Abstract
The rough endoplasmic reticulum-resident protein complex consisting of prolyl 3-hydroxylase 1 (P3H1), cartilage-associated protein (CRTAP), and cyclophilin B (CypB) can be isolated from chick embryos on a gelatin-Sepharose column, indicating some involvement in the biosynthesis of procollagens. Prolyl 3-hydroxylase 1 modifies a single proline residue in the α chains of type I, II, and III collagens to (3S)-hydroxyproline. The peptidyl-prolyl cis-trans isomerase activity of cyclophilin B was shown previously to catalyze the rate of triple helix formation. Here we show that cyclophilin B in the complex shows peptidyl-prolyl cis-trans isomerase activity and that the P3H1·CRTAP·CypB complex has another important function: it acts as a chaperone molecule when tested with two classical chaperone assays. The P3H1·CRTAP·CypB complex inhibited the thermal aggregation of citrate synthase and was active in the denatured rhodanese refolding and aggregation assay. The chaperone activity of the complex was higher than that of protein-disulfide isomerase, a well characterized chaperone. The P3H1·CRTAP·CypB complex also delayed the in vitro fibril formation of type I collagen, indicating that this complex is also able to interact with triple helical collagen and acts as a collagen chaperone.
Introduction
Procollagen biosynthesis occurs in the rough endoplasmic reticulum of cells and requires a large number of posttranslational modifications (1). Although it has been known for a long time that type I collagen contains 3-hydroxyproline residues (2), the enzyme activity catalyzing this reaction was only partially characterized (3–5) until recently (6). Prolyl 3-hydroxylase 1 (P3H1)2 modifies a single proline residue in the Xaa position of the Gly-Xaa-Hyp repeating sequence into (3S)-hydroxyproline (3-Hyp) in the α1 chains of type I, II, and III collagens. P3H1 extracted from chick embryos forms a multiprotein complex with CRTAP (previously described as Casp) and cyclophilin B (CypB) (6, 7). Laser light scattering and velocity sedimentation analysis shows that the three proteins form a 1:1:1 complex (7). P3H1 consists of two major domains: a carboxyl-terminal dioxygenase domain, which is similar to the α-subunit of prolyl 4-hydroxylase and lysyl hydroxylases, and a unique amino-terminal domain. The amino-terminal domain contains four cysteine (CXXXC) repeats and is homologous to CRTAP. This domain is found in five proteins of the human genome: P3H1, P3H2, P3H3, CRTAP, and SC65 (6). The function of this domain has not been established, but it also contains multiple tetratricopeptide repeat domains that are known to be important in protein-protein interactions (8, 9).
P3H1 was immunolocalized to cells in tissues that contain fibrillar collagens (6). Cells in tissues rich in basement membrane collagens did not stain with the monoclonal antibody against P3H1, and it is therefore likely that P3H2 (10) or P3H3 3-hydroxylates proline residues in these collagens.
To understand the potential roles of the P3H1·CRTAP·CypB complex during collagen biosynthesis a brief review of the known steps follows. After the translocation of the growing polypeptide chains of procollagens into the rER, proline residues become 4-hydroxylated by prolyl 4-hydroxlase. 4-Hydroxylation of proline residues increases the stability of the triple helix and is a key element in the folding of the triple helix. Prolyl 4-hydroxylase requires an unfolded chain as a substrate. The chain selection and association for triple helix formation are determined by the carboxyl-terminal propeptides in fibrillar collagens. Premature association between procollagen chains is thought to be prevented by chaperones such as protein-disulfide isomerase, BiP/GRP78, GRP94, HSP47, and FKBP65 and collagen modifying enzymes until the biosynthesis of the individual chain is completed. Additional modifications are the 3-hydroxylation of proline residues by the P3H1·CRTAP·CypB complex, the hydroxylation of lysine residues by lysyl hydroxylases, and glycosylation. The chains are then selected, and trimers are formed by association of the carboxyl-terminal propeptides. Disulfide bonds between the chains are formed, and this formation is most likely catalyzed by protein-disulfide isomerase. Triple helix formation proceeds from the carboxyl-terminal end toward the amino-terminal end in a zipper-like fashion. The rate-limiting step in this process is the cis-trans isomerization of peptide bonds. This process is catalyzed by peptidyl-prolyl cis-trans isomerase (CypB). Because procollagen molecules are only marginally stable, it was proposed that folding of procollagen molecules inside cells requires special chaperones (11) with HSP47 and FKBP65 as a potential candidates. Given the complexity of this process, it is not surprising that so many different proteins in the rER are involved and that these proteins interact and function as “folding machines.” Additionally collagen molecules have a strong tendency to aggregate. This process must be inhibited inside the cells and allowed only after secretion into the extracellular space. The functional importance of collagen chaperones is further shown by the embryonic lethality of HSP47 knock-out mice (12) and the accumulation of BiP/GRP78 in cells with mutations in the collagen chains (13).
The importance of the P3H1·CRTAP·CypB complex was recently demonstrated in CRTAP knock-out mice (7). These mice show osteochondrodysplasia characterized by severe osteoporosis and decreased osteoid production. They lack the 3-Hyp in type I and II collagens. In addition, human mutations in P3H1 and CRTAP lead to a lethal form of recessive osteogenesis imperfecta (14–16). In this report we show that the P3H1·CRTAP·CypB complex is not only responsible for the 3-hydroxylation of proline residues, but it is also a potent molecular chaperone.
EXPERIMENTAL PROCEDURES
Extraction and Purification of the Chicken P3H1·CRTAP·CypB Complex
The chicken P3H1·CRTAP·CypB complex was isolated from 15–17-day-old chick embryos following the previously published protocol (17, 18) with modifications to improve both yield and purity of the complex. 12 dozen chick embryos were mixed with an equal volume of 10 mm Tris/HCl buffer, pH 7.5, containing 0.25 m sucrose and protease inhibitors (5 mm EDTA, 2 mm phenylmethylsulfonyl fluoride, 2 mm N-ethylmaleimide, 1 μg/ml pepstatin A, and 1 μg/ml leupeptin). Homogenization was carried out in a Waring blender at maximum speed for 3 min. This and all subsequent steps were performed at 4 °C. The homogenate was centrifuged at 3,000 × g for 15 min in an H-6000A rotor (Thermo Fisher Scientific Inc., Waltham, MA). The supernatant was then centrifuged at 125,000 × g for 1.5 h in a 45 Ti rotor (Beckman Coulter, Inc., Fullerton, CA). The pellets from this step were resuspended in twice the volume of 50 mm Tris/HCl buffer, pH 7.5, containing 0.1% Tween 20, 0.2 m NaCl, and the same protease inhibitors as described above, treated with 1 μl/ml diisopropyl fluorophosphate, and gently stirred for 4 h on ice. The extract was centrifuged at 125,000 × g for 1.5 h, filtered through Miracloth, and run over a gelatin-Sepharose 4B column (2.6 × 30 cm; GE Healthcare) equilibrated in buffer A (50 mm Tris/HOAc buffer, pH 7.5, containing 0.2 m NaCl and 0.05% (v/v) Tween 20). The column was washed with at least 2 bed volumes of buffer A and then with 1 bed volume of 50 mm Tris/HOAc buffer, pH 7.5, containing 1 m NaCl and 0.05% Tween 20 followed by another bed volume of buffer A. Elution was performed using a pH gradient from 7.5 to 5.0 with buffer A. Peak fractions containing P3H1 complex and collagen-related proteins were pooled, dialyzed into phosphate-buffered saline at 4 °C, and filtrated through a 0.45-μm filter prior to being loaded onto the P3H1 monoclonal antibody affinity column. The column was washed with at least 5 bed volume of phosphate-buffered saline and then eluted by 50 mm glycine/HCl, pH 2.5, 150 mm NaCl, and 0.1% Triton X-100. P3H1 complex was pooled and dialyzed against each enzyme assay and reaction buffers.
Expression and Purification of Chicken CypB
The cDNA encoding chick CypB without the signal sequence was amplified by PCR from chick fibroblast cDNA using primers with the 5′ side containing an NcoI site and the 3′ side containing a SalI site after the stop codon. This DNA was inserted between the NcoI and SalI restriction sites of a pET30a(+) expression vector (Invitrogen). The expression vector was transformed into Escherichia coli BL21(DE3) and grown at 37 °C to an optical density of 0.6 at 600 nm, and isopropyl 1-thio-β-d-galactopyranoside was added (final concentration, 1 mm) to induce expression of CypB. After incubation at 30 °C overnight, cells were harvested by centrifugation and resuspended in B-PER (Thermo Fisher Scientific Inc.). Insoluble material was removed by centrifugation, and ammonium sulfate (final concentration, 30% (w/v)) was added to the supernatant, and it was incubated at room temperature for 1 h and then centrifuged to remove the precipitated materials. The supernatant from the ammonium sulfate precipitation was passed through a 0.22-μm filter and applied onto a Co2+-chelating resin column, and CypB was eluted by 20 mm sodium phosphate buffer, pH 7.5, containing 0.5 m NaCl and 0.5 m imidazole after washing with 20 mm sodium phosphate buffer, pH 7.5, containing 0.5 m NaCl and 0.05 m imidazole for at least 10 column volumes. The fractions containing CypB were dialyzed into enterokinase cleavage buffer (50 mm Tris/HCl, pH 8.0, containing 1 mm CaCl2 and 0.1% Tween 20), and enterokinase (0.1 unit/200-μl reaction volume) (Invitrogen) was added at 4 °C overnight to cleave the His tag. The reaction mixture was incubated with DEAE-Sepharose (100 μl/1.5-ml reaction volume) to remove enterokinase at room temperature for 1 h. The reaction mixture was spun down, the supernatant was applied onto a Co2+-chelating resin column, and CypB was eluted by 20 mm sodium phosphate buffer, pH 7.5, containing 0.5 m NaCl. The purified CypB was dialyzed against each enzyme assay and reaction buffer.
Citrate Synthase Thermal Aggregation Assay
The aggregation of citrate synthase upon thermal denaturation (19) was measured by the method of Shao et al. (20). Citrate synthase (Sigma-Aldrich) was diluted 200-fold to a final concentration of 0.15 μm into prewarmed 40 mm Hepes buffer, pH 7.4, at 43 °C. The aggregation of citrate synthase was monitored by absorbance at 500 nm in a Cary4 spectrophotometer (Varian Inc., Palo Alto, CA). All enzyme concentrations were determined by amino acid analysis. A stock solution of 0.5 mm cyclosporine A (EMD Chemicals, Inc., Gibbstown, NJ) was prepared in DMSO and further diluted to a final concentration of 1 μm (0.2% DMSO). Before measurement the P3H1·CRTAP·CypB complex was incubated with Cyclosporine A for 1 h at 4 °C.
Denatured Rhodanese Refolding and Aggregation Assay
Another frequently used assay for chaperone activity is the inhibition of aggregation of chemically denatured rhodanese (21, 22). Bovine rhodanese (Sigma-Aldrich) was denatured in 30 mm Tris/HCl buffer, pH 7.4, containing 6 m guanidine hydrochloride and 1 mm dithiothreitol at 25 °C for 1 h and then diluted 100-fold to a final concentration of 0.3 μm in 30 mm Tris/HCl buffer, pH 7.2, containing 50 mm KCl. The aggregation of denatured rhodanese was monitored at 320 nm with a Cary4 spectrophotometer. All protein concentrations were determined by amino acid analysis.
Enzyme Assays and Inhibition with Cyclosporine A
Measurements of the catalytic efficiency (kcat/Km) for the isomerization reaction were performed as described previously (23) based on the α-chymotrypsin assay (24) with the following modifications. Stock solutions of substrates were prepared in DMSO at a concentration of 2.5 mm for Suc-Ala-Ala-Pro-Phe-MCA. The final DMSO concentration in the assay was 0.352% for kcat/Km measurements and 0.602% for the inhibition studies. Kinetic measurements were made at 5 °C to minimize the non-enzymatic isomerization reaction in 35 mm Hepes/NaOH buffer, pH 7.8. Final substrate and chymotrypsin concentrations were 8.8 and 12.8 μm, respectively. Fluorescence changes were monitored at 380 nm with a HiTech stopped-flow spectrophotometer (TgK Scientific Ltd., Bradford-on-Avon, UK). The assay was started by mixing chymotrypsin and substrate. Progression curves were analyzed by fitting to a second-order exponential decay function with Origin (OriginLab Corp., Northampton, MA). Inhibition measurements were performed in a similar manner. A stock solution of cyclosporine A (0.1 mm) was prepared in DMSO and further diluted with 35 mm Hepes/NaOH buffer, pH 7.8. After preincubation of the inhibitor with the P3H1 complex at 4 °C for 1 h, the assay was started. Values for kcat/Km were calculated according to kcat/Km = (kobs − ku)/[E] where ku is the rate constant for the unanalyzed isomerization reaction and kobs is the rate constant for the catalyzed reaction in the presence of enzymes at a given concentration of [E]. k values were calculated using Origin.
Thermal Stability of Type I and Type III Collagens and Refolding of Type III Collagen Measured by Optical Rotary Dispersion
The thermal stability of bovine type I and III collagens was monitored at 365 nm using a 341MC polarimeter (PerkinElmer Life Sciences) with a 10-cm-path length thermostatted cell. The temperature was controlled by a circulating water bath and programmable temperature controller (RCS, Lauda Division, Brinkmann Instruments) and measured with a digital thermometer (Omega Engineering, Inc., Stamford, CT) and a thermistor inserted into the cell. Both the temperature and the optical rotatory dispersion signals were recorded and digitized on an HP9070A measurement and plotting system (Hewlett-Packard, Palo Alto, CA). The temperature was increased from 25 to 50 °C at a rate of 10 °C/h. Stock solutions of type I and type III collagens (final concentration, 0.025 μm) in 10 mm acetic acid were diluted into 50 mm Tris/HCl buffer, pH 7.5, containing 0.4 m NaCl. Refolding of pN type III collagen (0.125 μm) in 50 mm Tris/HCl buffer, pH 7.5, containing 0.2 m NaCl was monitored at 365 nm. The sample was denatured for 10 min at 45 °C and refolded at 25 °C by switching between two water baths.
Type I Collagen Fibril Formation Assay
A stock solution of type I collagen in 50 mm acetic acid was diluted to a final concentration of 0.3 μm in 150 mm sodium phosphate buffer, pH 7.8, containing 150 mm NaCl. The absorbance (light scattering) was monitored at 313 nm as a function of time at 34 °C. All protein concentrations were determined by amino acid analysis.
Surface Plasmon Resonance Analysis
Surface plasmon resonance experiments were carried out using a BIACore X instrument (GE Healthcare). Purified native bovine type I collagen was immobilized at a concentration of about 6 ng/mm2 (6,000 resonance units) on a CM5 sensor chip by amide coupling. The experiments were carried out at 25 °C in 20 mm Hepes buffer, pH 7.4, containing 150 mm NaCl, 5 mm EDTA, and 0.05% Tween 20. Various concentrations of P3H1·CRTAP·CypB complex (from 0.35 to 0.075 μm) were injected at a flow rate of 10 μl/min.
RESULTS
Purification of the P3H1·CRTAP·CypB Complex and Expression of Chicken CypB
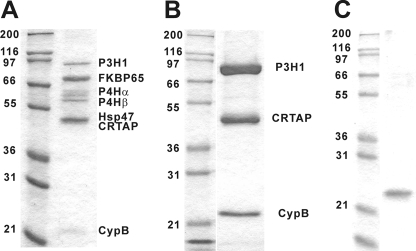
The P3H1·CRTAP·CypB complex was isolated from chick embryos and purified on a gelatin-Sepharose column followed by an antibody column loaded with a monoclonal antibody against chicken P3H1. Fig. 1 shows an SDS-polyacrylamide gel of the elution of the gelatin-Sepharose column (Fig. 1A) and the elution from the antibody column (Fig. 1B). To compare the biochemical activities of the P3H1·CRTAP·CypB complex and free CypB, we recombinantly expressed chicken CypB in E. coli. An SDS-polyacrylamide gel of the purified chicken CypB is shown in Fig. 1C. These protein preparations were used for the characterization of the biochemical activities.
FIGURE 1.
SDS-polyacrylamide gel electrophoresis of purified chicken P3H1·CRTAP·CypB complex and recombinant cyclophilin B. A, chicken rER proteins eluted from the gelatin-Sepharose column by low pH buffer. The proteins were identified by amino acid sequencing. This fraction was applied to a P3H1 antibody column and eluted with a pH 2.5 elution buffer. B, the fractions containing the purified P3H1 complex were run on an SDS, 10% polyacrylamide gel under reducing conditions and stained with GelCode Blue Stain Reagent. C, the purified chick CypB from E. coli after cleavage of the His tag by enterokinase was run on an SDS, 10% polyacrylamide gel under reducing conditions and stained with GelCode Blue Stain Reagent. The numbers on the left of each gel indicate the molecular masses of marker proteins in kDa. P4H, prolyl 4-hydroxylase.
Chaperone Activity of the P3H1·CRTAP·CypB Complex Using the Thermal Aggregation of Citrate Synthase
The P3H1·CRTAP·CypB complex exhibited potent chaperone activity in this assay. Fig. 2A shows the concentration-dependent inhibition of the thermal aggregation of citrate synthase. Compared with BSA as a negative control and 0.15 μm protein-disulfide isomerase, an established chaperone, the P3H1·CRTAP·CypB complex showed much stronger aggregation inhibition activity at only 0.01 μm concentration (Fig. 2B). Chicken CypB or cyclosporine A alone did not show any chaperone activity, and the addition of cyclosporine A to the P3H1·CRTAP·CypB complex did not alter its chaperone activity (Fig. 2C).
FIGURE 2.
Chaperone activity of P3H1·CRTAP·CypB complex using citrate synthase as a substrate. The inhibition of the thermal aggregation of citrate synthase by the P3H1·CRTAP·CypB complex was monitored at 500 nm. A 30 μm citrate synthase solution was diluted 200-fold into prewarmed 40 mm Hepes buffer, pH 7.5, at 43 °C. A, in the absence (black) and presence of 0.001 (red), 0.0025 (green), 0.005 (blue), and 0.01 μm (orange) P3H1·CRTAP·CypB complex. B, in the absence (black) and presence of 0.01 μm complex (red), 0.15 μm protein-disulfide isomerase (blue), and 0.15 μm bovine serum albumin (green). C, in the absence (black) and presence of 1 μm cyclosporine A (red) with 0.01 μm CypB (blue), with 0.005 μm P3H1·CRTAP·CypB complex (green), and with 0.005 μm P3H1·CRTAP·CypB and 1 μm cyclosporine A (orange).
Chaperone Activity of the P3H1·CRTAP·CypB Complex Using the Aggregation and Refolding of Chemically Denatured Rhodanese
The concentration-dependent inhibition of the aggregation and refolding of chemically denatured rhodanese by the P3H1·CRTAP·CypB complex is shown in Fig. 3A. The P3H1·CRTAP·CypB complex at a concentration of 0.08 μm showed a much higher chaperone activity than protein-disulfide isomerase at a concentration of 2 μm (Fig. 3B). Free CypB did not contribute to the chaperone effect in this assay as shown previously (25).
FIGURE 3.
Influence of P3H1·CRTAP·CypB complex on the aggregation and refolding of chemically denatured rhodanese. Chemically denatured rhodanese was diluted 100-fold (0.3 μm final concentration) into 30 mm Tris/HCl buffer, pH 7.2, containing 50 mm KCl. Absorbance (light scattering) was monitored at 320 nm. A, in the absence (black) and presence of 0.02 (red), 0.04 (blue), or 0.08 μm (green) P3H1·CRTAP·CypB complex. B, in the absence (black) and presence of 0.08 μm P3H1·CRTAP·CypB complex (red). 2 μm protein-disulfide isomerase (green) and bovine serum albumin (blue) were positive and negative controls, respectively.
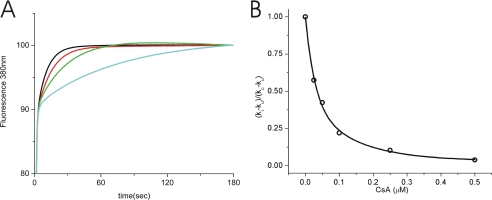
PPIase Activity of the P3H1·CRTAP·CypB Complex and Inhibition by Cyclosporine A
The P3H1·CRTAP·CypB complex was active as a PPIase when analyzed with a synthetic peptide substrate. Fig. 4A shows the PPIase activity using Suc-Ala-Ala-Pro-Phe-MCA as a substrate. The catalytic efficiency (kcat/Km 18,800 ± 1000 mm−1 s−1) of the complex is a little higher than that of chicken CypB (11,600 ± 1000 mm−1 s−1) (data not shown). The PPIase activity of the P3H1·CRTAP·CypB complex could be inhibited by cyclosporine A in a manner similar to that of the inhibition of CypB (Fig. 4B). This indicates that CypB in this complex is fully functional.
FIGURE 4.
PPIase activity of P3H1·CRTAP·CypB complex and inhibition by cyclosporine A. The PPIase activity of the P3H1·CRTAP·CypB complex was measured in 35 mm Hepes/NaOH, pH 7.8, at 5 °C using Suc-Ala-Ala-Pro-Phe-MCA as the substrate. A, the activity was monitored by the fluorescence at 380 nm of free MCA that was cleaved by chymotrypsin. In the absence (blue) and presence of 0.0016 (green), 0.0032 (red), and 0.0064 μm (black) P3H1 complex, respectively. B, the inhibition of the PPIase activity of the P3H1·CRTAP·CypB complex by cyclosporine A (CsA) was determined by the percentage of activity remaining upon increasing concentrations of cyclosporine A. The activity ((ki − ku)/(kc − ku)) was calculated using the rate constant ki in the presence of inhibitor, uninhibited rate constant kc, and ku, the rate constant of the uncatalyzed reaction. The final P3H1·CRTAP·CypB complex concentration was 0.013 μm.
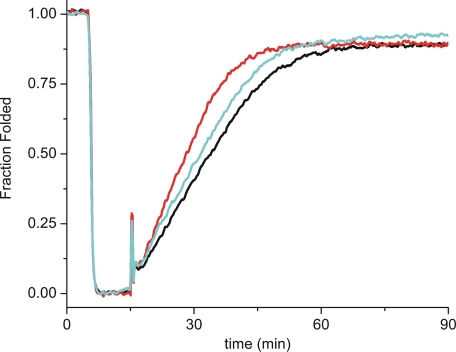
The refolding of type III collagen was rate-limited by cis-trans isomerizations of proline residues, and this rate could be catalyzed by CypB. Fig. 5 shows the refolding of thermally denatured pN type III collagen. The P3H1·CRTAP·CypB complex increased the rate of refolding of pN type III collagen; however, an equimolar amount of free CypB was a better catalyst in this reaction despite the higher catalytic efficiency of the complex against peptide substrates.
FIGURE 5.
Refolding of pN type III collagen in the presence of P3H1·CRTAP·CypB complex. Refolding was monitored by optical rotatory dispersion at 365 nm. The signal from pN type III collagen was observed for 5 min at 25 °C, and then native collagen was denatured for 10 min at 45 °C to unfold the triple helix. Refolding was initiated by quickly lowering the temperature back to 25 °C. Refolding curves are shown in the absence (black) and presence of 0.05 μm P3H1·CRTAP·CypB complex (blue) or 0.05 μm chicken CypB (red). The increased slope of the linear refolding phase indicates catalysis of cis-trans isomerizations. The final concentration of pN type III collagen was 0.125 μm.
The P3H1·CRTAP·CypB Complex Does Not Stabilize the Collagen Triple Helix
In contrast to the recently described FKBP65, the P3H1·CRTAP·CypB complex or free CypB did not stabilize the collagen triple helix. Fig. 6 shows the melting curves for type I collagen (Fig. 6A) and type III collagen (Fig. 6B). In both cases the curves are very close in the presence or absence of a 4-fold molar excess of P3H1·CRTAP·CypB or CypB.
FIGURE 6.
Melting curves of type I and III collagens in the presence of P3H1·CRTAP·CypB complex. Optical rotary dispersion was measured at 365 nm, and the temperature was increased at a rate of 10 °C/h. A stock solution of type I collagen and type III collagen (final concentration, 0.025 μm) in 50 mm acetic acid was diluted into 50 mm Tris/HCl buffer, pH 7.5, containing 0.4 m NaCl. The final concentration of both P3H1·CRTAP·CypB complex and CypB was 0.1 μm. Denaturation curves in the absence (black) and presence of P3H1·CRTAP·CypB complex (blue) or CypB (red) are shown for type I collagen (A) and type III collagen (B).
Inhibition of Collagen Fibril Formation by the P3H1·CRTAP·CypB Complex
The P3H1·CRTAP·CypB complex also interacted with folded triple helical collagen as shown by its activity in inhibiting the formation of type I collagen fibrils. Fig. 7 shows the concentration-dependent inhibition of type I collagen fibril formation by the P3H1·CRTAP·CypB complex and compares this activity with the known inhibitor HSP47. When equimolar amounts of HSP47 and P3H1·CRTAP·CypB were used HSP47 showed a lower activity in inhibiting fibril formation. Free CypB did not show any activity in this assay (data not shown).
FIGURE 7.
Inhibition of fibril formation of type I collagen in the presence of P3H1·CRTAP·CypB complex. A stock solution of type I collagen in 50 mm acetic acid was diluted into 150 mm sodium phosphate buffer, pH 7.8, containing 150 mm NaCl and measured at 34 °C. The absorbance (light scattering) was monitored at 313 nm. The final concentration of type I collagen was 0.3 μm. The following curves are shown: in the absence (black) and presence of 0.01 (red), 0.025 (green), or 0.05 μm (orange) P3H1·CRTAP·CypB complex and 0.05 μm HSP47 (blue) as a positive control.
Quantitation of Binding of the P3H1·CRTAP·CypB Complex to Native Type I Collagen
The dissociation constant of the type I collagen P3H1·CRTAP·CypB complex was determined to be 7.4 μm (Table 1). This value indicates a weaker interaction as compared with HSP47, which was used as a control (Kd = 2.3 μm). The difference in Kd was the result of a 5–10-fold faster dissociation rate of the complex compared with HSP47.
TABLE 1.
Dissociation and rate constants of binding of native bovine type I collagen to the P3H1·CRTAP·CypB complex and HSP47
| Kd | ka | kd | |
|---|---|---|---|
| 10−6m | 1/m s | 1/s | |
| P3H1·CRTAP·CypB | 7.4 ± 2.8 | 2.5 ± 0.4·104 | 2.0 ± 0.5·10−1 |
| HSP47 | 2.3 ± 0.6 | 2.8 ± 0.5·104 | 6.2 ± 1.0·10−2 |
| HSP47a | 1.1 | 2.08·104 | 2.36·10−2 |
a Taken from Ref. (27).
DISCUSSION
P3H1 catalyzes the hydroxylation of the Pro-986 to (3S)-Hyp of the α1 chains of type I collagen in vitro (6). However, when P3H1 is extracted from chick embryos, P3H1 is present in a complex with CRTAP and cyclophilin B. For the in vivo hydroxylation of the single Pro residue in the α1 chains of type I, II, and III collagens the P3H1·CRTAP·CypB complex seems to be required. The CRTAP knock-out mice and human mutations in CRTAP and P3H1 show a dramatic decrease in the level of 3-hydroxylation (7, 14–16). Assuming that the hydroxylase activity is in the carboxyl-terminal dioxygenase domain of P3H1, why are CRTAP and CypB required for the 3-hydroxylation of proline residues in vivo?
The results shown here indicate that the P3H1·CRTAP·CypB complex is not only responsible for the 3-hydroxylation of specific proline residues in the α chains of fibrillar collagens, but it also acts as a potent molecular chaperone. The complex was active in the inhibition of the thermal aggregation of citrate synthase and in the refolding and aggregation assay of chemically denatured rhodanese. Free CypB had no activity in these assays, and inhibition of the peptidyl-prolyl cis-trans isomerase activity of CypB in the complex did not change this activity. The CypB in the complex was active as a PPIase, and this activity could be inhibited with cyclosporine A. The catalytic efficiency of CypB in the complex was higher than that of free CypB against a peptide substrate. The catalytic efficiency of the complex was lower against pN type III collagen, but a slightly higher amount of collagen refolded compared with control or free CypB. The chaperone activity of the complex might help in the recognition of substrates, and therefore the complex combines chaperone activity and PPIase activity during the refolding of collagen triple helices. It was recently shown that the insertion of a chaperone domain into FKBP12 (a PPIase) increased the catalytic efficiency of protein refolding (26).
It has been suggested that the chaperones HSP47 and FKBP65 stabilize the collagen triple helix in the rER (11, 17). The P3H1·CRTAP·CypB complex had no effect on the stability of type III collagen. A slight decrease in the stability of type I collagen could be observed at a 4-fold molar excess of complex. This indicates that HSP47 and FKBP65 are the likely chaperones that stabilize the triple helix of these collagens in the rER. The P3H1·CRTAP·CypB complex also interacts with folded triple helical collagens as indicated by the inhibition of fibril formation of type I collagen. The complex inhibited the fibril formation of type I collagen as efficiently as HSP47 and FKBP65 (16). It therefore contributes in the prevention of premature aggregation of collagen chains and molecules in the rER.
The P3H1·CRTAP·CypB complex bound folded type I collagen less efficiently than did HSP47. The association rate constants were similar, but the complex showed a higher dissociation rate constant. Binding to unfolded collagen chains is likely to be very complex. The complex will bind to unfolded collagen chains for the 3-hydroxylation activity, the cis-trans isomerase activity, and likely for the chaperone activity. One can speculate that the complex preferentially binds to unfolded chains and catalyzes the folding of the triple helix. The complex can then interact at this site with the triple helical structure and temporarily stabilize this junction. The fast dissociation is then used to replace the complex with HSP47 or other helix-stabilizing chaperones.
The P3H1·CRTAP·CypB complex therefore has three distinct activities: it is a prolyl 3-hydroxylase, a PPIase, and a molecular chaperone. The observed phenotypes in the CRTAP-null mice and human mutations in CRTAP and P3H1 may not only result from the lack of the 3-Hyp residues in the type I, II, or III collagens but also from a disturbance in the additional function of the P3H1·CRTAP·CypB complex as a chaperone and PPIase. CRTAP-null mice show overmodified α chains on SDS-polyacrylamide gels (7). Overmodified α chains in collagens of fibroblasts of patients with CRTAP and P3H1 mutations were also observed, and in addition a slower rate of secretion of type I collagen was found in these cells (14–16). This indicates that the chaperone function of the P3H1·CRTAP·CypB complex could contribute significantly to the observed phenotypes.
Acknowledgments
We thank Kerry Maddox for amino acid analyses and Lauren Hayashi and Elena Pokidysheva for help with the extraction of chick embryos.
This work was supported by a grant from the Shriners Hospitals for Children (to H. P. B.) and in part by a Michael Geisman Research Fellowship from the Osteogenesis Imperfecta Foundation (to J. A. V.).
- P3H
- prolyl 3-hydroxylase
- PPIase
- peptidyl-prolyl cis-trans isomerase
- CypB
- cyclophilin B
- CRTAP
- cartilage-associated protein (Casp)
- pN type III collagen
- type III collagen containing the amino-terminal propeptide
- MCA
- methylcoumarylamide
- Suc
- succinyl
- Hyp
- hydroxyproline
- rER
- rough endoplasmic reticulum.
REFERENCES
- 1.Myllyharju J., Kivirikko K. I. ( 2004) Trends Genet. 20, 33– 43 [DOI] [PubMed] [Google Scholar]
- 2.Ogle J. D., Arlinghaus R. B., Lgan M. A. ( 1962) J. Biol. Chem. 237, 3667– 3673 [PubMed] [Google Scholar]
- 3.Risteli J., Tryggvason K., Kivirikko K. I. ( 1977) Eur. J. Biochem. 73, 485– 492 [DOI] [PubMed] [Google Scholar]
- 4.Risteli J., Tryggvason K., Kivirikko K. I. ( 1978) Anal. Biochem. 84, 423– 431 [DOI] [PubMed] [Google Scholar]
- 5.Tryggvason K., Majamaa K., Risteli J., Kivirikko K. I. ( 1979) Biochem. J. 183, 303– 307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vranka J. A., Sakai L. Y., Bächinger H. P. ( 2004) J. Biol. Chem. 279, 23615– 23621 [DOI] [PubMed] [Google Scholar]
- 7.Morello R., Bertin T. K., Chen Y., Hicks J., Tonachini L., Monticone M., Castagnola P., Rauch F., Glorieux F. H., Vranka J., Bächinger H. P., Pace J. M., Schwarze U., Byers P. H., Weis M., Fernandes R. J., Eyre D. R., Yao Z., Boyce B. F., Lee B. ( 2006) Cell 127, 291– 304 [DOI] [PubMed] [Google Scholar]
- 8.Blatch G. L., Lässle M. ( 1999) BioEssays 21, 932– 939 [DOI] [PubMed] [Google Scholar]
- 9.D'Andrea L. D., Regan L. ( 2003) Trends Biochem. Sci. 28, 655– 662 [DOI] [PubMed] [Google Scholar]
- 10.Tiainen P., Pasanen A., Sormunen R., Myllyharju J. ( 2008) J. Biol. Chem. 283, 19432– 19439 [DOI] [PubMed] [Google Scholar]
- 11.Makareeva E., Leikin S. ( 2007) PLoS ONE 2, e1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai N., Hosokawa M., Itohara S., Adachi E., Matsushita T., Hosokawa N., Nagata K. ( 2000) J. Cell Biol. 150, 1499– 1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chessler S. D., Byers P. H. ( 1993) J. Biol. Chem. 268, 18226– 18233 [PubMed] [Google Scholar]
- 14.Barnes A. M., Chang W., Morello R., Cabral W. A., Weis M., Eyre D. R., Leikin S., Makareeva E., Kuznetsova N., Uveges T. E., Ashok A., Flor A. W., Mulvihill J. J., Wilson P. L., Sundaram U. T., Lee B., Marini J. C. ( 2006) N. Engl. J. Med. 355, 2757– 2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabral W. A., Chang W., Barnes A. M., Weis M., Scott M. A., Leikin S., Makareeva E., Kuznetsova N. V., Rosenbaum K. N., Tifft C. J., Bulas D. I., Kozma C., Smith P. A., Eyre D. R., Marini J. C. ( 2007) Nat. Genet. 39, 359– 365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldridge D., Schwarze U., Morello R., Lennington J., Bertin T. K., Pace J. M., Pepin M. G., Weis M., Eyre D. R., Walsh J., Lambert D., Green A., Robinson H., Michelson M., Houge G., Lindman C., Martin J., Ward J., Lemyre E., Mitchell J. J., Krakow D., Rimoin D. L., Cohn D. H., Byers P. H., Lee B. ( 2008) Hum. Mutat. 29, 1435– 1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa Y., Vranka J., Wirz J., Nagata K., Bächinger H. P. ( 2008) J. Biol. Chem. 283, 31584– 31590 [DOI] [PubMed] [Google Scholar]
- 18.Zeng B., MacDonald J. R., Bann J. G., Beck K., Gambee J. E., Boswell B. A., Bächinger H. P. ( 1998) Biochem. J. 330, 109– 114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchner J., Schmidt M., Fuchs M., Jaenicke R., Rudolph R., Schmid F. X., Kiefhaber T. ( 1991) Biochemistry 30, 1586– 1591 [DOI] [PubMed] [Google Scholar]
- 20.Shao F., Bader M. W., Jakob U., Bardwell J. C. ( 2000) J. Biol. Chem. 275, 13349– 13352 [DOI] [PubMed] [Google Scholar]
- 21.Martin J., Langer T., Boteva R., Schramel A., Horwich A. L., Hartl F. U. ( 1991) Nature 352, 36– 42 [DOI] [PubMed] [Google Scholar]
- 22.Mendoza J. A., Rogers E., Lorimer G. H., Horowitz P. M. ( 1991) J. Biol. Chem. 266, 13044– 13049 [PubMed] [Google Scholar]
- 23.Harrison R. K., Stein R. L. ( 1990) Biochemistry 29, 1684– 1689 [DOI] [PubMed] [Google Scholar]
- 24.Fischer G., Bang H., Mech C. ( 1984) Biomed. Biochim. Acta 43, 1101– 1111 [PubMed] [Google Scholar]
- 25.Horibe T., Yosho C., Okada S., Tsukamoto M., Nagai H., Hagiwara Y., Tujimoto Y., Kikuchi M. ( 2002) J. Biochem. 132, 401– 407 [DOI] [PubMed] [Google Scholar]
- 26.Knappe T. A., Eckert B., Schaarschmidt P., Scholz C., Schmid F. X. ( 2007) J. Mol. Biol. 368, 1458– 1468 [DOI] [PubMed] [Google Scholar]
- 27.Natsume T., Koide T., Yokota S., Hirayoshi K., Nagata K. ( 1994) J. Biol. Chem. 269, 31224– 31228 [PubMed] [Google Scholar]