Abstract
Engineered overexpression of protein kinase Cα (PKCα) was previously shown to endow nonmotile MCF-10A human breast cells with aggressive motility. A traceable mutant of PKCα (Abeyweera, T. P., and Rotenberg, S. A. (2007) Biochemistry 46, 2364–2370) revealed that α6-tubulin is phosphorylated in cells expressing traceable PKCα and in vitro by wild type PKCα. Gain-of-function, single site mutations (Ser → Asp) were constructed at each PKC consensus site in α6-tubulin (Ser158, Ser165, Ser241, and Thr337) to simulate phosphorylation. Following expression of each construct in MCF-10A cells, motility assays identified Ser165 as the only site in α6-tubulin whose pseudophosphorylation reproduced the motile behavior engendered by PKCα. Expression of a phosphorylation-resistant mutant (S165N-α6-tubulin) resulted in suppression of MCF-10A cell motility stimulated either by expression of PKCα or by treatment with PKCα-selective activator diacylglycerol-lactone. MCF-10A cells treated with diacylglycerol-lactone showed strong phosphorylation of endogenous α-tubulin that could be blocked when S165N-α6-tubulin was expressed. The S165N mutant also inhibited intrinsically motile human breast tumor cells that express high endogenous PKCα levels (MDA-MB-231 cells) or lack PKCα and other conventional isoforms (MDA-MB-468 cells). Comparison of Myc-tagged wild type α6-tubulin and S165N-α6-tubulin expressed in MDA-MB-468 cells demonstrated that Ser165 is also a major site of phosphorylation for endogenously active, nonconventional PKC isoforms. PKC-stimulated motility of MCF-10A cells was nocodazole-sensitive, thereby implicating microtubule elongation in the mechanism. These findings support a model in which PKC phosphorylates α-tubulin at Ser165, leading to microtubule elongation and motility.
Introduction
For many years, attention has been focused on protein kinase Cα (PKCα)3 as an upstream element of signaling pathways governing cell adhesion and migration of cancer cells. In light of the role of this PKC isoform in cytoskeletal events that drive the metastatic phenotype (1), identification of its protein substrates would offer attractive targets for design of anti-metastasis drugs. Despite the large number of PKC substrates that undergo phosphorylation in vitro, there is at present a short list of proteins that have been demonstrated to serve as intracellular PKC substrates. These substrates have been found to associate with the actin cytoskeleton and cell-cell contacts (2, 3). Upon phosphorylation by PKCα, these proteins impact cytoskeletal dynamics that promote adhesion and movement (1).
The MCF-10A cell line provides a valuable model for probing PKCα-specific pathways in human breast cells. This nontransformed, nontumorigenic cell line expresses very low levels of PKCα and therefore offers a low background for experiments seeking to correlate overexpression of PKCα with its phenotypic consequences. Of the diacylglycerol (DAG)-sensitive isoforms expressed in these cells, PKCα is the only Ca2+/DAG-dependent (conventional) isoform. This laboratory previously reported that stable overexpression of PKCα endowed these nonmotile cells with a high level of motility, decreased proliferation, loss of E-cadherin expression, and altered cell morphology (4).
A distinct advantage of MCF-10A cells is that following engineered overexpression of PKCα, they require no treatment with PKC activators (such as DAG or the tumor promoter tetradecanoyl-12,13-phorbol acetate) in order to observe PKCα-related phenotypes (4). This condition is related to the MCF-10A cell environment, since previous use of the same PKCα-encoding plasmid with NIH 3T3 cells did not give rise to a constitutively active protein (5). The probable source of this activation is the presence of epidermal growth factor (EGF) in the culture medium, a requirement for MCF-10A cell growth. Following its expression, recombinant PKCα is activated by EGF-induced levels of intracellular DAG and Ca2+, resulting in motile behavior. In parental MCF-10A cells, which are only weakly motile, endogenous PKC activity is apparently unresponsive to extracellular EGF. However, treatment of MCF-10A cells with a cell-permeable and PKCα-selective DAG-lactone produces dramatic motility (6, 7). These observations suggest that nonmotile MCF-10A cells express an inhibitor of endogenous PKC that prevents its activation by (EGF-generated) DAG but that can be either displaced by micromolar concentrations of DAG-lactone or titrated by high dosage expression of recombinant PKCα. An alternative but less probable scenario is that the engineered overexpression of PKCα results in formation of the constitutively active 50-kDa catalytic fragment. However, our previous studies argue against this possibility (4). First, in MCF-10A cells expressing the recombinant PKCα, the 80-kDa protein was observed to translocate to the membrane fraction, thus signifying endogenous activation of the intact enzyme. Second, calphostin C, a PKC inhibitor that binds the DAG binding site in the regulatory domain and therefore requires the intact enzyme for inhibition of catalytic activity, was observed to block PKCα-induced motility of MCF-10A cells.
The idea that cancer-related phenotypes of MCF-10A cells arise from aberrant EGF-mediated signaling is further supported by evidence that many PKC-driven phenotypes of this cell line (including motility) can be recapitulated by ErbB-2 overexpression (8, 9). This EGF receptor-related oncogene is frequently overexpressed in breast tumors and is thought to act through PKCα to engender breast cancer invasiveness (10). Because the MCF-10A cell line exhibits a low background of endogenous PKCα activity, it offers a substrate-rich environment in which to screen and identify protein substrates that mediate PKCα-induced phenotypes. Any candidate substrate can be subsequently validated as a PKC target in human breast tumor cells or other cancer cells.
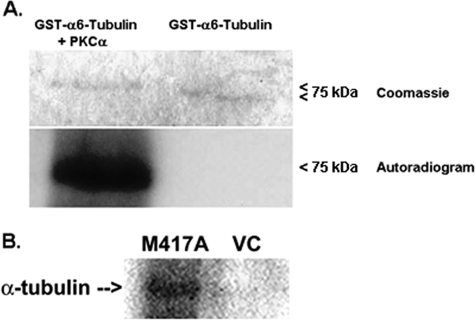
Until recently, identification of PKCα substrates was not possible due to the inherent difficulties of linking a phosphoprotein directly to the specific protein kinase that produced it. Initially developed by the Shokat laboratory (11–14), the traceable kinase method offers a chemical-genetic approach to identify the immediate substrate(s) of any protein kinase (15–17). This method entails site-directed mutagenesis at the ATP binding domain in the vicinity of the adenosine N6-amino group of bound ATP. Mutation at this site replaces an amino acid residue bearing a long aliphatic side chain (e.g. methionine) with a residue containing a shorter side chain, such as alanine, thereby removing steric hindrance near the N6-amino group. This mutant productively binds a bulky analogue of ATP that is derivatized at the N6-amino group (e.g. N6-phenyl-ATP). Phosphoproteins that are produced when the ATP analogue is added to cell lysates containing the mutant protein kinase are the result of the mutant activity. Guided by the x-ray crystal structure of cAMP-dependent protein kinase that contained bound ATP, this laboratory recently developed a traceable mutant of PKCα (18). This mutant (M417A-PKCα) was effective in identifying proteins already known to be PKC substrates and in promoting the motility phenotype in MCF-10A cells that was previously ascribed to wild type PKCα (4).
In the present work, the potential of the traceable kinase method is fully realized. Phosphoproteins identified as substrates of M417A-PKCα led us to discover α6-tubulin as a new PKCα substrate in MCF-10A cells. Because heterodimers of α- and β-tubulin are the building blocks of microtubules whose assembly dynamics are critical during cell movement, we hypothesized that its phosphorylation by PKCα engenders motile behavior. By use of gain-of-function and loss-of-function mutants, our results implicate a single site in α6-tubulin whose phosphorylation by PKCα reproduces the motility phenotype of MCF-10A cells that was previously ascribed to PKCα.
MATERIALS AND METHODS
Cell culture serum, growth factors, media, DNA sequencing primers, Alexa fluor antibodies, and pcDNA3.1 vector were purchased from Invitrogen. The QuikChange mutagenesis kit and pCMV4 vector were purchased from Stratagene (La Jolla, CA). A plasmid encoding α6-tubulin was obtained from ATCC (Manassas, VA). All restriction enzymes were acquired from New England Biolabs (Ipswich, MA). Phosphoserine protein phosphatase inhibitor mixture, protease inhibitors, saponin, nocodazole, and α-tubulin monoclonal antibody (DM1A) were purchased from Sigma. bis-Indoleylmaleimide-1 (BIM) was purchased from EMD-Calbiochem. Rabbit polyclonal α-tubulin antibody (E-19), Protein A/G-agarose beads, horseradish peroxidase-conjugated secondary antibody, mouse IgG-agarose, and radioimmune precipitation buffer were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Fugene 6 was acquired from Roche Applied Science; Gelcode Blue and chemiluminescence reagents (Supersignal West Pico) were purchased from Pierce; Duracryl was obtained from NextGen Sciences (Ann Arbor, MI); and Immobilon-P transfer membranes and mouse monoclonal anti-Myc were purchased from Millipore Corp. (Bedford, MA). Recombinant human GST (Mu)-α6-tubulin was purchased from Abnova Corp. (Taiwan), GST-PKCα and the antibody recognizing phosphorylated PKC substrates were obtained from Cell Signaling Technology (Beverly, MA), and GST-Mu protein standard was purchased from Alpha Diagnostic International, Inc. (San Antonio, TX). Radiochemicals were obtained from PerkinElmer Life Sciences.
Mutagenesis of α6-Tubulin
cDNA encoding human α6-tubulin was subcloned into the pCMV4 vector at BamHI and XhoI restriction sites. Expression from this vector produced a protein that was FLAG-tagged at the COOH terminus. Substitution of either Asp or Asn codons (underlined) was carried out by the QuikChange method. The following primers were used to produce the mutations at Ser165: 5′-CGT CTC TCA GTT GAT TAT GGC AAG AAG GAC AAG CTG GAG TTC TCC-3′ for S165D and 5′-CGT CTC TCA GTT GAT TAT GGC AAG AAG AAC AAG CTG GAG TTC TCC-3′ for S165N. To produce S158D, S241D, and T337D, the following primers were used: 5′-GAA CGT CTC GAC GTT GAT TAT GGC AAG AAG TCC AAG CTG GAG-3′ for S158D, 5′-GTG TCC TCC ATC ACT GCT GAC CTG AGA TTT GAT GGA GC-3′ for S241D, and 5′-GCC ATT GCC ACC ATC AAA GAC AAG CGT ACC ATC CAG-3′ for T337D. To verify the presence of each site-specific mutation, DNA sequence analysis of the entire open reading frame was performed (Macrogen (Seoul, Korea)).
Myc fusion proteins were constructed for the wild type (WT) α6-tubulin and for the S165D and S165N mutants of α6-tubulin by subcloning the cDNA from the corresponding pCMV4 construct into a pcDNA3.1/Myc-His A vector at BamHI and XbaI sites. The sequence of the entire open reading frame was verified for each construct (Macrogen, Rockville, MD). Upon expression, the Myc tag sequence was conferred at the carboxyl terminus. High level expression of the 50-kDa Myc-tagged fusion proteins in MCF-10A cells was verified by Western blot with rabbit polyclonal Myc antibody (Millipore Corp.).
Construction of GFP-α6-Tubulin
Using a standard PCR-based approach, humanized Renilla reniformis green fluorescent protein (GFP) (Stratagene) was fused at the 5′-end of α6-tubulin cDNA (wild type or mutant) via a six-Gly linker. The fusion construct was inserted into a pCMV4 vector at SacII and XhoI restriction sites. The sequence of the entire open reading frame was verified for each construct (Macrogen, Seoul, Korea).
Cell Culture and Transfection
Midpassage MCF-10A human breast epithelial cells (19) were obtained from the Barbara Ann Karmanos Cancer Institute (Detroit, MI). MCF-10A cells were cultured on 10-cm plates (Falcon), as previously described (4). MDA-MB-231 cells were cultured in Iscove's modified Dulbecco's medium with l-glutamine, 10% fetal bovine serum, and antibiotics (1% penicillin/streptomycin and 0.5 μg/ml fungizone). MDA-MB-468 cells were cultured in RPMI containing glutamine, 10% fetal bovine serum, and antibiotics. Transient transfection of cells was carried out with 4 μg of cDNA complexed with Fugene 6 for 6 h in serum-free medium, followed by the addition of serum and incubation at 37 °C, 5% CO2. After 48 h, the cells were harvested. Based on an experiment with a GFP-encoding vector, the efficiency of transfection was typically 70% or higher.
Motility Assay
The cells were applied to a 10-well slide through a 10-hole manifold (CSM, Inc., Phoenix, AZ) that restricts sedimentation of cells to a small, circumscribed area. Upon removal of the manifold, the cells radiated outwardly over an 8-h period. Cell motility was analyzed by a digital camera (Moticam 2000) attached to a computer and an inverted Nikon Diaphot microscope. The extent of movement was determined by measuring the change in total area (in μm2) occupied by the cells using Motic Images Plus 2.0 software. Between measurements, cells were incubated at 37 °C, 5% CO2. In experiments employing DAG-lactone or nocodazole, the reagent (or 0.1% (v/v) DMSO as vehicle control) was added immediately after the removal of the manifold (t = 0) and was present for the duration of the assay. Each reported value is the average of triplicate measurements with the corresponding standard deviation value (S.D.).
Radiolabeling of Permeabilized MCF-10A Cells
Transfected MCF-10A cells were treated for 5 min with saponin (50 μg/ml) in isotonic sucrose buffer (130 mm sucrose, 30 mm potassium chloride, 30 mm potassium acetate, 20 mm magnesium acetate, 1 mm calcium chloride, 20 mm HEPES, pH 7.4) containing phosphoserine phosphatase inhibitors, followed by the addition of [γ-32P]N6-phenyl-ATP to a final concentration of 100 μm (∼800 cpm/pmol). The synthesis of this reagent has been described elsewhere (18). Cells were incubated with occasional mixing for 1 h at 30 °C and pelleted by centrifugation. The cell pellet was lysed by sonication and centrifuged at 8000 × g, and the resulting supernatant was subjected to immunoprecipitation with α-tubulin antibody and protein A/G-agarose, as described below. The immunopellet was treated with Laemmli sample buffer and heated for 5 min at 95 °C prior to performing 8% SDS-PAGE.
Sample Preparation and Immunoprecipitation
MCF-10A cells were harvested and dissociated by trypsinization, followed by two washes with complete medium and a final wash with serum-free medium. Prior to lysis, the cells were resuspended and transferred to a new Eppendorf tube. These steps facilitated cell dissociation and disruption and proved superior to simple cell scraping. To prepare cell lysates directly for Western blot, cells were lysed in 50 mm Tris (pH 7.4), 5 mm EDTA, 5 mm EGTA, 15 mm 2-mercaptoethanol, 1% (v/v) Triton X-100, protease inhibitors, and phosphatase inhibitors, as previously described (18). The protein concentration of each sample was measured with a colorimetric protein reagent (Bio-Rad) with bovine serum albumin as a standard.
Preparation of the soluble fraction (containing cytosolic α-tubulin) was carried out as previously described (20). MCF-10A cells were harvested, pelleted by centrifugation (1000 × g for 5 min at room temperature), and lysed at room temperature for 20 min in 0.2 ml of stabilization buffer (0.1 m PIPES, pH 6.9, 30% glycerol, 5% (v/v) DMSO, 1 mm MgSO4, 1 mm EGTA, protease inhibitor mixture, and phosphatase inhibitors), containing 1.0% Triton X-100. The addition of 10 μm BIM (a pan-PKC inhibitor) to the lysis buffer greatly decreased the background, since it prevented adventitious phosphorylation during the isolation procedure. Lysates were transferred to fresh tubes and centrifuged at 100,005 × g for 45 min at room temperature. Supernatants containing soluble tubulin (0.2 ml) were normalized for total protein (typically 200–300 μg), and the sample volumes were adjusted to 1.0 ml with radioimmune precipitation buffer. To isolate α-tubulin from each soluble fraction, immunoprecipitation was performed with monoclonal anti-α-tubulin (2 μg/ml), as described below.
For immunoprecipitation, lysates were prepared from transfected cells in buffer (specified in the legends for Figs. 3 and 4) containing protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 10 ng/ml leupeptin, 10 ng/ml soybean trypsin inhibitor) and phosphoserine phosphatase inhibitors. After normalizing samples for total protein and preclearing for 30 min with rotation at 4 °C with mouse IgG-agarose, immunoprecipitation with rotation was carried out at 4 °C with the specified antibody, followed by treatment with protein A/G-agarose for 1 h. Immunocomplexes were collected by centrifugation at 800 × g at 4 °C for 5 min. Pellets were washed three times with resuspension in 0.5 ml of detergent-free intraperitoneal buffer and rotated at 4 °C for 5 min and centrifuged at 800 × g for 5 min.
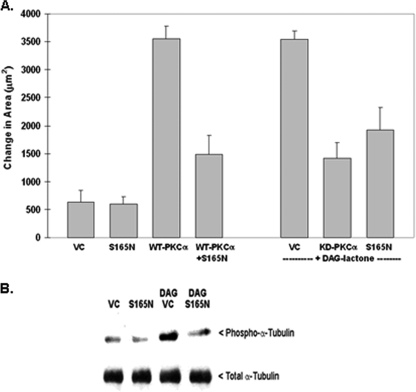
FIGURE 3.
Phosphorylation-resistant α6-tubulin mutant (S165N) inhibits motility of MCF-10A cells and decreases phosphorylation of endogenous α-tubulin. A, S165N-α6-tubulin suppresses motile behavior of stimulated MCF-10A cells. Following expression of the S165N mutant (GFP-(S165N)-α6-tubulin), cells were tested for a dominant negative effect on motility that had been induced by either WT PKCα overexpression or by treatment with 5 μm DAG-lactone for the duration of the experiment. The ability of the S165N mutant to impede motility was compared with that of the kinase-defective mutant of PKCα (KD). An equivalent amount of plasmid DNA (4 μg) was used for all transfections. B, S165N-α6-tubulin suppresses DAG-lactone-stimulated phosphorylation of endogenous α-tubulin in MCF-10A cells. Cells were transfected with 4 μg of S165N mutant or the VC. Identical transfectants were treated with 5 μm DAG-lactone or DMSO (0.1%, v/v) for 30 min at 37 °C prior to lysis. Lysates were prepared in 0.2 ml of microtubule stabilization-extraction buffer (0.1 m PIPES, pH 6.9, 30% glycerol, 5% DMSO, 1 mm MgSO4, 1 mm EGTA, and 1% Triton X-100) containing protease inhibitors and 10 μm BIM. A soluble fraction containing unincorporated tubulin was prepared from lysates (see “Materials and Methods”) and normalized for total protein content (280 μg), and each sample volume was adjusted to 1.0 ml with radioimmune precipitation buffer. The samples were precleared with mouse IgG-agarose and immunoprecipitated with 2 μg of mouse anti-α-tubulin for 15 h with rotation at 4 °C. Immunopellets were divided into duplicate blots and probed in parallel with either rabbit polyclonal phospho-PKC substrate antibody (1:500 dilution) or rabbit anti-α-tubulin (1:2500 dilution). The results are representative of three independent experiments.
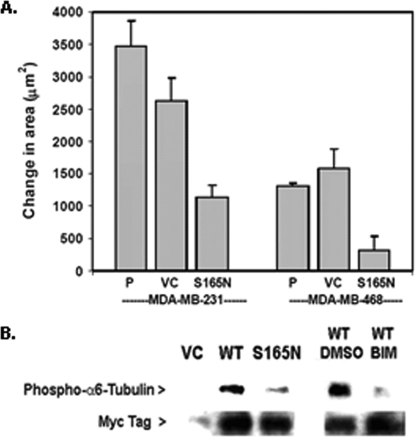
FIGURE 4.
Phosphorylation-resistant α6-tubulin mutant (S165N) inhibits motility of human breast tumor cells. A, dominant-negative effect on motility following expression of GFP-(S165N)-α6-tubulin (4 μg of plasmid) in human breast tumor cell lines that express very high levels of PKCα (MDA-MB-231) or none at all (MDA-MB-468). Each value is the average of triplicate measurements ± S.D., and the results are representative of three independent experiments. P, parental cells. B, phosphorylation of Myc-tagged α6-tubulin reporter constructs in MDA-MB-468 cells. Cells were transfected for 48 h with 4 μg of plasmid encoding the Myc-tagged WT α6-tubulin, S165N mutant, or VC. Where indicated, cells were treated with either 1 μm BIM or DMSO (0.1%, v/v) and incubated at 37 °C for 30 min prior to cell lysis. Whole cell lysates were prepared in 0.2 ml of buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EGTA, 10 μm BIM, and 0.5% Nonidet P-40. After normalizing for total protein content (550 μg), lysates were diluted 5-fold in detergent-free buffer, followed by preclearing with mouse IgG-agarose and immunoprecipitation with 5 μg of mouse anti-Myc (6 h with rotation, 4 °C). Myc-tagged α6-tubulin (50 kDa band) was detected in a Western blot that was probed sequentially with rabbit anti-phospho-PKC substrates (1:500) and rabbit anti-Myc (1:1000). The results are representative of three independent experiments.
Western Blot Analysis
Samples of known protein concentration were denatured in sample buffer, as previously described (18) and subjected to 8% SDS-PAGE on Duracryl followed by electrophoretic transfer to a 15 × 15-cm polyvinylidene difluoride membrane (Immobilon-P), and blocked in 5% powdered milk. An immunochemical assay was carried out with primary and horseradish peroxidase-conjugated secondary antisera and detected by chemiluminescence.
In Vitro Phosphorylation of Recombinant α6-Tubulin
To carry out phosphorylation in vitro, 1 μg each of highly pure recombinant GST-α6-tubulin (Abnova Corp.) and GST-PKCα (Cell Signaling Technology, Inc.) were combined in a reaction medium containing 25 mm Tris, pH 7.4, 10 mm magnesium acetate, 0.5 mm Ca2+, phosphatidylserine (0.1 mg/ml), and 0.5 mm dithiothreitol. Following the addition of 50 μm ATP or [γ-32P]ATP (∼200 cpm/pmol), the reaction was carried out for 30 min at 30 °C. Each reaction was quenched by the addition of Laemmli sample buffer and heated for 5 min at 95 °C prior to performing 6% SDS-PAGE and staining with Gelcode Blue. Samples to be used for mass spectrometry were resolved on a 6% SDS-PAGE gel having a thickness of 0.75 mm.
Protein Identification by Mass Spectrometry
Identification of α6-tubulin was performed at the Keck Foundation Mass Spectrometry Resource Laboratory at Yale Cancer Center. Gel-resolved proteins were digested in situ with trypsin (Promega) and batch-purified on a reversed-phase microtip, and resulting peptide pools were individually analyzed by liquid chromatography-tandem mass spectrometry on a Waters Q-TOF mass spectrometer, as previously described (18). All tandem mass spectrometry spectra were searched using the automated MASCOT algorithm. Identification required that two or more spectra matched the same protein entry in the data base.
RESULTS
Identification of α6-Tubulin as an Intracellular PKCα Substrate
Identification of α6-tubulin was initially suggested by its co-immunoprecipitation with M417A-PKCα from MCF-10A cell lysates. This traceable mutant of PKCα was expressed as a FLAG-tagged product and immunoprecipitated from detergent-free cell lysates with FLAG antibody in order to “pull down” any high affinity substrate proteins bound to it. Reaction of each immunopellet with [γ-32P]phenyl-ATP produced several 32P-labeled products that were resolved by one-dimensional SDS-PAGE and analyzed by autoradiography. In a typical experiment (18), a strong band at 50–55 kDa was produced by M417A-PKCα but was absent in the banding patterns observed with either WT PKCα or the vector control. The finding of radiolabeled bands unique to the traceable kinase of PKCα indicated that these proteins were potential substrates. When analyzed by mass spectrometry, the 50–55 kDa band was found to contain α6-tubulin. Whether α6-tubulin was indeed phosphorylated remained to be demonstrated. In view of the well established importance of microtubules to the mechanics of cell movement, the novel possibility that α6-tubulin serves as a direct PKCα substrate in the intact cell environment was further investigated.
To establish that PKCα directly phosphorylates α6-tubulin, a reaction was performed in vitro with recombinant human GST-α6-tubulin, recombinant human GST-PKCα, and [γ-32P]ATP, as described under “Materials and Methods.” The GST-α6-tubulin protein (75-kDa) used for this analysis was produced in a eukaryotic system (wheat germ cell-free system) that ensured that the recombinant protein was properly folded. In the experiment shown in Fig. 1A, substantial incorporation of 32P into α6-tubulin protein could be demonstrated by autoradiography. Gelcode Blue staining revealed that, relative to untreated α6-tubulin, the phosphoprotein had a detectably higher molecular mass. A control experiment (not shown) performed in vitro with pure, recombinant GST protein (GST-Mu) verified that this tag was not a substrate for PKCα.
FIGURE 1.
In vitro and intracellular phosphorylation of α6-tubulin. A, direct phosphorylation of recombinant 75-kDa GST-α6-tubulin (1 μg) by recombinant GST-PKCα (1 μg), was carried out in the presence of 50 μm [γ-32P]ATP plus activators (0.5 mm Ca2+, phosphatidylserine (0.1 mg/ml)), for 30 min at 30 °C, as described under “Methods.” The products were resolved by 6% SDS-PAGE, followed by staining with Gelcode Blue and autoradiography at −20 °C for 2 days. The results are representative of three independent experiments. B, intracellular phosphorylation of α-tubulin in permeabilized MCF-10A transfectants. Cells that had been transfected with M417A-PKCα or VC were harvested, lysed in hypotonic, detergent-free buffer (20 mm Tris, pH 7.4, 2 mm EGTA, 2 mm MgCl2, 1 mm dithiothreitol), permeabilized with saponin (50 μg/ml), and treated with 100 μm [γ-32P]N6-phenyl-ATP, as described under “Methods.” Cell lysates were immunoprecipitated with anti-α-tubulin, and the immunopellets were analyzed by 8% SDS-PAGE and autoradiography for 1 week at −20 °C. The experiment was performed twice with similar results.
To demonstrate that α6-tubulin is also phosphorylated by PKCα in intact MCF-10A cells, endogenous α-tubulin was subjected to an intracellular reaction with the traceable mutant of PKCα (M417A-PKCα) in the presence of [γ-32P]phenyl-ATP. In cells expressing M417A-PKCα, the reaction was initiated by permeabilizing the cells with saponin so as to facilitate entry of 100 μm [γ-32P]phenyl-ATP (21). This ATP analogue was expected to bind preferentially to the mutant PKCα in the presence of endogenous ATP, since the mutant exhibits a 10-fold lower affinity for natural ATP (18). Following a 30-min intracellular reaction, lysates were prepared, and α-tubulin was immunoprecipitated with a monoclonal antibody. In the autoradiogram shown in Fig. 1B, it was observed that α-tubulin underwent radiolabeling in intact cells expressing M417A-PKCα, in contrast with cells that received only the control vector.
Development and Characterization of α6-Tubulin Proteins Mutated at Consensus Sites for PKC Phosphorylation
The primary sequence of human α6-tubulin4 was examined for the presence of the PKC consensus sequence, which consists of a Ser or Thr flanked by basic residues (22). Four potential sites were found. One site, Ser165, was nested within a full consensus sequence, whereas the other three sites, namely Ser158, Ser241, and Thr337, contained a partial consensus sequence. It is noted that the four sites are present in all known isoforms of human α-tubulin but are entirely absent in β-tubulin. To determine whether α6-tubulin phosphorylation by PKCα has functional significance, single site mutations were introduced at each potential site of phosphorylation: Ser158 or Ser165 (RLS158VDYGKKS165K), Ser241 (S241LR), or Thr337 (KT337KR). Substitution of Ser or Thr with an Asp residue was performed in order to simulate the presence of phosphate (“pseudophosphate”). Initially, these mutants were developed with a COOH-terminal FLAG tag, which consists of a short segment of acidic residues. However, because α6-tubulin possesses multiple acidic residues at its COOH terminus that are also recognized by this antibody, the FLAG antibody could not be used to discriminate between native and mutant proteins. To detect mutant α6-tubulin, an amino-terminal fusion of α6-tubulin (WT or mutant) with a GFP tag was prepared (see “Materials and Methods”). A previous study of a similar fusion of GFP to α-tubulin indicated that the GFP moiety did not interfere with its incorporation into microtubules (23).
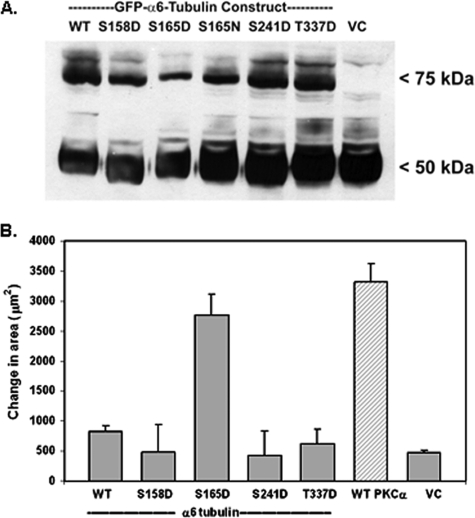
Following transfection of each construct into MCF-10A cells, stable expression of each GFP-α6-tubulin mutant protein (75 kDa) was verified by Western blot with α-tubulin antibody (Fig. 2A) that detected both the GFP-fusion proteins (75 kDa band) and endogenous α-tubulin (50 kDa band). The GFP constructs were stably expressed at a high and equivalent level.
FIGURE 2.
Effect of GFP-α6-tubulin pseudophosphorylated mutants on motility of MCF-10A cells. A, lysates from MCF-10A transfectants (25 μg/lane) expressing recombinant WT GFP-α6-tubulin, pseudophosphorylated GFP mutants (S165D, S158D, S241D, or T337D), or VC were analyzed by Western blot using α-tubulin monoclonal antibody to detect the GFP fusion proteins (75 kDa) and endogenous α-tubulin (50 kDa). Note that the phosphorylation-resistant mutant S165N was included for comparison. The results were observed in three independent experiments. B, measurement of motility was carried out with MCF-10A cells that had been transfected with single site-pseudophosphorylated mutants of GFP-α6-tubulin, WT PKCα, or the vector control. Each value is the average of triplicate measurements ± S.D. The results are representative of three independent experiments.
To determine whether expression of any of the pseudophosphorylated α6-tubulin mutants recapitulated the effect of PKCα expression on cell motility (4), each α6-tubulin mutant was expressed in MCF-10A cells and then tested for an effect on motile behavior. As shown in Fig. 2B, cells showing the strongest motile behavior were those that expressed α6-tubulin bearing the S165D mutation. Importantly, this level of motility was equivalent to that produced in cells that had been transfected with WT PKCα. By contrast, expression of the S158D, S241D, or T337D mutants did not produce motilities above that observed with either the WT α6-tubulin or vector control (VC). Identical effects on motility (or absence thereof) were observed with three different clones of each construct, thus confirming that clonal variation was not a factor. It is further noted that the motile behaviors measured with GFP-tagged α6-tubulin mutants were identical to that of cells transfected with the analogous FLAG-tagged constructs (data not shown).
We considered the possibility that PKCα is phosphorylating sites in α6-tubulin that are not consensus sites. Mass spectrometry of tryptic peptides derived from pure, recombinant α6-tubulin that had been phosphorylated by pure, recombinant PKCα in vitro revealed only Ser158 as a phosphorylation site (data not shown). However, pseudophosphorylation of this site (S158D) was found to have no impact on motility (Fig. 2B). It is likely that phospho-Ser165 was not detected, since it is flanked by lysine residues that would have been cleaved by trypsin, thus yielding Ser165 as part of a dipeptide. Furthermore, because the mass spectrometry measurement was based on only 27% coverage of the protein, additional sites could easily have eluded this analysis.
A Phosphorylation-resistant Ser165 Mutant of α6-Tubulin Suppresses α-Tubulin Phosphorylation and Motility of Stimulated MCF-10A Cells and Metastatic Human Breast Tumor Cells
To explore the functional significance of phosphorylation at Ser165, a phosphorylation-resistant mutant of α6-tubulin was prepared by substituting Ser165 with an Asn residue (S165N), so as to provide an isosteric control for the S165D mutant. The stable expression of S165N-α6-tubulin in MCF-10A cells was confirmed by Western blot (Fig. 2A). The next step was to test whether this mutant has a dominant-negative effect on motility of MCF-10A cells whose motility is induced by either engineered expression of WT PKCα (constitutively active in MCF-10A cells) or by treatment of parental cells with DAG-lactone, a membrane-permeable analogue of DAG that selectively activates endogenous PKCα (6). To test for a dominant-negative effect on motility, MCF-10A cells were either co-transfected with WT PKCα and S165N-α-tubulin or transfected only with the S165N mutant and then treated with DAG-lactone. As shown in Fig. 3A, the S165N mutant inhibited WT PKCα-induced motility by 72%. Similarly, when S165N-transfected cells were activated with DAG-lactone, the resulting cell movement was suppressed by 44% as compared with vector control cells. Motility of cells treated with DAG-lactone was inhibited to a similar degree by engineered expression of kinase-dead PKCα.
To assess whether Ser165 in α-tubulin is a major target of PKC-mediated phosphorylation, expression of S165N-α6-tubulin (or the vector control) was performed. If Ser165 is a preferred site of phosphorylation, then it should block phosphorylation of endogenous α-tubulin that is stimulated by DAG-lactone. MCF-10A transfectants that had been treated with or without DAG-lactone were lysed, and the soluble fraction (containing cytosolic tubulin) was prepared. Immunoprecipitation of endogenous α-tubulin was carried out, and the immunopellets were analyzed by Western blot. Phospho-α-tubulin was detected with an antibody that recognizes the phosphorylated PKC consensus site (PKC substrates antibody). The results (Fig. 3B) show that endogenous α-tubulin protein (50 kDa band) was strongly phosphorylated when cells were stimulated by treatment with DAG-lactone. However, compared with the unstimulated control, this phosphorylation signal could be substantially decreased when DAG-lactone-stimulated cells also expressed the S165N mutant of α6-tubulin. Therefore, Ser165 in α-tubulin appears to be a major target site of phosphorylation by PKCα.
Expression of the S165N mutant had a similar inhibitory effect on the intrinsic motility of metastatic human breast tumor cells. A comparison was made of MDA-MB-231 cells, which express elevated levels of PKCα (24), and MDA-MB-468 cells, which do not express any of the conventional isoforms (including PKCα) (25) but do express other DAG-sensitive isoforms (e.g. PKCδ (most abundant), PKCϵ, and PKCη (25)). Fig. 4A demonstrates that engineered overexpression of the S165N mutant in MDA-MB-231 and MDA-MB-468 cells inhibited their intrinsic motilities by 60 and 75%, respectively.
Since MDA-MB-468 cells do not express conventional PKC isoforms, phosphorylation of ectopically expressed Myc-WT α-tubulin or Myc-S165N mutant was compared in these cells. Each construct was expressed and immunoprecipitated from whole cell lysates with mouse anti-Myc. A Western blot was prepared from the immunopellets and probed sequentially with the PKC substrates antibody and anti-Myc (Fig. 4B). The analysis showed that MDA-MB-468 cells expressing the reporter WT α-tubulin (50 kDa) exhibited a strong level of phosphorylation, whereas cells expressing WT α6-tubulin and treated with bis-indolylmaleimide-1 (a pan-PKC inhibitor) resulted in very weak phosphorylation. This finding indicated that the site is primarily phosphorylated by PKC isoforms. Importantly, expression of Myc-tagged S165N-α6-tubulin led to a substantial decrease in its phosphorylation. That there was some evidence of phosphorylation in the S165N construct revealed the possibility of an additional phosphorylation site(s) (such as Ser158). Nonetheless, these results with breast tumor cells implicate Ser165 as the major site of phosphorylation by nonconventional PKC isoforms that correlates well with motility behavior.
Phosphorylation of α-Tubulin and Microtubule Elongation
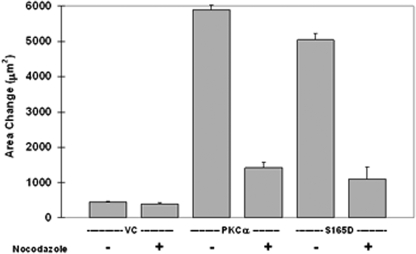
The possibility that motile behavior produced by α-tubulin phosphorylation is in fact mediated by microtubules was addressed by testing the effect on motility by nocodazole. This reagent inhibits microtubule elongation and shortening velocities and results in reduced dynamic instability (26). Highly motile MCF-10A cells expressing either recombinant PKCα or S165D-α6-tubulin, were inhibited by nearly 80% when subjected to continuous treatment (8 h) with 5 μm nocodazole (Fig. 5). That cell motility was nocodazole-sensitive is consistent with a requirement for elongating microtubules. Overall, these findings support a model in which PKC phosphorylates Ser165 and thereby promotes microtubule elongation and motile behavior.
FIGURE 5.
Motility of MCF-10A cells engendered by PKCα or S165D-α6-tubulin is eliminated by treatment with nocodazole. Cells were transfected with 4 μg of plasmid DNA encoding PKCα, S165D-α6-tubulin, or the vector control, as described under “Materials and Methods.” After 48 h, cells were plated onto 10-well slides, and, following incubation at 37 °C and 5% CO2 overnight, the cells were treated (t = 0) with 5 μm nocodazole (or 0.1% (v/v) DMSO) and analyzed for motility after 8 h. Each measurement is the average of triplicate measurements ± S.D., and the results are representative of three independent experiments.
DISCUSSION
In a previous study from this laboratory, the acquisition of motile behavior resulted from engineered overexpression of WT PKCα in nonmotile MCF-10A cells (4). In the present work, application of the traceable kinase method led us to identify α6-tubulin as a substrate of PKCα in MCF-10A cells that also engenders the motility phenotype. Construction of pseudophosphorylated mutants of α6-tubulin at PKC consensus sites provided the molecular tools by which to identify Ser165 as a key site of phosphorylation. Simulated phosphorylation at Ser165 (S165D-α6-tubulin) endowed transfectant cells with nocodazole-sensitive motility. In complementary experiments, expression of a phosphorylation-resistant mutant (S165N-α6-tubulin) had a dominant-negative effect on the motility of metastatic human breast tumor cells. These results further implicated α6-tubulin and its phosphorylation at Ser165 by PKC in the regulation of microtubule structure and motile behavior of metastatic breast cells.
That α6-tubulin serves as an intracellular PKCα substrate is a new development in our understanding of post-translational modifications that occur on tubulin proteins (27). Unlike other known tubulin modifications, such as tyrosination, glutamylation, and acetylation, phosphorylation of tubulin has received only limited attention. In this regard, phosphorylation of the carboxyl terminus of α-tubulin has been observed (28), and phosphorylation of β-tubulin by PKCθ (29) and by G-protein-coupled receptor kinase 2 (30) has been reported. The most complete investigation to date examined cyclin-dependent kinase Cdk1-mediated phosphorylation of intracellular β-tubulin, whose phosphorylation promoted disassembly of existing polymers (31, 32). A more recent study by Fourest-Lieuvin et al. (33) demonstrated that β-tubulin undergoes direct phosphorylation by Cdk1 at Ser172 (colored brown in Fig. 6). This site, which is absent in α-tubulin, occurs within the GTP binding domain at the interface of a β-subunit and the α-subunit of a second heterodimer. In that study, pseudophosphorylated GFP-β3-tubulin (with gain-of-function mutants S172D or S172E) was poorly incorporated into microtubules, especially during mitosis. These findings suggested that phosphorylation by Cdk1 at Ser172 in β-tubulin acts to prevent and/or reverse polymerization into microtubules, possibly by impairing GTP binding or interactions between heterodimers. Fourest-Lieuvin et al. (33) observed that Cdk1-mediated β-tubulin phosphorylation accompanies mitosis. In contrast with their findings, we have shown that when PKCα is overexpressed in MCF-10A cells, there is a dramatic decrease in the rate of proliferation due to slower progress through G1 (4). Irrespective of the other targets of these two enzymes, we speculate that Cdk1 and PKCα interact with tubulin subunits in distinct ways and perhaps at different points in the cell cycle to produce their characteristic effects on microtubule structure.
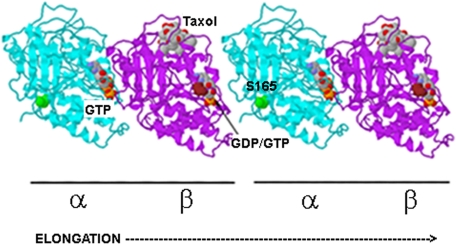
FIGURE 6.
Structural model of assembling tubulin heterodimers. The model shows that Ser165 (green) in the α-tubulin subunit (blue) lies at the interface of two polymerizing α/β-heterodimers. Ser165 participates in longitudinal contacts with a β-tubulin subunit at the plus end of a growing polymer and may influence GTP hydrolysis or GDP/GTP exchange on β-tubulin. The exchangeable GTP/GDP on β-tubulin and the nonexchangeable GTP on α-tubulin are indicated. Highlighted in brown is the site in β-tubulin phosphorylated by Cdk1 (Ser174 in this model) (33). Molecular modeling of the α/β-heterodimer (Protein Data Bank entry 1TUB) was performed with Protein Explorer, version 2.80.
It is notable that Ser165 of α-tubulin and Ser172 in β-tubulin lie in close juxtaposition near the interface of two assembling heterodimers. As depicted in Fig. 6, Ser165 (green) lies at the surface of α-tubulin. This site is close to the T5 loop (amino acids 175–184), which collaborates in longitudinal contacts between assembling heterodimers, especially in the vicinity of the exchangeable GTP (GTP cap) on the β-tubulin subunit at the growing end (“plus end”) (34, 35). During polymerization of tubulin heterodimers into protofilaments, the α-tubulin of an incoming dimer makes contact with the plus end β-tubulin and consequently causes hydrolysis of the exchangeable GTP to GDP; this reaction is thought to involve catalytic residues in the T7 loop of α-tubulin (35). It is possible that the presence of a negatively charged phosphoryl group at Ser165 in α-tubulin promotes tubulin polymerization by (i) increasing contacts between α-tubulin and a plus end β-tubulin, (ii) increasing hydrolysis of GTP to GDP on β-tubulin at the plus end, or (iii) facilitating exchange via guanine-nucleotide exchange factors of the resulting GDP for new GTP.
During cell movement, microtubules undergo dynamic instability that is defined by alternating phases of elongation and shortening and is thought to be regulated by proteins (e.g. EB-1, CLIP-170, or APC) that bind at plus ends (36–38). Several laboratories have used GFP-EB1 as a tool to track the dynamic formation of plus ends as indicative of microtubule elongation activity (37, 38). The alternating phases of elongation and shrinkage of microtubules are thought to be influenced by downstream events that include the Rho GTPases (e.g. Rac 1, Cdc42, and RhoA) and stathmin/Op18 phosphorylation (36, 39–46). In this regard, the plus ends of growing microtubules are captured and stabilized at the leading edge by Rac 1 and Cdc42, whose activation is highest in this region of a motile cell. In a model advanced by others, it was suggested that elongating microtubules, acting as positive feedback, can promote Rho GTPase activity (39, 42) and stathmin/Op18 phosphorylation (47), two events associated with microtubule dynamics and directional cell movement (38). In MCF-10A cells that are treated with DAG-lactone or that express the S165D mutant of α6-tubulin, we observed that the resulting motility could be inhibited by >80% by Rac 1 inhibitor NSC23766,5 thereby supporting a downstream role for Rac 1, as initially suggested by our earlier study with dominant negative Rac 1 (4). However, Western blot analysis with phosphostathmin antibodies, each recognizing one of the three known phosphorylation sites (Ser24, Ser37, or Ser63) (47), revealed no increased phosphorylation in PKCα-stimulated or DAG-lactone-treated MCF-10A cells.5 The precise mechanism by which phosphorylation of α-tubulin directs the structural dynamics of microtubules provides a new direction for further study.
These studies implicate Ser165 in α-tubulin as a target site for PKCα and other DAG-sensitive PKC isoforms that would include the conventional isoforms (α, βI, βII, and γ), which are Ca2+- and DAG-dependent, and the novel isoforms (δ, ϵ, η, and θ), which are DAG-stimulated and Ca2+-independent. In this regard, the intrinsic motility of MDA-MB-468 cells (Fig. 4A) reflected the activity of nonconventional PKC isoforms (25). Nevertheless, inhibition of α-tubulin phosphorylation by the PKC-selective inhibitor bis-indolelymaleimide-1 confirmed that PKC isoforms rather than unidentified protein kinases were responsible for phosphorylating the reporter WT α6-tubulin. Furthermore, blockade of Ser165 by expression of S165N-α6-tubulin produced both inhibition of motility (Fig. 4A) and loss of phosphorylation of the reporter construct (Fig. 4B). These findings strongly imply that nonconventional PKC isoforms also recognize Ser165 as a major common target site in α-tubulin.
In a related study in which MCF-10A cells were treated with siRNA reagents targeted to genes involved in migration and adhesion (9), two novel PKC isoforms were found to have opposing effects (i.e. PKCϵ activity was judged to have a pro-motility effect, whereas PKCη opposed migration). Endogenous PKCα was also linked to migration but only if MCF-10A cells overexpressed ErbB2 in order to activate PKCα as a component of the EGF receptor pathway (see supplemental materials of Ref. 9). In other studies performed with intestinal epithelial Caco-2 cells (29), the effects of specific PKC isoforms on microtubule structure were investigated. Here, PKCθ, a novel isoform, was shown to phosphorylate β-tubulin (at unspecified Thr site(s)), and its activity was correlated with increased microtubule polymerization. In contrast, the novel isoform PKCδ and atypical isoform PKCλ (insensitive to both DAG and Ca2+) promoted microtubule disassembly (48). Although these studies suggest that PKC isoforms produce differential effects on microtubule dynamics, the target sites were not determined.
The PKCα-stimulated signaling pathway described here need not be mediated exclusively by α6-tubulin, since multiple isoforms also exist for this protein. Six α-tubulin isotypes have been isolated, all of which contain 90–98% sequence homology that includes all PKC consensus sites tested here, including Ser165. However, these consensus sites are entirely absent in the other members of the tubulin superfamily: β, γ, δ, ϵ, and ζ (49). Of the six human α-isotypes known to exist, only α6- and Kα1-tubulin have been detected in human carcinoma cancer cells (including MDA-MB-231 cells) by mass spectrometry (50–52) or by real time quantitative reverse transcription PCR (53).
Acknowledgments
We thank Professors Corinne Michels and Karl Fath, Dr. Areti Tsiola, and Dr. Saima Cheema (Queens College) for expert advice and helpful discussion. Mass spectrometry was carried out at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University Medical Center. The DAG-lactone reagent (JH-131E-153), a gift from Dr. Victor Marquez (NCI-Frederick, National Institutes of Health), was the pure R-enantiomer contained in the previously reported racemic mixture (HK654 in Ref. 6).
This work was supported, in whole or in part, by National Institutes of Health Grant CA125632 (to S. A. R.) from NCI. This work was also supported by The Breast Cancer Alliance (Greenwich, CT) (to S. A. R.) and the Professional Staff Congress of The City University of New York. This work was carried out by T. P. A. and X. C. in partial fulfillment of the requirements for the Ph.D. degree in Biochemistry administered by The City University of New York.
- PKC
- protein kinase C
- WT
- wild type
- VC
- vector control
- GFP
- green fluorescent protein
- DAG
- diacylglycerol
- GST
- glutathione S-transferase
- BIM
- bis-indoleylmaleimide
- EGF
- epidermal growth factor
- PIPES
- 1,4-piperazinediethanesulfonic acid.
4 The amino acid sequence for human α6-tubulin can be accessed through NCBI protein data base under NCBI accession number AAH04949 (54). The nucleotide sequence of this protein has been deposited in the GenBankTM data base under GenBankTM accession number 84790 (54). The atomic coordinates for the crystal structure of α-β-tubulin heterodimer are available in the Molecular Modeling Database (available on the World Wide Web) under MMDB accession number 8900 (Protein Data Bank entry 1TUB) (55).
5 X. Chen and S. A. Rotenberg, unpublished data.
REFERENCES
- 1.Larsson C. ( 2006) Cell. Signal. 18, 276– 284 [DOI] [PubMed] [Google Scholar]
- 2.Jaken S., Parker P. J. ( 2000) BioEssays 22, 245– 254 [DOI] [PubMed] [Google Scholar]
- 3.Rabinovitz I., Tsomo L., Mercurio A. M. ( 2004) Mol. Cell Biol. 24, 4351– 4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X. G., Rotenberg S. A. ( 1999) Cell Growth Differ. 10, 343– 352 [PubMed] [Google Scholar]
- 5.Goodnight J. A., Mischak H., Kolch W., Mushinski J. F. ( 1995) J. Biol. Chem. 270, 9991– 10001 [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Bermejo M. L., Leskow F. C., Fuji T., Wang Q., Blumberg P. M., Ohba M., Kurok T., Han K. C., Lee J., Marquez V. E., Kazanietz M. G. ( 2002) J. Biol. Chem. 277, 645– 655; Correction (2004) J. Biol. Chem.279, 23846 [DOI] [PubMed] [Google Scholar]
- 7.Duan D., Sigano D. M., Kelley J. A., Lai C. C., Lewin N. E., Kedei N., Peach M. L., Lee J., Abeyweera T. P., Rotenberg S. A., Kim H., Kim Y. H., El-Kazzouli S., Chung J. U., Young H. A., Young M. R., Baker A., Colburn N. H., Haimovitz-Friedman A., Truman J. P., Parrish D. A., Deschamps J. R., Perry N. A., Surawski R. J., Blumberg P. M., Marquez V. E. ( 2008) J. Med. Chem. 51, 5198– 5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu B. W., Bowers M., Witkowski G., Huang M., Ramachandran S. ( 2002) Breast Cancer Res. Treat. 76, 181– 193 [DOI] [PubMed] [Google Scholar]
- 9.Simpson K. J., Selfors L. M., Bui J., Reynolds A., Leake D., Khvorova A., Brugge J. S. ( 2008) Nat. Cell Biol. 10, 1027– 1038 [DOI] [PubMed] [Google Scholar]
- 10.Tan M., Li P., Sun M., Yin G., Yu D. ( 2006) Oncogene 25, 3286– 3295 [DOI] [PubMed] [Google Scholar]
- 11.Shah K., Liu Y., Deirmengian C., Shokat K. M. ( 1997) Proc. Natl. Acad. Sci. U. S. A. 94, 3565– 3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ting A. Y., Witte K., Shah K., Kraybill B., Shokat K. M., Schultz P. G. ( 2001) Biopolymers 60, 220– 228 [DOI] [PubMed] [Google Scholar]
- 13.Shah K., Shokat K. M. ( 2002) Chem. Biol. 9, 35– 47 [DOI] [PubMed] [Google Scholar]
- 14.Witucki L. A., Huang X., Shah K., Liu Y., Kyin S., Eck M. J., Shokat K. M. ( 2002) Chem. Biol. 9, 25– 33 [DOI] [PubMed] [Google Scholar]
- 15.Habelhah H., Shah K., Huang L., Burlingame A. L., Shokat K. M., Ronai Z. ( 2001) J. Biol. Chem. 276, 18090– 18095 [DOI] [PubMed] [Google Scholar]
- 16.Eblen S. T., Kumar N. V., Shah K., Henderson M. J., Watts C. K., Shokat K. M., Weber M. J. ( 2003) J. Biol. Chem. 278, 14926– 14935 [DOI] [PubMed] [Google Scholar]
- 17.Hindley A. D., Park S., Wang L., Shah K., Wang Y., Hu X., Shokat K. M., Kolch W., Sedivy J. M., Yeung K. C. ( 2004) FEBS Lett. 556, 26– 34 [DOI] [PubMed] [Google Scholar]
- 18.Abeyweera T. P., Rotenberg S. A. ( 2007) Biochemistry 46, 2364– 2370 [DOI] [PubMed] [Google Scholar]
- 19.Soule H. D., Maloney T. M., Wolman S. R., Peterson W. D., Jr., Brenz R., McGrath C. M., Russo J., Pauley R. J., Jones R. F., Brooks S. C. ( 1990) Cancer Res. 50, 6075– 6086 [PubMed] [Google Scholar]
- 20.Banan A., Smith G. S., Rieckenberg C. L., Kokoska E. R., Miller T. A. ( 1998) Am. J. Physiol. Gastrointest. Liver Physiol. 274, G111– G121 [DOI] [PubMed] [Google Scholar]
- 21.Hudder A., Nathanson L., Deutscher M. P. ( 2003) Mol. Cell. Biol. 23, 9318– 9326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennelly P. J., Krebs E. G. ( 1991) J. Biol. Chem. 266, 15555– 15558 [PubMed] [Google Scholar]
- 23.Rusan N. M., Fagerstrom C. J., Yvon A. M., Wadsworth P. ( 2001) Mol. Biol. Cell 12, 971– 980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauthier M. L., Torretto C., Ly J., Francescutti V., O'Day D. H. ( 2003) Biochem. Biophys. Res. Commun. 307, 839– 846 [DOI] [PubMed] [Google Scholar]
- 25.Mao M., Fang X., Lu Y., Lapushin R., Bast R. C., Mills G. B. ( 2000) Biochem. J. 352, 475– 482 [PMC free article] [PubMed] [Google Scholar]
- 26.Vasquez R. J., Howell B., Yvon A. M., Wadsworth P., Cassimeris L. ( 1997) Mol. Biol. Cell 8, 973– 985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westermann S., Weber K. ( 2003) Nat. Rev. Mol. Cell Biol. 4, 938– 947 [DOI] [PubMed] [Google Scholar]
- 28.Wandosell F., Serrano L., Avila J. ( 1987) J. Biol. Chem. 262, 8268– 8273 [PubMed] [Google Scholar]
- 29.Banan A., Zhang L. J., Shaikh M., Fields J. Z., Farhadi A., Keshavarzian A. ( 2004) Am. J. Physiol. Cell Physiol. 287, C218– 234 [DOI] [PubMed] [Google Scholar]
- 30.Yoshida N., Haga K., Haga T. ( 2003) Eur. J. Biochem. 270, 1154– 1163 [DOI] [PubMed] [Google Scholar]
- 31.Lamb N. J., Fernandez A., Watrin A., Labbé J. C., Cavadore J. C. ( 1990) Cell 60, 151– 165 [DOI] [PubMed] [Google Scholar]
- 32.Lieuvin A., Labbé J. C., Dorée M., Job D. ( 1994) J. Cell Biol. 124, 985– 996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fourest-Lieuvin A., Peris L., Gache V., Garcia-Saez I., Juillan-Binard C., Lantez V., Job D. ( 2006) Mol. Biol. Cell 17, 1041– 1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Downing K. H., Nogales E. ( 1998) Curr. Opin. Cell Biol. 10, 16– 22 [DOI] [PubMed] [Google Scholar]
- 35.Löwe J., Li H., Downing K. H., Nogales E. ( 2001) J. Mol. Biol. 313, 1045– 1057 [DOI] [PubMed] [Google Scholar]
- 36.Watanabe T., Noritake J., Kaibuchi K. ( 2005) Trends Cell Biol. 15, 76– 83 [DOI] [PubMed] [Google Scholar]
- 37.Piehl M., Cassimeris L. ( 2003) Mol. Biol. Cell 14, 916– 925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salaycik K. J., Fagerstrom C. J., Murthy K., Tulu U. S., Wadsworth P. ( 2005) J. Cell Sci. 118, 4113– 4122 [DOI] [PubMed] [Google Scholar]
- 39.Wittmann T., Bokoch G. M., Waterman-Storer C. M. ( 2004) J. Biol. Chem. 279, 6196– 6203 [DOI] [PubMed] [Google Scholar]
- 40.Ballestrem C., Wehrle-Haller B., Hinz B., Imhof B. A. ( 2000) Mol. Biol. Cell 11, 2999– 3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y. L., Li S., Miao H., Tsou T. C., del Pozo M. A., Chien S. ( 2002) J. Vasc. Res. 39, 465– 476 [DOI] [PubMed] [Google Scholar]
- 42.Waterman-Storer C. M., Worthylake R. A., Liu B. P., Burridge K., Salmon E. D. ( 1999) Nat. Cell Biol. 1, 45– 50 [DOI] [PubMed] [Google Scholar]
- 43.Küntziger T., Gavet O., Manceau V., Sobel A., Bornens M. ( 2001) Mol. Biol. Cell 12, 437– 448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wittmann T., Waterman-Storer C. M. ( 2001) J. Cell Sci. 114, 3795– 3803 [DOI] [PubMed] [Google Scholar]
- 45.Wittmann T., Bokoch G. M., Waterman-Storer C. M. ( 2003) J. Cell Biol. 161, 845– 851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lansbergen G., Akhmanova A. ( 2006) Traffic 7, 499– 507 [DOI] [PubMed] [Google Scholar]
- 47.Cassimeris L. ( 2002) Curr. Opin. Cell Biol. 14, 18– 24 [DOI] [PubMed] [Google Scholar]
- 48.Farhadi A., Keshavarzian A., Ranjbaran Z., Fields J. Z., Banan A. ( 2006) J. Pharmacol. Exp. Ther. 316, 1– 7 [DOI] [PubMed] [Google Scholar]
- 49.McKean P. G., Vaughan S., Gull K. ( 2001) J. Cell Sci. 114, 2723– 2733 [DOI] [PubMed] [Google Scholar]
- 50.Rao S., Aberg F., Nieves E., Band, Horwitz S., Orr G. A. ( 2001) Biochemistry 40, 2096– 2103 [DOI] [PubMed] [Google Scholar]
- 51.Verdier-Pinard P., Wang F., Martello L., Burd B., Orr G. A., Horwitz S. B. ( 2003) Biochemistry 42, 5349– 5357 [DOI] [PubMed] [Google Scholar]
- 52.Verdier-Pinard P., Wang F., Burd B., Angeletti R. H., Horwitz S. B., Orr G. A. ( 2003) Biochemistry 42, 12019– 12027 [DOI] [PubMed] [Google Scholar]
- 53.Takano T., Hasegawa Y., Miyauchi A., Matsuzuka F., Yoshida H., Kuma K., Amino N. ( 2001) Cancer Lett. 168, 51– 55 [DOI] [PubMed] [Google Scholar]
- 54.Mammalian Gene Collection Program Team ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 16899– 1690312477932 [Google Scholar]
- 55.Nogales E., Wolf S. G., Downing K. H. ( 1998) Nature 391, 199– 203 [DOI] [PubMed] [Google Scholar]