 × 108 bp, with 27,000 predicted genes (43). A blast search of approximately 4.25% of the sea urchin genome that has been covered by sequencing both ends of BAC inserts (68,006 sequences of about 500 bp) revealed 51 unique SRCR sequences, which is the equivalent of 1,200 SRCR domains in the whole genome. The number of SRCR domains in each gene is unknown but averages to seven in the sample of six complete sequences available thus far. Thus, the most conservative estimate for the number of SRCR genes in the sea urchin genome would be around 150 genes (1,200 ÷ 7 = 171).
Fingerprinting Expression of Coelomocyte SRCR Genes.
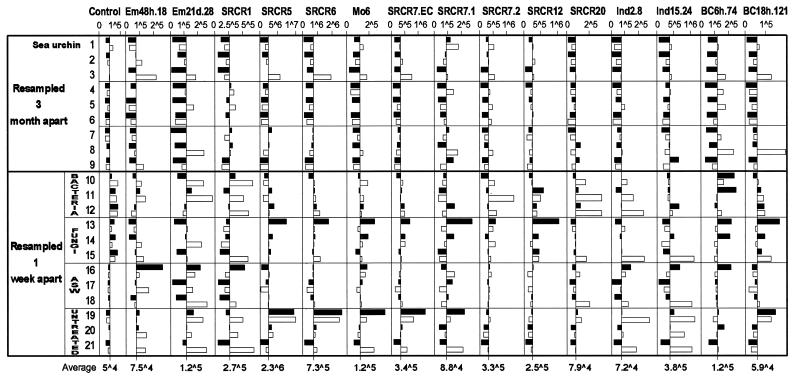
An SRCR array was used for simultaneous detection of transcription levels of multiple SRCR genes. The array consisted of 87 unique sequence SRCR gene fragments that were cloned by PCR with degenerate SRCR primers. Of these markers, 44 represent previously unknown SRCR domains, and the remaining 43 are segments of the SRCR clones shown in Fig. 1. Probes for hybridization were synthesized from mRNA of on average 3.5 × 106 coelomocytes, drawn with a fine-needle syringe in 0.5 ml of coelomic fluid. Repeated coelomocyte samples were collected 1 week or 3 months apart from the same animals with minimal disturbance. To test reproducibility of hybridization patterns, probes were synthesized from five samples that were split on collection. These five pairs of coelomocyte replicas hybridized in highly reproducible patterns (0.97–0.99 pairwise correlation). Next, 21 sea urchins were sampled twice, either 1 week or 3 months apart. Their hybridization patterns are presented in Fig. 5. Values shown are for 15 markers that produced strong hybridization signals representing SRCR types 1, 5, 6, 7, 12, and 20; the two C-terminal variants of SpSRCR7; and seven incomplete previously unidentified sequences. Several other SRCR markers revealed little variation in hybridization values among the samples, and one example is shown as a control demonstrating small fluctuations in gene expression. For the control SRCR marker (BC12h.77), the average from all 42 samples is 49,800 arbitrary PhosphorImager units (range: 29,200–88,500), and the ratio between first and second samples from 21 animals averaged to 1.0 (range: 0.48–2.04). Because alterations of up to 2-fold were detected in the hybridization levels of the control marker, a threshold of at least 3-fold alteration in expression values was chosen as significant in interpreting the array hybridizations.
Figure 5.
Graphic presentation of hybridization values for 16 SRCR gene markers in repeated samples from 21 sea urchins (black bar for the first sample, white for the second). Data bars represent distance from the average value calculated for that marker (x axis crosses at the average value, which is shown at the bottom). Nine sea urchins were each sampled twice 3 months apart (nos. 1–9). Another group of 12 sea urchins were sampled before, and 1 week after treatment. The coelomic cavities of these animals were injected with 1 ml of OD600 = 1 of live sea urchin surface bacteria (nos. 10–12) or with 1 ml of OD600 = 1 of live sea urchin surface fungi (nos. 14–16) or with 1 ml of artificial seawater (ASW; nos. 17–19), and three animals were untreated (nos. 19–21). Arbitrary PhosphorImager values of hybridization are shown for markers representing SRCR types 1, 5, 6, 7, 12, and 20, the two C-terminal variants of SpSRCR7, and seven incomplete previously unknown sequences. Control, hybridization values for SRCR marker BC12h.77 that show little variation in gene expression. In the numerical values, ∧ represents exponent of 10.
All of the 15 SRCR markers, other than the control, detected diverse patterns of SRCR transcripts. For instance, the average levels of expression of SRCR5 in samples from animal 19 were 73-fold higher than those of animal 9, and 14-fold higher for SRCR6.
Modulation of Expressed Coelomocyte SRCR Repertoires over a 3-Month Period.
To learn whether SRCR repertoires of individual sea urchins are fixed or transient, coelomocytes were collected twice 3 months apart from nine animals that were housed in individual cages (nos. 1–9 in Fig. 5). All samples repeated 3 months apart from nine sea urchins revealed alterations greater than 3-fold in SRCR profiles. For example, sea urchin 3 up-regulated 11 markers in the range of 4.2- to 30-fold: 30.5-fold for marker Em21d.28, 13.1-fold for SRCR7.IC2, and 11.2-fold for marker Ind.2.8. Only animal 7 retained its SRCR repertoire with only minor fluctuations, except for a 21.5-fold up-regulation of marker Em21d.28. In total, pairwise correlation between hybridization values ranged 0.42–0.89 (average 0.78) among repeated samples collected 3 months apart from the same animals.
Expression Fingerprints of Traumatized Sea Urchins.
To examine the possible effect of pathogen challenge and injury on transcription of SRCR genes, 12 animals were sampled twice 1 week apart, once before and once after treatment. Sea urchins were injected with 1 ml of OD600 = 1 of live bacteria (Fig. 5, nos. 10–12) or injected with 1 ml of OD600 = 1 of live fungi (Fig. 5, nos. 13–15) or injected with 1 ml of sterile artificial seawater (Fig. 5, nos. 16–18). Three animals were left untreated as a control (Fig. 5, nos. 19–21). In 1 week, all 12 sea urchins displayed altered SRCR profiles, which varied among the treated animals. For example, sea urchin 13, which was injected with fungi, down-regulated four markers 3.1- to 5.6-fold and up-regulated three markers 3.6- to 5.2-fold, whereas animal 10, which was injected with bacteria, up-regulated three markers 3- to 10.1-fold. Inspection of hybridization profiles of untreated sea urchins revealed that animal 20 retained its repertoire and that animal 19 up-regulated marker Ind2.8 3.7-fold. In contrast, the third untreated animal, 21, up-regulated 10 of these markers 3.9- to 13.2-fold: 13.2-fold for SRCR1 and 9.7-fold for marker Ind15.24. This unexpected response may indicate an inadvertent internal injury (4, 44).
Discussion
Coelomocytes of individual sea urchins express multiple SRCR genes from among a multigene family in diverse transcription profiles. It is estimated that the S. purpuratus genome encodes about 1,200 SRCR domains, which are components of at least 150 genes, and this large number can attest to the importance of SRCR genes in the biology of sea urchins. Genes encoding coelomocyte SRCR transcripts exist in all tested animals, whether messages of these genes were detectable in their coelomocytes. Hence, variability in expression of SRCR genes reflects animal-to-animal differences in levels of coelomocyte transcripts from the same or closely related SRCR genes. The genome of S. purpuratus encodes multiple variants of all tested SRCR gene types, as evident from the complex genome blots corresponding to these loci. Type 1 SRCR genes are most likely encoded in a large locus, arranged as a gene cluster. Furthermore, sequence analysis of the 5′ flanking and the beginning of the coding region in three genomic SRCR type 1 clones revealed conserved as well as completely diverged regions, which suggests a high rate of divergence among alleles and duplicated genes. The identity in 5′ gene regulatory regions may indicate concerted regulation of transcription of these genes.
Coelomocyte SRCR expression profiles change over time in individual sea urchins. Array hybridizations with coelomocyte messages from repeated samples collected 3 months apart revealed individual-specific alterations in the SRCR repertoires of all sea urchins. During this period, expression values for specific SRCR genes changed in several animals on the order of 20- to 30-fold. Furthermore, animals traumatized by pathogen challenges or injection of artificial seawater and even an untreated sea urchin displayed substantially altered SRCR profiles within a week. The SRCR repertoires of challenged sea urchins changed in an inconsistent pattern among the animals. It is possible that consistent alterations of particular SRCR genes can be observed only shortly after challenge. For instance, the kinetics of coelomocyte response to bacterial challenge were shown to peak at 6 h for SpNFkB, SpRunt-1, and SpGATAc, resuming steady-state mRNA levels within 18–24 h (4). A rapid response was reported in mice for the SRCR protein MARCO, which is up-regulated in macrophages within 45 min of injection of bacteria (26). However, we could not carry out experiments on a similar short-term scale in sea urchins because of the stress induced by frequent collection of coelomocyte samples. On the other hand, it is possible that these coelomocyte SRCR gene products are not components of an antipathogen system. Vertebrate SRCR proteins are implicated in development of the immune system and in regulation of immune responses, mediating growth, differentiation, and activation of immune cells as well as other cells of the body most likely via protein–protein interactions (9, 15, 16). In view of the broad range of functions attributed to vertebrate immune-related SRCR genes, it might be reasonable to assume multiple functions for similar gene products in sea urchin coelomocytes.
Elucidation of the function of coelomocyte SRCR gene products awaits further research, but their magnitude is presented here, in an invertebrate, in terms of gene numbers, complex genomic organization, and dynamic transcription profiles. Irrespective of their function, as biological markers, these SRCRs can be used for identification of subsets of the coelomocytes, as were classical cluster of differentiation antigens for the development of vertebrate cellular immunology. Such markers may provide us with powerful tools to study the evolution of multilineages of putative immune cells in invertebrate deuterostomes. The sea urchin SRCR gene system is fundamentally different from the rearranging Ig genes in vertebrate adaptive immunity and yet may reveal aspects of molecular and cellular diversity heretofore never considered in any invertebrate.
Acknowledgments
Special gratitude is expressed to Prof. Eric H. Davidson for encouragement and supervision throughout this project. I am grateful to my colleagues here at California Institute of Technology, Profs. Jose Alberola-Ila and Ellen Rothenberg and Drs. Mark P. Boldin, Alexander Hoffmann, Carlos Lois, Jonathan P. Rast, Luk Van Parijs, and Xiao-Feng Qin, for advice and discussion of this manuscript. Dr. Andrew R. Cameron kindly supplied data from the genome project sequence database. Miki Yun provided invaluable assistance in the analysis of the many previously unidentified clones described in this work, and Patrick Leahy was of enormous assistance in handling the sea urchins and in collection of samples for all these experiments. The research was supported by Human Frontier Science Program Organization Grant RG-333/96; Z.P. was supported by National Institutes of Health Training Grant HD-07257.
Abbreviations
- SRCR
scavenger receptor cysteine-rich
- BAC
bacterial artificial chromosome
- UTR
untranslated region
- vWF
von Willebrand factor
- ECM
extracellular matrix
- EGF
epidermal growth factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF064259 and AF228823–AF228878).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230096397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230096397
References
- 1.Smith L C, Chang L, Britten R J, Davidson E H. J Immunol. 1996;156:593–602. [PubMed] [Google Scholar]
- 2.Al-Sharif W Z, Sunyer J O, Lambris J D, Smith L C. J Immunol. 1998;160:2983–2997. [PubMed] [Google Scholar]
- 3.Smith L C, Shih C-S, Dachenhausen S G. J Immunol. 1998;161:6784–6793. [PubMed] [Google Scholar]
- 4.Pancer Z, Rast J P, Davidson E H. Immunogenetics. 1999;49:773–786. doi: 10.1007/s002510050551. [DOI] [PubMed] [Google Scholar]
- 5.Smith L C, Davidson E H. Ann NY Acad Sci. 1994;712:213–226. doi: 10.1111/j.1749-6632.1994.tb33575.x. [DOI] [PubMed] [Google Scholar]
- 6.Smith L C, Davidson E H. Immunol Today. 1992;13:356–361. doi: 10.1016/0167-5699(92)90172-4. [DOI] [PubMed] [Google Scholar]
- 7.Klein J. Immunogenetics. 1999;50:116–123. doi: 10.1007/s002510050587. [DOI] [PubMed] [Google Scholar]
- 8.Smith L C, Britten R J, Davidson E H. Dev Comp Immunol. 1995;19:217–224. doi: 10.1016/0145-305x(95)00009-i. [DOI] [PubMed] [Google Scholar]
- 9.Resnick D, Pearson A, Krieger M. Trends Biochem Sci. 1994;19:5–8. doi: 10.1016/0968-0004(94)90165-1. [DOI] [PubMed] [Google Scholar]
- 10.Hohenester E, Sasaki T, Timpl R. Nat Struct Biol. 1999;6:228–232. doi: 10.1038/6669. [DOI] [PubMed] [Google Scholar]
- 11.Pancer Z, Münkner J, Müller I, Müller W E G. Gene. 1997;193:211–218. doi: 10.1016/s0378-1119(97)00135-2. [DOI] [PubMed] [Google Scholar]
- 12.Blumbach B, Pancer Z, Diehl-Seifert B, Steffen R, Münkner J, Müller I, Müller W E G. J Cell Sci. 1998;111:2635–2644. doi: 10.1242/jcs.111.17.2635. [DOI] [PubMed] [Google Scholar]
- 13.Dangott L J, Jordan J E, Bellet R A, Garbers D L. Proc Natl Acad Sci USA. 1989;86:2128–2132. doi: 10.1073/pnas.86.7.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer W E, Tichy H. Gene. 1995;164:267–271. doi: 10.1016/0378-1119(95)94092-z. [DOI] [PubMed] [Google Scholar]
- 15.Aruffo A, Bowen M A, Patel D D, Hynes B F, Starling G C, Gebe J A, Bajorath J. Immunol Today. 1997;18:498–504. doi: 10.1016/s0167-5699(97)01130-4. [DOI] [PubMed] [Google Scholar]
- 16.O'Keeffe M A, Metcalfe S A, Cunningham C P, Walker I D. Immunogenetics. 1999;49:45–55. doi: 10.1007/s002510050462. [DOI] [PubMed] [Google Scholar]
- 17.Yu Q, Reichert M, Brousseau T, Cleuter Y, Burny A, Kettmann R. Nucleic Acids Res. 1990;18:5296. doi: 10.1093/nar/18.17.5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aruffo A, Melnick M B, Linsley P S, Seed B. J Exp Med. 1991;174:949–952. doi: 10.1084/jem.174.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pena-Rossi C, Zuckerman L A, Strong J, Kwan J, Ferris W, Chan S, Tarakhovsky A, Beyers A D, Killeen N. J Immunol. 1999;163:6494–6501. [PubMed] [Google Scholar]
- 20.Kobarg J, Whitney G S, Palmer D, Aruffo A, Bowen M A. Eur J Immunol. 1997;27:2971–2980. doi: 10.1002/eji.1830271133. [DOI] [PubMed] [Google Scholar]
- 21.Miyazaki T, Hirokami Y, Matsuhashi N, Takatsuka H, Naito M. J Exp Med. 1999;2:413–422. doi: 10.1084/jem.189.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yusa S, Ohnishi S, Onodera T, Miyazaki T. Eur J Immunol. 1999;29:1086–1093. doi: 10.1002/(SICI)1521-4141(199904)29:04<1086::AID-IMMU1086>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 23.Ritter M, Buechler C, Langmann T, Schmitz G. Biochem Biophys Res Commun. 1999;260:466–474. doi: 10.1006/bbrc.1999.0866. [DOI] [PubMed] [Google Scholar]
- 24.Trahey M, Weissman I L. Proc Natl Acad Sci USA. 1999;96:3006–3011. doi: 10.1073/pnas.96.6.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elomaa O, Kangas M, Sahlberg C, Tuukkanen J, Sormunen R, Liakka A, Thesleff I, Kraal G, Tryggvason K. Cell. 1995;80:603–609. doi: 10.1016/0092-8674(95)90514-6. [DOI] [PubMed] [Google Scholar]
- 26.Van der Laan L J W, Döpp E A, Haworth R, Pikkarainen T, Kangas M, Elomaa O, Dijkstra C D, Gordon S, Tryggvason K, Kraal G. J Immunol. 1999;162:939–947. [PubMed] [Google Scholar]
- 27.Thomas C A, Li Y M, Kodama T, Suzuki H, Silverstein S C, El Khoury J. J Exp Med. 2000;191:147–156. doi: 10.1084/jem.191.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haworth R, Platt N, Keshav S, Hughes D, Darley E, Suzuki H, Kurihara Y, Kodama T, Gordon S. J Exp Med. 1997;186:1431–1439. doi: 10.1084/jem.186.9.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenberg J W, Fischer W, Joiner K A. Infect Immun. 1996;64:3318–3325. doi: 10.1128/iai.64.8.3318-3325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunne D W, Resnick D, Greenberg J, Krieger M, Joiner K A. Proc Natl Acad Sci USA. 1994;91:1863–1867. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmskov U, Mollenhauer J, Madsen J, Vitved L, Grønlund J, Torøne I, Kliem A, Reid K B M, Poustka A, Skjødt K. Proc Natl Acad Sci USA. 1999;96:10794–10799. doi: 10.1073/pnas.96.19.10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freeman M, Ashkenas J, Rees D J, Kingsley D M, Copeland N G, Jenkins N A, Krieger M. Proc Natl Acad Sci USA. 1990;87:8810–8814. doi: 10.1073/pnas.87.22.8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldberger G, Bruns G A, Rits M, Edge M D, Kwiatkowski D J. J Biol Chem. 1987;262:10065–10071. [PubMed] [Google Scholar]
- 34.Wijngaard P L, Metzelaar M J, MacHugh N D, Morrison W I, Clevers H C. J Immunol. 1992;149:3273–3277. [PubMed] [Google Scholar]
- 35.Kanan J H C, Nayeem N, Binns R M, Chain B M. Immunogenetics. 1997;46:276–282. doi: 10.1007/s002510050273. [DOI] [PubMed] [Google Scholar]
- 36.Walker I D, Glew M D, O'Keeffe M A, Metclafe S A, Clevers H C, Wijngaard P L J, Adams T E, Hein W R. J Immunol. 1994;83:517–523. [PMC free article] [PubMed] [Google Scholar]
- 37.Kirkham P A, Takamatsu H-H, Parkhouse R M E. Eur J Immunol. 1997;27:717–725. doi: 10.1002/eji.1830270321. [DOI] [PubMed] [Google Scholar]
- 38.Takamatsu H-H, Kirkham P A, Parkhouse R M E. Eur J Immunol. 1997;27:105–110. doi: 10.1002/eji.1830270116. [DOI] [PubMed] [Google Scholar]
- 39.Leahy P S, Tutschulte T C, Britten R J, Davidson E H. J Exp Zool. 1978;204:369–380. doi: 10.1002/jez.1402040308. [DOI] [PubMed] [Google Scholar]
- 40.Martinez P, Rast J P, Arenas-Mena C, Davidson E H. Proc Natl Acad Sci USA. 1999;96:1469–1474. doi: 10.1073/pnas.96.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinnett D, Richer C, Baccichet A. BioTechniques. 1998;24:752–754. doi: 10.2144/98245bm12. [DOI] [PubMed] [Google Scholar]
- 42.Cameron R A, Leahy P S, Britten R J, Davidson E H. Dev Biol. 1999;208:255–264. doi: 10.1006/dbio.1999.9224. [DOI] [PubMed] [Google Scholar]
- 43.Cameron R A, Mahairas G, Rast J P, Martinez P, Biondi T R, Swartzell S, Wallace J C, Poustka A J, Livingston B T, Wray G A, et al. Proc Natl Acad Sci USA. 2000;97:9514–9518. doi: 10.1073/pnas.160261897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith L C, Britten R J, Davidson E H. Mol Biol Cell. 1992;3:403–414. doi: 10.1091/mbc.3.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]