Abstract
Induced pluripotent stem cell (iPS) technology appears to be a general strategy to generate pluripotent stem cells from any given mammalian species. So far, iPS cells have been reported for mouse, human, rat, and monkey. These four species have also established embryonic stem cell (ESC) lines that serve as the gold standard for pluripotency comparisons. Attempts have been made to generate porcine ESC by various means without success. Here we report the successful generation of pluripotent stem cells from fibroblasts isolated from the Tibetan miniature pig using a modified iPS protocol. The resulting iPS cell lines more closely resemble human ESC than cells from other species, have normal karyotype, stain positive for alkaline phosphatase, express high levels of ESC-like markers (Nanog, Rex1, Lin28, and SSEA4), and can differentiate into teratomas composed of the three germ layers. Because porcine physiology closely resembles human, the iPS cells reported here provide an attractive model to study certain human diseases or assess therapeutic applications of iPS in a large animal model.
Introduction
Induced nuclear reprogramming through induced pluripotent stem cell (iPS)2 technology is an amazing achievement full of challenge to the intellect and important practical implications (1, 2). Overexpression of exogenous factors that are highly enriched in embryonic stem cell (ESC) can rearrange the genetic program of different cell types, including somatic and adult stem cells, and induce a long lasting ESC-like pluripotent state (3–7). The repercussions of iPS technology are vast: it provides a way to create patient-specific stem cells that bypasses ethical and technical issues surrounding human ESC derivation and somatic cell nuclear transfer (8, 9), a state of the art model for studying genetic diseases in vitro (10, 11), and an incredible backwards route that can crystallize our current understanding of developmental and stem cell biology. Many questions, especially mechanistic, remain unanswered, but the current rhythm of research may bring iPS to clinical application sooner than expected. However, before jumping onto such extraordinary endeavor, safety must be scrupulously tested in an animal model close enough to humans. Nowadays that iPS technology is expanding, with improved delivery systems, chemical additions, new tissue culture conditions, and multiple cell sources being reported regularly, such animal model is essential to set up quality standards (12–18). Mice, and maybe rats, will possibly continue unrivalled as the easier ways to learn about reprogramming machinery and improve methodology, but their size, physiology, and reduced lifespan are handicaps for making serious assumptions regarding safety in humans. Given philogenetic similarity, monkeys are theoretically an excellent alternative, but in practice ethical concerns remain to at least some extent, and they are neither easy to maintain nor to breed. Swine, a regular source of food whose farming humans have adapted over myriads of years and whose physiology is remarkably similar to ours, stands up as arguably the most attractive model for preclinical iPS. Notably, insulin obtained from pigs is widely used to treat diabetes, whereas pig heart valves and skin have been, respectively, transplanted and applied to human burn victims for decades (19). Pig organs have also raised enormous interest for xenotransplantation: use of transgenic pigs lacking α(1–3)-galactosyltransferase gene, a major xenoantigen involved in acute rejection, holds optimism regarding effectiveness of more comprehensive genetic manipulations (20). Isolation of fully competent ESC from pigs or animal species apart from mouse, human, monkey, and more recently the rat, has proven impossible despite years of maintained effort (21, 22). Consequently, porcine genetic manipulation can only be achieved through laborious and inefficient somatic cell nuclear transfer (23). Moreover, porcine embryonic fibroblasts (PEF), the cell choice for somatic cell nuclear transfer, have a limited lifespan that complicates homologous recombination techniques (24). Genetic manipulation of pig iPS cell lines could provide an outstanding supply of tissues for xenotransplantation. From a different perspective, knowledge derived from porcine iPS may as well accelerate the isolation of bona fide ESC.
EXPERIMENTAL PROCEDURES
Cells and Tissue Culture
For isolating PEF, 37-day-old fetuses were minced with a scalpel blade in HEPES-buffered medium, and digested for 4 h in collagenase (Invitrogen) at 39 °C. The solution was mixed multiple times with a pipette and diluted 1/1 in PEF culture medium: Dulbecco's modified Eagle's medium, high glucose with penicillin/streptomycin (Invitrogen) and 10% fetal bovine serum (Hyclone). Samples were centrifuged at 700 × g for 10 min and the pellet resuspended in PEF medium. Only early passages were used for iPS generation. Mouse embryonic fibroblasts treated with mitomycin C were used as feeder cells. HEK293T cells were used as the packaging cell line for retroviral production. All 3 cell types were cultured in the same medium as PEF. iPS cell lines were successfully generated using Dulbecco's modified Eagle's medium, high glucose with antibiotics, glutamine (2 mm, Invitrogen), pyruvate (2 mm, Invitrogen), nonessential amino acids (1%, Invitrogen), β-mercaptoethanol (0.1 mm, Sigma), basic fibroblast growth factor (4 ng/ml, Invitrogen), and 10% defined fetal bovine serum (Hyclone); mouse ESC medium also contained mouse LIF (1000 units/ml, Millipore) but not basic fibroblast growth factor, and fetal bovine serum (15%) was from Invitrogen. All cells apart from HEK293T were cultured at all times (except for retroviral infection) at 39 °C. iPS colonies were first passaged mechanically using a Pasteur pipette, and afterward using dispase (1 mg/ml, Invitrogen). 5-Azacytidine (20 μm, Sigma) was maintained for 3 passages (around 12 days) before RNA was extracted. For iPS production and maintenance culture media were renewed daily.
Retroviral Transduction
pMX plasmids containing epidermal growth factor protein or mouse Sox2, Klf4, Oct4, and c-Myc, have been described by us previously (25); those containing human factors were purchased from Adgene. HEK293T were transfected with Lipofectamine 2000 (Invitrogen) following the instructions of the manufacturer. Two rounds (24 h each) of supernatants were collected after stopping the transfection, filtered (0.45 μm pore size), and added onto sparse PEF split the day before; Polybrene (8 μg/ml, Sigma) was added to increase infection efficiency.
Alkaline Phosphatase (AP) Staining and Immunofluorescence Microscopy
AP staining was performed as previously described (25). For immunofluorescence, iPS cell lines were grown for 2–3 days on coverslips coated with feeder cells before fixing with 4% paraformaldehyde. Coverslips were permeabilized with Triton X-100 before 30 min incubation with blocking reagent (5% fetal bovine serum in phosphate-buffered saline). Antibodies against SSEA4 and secondary antibodies (goat anti-mouse TRITC) were purchased from Invitrogen and Zhongshan Goldenbridge Biotechnology, respectively, antibodies against Nanog and Rex1 were made by us and have been described previously (25). Primary antibodies were incubated for at least 1 h at room temperature before washing, secondary antibodies for 1 h or less. 4′,6-Diamidino-2-phenylindole was purchased from Sigma. Coverslips were mounted on a slide using glycerol and sealed with nail polish; a conventional fluorescence microscope (Carl Zeiss) was used for visualization.
Semiquantitative RT-PCR and Real Time PCR
RNA was extracted using TRIzol (MRC). Semiquantitative PCR of retrotranscribed samples was performed using a touchdown protocol and LA Taq polymerase (Takara). Real time PCR was performed using SYBR Green (Takara) and a ABI 7300 machine; samples were normalized on the basis of ribosomal 18 S RNA values. Primers for porcine sequences were as follows: endogenous Sox2 (NCBI accession number NM_001123197) forward, 5′-GGTTACCTCTTCTTCCCACTCCA-3′ and reverse, 5′-CAAAAATAGTCCCCCCAAAAGAAG-3′; Nanog (NM_001129971) forward, 5′-CTTATTCAGGACAGCCCTGATTCTTC-3′ and reverse, 5′-AAGACGGCCTCCAAATCACTG-3′; Lin28 (NM_001123133) forward, 5′-TCAACGTGCGCATGGGGTTCGGCTTCCTGT-3′ and reverse, 5′-GTGGACGTCTTTGTGCACCAGAGTAAGCTG-3′; β-actin (AY550069) forward, 5′-CCGTGAGAAGATGACCCAGATCATGT-3′ and reverse, 5′-CGTGATCTCCTTCTGCATCCTGTC-3′; reverse telomerase-reverse transcriptase (NCBI accession number AY785158) forward, 5′- TGCTCGCCAACGTTTACA-3′ and reverse, 5′-CAAGCCGGAGGAAAAATG-3′, 18S (NR_002170) forward, 5′-ACCCACGGAATCGAGAAA-3′ and reverse, 5′-GCCTGCGGCTTAATTTGA-3′. Amplicons for Sox2, Lin28, and Nanog were cloned into pMD18-T (TAKARA) and sequenced. Identity with the predicted sequences was 100%; eSox2 and Lin28 did not match any other DNA sequence available in NCBI and Sanger Institute pig genome databases. Primers for measuring the degree of silencing of exogenous transgenes were: pMX vector forward, 5′-GCCGACACCAGACTAAGAACCTAGAACCTC-3′; mouse Sox2 reverse, 5′-GCTTCAGCTCCGTCTCCATCATGTTATACAT-3′; mouse Oct4 reverse, 5′-AGTATGCCATCCCTCCGCAGAACTCGTATG-3′; mouse Klf4 reverse, 5′-AGGATAAAGTCTAGGTCCAGGAGGTCGTTG-3′; mouse c-Myc reverse, 5′-AGTCGTAGTCGAGGTCATAGTTCCTGTTGG-3′; human Sox2 reverse, 5′-TGACCACCGAACCCATGGAGCCAAGAG-3′; human Oct4 reverse, 5′-GTTGCTCTCCACCCCGACTCCTGCTTC-3′, human Klf4 reverse, 5′-GGAGGATGGGTCAGCGAATTGGAGAGA-3′; human c-Myc reverse, 5′-AGGACGGAGAGAAGGCGCTGGAGTCTTG-3′. Semiquantitative RT-PCRs were performed using retrotranscribed samples as above. DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega). Primers for detecting integration of mouse exogenous factors into the genome were similar to those used for measuring the silencing. Primers used for human factors were as follows: pMX vector forward, 5′-GCCGACACCAGACTAAGAACCTAGAACCTC-3′; human Sox2 forward, 5′-CTTGGCTCCATGGGTTCG-3′; human Oct4 forward, 5′-GAGAACCGAGTGAGAGGCAAC-3′; human Klf4 forward, 5′-TCTCTTCGTGCACCCACTTG-3′; human c-Myc forward, 5′-AGAGTCTGGATCACCTTCTGCTG-3′.
Karyotype Analysis
Cells were grown sparse in T flasks (Corning) and demecolcine (Dahui Biotech) was added to a final concentration of 50 μg/ml for 1 h. Cells were then trypsinized, pelleted by centrifugation at 2,000 × g for 5 min, resuspended in 8 ml of 0.075 m KCl, and incubated for 20 min at 37 °C. Fixative solution composed of 1 part of acetic acid and 3 parts methanol was added to a final volume of 10 ml, mixed gently, and incubated for 10 min at 37 °C. After further centrifugation, the supernatant was removed, and ice-cold fixative solution composed of 1 part acetic acid and 3 parts methanol were added to a final volume of 10 ml. Cells were dropped on a cold slide and incubated at 75 °C for 3 h. Belts were treated with trypsin and colorant, and metaphases analyzed on a Olympus BX51 microscope.
Teratoma Formation
Pig iPS cells were harvested using dispase and one million cells were injected into the flanks of nude mice subcutaneously. After 9 weeks, mice were sacrificed, tumors were embedded in paraffin, and sections stained with hematoxilin/eosin and histologically analyzed.
RESULTS AND DISCUSSION
We chose the Tibet miniature pig as a source for generating iPS. A significant advantage of this strain over the farm pig (Sus scrofa) is their reduced size and subsequent easier maintenance and experimentation (Fig. 1A) (26). Retroviral overexpression of Sox2, Klf4, Oct4, and c-Myc (SKOM) remains yet the most standard approach to induce iPS, and has been successfully used in mouse, human, monkey, and rat cells (4, 5, 28, 29). Despite cross-species differences, mouse SKOM factors can reprogram human cells efficiently (30). We employed both mouse and human factors delivered by means of retroviral transduction, and used PEF as a target (Fig. 1). Infection efficiency, measured with control GFP retroviruses, was close to 100% (Fig. 1, bottom). Three culture conditions (see “Experimental Procedures” for further details) were tested: standard mouse ESC medium (containing 15% serum and mouse LIF), Dulbecco's modified Eagle's medium, high glucose with 15% of defined (human ESC tested) serum (hereafter named as defined medium), and no LIF but with basic fibroblast growth factor (Fig. 1), and a half and half mixture of the two. Except for the infection, cells were maintained at all times in a CO2 incubator set at 39 °C, as this is the physiological body temperature in pigs and we considered it might affect the reprogramming. Early morphological changes comparable with those seen during mouse or human iPS generation were detected in all 3 media starting at day 5–6 post-infection (Fig. 1), after which 104 cells were split onto feeder monolayers. Colonies with human ESC-like morphology (clear-cut borders and with flat cells), only more compacted, appeared on the feeders in all 3 media around day 8–10 post-infection and irrespective of the factors mixture. More irregular non-ESC-like cell clusters were also abundant and tended to take over more standard colonies progressively (Fig. 2A). At day 16 discernible colonies with human ESC-like characteristics remained only in the defined medium, and were picked mechanically (Figs. 1 and 2A). 7 of 20 picked colonies in the mouse factor combination, and 11 of 24 in the human, survived the initial passage and stained positive for AP. These cell lines could be routinely passaged on feeders (after dispase digestion) without losing their characteristics, but trypsinization or splitting without feeders induced quick differentiation (not shown). Selected colonies were expanded and further characterized. Expanded colonies retained the original morphology, displayed a high nuclear/cytoplasmic ratio with big nucleoli characteristic of human and monkey ESC/iPS (28), and were AP positive (Fig. 2B). Semiquantitative RT-PCR demonstrated integration of the 4 transgenes into the genome of all tested iPS cell lines (Fig. 2C), whereas reverse telomerase-reverse transcriptase expression, an indication of high replication potential, was low or absent in PEF and high in iPS clones (Fig. 2D). Indicative of acquisition of pluripotency characteristics: porcine iPS colonies stained positive for the human and monkey ESC-specific glycoprotein SSEA4 and transcription factors Nanog and Rex1 (Zfp42) (Fig. 3A). No clear differences in morphology, AP, or immunofluorescence staining were detected between iPS cell lines resulting from mouse or human factor combinations, or after repeated passages (over 25 in this study). Semiquantitative RT-PCR demonstrated high expression of endogenous Sox2 (eSox2), detected with primers that cannot amplify the overexpressed transgene, and also of Nanog and Lin28 (Fig. 3B). mRNA products for the expressed transgenes were not silenced (Fig. 3C). Incomplete transgene silencing has also been described by others in human and rat iPS cell lines (6, 31). Specific chemicals can allow the transformation of incompletely reprogrammed mouse iPS cell lines into full iPS (32, 33). Treatment of pig iPS cell lines with 5′-azad over a period of 2 weeks did not produce any change in cell morphology (not shown) or ESC markers (Fig. 3B). Addition of ERK (PD0325901) and GSK3b (CHIR99021) inhibitors enhanced compaction and increased proliferation, but did not affect expression levels of the tested ESC markers (data not shown). To demonstrate multilineage differentiation, pig iPS cell lines were injected subcutaneously into the flanks of nude mice, which after 9 weeks resulted in teratomas comprising tissues derived from the three germ layers (Fig. 4A, a-b and g-h, mesoderm-derived striated muscle and adipose tissue; c-d, endoderm-derived gland like structures; e-f, ectoderm-derived neural epithelium). Noteworthy, karyotype analysis of these pig iPS cell lines showed that their pluripotent characteristics were not associated with accumulation of chromosomal abnormalities (Fig. 4B). Both fetal and born live chimeras have been obtained after injection of freshly isolated porcine inner mass cells into blastocysts (19). During the course of our study we injected pig iPS cell lines into blastocysts from farm pigs (white), and deposited them into pseudopregnant recipient females. The outcome of such experiment is still waiting. To our knowledge only one case of teratoma, using pig ESC that had been expanded for only 8 weeks (equivalent to 12–14 passages of pig iPS clones), and one describing the birth of chimeric piglets from pig ESC has been reported (19).
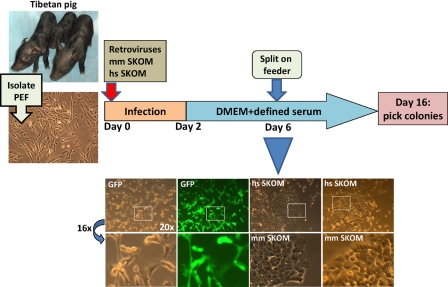
FIGURE 1.
Scheme depicting the generation of porcine iPS cell lines from the moment of viral transduction to colony picking. Captures of Tibetan pigs (5 days old), similar to the strain used for our experiments, are shown on the left. PEF (capture shown on the left) were isolated as described under “Experimental Procedures” and infected with retroviruses coding mouse or human factors. At the bottom of the scheme, captures at day 6 post-infection of PEF transduced with either GFP control retroviruses or SKOM mixtures and culture with defined medium are shown. GFP detected with a fluorescence filter showed almost 100% infection efficiency. Notice the early classical (similar to mouse or human iPS generation) morphology changes (cells becoming rounded and aggregating) only in the SKOM-infected pools. DMEM, Dulbecco's modified Eagle's medium. hs, Homo sapiens; mm, Mus musculus.
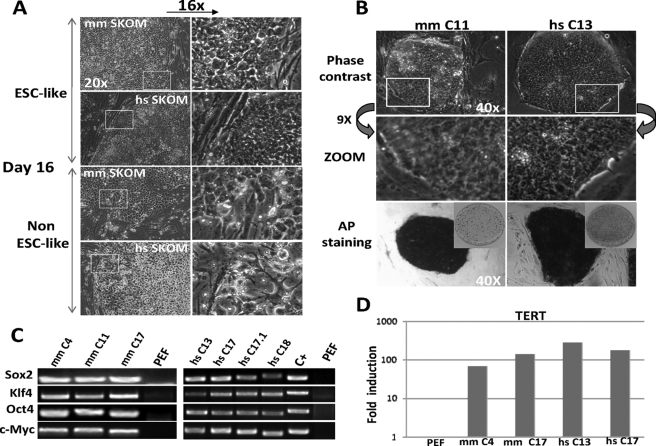
FIGURE 2.
Isolation of porcine iPS cell lines. A, colonies with ESC-like characteristics were picked at day 16. Colonies with a variable non-ESC-like morphology were abundant. Magnifications are indicated. B, expanded pig iPS colonies (passage 10 is shown) maintained the original morphology and stained positive for AP after repeated passages. AP staining on 3-cm dishes is shown in reduced size. C, semiquantitative RT-PCR with specific primers shows integration of exogenous mouse or human factors into the genomic DNA of pig iPS clones. Positive (C+) control corresponding to pMX-Sox2 plasmid and negative (uninfected PEF) were included. D, real-time RT-PCR for the reverse telomerase-reverse transcriptase gene shows high expression in selected PEF iPS clones compared with uninfected PEF. Values were normalized with 18S. Pig iPS colonies at passages 9 (mouse SKOM) and 15 (human SKOM) were used. mm, M. musculus; hs, H. sapiens.
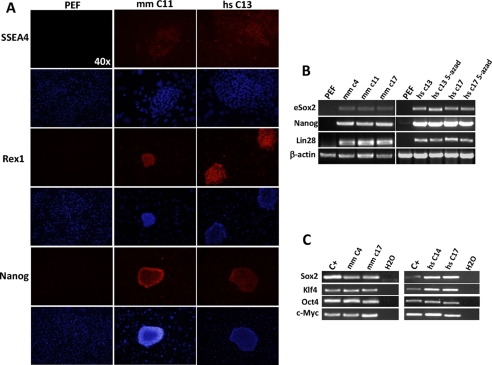
FIGURE 3.
Characterization of porcine iPS cell lines. A, immunofluorescence microscopy shows activation of the endogenous ESC program. PEF are shown on the left and stained negative for all markers. Pig iPS colonies at passage 10 were used, note that feeder layers stained negative and serve as an internal comparison. Magnifications are indicated. B, semiquantitative RT-PCR of selected PEF iPS cell lines. eSox2 indicates the endogenous gene; Nanog and Lin28 are also included and β-actin was used as loading control. Uninfected PEF were used as negative control. Treatment of human factors C13 and C17 iPS clones with 5-azacytidine did not further increase expression of ESC markers (right panel). Pig iPS colonies at passages 9 (mouse SKOM) and 15 (human SKOM) were used. C, semiquantitative RT-PCR with primers that specifically amplify the mRNA product of the integrated transgenes shows no silencing in selected iPS cell lines compared with mRNA extracted from infected control cells at day 8 after viral transduction. Water was used as negative control for the PCR. mm, M. musculus; hs, H. sapiens.
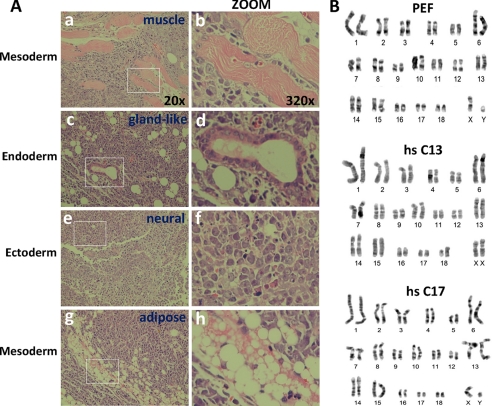
FIGURE 4.
Pig iPS cell lines are pluripotent. A, teratoma formation in immunodeficient mice demonstrates differentiation into the three germ layers. Results from the hs SKOM C13 clone (passage 16) are displayed, a similar pattern was observed with hs SKOM C17 (passage 16) (not shown). Magnified pictures are shown on the right, and magnification is indicated for both panels. Mesoderm-derived muscle and fat are shown in a-b and g-h, respectively, a gland-like structure (endoderm derived) in c-d, and neural-like tissue is shown in e-f. B, karyotype analysis demonstrates an equal number of chromosomes (19 pairs) in two different human SKOM iPS cell lines (passage 18) compared with control PEF. Note that the two iPS cell lines correspond to different genders. hs, H. sapiens.
In summary, herein we explain a method for reprogramming PEF into iPS cells, and provide tools for their characterization. The consequences of lack of silencing for the endogenous transgenes in our cell lines are uncertain. Conceivably, this may have an effect on the readiness of our cell lines to differentiate into different tissues, and this may explain the long time (9 weeks) needed for teratoma formation. The use of different cell types other than fibroblasts has a dramatic impact on iPS generation (25, 34, 35). A more systematic analysis of susceptibility to iPS among different porcine tissues would be important and might allow full transgene silencing by increasing the extent of the reprogramming. Besides, shifting away from PEF toward an easily obtained cell that does not involve sacrificing the animal will be needed for autologous transplantation experiments. Use of loxP flanked polycistronic exogenous factors would also allow elimination of the exogenous DNA insertions, and we are now setting this system up for porcine iPS (27). Models of lineage/tissue-specific differentiation will as well need to be validated in pig iPS. Such models may require variations from established mouse and human models, and in those cases in which cytokines are needed the cross-species jump may be problematic. The same can be argued regarding antibodies or other reagents needed for the characterization. In addition, Tibetan pigs, whereas having remarkable advantages over farm pigs in terms of their handling, have evolved for thousands of years in a restricted environment and this could have imposed evolutionary changes that affect their susceptibility to iPS generation. Systematic analysis of cells from other pig strains will thus be important as well. We are currently working on disease models using pigs in which iPS cell lines will be tested. Given the long life span of pigs (18–25 years), time consuming iPS generation is not an issue like it is now in mice. The latter implies the in vivo stability of iPS-derived lineages can be more rigorously monitored. Rather than all the above mentioned seeming incapacitating obstacles, and having in mind the creation of an outstanding model for preclinical testing, porcine iPS research is exciting and will likely move fast.
Acknowledgment
We thank Professor Gang Pei of the Shanghai Institutes for Biological Sciences for encouragement, support, and suggestions during the entire course of this study.
This work was supported by National Natural Science Foundation of China Grants 30725012 and 30630039, Chinese Academy of Sciences Grant KSCX2-YW-R-48, Guangzhou Science and Technology Grant 2006A50104002, Ministry of Science and Technology 973 Grants 2006CB701504, 2006CB943600, 2007CB948002, 2007CB947804, and 2009CB941000, and National High Technology Project 863 Grant 2005AA210930.
- iPS
- induced pluripotent stem cell
- ESC
- embryonic stem cell
- PEF
- porcine embryonic fibroblasts
- AP
- alkaline phosphatase
- TRITC
- tetramethylrhodamine isothiocyanate
- RT
- reverse transcriptase.
REFERENCES
- 1.Tweedell K. S. ( 2008) Curr. Stem Cell Res. Ther. 3, 151– 162 [DOI] [PubMed] [Google Scholar]
- 2.Pei D. ( 2008) Cell Res. 18, 221– 223 [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K., Yamanaka S. ( 2006) Cell 126, 663– 676 [DOI] [PubMed] [Google Scholar]
- 4.Okita K., Ichisaka T., Yamanaka S. ( 2007) Nature 448, 313– 317 [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. ( 2007) Cell 131, 861– 872 [DOI] [PubMed] [Google Scholar]
- 6.Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. ( 2007) Science 318, 1917– 1920 [DOI] [PubMed] [Google Scholar]
- 7.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. ( 2007) Nature 448, 318– 324 [DOI] [PubMed] [Google Scholar]
- 8.Thomson J. A., Kalishman J., Golos T. G., Durning M., Harris C. P., Becker R. A., Hearn J. P. ( 1995) Proc. Natl. Acad. Sci. U. S. A. 92, 7844– 7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilmut I., Schnieke A. E., McWhir J., Kind A. J., Campbell K. H. ( 1997) Nature 385, 810– 813 [DOI] [PubMed] [Google Scholar]
- 10.Park I. H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M. W., Cowan C., Hochedlinger K., Daley G. Q. ( 2008) Cell 134, 877– 886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebert A. D., Yu J., Rose F. F., Jr., Mattis V. B., Lorson C. L., Thomson J. A., Svendsen C. N. ( 2009) Nature 457, 277– 280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. ( 2009) Nature 458, 771– 775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woltjen K., Michael I. P., Mohseni P., Desai R., Mileikovsky M., Hämäläinen R., Cowling R., Wang W., Liu P., Gertsenstein M., Kaji K., Sung H. K., Nagy A. ( 2009) Nature 458, 766– 770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. ( 2008) Science 322, 949– 953 [DOI] [PubMed] [Google Scholar]
- 15.Stadtfeld M., Nagaya M., Utikal J., Weir G., Hochedlinger K. ( 2008) Science 322, 945– 949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoi T., Yae K., Nakagawa M., Ichisaka T., Okita K., Takahashi K., Chiba T., Yamanaka S. ( 2008) Science 321, 699– 702 [DOI] [PubMed] [Google Scholar]
- 17.Hanna J., Markoulaki S., Schorderet P., Carey B. W., Beard C., Wernig M., Creyghton M. P., Steine E. J., Cassady J. P., Foreman R., Lengner C. J., Dausman J. A., Jaenisch R. ( 2008) Cell 133, 250– 264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y., Do J. T., Desponts C., Hahm H. S., Schöler H. R., Ding S. ( 2008) Cell Stem Cell 2, 525– 528 [DOI] [PubMed] [Google Scholar]
- 19.Hall V. ( 2008) Stem Cell Rev. 4, 275– 282 [DOI] [PubMed] [Google Scholar]
- 20.Kolber-Simonds D., Lai L., Watt S. R., Denaro M., Arn S., Augenstein M. L., Betthauser J., Carter D. B., Greenstein J. L., Hao Y., Im G. S., Liu Z., Mell G. D., Murphy C. N., Park K. W., Rieke A., Ryan D. J., Sachs D. H., Forsberg E. J., Prather R. S., Hawley R. J. ( 2004) Proc. Natl. Acad. Sci. U. S. A. 101, 7335– 7340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talbot N. C., Blomberg le A. ( 2008) Stem Cell Rev. 4, 235– 254 [DOI] [PubMed] [Google Scholar]
- 22.Vackova I., Ungrova A., Lopes F. ( 2007) J. Reprod. Dev. 53, 1137– 1149 [DOI] [PubMed] [Google Scholar]
- 23.Lai L., Kolber-Simonds D., Park K. W., Cheong H. T., Greenstein J. L., Im G. S., Samuel M., Bonk A., Rieke A., Day B. N., Murphy C. N., Carter D. B., Hawley R. J., Prather R. S. ( 2002) Science 295, 1089– 1092 [DOI] [PubMed] [Google Scholar]
- 24.Hao Y., Wax D., Zhong Z., Murphy C., Ross J. W., Rieke A., Samuel M., Spate L., Dyce P., Li J., Sutovsky P., Prather R. S. ( 2009) Cloning Stem Cells 11, 101– 110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin D., Gan Y., Shao K., Wang H., Li W., Wang T., He W., Xu J., Zhang Y., Kou Z., Zeng L., Sheng G., Esteban M. A., Gao S., Pei D. ( 2008) J. Biol. Chem. 283, 33730– 33735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X. M., Liu B., Zhao S. H., Fan B., Zhu M. J., Yu M., Xiong T. A., Li K. ( 2006) Anim. Biotechnol. 17, 99– 107 [DOI] [PubMed] [Google Scholar]
- 27.Soldner F., Hockemeyer D., Beard C., Gao Q., Bell G. W., Cook E. G., Hargus G., Blak A., Cooper O., Mitalipova M., Isacson O., Jaenisch R. ( 2009) Cell 136, 964– 977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H., Zhu F., Yong J., Zhang P., Hou P., Li H., Jiang W., Cai J., Liu M., Cui K., Qu X., Xiang T., Lu D., Chi X., Gao G., Ji W., Ding M., Deng H. ( 2008) Cell Stem Cell 3, 587– 590 [DOI] [PubMed] [Google Scholar]
- 29.Li W., Wei W., Zhu S., Zhu J., Shi Y., Lin T., Hao E., Hayek A., Deng H., Ding S. ( 2009) Cell Stem Cell 4, 16– 19 [DOI] [PubMed] [Google Scholar]
- 30.Mali P., Ye Z., Hommond H. H., Yu X., Lin J., Chen G., Zou J., Cheng L. ( 2008) Stem Cells 26, 1998– 2005 [DOI] [PubMed] [Google Scholar]
- 31.Liao J., Cui C., Chen S., Ren J., Chen J., Gao Y., Li H., Jia N., Cheng L., Xiao H., Xiao L. ( 2009) Cell Stem Cell 4, 11– 15 [DOI] [PubMed] [Google Scholar]
- 32.Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., Meissner A. ( 2008) Nature 454, 49– 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T. W., Smith A. ( 2008) PLoS Biol. 6, e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J. B., Sebastiano V., Wu G., Araúzo-Bravo M. J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., Meyer J., Hübner K., Bernemann C., Ortmeier C., Zenke M., Fleischmann B. K., Zaehres H., Schöler H. R. ( 2009) Cell 136, 411– 419 [DOI] [PubMed] [Google Scholar]
- 35.Eminli S., Utikal J., Arnold K., Jaenisch R., Hochedlinger K. ( 2008) Stem Cells 26, 2467– 2474 [DOI] [PubMed] [Google Scholar]