Abstract
Hepatitis C virus (HCV) is an enveloped, positive strand RNA virus of about 9.6 kb. Like all enveloped viruses, the HCV membrane fuses with the host cell membrane during the entry process and thereby releases the genome into the cytoplasm, initiating the viral replication cycle. To investigate the features of HCV membrane fusion, we developed an in vitro fusion assay using cell culture-produced HCV and fluorescently labeled liposomes. With this model we could show that HCV-mediated fusion can be triggered in a receptor-independent but pH-dependent manner and that fusion of the HCV particles with liposomes is dependent on the viral dose and on the lipid composition of the target membranes. In addition CBH-5, an HCV E2-specific antibody, inhibited fusion in a dose-dependent manner. Interestingly, point mutations in E2, known to abrogate HCV glycoprotein-mediated fusion in a cell-based assay, altered or even abolished fusion in the liposome-based assay. When assaying the fusion properties of HCV particles with different buoyant density, we noted higher fusogenicity of particles with lower density. This could be attributable to inherently different properties of low density particles, to association of these particles with factors stimulating fusion, or to co-floatation of factors enhancing fusion activity in trans. Taken together, these data show the important role of lipids of both the viral and target membranes in HCV-mediated fusion, point to a crucial role played by the E2 glycoprotein in the process of HCV fusion, and reveal an important behavior of HCV of different densities with regard to fusion.
Introduction
Hepatitis C virus (HCV)4 is an important public health concern worldwide as it is a major cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma. HCV is an enveloped virus that belongs to the Hepacivirus genus of the Flaviviridae family (1). Based on sequence comparison, patient isolates are classified into seven genotypes, differing in their nucleotide sequence by 30–35% (2–5). The two viral surface proteins, E1 (residues 192–383) and E2 (residues 384–746), are processed by signal peptidases of the endoplasmic reticulum from a 3,000-amino acid-long polyprotein encoded by the HCV genome (reviewed in Ref. 2). The E1 (∼31 kDa) and E2 (∼70 kDa) proteins are glycosylated in their large amino-terminal ectodomains (6) and are anchored in the viral membrane by their carboxyl-terminal transmembrane domains. E1 and E2 form a heterodimer stabilized by noncovalent interactions. This oligomer is thought to be present at the surface of HCV particles (7) and to be involved in viral entry. Carboxyl-terminally truncated soluble E2 protein is known to specifically bind to crucial HCV entry factors like glycosaminoglycans, the tetraspanin CD81, and the scavenger receptor BI (8–12). Thus, virus-associated E2 is likely directly involved in interactions important for virus attachment and productive infection (reviewed in Refs. 13, 14).
Both HCV envelope glycoproteins are the targets for virus-neutralizing antibodies (7, 15–19). In E2, one important neutralizing epitope is the so-called hypervariable region 1, which includes the 27 amino-terminal residues of E2 (20–22). Other neutralizing epitopes lie within or encompass the regions implicated in CD81 binding of E2 (23). Therefore, antibodies recognizing such epitopes may prevent infection by way of a neutralization-of-binding (NOB) activity with respect to CD81. In addition, they are valuable reagents to characterize at the molecular level the implication of targeted regions of E1 or E2 in the fusion process. In fact, both E1 and E2 have been reported to contain fusion determinants or fusion peptide candidates (24, 25), suggesting that distinct regions in both E1 and E2 may cooperate to complete the fusion process (25). However, little is known at the molecular level about the events mediating HCV membrane fusion. In recent times, significant progress has been made with the development of robust assays for the analysis of productive HCV infection in tissue culture. These models are based on the following: (i) so-called HCV pseudotyped particles (HCVpp), consisting of unmodified HCV E1E2 glycoproteins assembled onto retroviral nucleocapsids (26–28), and more recently (ii) cell culture-derived HCV particles (HCVcc) of the infectious HCV clone JFH1 (genotype 2a), able to replicate and produce viral particles in cell culture (29–31). Extensive characterization of HCVpp established that these particles mimic the early steps of the HCV replication cycle (reviewed in Refs. 13, 14). Infection assays and our in vitro liposome fusion assays based on HCVpp have established that HCV entry and fusion is pH-dependent (28, 32, 33). This was confirmed by cell-cell fusion assays (34) and by using HCVcc particles (35–37). Furthermore, low pH treatment of HCVpp led to the exposure of new epitopes in E2 (7), suggesting that low pH induces conformational rearrangements in HCV glycoproteins that eventually trigger fusion with the endosome membrane.
The majority of HCV circulating in blood was found associated with β-lipoproteins and very low and low density lipoproteins (38–40), and the low density lipoprotein receptor has been reported as a receptor for HCV (41). Moreover, lipids associated with the virion such as cholesterol and sphingomyelin, together with cholesterol of the target membranes, were found to play a critical role in the cellular entry, fusion, and overall infectivity of HCV (33, 42, 43). Interestingly, serum-derived HCV displays a highly heterogeneous density, of which low density particles are more infectious for chimpanzees than viruses with higher density (44). Similarly, HCVcc particles with low density (1.09–1.10 g/ml) display the highest specific infectivity (29). Taken together, these data suggest a key role of lipids and/or lipoprotein-associated lipids for productive infection by HCV, which may be related to facilitated virus binding, entry, and/or fusion.
To improve our knowledge about the requirements for HCV glycoprotein-dependent fusion and infection, we have developed in this work an HCV fusion assay. Using this model based on fluorescently labeled liposomes and infectious HCVcc particles of the Jc1 chimera (45), we define basic parameters of the HCV fusion process. We demonstrate the dependence of HCV fusion upon low pH and the envelope glycoproteins. Our studies point to a crucial role played by E2 in the fusion process per se and shed light on the influence of lipids of both viral and target membranes in HCV infectivity and membrane fusion.
MATERIALS AND METHODS
Cell Culture and Cell Lines
Human hepatoma cells Huh-7.5 (46) were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen), supplemented with 2 mm l-glutamine, nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum (DMEM complete).
Constructs
The genotype 2a/2a chimera Jc1, the Luc-Jc1 reporter virus, and JFH1-ΔE1E2 have been described recently (30, 37, 45). Jc1 derivatives Jc1G418A and Jc1G418D were constructed by standard PCR-based cloning strategies using appropriate primers to introduce the respective point mutation at codon 418 of the Jc1 open reading frame. PCR-based inserts were sequenced to verify the introduced mutation and to exclude off-side mutations. Detailed cloning strategies and sequence information are available upon request.
In Vitro Transcription and Electroporation
In vitro transcripts were generated by linearizing 10 μg of the respective plasmid by digestion for 1 h with MluI. Plasmid DNA was extracted with phenol and chloroform and, after precipitation with ethanol, dissolved in RNase-free water. In vitro transcription reaction mixtures contained 80 mm HEPES, pH 7.5, 12 mm MgCl2, 2 mm spermidine, 40 mm dithiothreitol, a 3.125 mm concentration of each ribonucleoside triphosphate, 1 unit of RNasin (Promega, Mannheim, Germany) per μl, 0.1 μg of plasmid DNA/μl, and 0.6 unit of T7 RNA polymerase (Promega) per μl. After incubating the mixture for 2 h at 37 °C, an additional 0.3 unit of T7 RNA polymerase/μl was added, followed by another 2 h at 37 °C. Transcription was terminated by the addition of 1.2 units of RNase-free DNase (Promega) per μg of plasmid DNA and 30 min of incubation at 37 °C. The RNA was extracted with acidic phenol and chloroform, precipitated with isopropyl alcohol, and dissolved in RNase-free water. The concentration was determined by measurement of the absorbance at 260 nm.
For electroporation of HCV RNA into Huh-7.5 cells, single-cell suspensions were prepared by trypsinization of monolayers and subsequent resuspension with complete DMEM. Cells were washed with phosphate-buffered saline (PBS), counted, and resuspended at 1.5 × 107 cells per ml in Cytomix (47), containing 2 mm ATP and 5 mm glutathione. Five μg of in vitro transcribed RNA was mixed with 400 μl of cell suspension by pipetting and then electroporated with a Gene Pulser system (Bio-Rad) in a cuvette with a gap width of 0.4 cm (Bio-Rad) at 975 microfarads and 270 V. Cells were immediately transferred to 10 ml of complete DMEM, and 2 ml of the cell suspension were seeded per well of a 6-well plate.
Luciferase Infection Assay
For standard infection assays with Jc1 firefly luciferase reporter viruses Luc-Jc1 (37), Huh-7.5 cells were seeded at a density of 6 × 104 cells per well of a 12-well plate, 24 h prior to inoculation with 350 μl of reporter virus for 4 h. Infectivity was quantified 72 h after inoculation using luciferase assays. To this end, cells were washed once with PBS, lysed directly on the plate with 350 μl of ice-cold lysis buffer (0.1% Triton X-100, 25 mm glycylglycine, 15 mm MgSO4, 4 mm EGTA, and 1 mm dithiothreitol, pH 7.8), and frozen. After being thawed, lysates were resuspended by pipetting up and down. For each well, 100 μl of lysate were mixed with 360 μl of assay buffer (25 mm glycylglycine, 15 mm MgSO4, 4 mm EGTA, 1 mm dithiothreitol, 2 mm ATP, and 15 mm K2PO4, pH 7.8) and, after addition of 200 μl of a luciferin solution (200 μm luciferin, 25 mm glycylglycine, pH 8.0), measured for 20 s in a luminometer (Lumat LB9507; Berthold, Freiburg, Germany).
Titration of Viruses Using Immunohistochemistry
Virus titers were determined as described previously with minor modifications (29). In brief, cells were seeded in 96-well plates at a density of 1 × 104 cells per well 24 h prior to inoculation with dilutions of filtered cell culture supernatant (6 parallel wells were used for each dilution). After 72 h, cells were fixed for 20 min with ice-cold methanol at −20 °C, washed once with PBS, and then permeabilized for 5 min with 0.5% Triton X-100 in PBS. After three washes with PBS, NS5A was detected with a 1:2,000 dilution of hybridoma supernatant 9E10 (29) in PBS for 1 h at room temperature. Subsequently, cells were washed as described above, and bound 9E10 was detected by incubation with peroxidase-conjugated antibodies specific to murine IgG (Sigma) diluted at 1:200 in PBS. After a 1-h incubation at room temperature, cells were washed as specified above. Finally, peroxidase activity was detected by using carbazole substrate. To this end, 0.32% (w/v) of 3-amino-9-ethylcarbazole (Sigma) in N,N-dimethylformamide was diluted at a ratio of 1:3.3 with 15 mm acetic acid, 35 mm sodium acetate, pH 8.0, and 0.4% H2O2 and incubated with the cells for ∼15 min at room temperature until brown staining of infected cells was clearly visible. Subsequently, the carbazole substrate was aspirated, and cells were kept in distilled H2O. Virus titers (50% tissue culture infective dose [TCID50/ml]) were calculated based on the methods of Kärber (48) and Spearman (49).
Density Gradient Centrifugation
Cell culture-derived HCV particles were harvested 48 h after electroporation of appropriate HCV RNA into cells and passed through 0.45-μm pore-size filters. One milliliter of the preparation was layered under a 0–30% continuous iodixanol gradient (Optiprep; Axis-Shield, Oslo, Norway) prepared in a cell suspension medium containing 0.85% (w/v) NaCl and 10 mm Tricine-NaOH, pH 7.4. Gradients were centrifuged for 15–18 h at 154,000 × g in a TH-641 swing-out rotor at 4 °C using a Sorvall Ultra WX80 centrifuge. Twenty fractions of 0.5 ml each were collected from the bottom, and virus infectivity and the quantity of core protein were determined using a limiting dilution assay and a core-specific enzyme-linked immunosorbent assay, respectively. The density of each fraction was quantified by refractometry.
Quantitative Detection of HCV Core Protein
HCV core protein was measured using an HCV core antigen kit (Wako Chemicals, Neuss, Germany) according to the manufacturer's instructions. Cell culture medium was filtered through 0.45-μm pore-size filters and either directly used for enzyme-linked immunosorbent assay or diluted with PBS prior to measurement.
Cytotoxicity Assay
Cytotoxicity of arbidol was measured using a CytoTox-Glo cytotoxicity assay (Promega, Madison, WI). In brief, Huh-7.5 cells were seeded at a density of 1 × 104 cells per well in a 96-well plate 24 h prior to treatment. Cells were incubated with arbidol for 4 h, washed once with PBS, and cultured for 48 h. Cytotoxicity was measured using a plate luminometer Centro XS LB 960 (Berthold, Freiburg, Germany) according to the manufacturer's instructions.
Immunoprecipitation
Huh-7.5 cells were transfected with 5 μg of Jc1, Jc1G418A, or Jc1G418D RNA constructs. After 24 h cells were labeled with [35S]methionine/cysteine-containing culture fluid for 16 h. Cells were lysed with NPB buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 1 mm phenylmethylsulfonyl fluoride, 0.01 unit/ml aprotinin, and 0.02 unit/ml leupeptin), and the proteins were immunoprecipitated using a 1:1 mixture of protein A-agarose (Bio-Rad) and protein G-agarose (Roche Applied Science) coupled to the E2-specific CBH-5 antibody kindly provided by Steven Foung, Stanford University. Beads were washed three times with NPB buffer. Bound proteins were eluted by boiling the samples in SDS-PAGE sample buffer (150 mm Tris, pH 6.8, 1.2% SDS, 30% glycerol, 15% mercaptoethanol, and 18 mg/liter bromphenol blue). The proteins were resolved by 10% SDS-PAGE and detected by autoradiography and exposure to MS film (Eastman Kodak Co.).
Chemicals and Reagents
Phosphatidylcholine from egg yolk (PC, 99% pure), cholesterol (chol, 99% pure), sphingomyelin from bovine brain (SM, 99% pure), and Triton X-100 were from Avanti Polar Lipids and Sigma, respectively. Octadecyl rhodamine B chloride (R18) was from Invitrogen. The human monoclonal antibody CBH-5 (isotype IgG1κ) was a kind gift of Steven Foung and the antibody against green fluorescent protein (anti-GFP; IgG1) was from Invitrogen. Arbidol (ARB, 1H-indole-3-carboxylic acid, 6-bromo-4-[(dimethylamino)methyl]-5-hydroxy-1-methyl-2-[(phenylthio)-methyl]-, ethyl ester, monohydrochloride) was a kind gift from S. J. Polyak. ARB was dissolved in sterile twice-distilled water, up to 0.7 mg/ml stock solution.
Preparation of Liposomes
All liposomes were large unilamellar vesicles (100 nm), consisting of PC or PC:chol (70:30 mol %) or PC:SM (90:10 mol %) or PC:chol:SM (65:30:5 mol %). R18-labeled liposomes were obtained by mixing R18 and lipids as ethanol and chloroform solutions, respectively (5 mol % R18 final), and liposomes were prepared as described previously (33). R18 was used for liposome labeling because of its photostability and the relative insensitivity of its fluorescence to pH variations.
Fusion Assay
Lipid mixing was assessed essentially as described (33) and monitored as the dequenching of R18. Briefly, viruses suspended in CSM (Cell Suspension Medium: 0.85% (w/v) NaCl, 10 mm Tricine-NaOH, pH 7.4) were added to a cuvette containing R18-labeled liposomes (final lipid concentration, 15 μm). Unless otherwise indicated, liposomes consisted of PC and cholesterol. After temperature equilibration, fusion was initiated through acidification by adding an appropriate volume of diluted HCl to the cuvette, and kinetics were recorded using a dual-channel PicoFluor hand-held fluorimeter (Turner Biosystems, Sunnyvale, CA), operated under the “rhodamine” channel (excitation and emission wavelengths 540 ± 20 and >570 nm, respectively). Maximal R18 dequenching was measured after the addition of 0.1% Triton X-100 (final concentration) to the cuvette. Initial rates of fusion were taken as the value of the slope of the tangent, drawn to the steepest part of the fusion kinetics. Final extent of lipid mixing was the value obtained when fluorescence reached a plateau.
When performing fusion experiments on density-fractionated viruses, a control gradient centrifugation was run in parallel to the virus gradient. This control gradient was prepared using culture fluid from Huh-7.5 cells transfected with 5 μg of tRNA. Fusion parameters of each of the control fractions were subtracted from the fusion values of the corresponding Jc1 virus gradient fraction.
For fusion experiments in the presence of arbidol, increasing concentrations of arbidol were added to liposomes in buffer at neutral pH; after a 2-min equilibration, Jc1 virus was added, and lipid mixing was measured. When the human monoclonal antibody CBH-5 was used, Jc1 was incubated for 15 min in the presence of increasing concentrations of CBH-5 in CSM buffer at pH 7.4. Liposomes were then added, and lipid mixing was measured as described above.
RESULTS
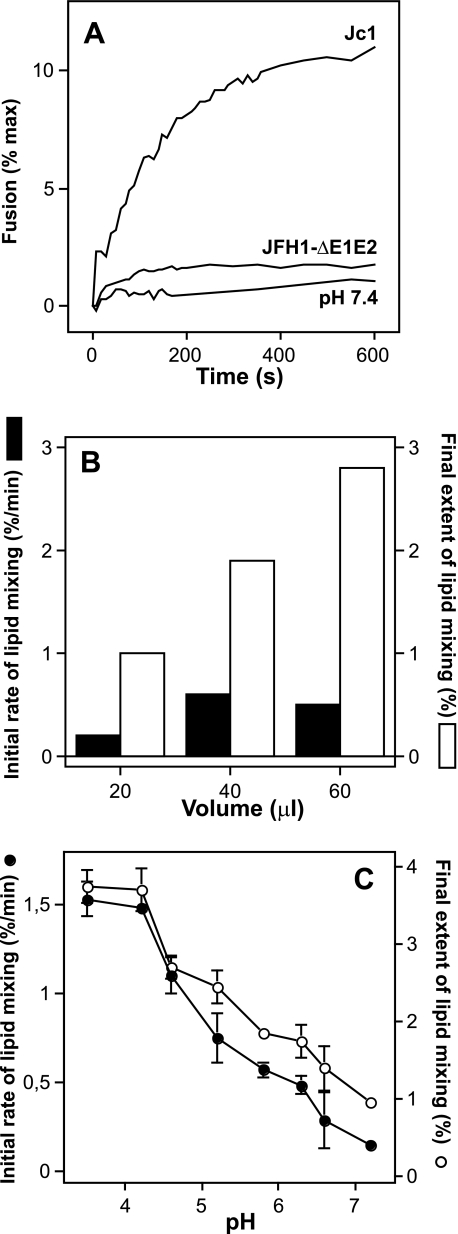
HCV fusion depends on E1 and E2, viral dose, and occurs within a specific pH range. The recent development of a fully permissive infection system based on the HCV clone JFH1 provides the opportunity to dissect the requirements for HCV infection using HCVcc that are infectious both in vitro and in animal models (29–31, 50). To reach maximal sensitivity, we took advantage of a virus chimera consisting of J6CF- and JFH1-derived segments and designated Jc1 which grows to high virus titers in tissue culture (45). In an initial experiment we assessed the fusion activity associated with a preparation of Jc1 particles harvested 48 h post-transfection of Huh-7.5 cells. As a control for background fusion activity, we used a virus mutant carrying a large in-frame deletion of viral E1 and E2 proteins (Fig. 1A, JFH1-ΔE1E2) (30). The Jc1 virus preparation exhibited an immediate lipid mixing with liposomes when the pH was decreased to 5.0, the pH threshold which we have recently defined to be optimal for fusion of HCVpp (33). Conversely, the preparation of the virus mutant lacking envelope glycoproteins (JFH1-ΔE1E2) or Jc1 left at pH 7.4 displayed only low or negligible fusion activity (Fig. 1A). This confirms that HCV membrane fusion is pH-dependent and relies on the E1 and E2 glycoproteins, as shown by us using a similar approach with HCV-pseudotyped particles (33). HCV fusion was specific, because increasing the viral dose increased the final extent of lipid mixing in a dose-dependent manner (Fig. 1B). However, the initial rate of lipid mixing leveled off for 60 μl of viruses. To determine whether any pH optimum could be observed for the fusion activation of Jc1-resident E1/E2, we mixed R18-labeled liposomes with viruses and followed the lipid mixing kinetics over a broad pH range. This was rendered possible by the robustness of R18 fluorescence toward low pH application. Analyzing the initial rates and final extents of lipid mixing demonstrated that Jc1 fusion occurred over a large range of pH (from below pH 4.0 to ∼6.3), with an optimum shifted to pH values below pH 5.0 (Fig. 1C). These results are partially in agreement with our previous data obtained with HCVpp (33) and with data of cell-cell fusion as induced by E1/E2 expressed at the surface of cells (34) (see “Discussion”).
FIGURE 1.
Characteristics of HCVcc-Jc1 membrane fusion. A, representative experiment of Jc1-mediated lipid mixing (100 μl) with R18-labeled PC:chol liposomes (15 μm final concentration; see “Materials and Methods”). Culture fluid of Jc1- or JFH1-ΔE1E2-transfected Huh-7.5 cells was harvested 48 h post-transfection, concentrated (10-fold), and partially purified by ultracentrifugation through a 20% sucrose cushion. The respective pellets were resuspended in PBS and directly applied to the fusion assay. At time 0, fusion was initiated by acidifying the medium to pH 5.0 through addition of diluted HCl to the cuvette containing PBS. The value for complete lipid mixing, which corresponds to 100% fluorescence, was obtained by adding 0.1% (v/v, final) Triton X-100 to the suspension. B, representative experiment of the influence of viral dose on Jc1 fusion. Increasing amounts of Jc1 viruses (in μl) were added to the cuvette containing fluorescent liposomes in PBS, and lipid mixing was recorded at pH 5.0; solid bars, initial rate; open bars, final extent of lipid mixing. C, pH dependence of Jc1 lipid mixing. Lipid mixing was recorded at the indicated pH; initial rates were determined for each pH from the tangents to the steepest part of the fusion curves (closed symbols). The final extent of lipid mixing is the value of fluorescence for each pH at the 10-min time point (open symbols). Each point represents the average value of three separate measurements.
HCV Membrane Fusion Is Inhibited by Arbidol
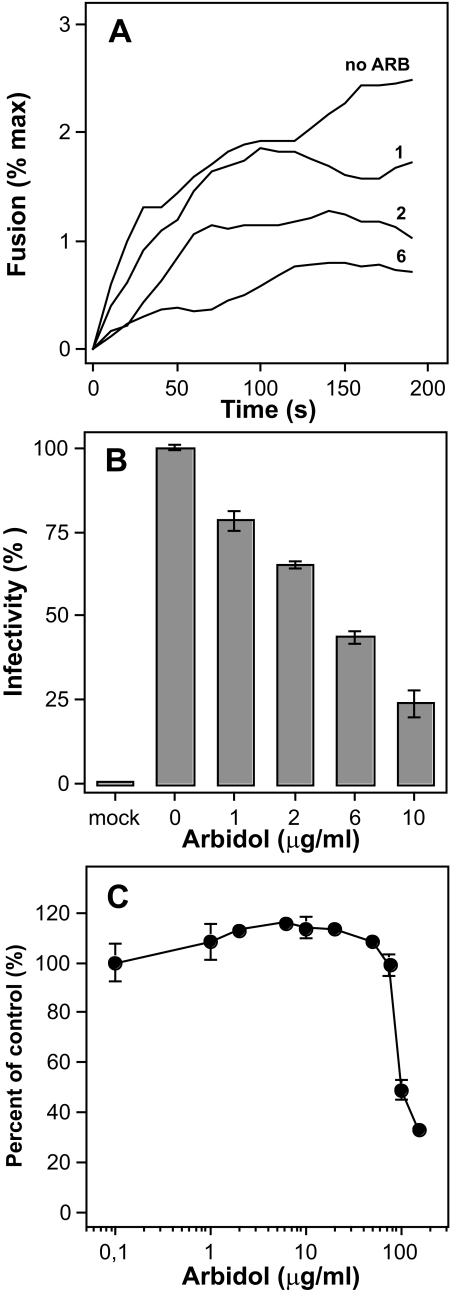
The broad-spectrum antiviral arbidol has been shown to inhibit the entry of several viruses into their target cells (51); in particular, arbidol is most efficient against the influenza virus, through the inhibition of the fusion process (52). We recently described an anti-HCV activity of arbidol that is based on inhibition of HCV cell entry and membrane fusion by the drug (53, 54). However, in our previous work the fusion inhibition exerted by arbidol was only assessed using HCV pseudoparticles. Here we tested whether arbidol would display any effect on HCVcc fusion and overall infectivity. Results presented in Fig. 2A indicate that arbidol can efficiently inhibit Jc1 virus membrane fusion, in a dose-dependent manner. The concentration of arbidol able to inhibit fusion by 50% (IC50) was ∼2 μg/ml, in close agreement with our previous data on HCVpp (53, 54). When the virus was preincubated with arbidol before infection of Huh-7.5 cells and inoculation was done in the presence of the drug, a dose-dependent reduction of infectivity was observed, with a 50% inhibition at ∼6 μg/ml arbidol (Fig. 2B). It should be noted that the concentration of arbidol toxic to 50% of cells (TC50) was found at a much higher concentration, i.e. 100 μg/ml (Fig. 2C). Having established the overall properties of HCVcc membrane fusion, we next investigated the involvement of E2 in HCV fusion.
FIGURE 2.
Inhibitory effect of arbidol on Jc1 fusion and infectivity. A, Jc1 virus was added to R18-labeled PC:chol liposomes in PBS, pH 7.4, with or without 1, 2, or 6 μg/ml arbidol (ARB). After a 2-min equilibration, lipid mixing was initiated by decreasing the pH to 5.0 (time 0). B, Huh-7.5 cells were inoculated with Luc-Jc1 virus preparations that had been preincubated with ARB at the given doses for 1 h at 37 °C. After 4 h, the medium containing virus and ARB was replaced by fresh medium without virus and inhibitor. Infectivity was determined 72 h after inoculation. Mean values and standard errors of quadruplicate experiments are shown. C, Huh-7.5 cells were cultured in medium containing arbidol at the given concentration for 4 h. Cells were then washed with PBS and cultured for an additional 48 h in medium without inhibitor. Cytotoxicity was measured according to manufacturer's instructions (see under “Experimental Procedures”). Mean values and standard errors of quadruplicate measurements are shown.
HCV Fusion Is Abolished by an HCV E2-specific Antibody
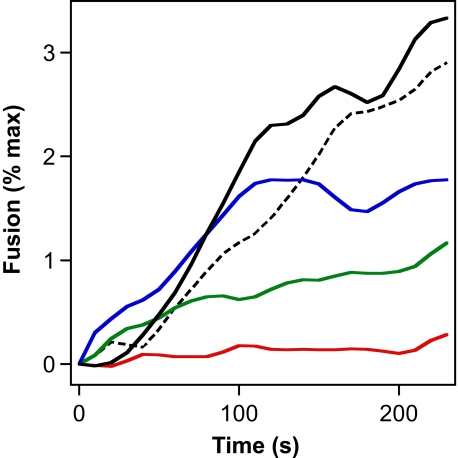
First, we analyzed if an E2-specific antibody specifically interferes with HCVcc fusion. For this series of experiments, we chose the human monoclonal antibody CBH-5, which neutralizes infectivity of HCVpp carrying diverse HCV genotypes (17, 55), via binding to an epitope within E2. The Jc1 virus was preincubated with increasing concentrations of CBH-5 at neutral pH, and the fusion of such mixtures was then assessed after acidification in our lipid mixing assay. As shown in Fig. 3, CBH-5 exerted a dose-dependent inhibition of Jc1 fusion, completely abrogating measurable fusion activity at a concentration of 25 μg/ml. An approximate half-maximal inhibition of fusion activity was achieved at a CBH-5 concentration of ∼5 μg/ml. Importantly, a control and isotype-matched antibody directed against GFP did not affect the final extent of Jc1 fusion activity, whereas the initial rate of fusion was slightly lowered, possibly due to the high protein content of the fusion mixture (Fig. 3, dotted curve). Taken together these data further confirm that the HCVcc-liposome fusion assay is dependent upon the HCV E2 glycoprotein.
FIGURE 3.
Specific inhibition of Jc1 fusion by the human monoclonal antibody CBH-5. Concentrated Jc1 virus particles suspended in CSM buffer were incubated for 15 min with increasing doses of CBH-5 at pH 7.4. The mixture was then transferred into a cuvette containing R18-labeled liposomes, and after a 1-min equilibration, pH was decreased to 5.0 and lipid mixing measured. Curves are as follows: black, Jc1 without CBH-5; blue, Jc1 with 1 μg/ml CBH-5; green, with 5 μg/ml CBH-5; red, with 25 μg/ml CBH-5; dashed black curve, Jc1 with anti-GFP antibody at 30 μg/ml final concentration, respectively.
Single Mutation in E2 Leads to a Severe Impairment of Jc1 Infectivity and Fusion
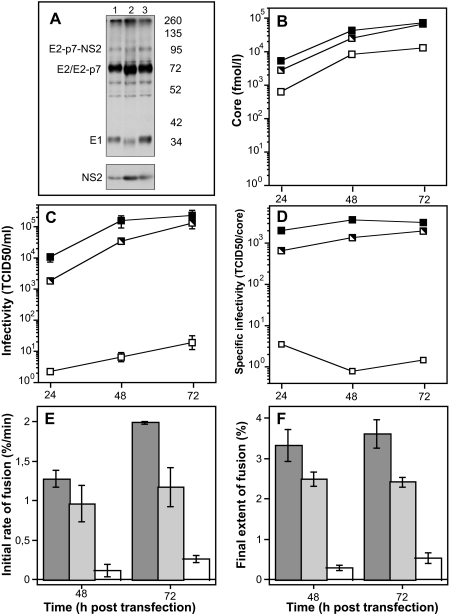
We recently showed that specific point mutations introduced in the sequence of E1 or E2 led to severe defects in HCVpp infectivity and membrane fusion without affecting HCVpp production and incorporation of E1-E2 complexes into HCVpp (25). These mutations reside within three discrete regions, one located in E1 and two located in E2. Interestingly, HCVpp harboring mutant E2 proteins with a single mutation of a highly conserved glycine at position 418 to alanine or aspartic acid (G418A; G418D) were heavily impaired with regard to infectivity and cell-cell fusion capacity, but they exhibited different fusion behaviors depending on whether Gly-418 had been mutated to Ala or Asp (25). We therefore introduced such mutations in the Jc1 context and analyzed glycoprotein processing and release of HCV particles, as well as their infectivity and fusion properties. When using the E2-specific CBH-5 monoclonal antibody to precipitate E1-E2 complexes, we observed comparable levels of E2 for Jc1 and both the Jc1G418A and the Jc1G418D mutants, demonstrating that neither mutation grossly affected reactivity toward this antibody (Fig. 4A). In the case of the G418D mutant, comparable quantities of mature E2, E1, and NS2 were detected, indicating that this mutation did not affect processing nor co-precipitation of E1 and NS2 with E2. In the case of the G418A mutant, however, we noted a slightly faster electrophoretic mobility of both E2 and E1, a decrease in E1 amount, as well as a minor increase of NS2 co-precipitation (Fig. 4A), suggesting that the overall structure of the E1-E2 complex may be modulated by this mutation, possibly resulting in differential glycosylation and interaction with NS2. The number of physical virus particles as determined by quantification of extracellular HCV core protein (Fig. 4B) obtained upon transfection of the Jc1G418D mutant was comparable with wild-type Jc1, indicating that the virus assembly and release process were only slightly affected by this mutation. Conversely, assembly and release of the Jc1G418A mutant were impaired with ∼10-fold lower levels of extracellular core protein detected between 24 and 72 h after transfection (Fig. 4B). The infectivity of Jc1G418D was decreased compared with Jc1 by only 2–5-fold (Fig. 4C). Strikingly, Jc1G418A infectivity was heavily impaired, with peak infectious titers ∼10,000-fold lower compared with wild-type Jc1 (Fig. 4C). Consequently, the specific infectivity, i.e. the infectivity associated with a given quantity of released core protein, was much lower for the G418A mutant and only slightly lower for the G418D mutant when compared with Jc1 (Fig. 4D). Analyzing the membrane fusion capacity of wild-type and mutant viruses, we noted that Jc1G418D fusion was only reduced by ∼15%, whereas fusion of Jc1G418A was almost totally abolished (Fig. 4, E and F). Taken together these data support an important function of the highly conserved Gly-418 within the ectodomain of E2 for assembly and release of infectious particles in general and for the fusion process in particular, thus lending further support to the idea that this region could be a fusion determinant.
FIGURE 4.
Effect of a single mutation in E2 on Jc1 glycoprotein processing, virus release, infectivity, and fusion properties of released particles. A, Huh-7.5 cells were transfected with 5 μg of Jc1 (lane 1), Jc1G418A (lane 2), or Jc1G418D (lane 3) RNA constructs. After 24 h cells were labeled with [35S]methionine/cysteine in culture fluid for 16 h. Cell lysates were immunoprecipitated with the E2-specific CBH-5 antibody. B and C, Huh-7.5 cells were transfected with 5 μg of the given Jc1 constructs (black symbol, wild type Jc1; black and white symbol, Jc1G418D; white symbol, Jc1G418A) and were seeded into replicate culture plates. After the indicated time points, the culture fluid of the transfected cells was harvested to determine the quantity of released HCV core protein (B) and the infectivity of released viruses (C). The specific infectivity, i.e. the quantity of TCID50/ml associated with 1 fmol of released core protein is depicted in D. E and F, preparations of the wild-type Jc1 chimera (dark gray bars), Jc1G418D, or Jc1G418A (light gray or open bars, respectively) were normalized for equal amount of core protein and were added to the cuvette containing R18-labeled liposomes. Fusion was initiated and measured as described previously. Initial rates (E) and final extents of lipid mixing (F) were determined as described in the legend to Fig. 1. Results are expressed as mean ± S.E. of three separate measurements.
Sphingomyelin, in Conjunction with Cholesterol, Enhances HCVcc Fusion
HCV is known to circulate in complex with lipoproteins. In addition, high density lipoprotein and oxidized low density lipoprotein, natural ligands of scavenger receptor BI, an essential host factor for productive infection by HCV, modulate HCV infection (56, 57).
Given the accumulating evidence that lipids and lipoproteins are important cofactors for the HCV infection process, we wished to directly assess the role of cholesterol (chol) and sphingomyelin (SM) in HCVcc fusion. To this end, Jc1 was mixed with R18 liposomes consisting of PC, PC:chol (70:30 mol %), PC:SM (90:10 mol %) or PC:chol:SM (65:30:5 mol %). Initial rates and final extents of lipid mixing obtained for each liposome condition and from three different batches of virus were assessed. Using this approach we observed higher fusion levels for all liposome compositions after lowering the pH to pH 5.0 (data not shown), establishing that in all cases fusion remained pH-dependent. Results presented in Fig. 5 indicate that, although incorporation of cholesterol to liposomes induced a 2–3-fold increase in initial rate and final extent of Jc1-mediated lipid mixing, incorporation of both chol and SM induced an ∼11-fold increase in initial rate and ∼8-fold increase in the final extent of lipid mixing. Conversely, incorporation of SM to PC liposomes did not enhance Jc1 fusion as compared with PC alone. This suggests that cholesterol and sphingomyelin could act in synergy to enhance HCV membrane fusion, and that specific microdomains enriched in chol/SM might play a role in the process of HCV fusion (see “Discussion”).
FIGURE 5.
Influence of the lipid composition of liposomes on Jc1 fusion. Liposomes of indicated composition were prepared (see under “Materials and Methods”) and added to a cuvette containing Jc1 viruses. Lipid mixing was then measured as described above; initial rates (solid bars) and final extents of lipid mixing (open bars) were determined as in Fig. 4. Results are the mean ± S.E. of four separate experiments.
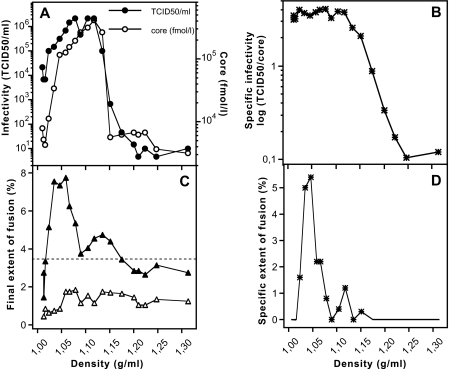
HCV Fractions of Lower Density Display Higher Fusogenicity
As reported earlier, the most infectious material in the sera of patients as well as in HCVcc preparations has low buoyant density (29, 58). Moreover, the HCV particle has the propensity to associate with β-lipoproteins (38, 40, 59–61). Therefore, we sought to investigate the relationship between HCVcc particle density, infectivity, and fusion activity. To this end, we separated Jc1 particles through a linear iodixanol density gradient and collected 20 fractions. These were each analyzed for their core levels, infectivity, and fusion activity (estimated from the final extent of fusion) (Fig. 6). To determine the background of the fusion assay, we prepared a control gradient that had been loaded with tissue culture fluid of tRNA-transfected Huh-7.5 cells and measured fusion activity of these fractions in parallel. Additionally, we calculated the specific infectivity and specific fusion activity, i.e. infectivity and fusion per given quantity of HCV core, by normalizing infectivity or fusion activity to the amount of core present in the respective fraction (Fig. 6, B and D, respectively). Most Jc1 particles displayed a density between ∼1.02 and 1.15 g/ml. Structures rich in core with a density between ∼1.02 and 1.12 displayed a very high infectivity, those with a density of ∼1.13–1.15 an intermediate infectivity, and those with higher density a very low infectivity (Fig. 6, A and B), in agreement with previous findings (29). Most interestingly, (specific) fusion activity was highest at low density peaking around 1.05 g/ml, intermediate in a density range between ∼1.12 and 1.15, and very low in fractions with higher density (Fig. 6, C and D). These data regarding fusogenicity suggest that these differences may be mediated in part by alternate fusion competence of the respective particles, and pinpoint a crucial role played by the lipids that could be associated to low density fractions (see under “Discussion”).
FIGURE 6.
Characterization of the infectivity and fusion properties of Jc1 particles with different buoyant density. Results are a representative experiment. A, culture fluid derived from transfected Huh-7.5 cells and containing Jc1 was layered under a linear iodixanol density gradient (0–30% iodixanol) and centrifuged for 16 h. Fractions of 0.5 ml were harvested from the bottom, and density, HCV core content (open circles), and infectivity (closed circles) were determined for each fraction. B, specific infectivity of each fraction in A was calculated as the infectivity per fmol of core and plotted against the gradient density. C, lipid mixing was assessed for each gradient fraction, and the final extent of fusion was determined as already described. Fusion activity is given for the control gradient loaded with culture fluid of tRNA-transfected cells (open triangles) and for a gradient with Jc1 virus (solid triangles, raw extents of fusion). Fusion activity was considered significant when exceeding a threshold (dashed line) defined as the mean value of the fusion signal of the control gradient plus twice the standard deviation of the mean. D, specific fusion activity of each fraction depicted in C (solid symbols), corrected from the extent of the corresponding fraction of the control gradient, was calculated as the fusion activity per fmol of core protein. It is expressed as specific extent of fusion. The lines without symbol correspond to fractions from C below the threshold of significance for fusion activity (9 fractions).
DISCUSSION
Increasing evidence indicates that HCV enters its host cells by clathrin-dependent endocytosis (36, 62), followed by fusion in the low pH compartment of endosomes (33–35). However, the molecular details of these steps remain elusive, and the fusion properties of HCVcc are unknown. Nevertheless, based on analogy with other viral fusion proteins, it is reasonable to assume that virus-receptor interactions and low pH could trigger glycoprotein rearrangements, thereby allowing the transition of the protein complex from a metastable pre-fusion state to a thermodynamically stable post-fusion structure (63). Therefore, in this study we aimed at further dissecting HCV membrane fusion using HCVcc, focusing on two essential questions as follows: the role played by the E2 glycoprotein and the involvement of lipids of the viral and target membranes in HCV fusion. To this end we used HCV Jc1 particles and fluorescently labeled liposomes as surrogate for the membrane of the host cell. Of note, liposomes lack known HCV entry factors (e.g. CD81), and the fusion reaction in this assay is therefore independent of host proteins. Nevertheless, because of the metastable conformation of the viral fusion machinery, it is likely that suboptimal triggers (e.g. low pH only) are sufficient to elicit rearrangements that mediate membrane fusion. Similar experimental systems have been successfully utilized to dissect basic parameters of viral fusion (64–66). Using the fusion assay described in this study, we observed a strictly pH-dependent fusion with liposomes, which relied on the presence of E1 and E2 glycoproteins at the viral surface. Together with the finding that increase of the viral dose enhanced the extent of fusion, our data indicate that our liposome-based assay is a quantitative measure of glycoprotein- and pH-dependent HCV fusion (Fig. 1). This is the first direct description of HCV fusion characteristics in a genotype 2a context using physio-pathologically relevant HCVcc particles. Moreover, assessing the interference of arbidol, a small fusion inhibitor molecule recently described by us as a potential HCV entry inhibitor (52–54), further confirmed the utility of the HCVcc fusion assay for the characterization of HCV fusion inhibitors (Fig. 2).
The pH range for HCVcc fusion was broad (from pH 6.3 to ∼4.0), as reported previously (33, 34), and interestingly its optimum was slightly shifted toward lower pH values (below pH 4.5; see Fig. 1C) than those reported previously. Because this was reproducibly observed, it is likely that HCVcc behave in a slightly different manner toward pH as compared with other HCV models. A plausible explanation would be that HCVcc fusion, to be optimal, would require additional factors or receptors that are lacking in our in vitro fusion assay; this lack could be compensated at lower pH values through a high protonation level. Also our actual knowledge on this particular topic is derived from experiments performed with E1 and E2 protein sequences of genotypes 1a and 1b, whereas Jc1 is a 2a/2a hybrid virus. This difference in genotype, and thus in envelope protein sequence, might therefore account for this pH shift, because point mutations in the primary sequence might lead to differences in the reactivity of fusion determinants to protonation, because of subtle modifications of their local surrounding. This could act together with the differences in overall physicochemical properties of HCVcc compared with HCVpp, notably the heterogeneity observed on density gradients (29) and the association with apolipoproteins (67), in agreement with the behavior observed for HCV isolated from the sera of patients (Refs. 38–40, 60 and references therein). Because the morphology of virions of different densities is unknown, we could only speculate that the global accessibility of protons to HCV envelope proteins might be affected by the “lipoprotein surrounding” of the viral particles (see also below).
A first set of experiments was then designed to address the role of E2 in HCV fusion. The human monoclonal antibody CBH-5, directed against a conformation-dependent epitope of E2 (7, 68), has been shown to neutralize HCV infectivity in the context of both HCVpp and HCVcc (6, 7, 17, 55), and especially in the context of genotype 2a HCVpp and HCVcc (69, 70). This latter remark is of importance, because only very few neutralizing antibodies in the context of genotype 2a HCV are available for experiments. It also displayed NOB activity toward CD81 (17, 68). CBH-5 exerted a dose-dependent inhibition of HCV fusion in our assay, with a complete abrogation of fusion at 25 μg/ml, a concentration consistent with data from the literature (Fig. 3). This inhibition was not because of a nonspecific competition between virus particles and a high amount of antibody, because a control antibody exerted no inhibition on fusion even at 30 μg/ml. Because our fusion assay relies on the use of plain lipid membranes as targets, CBH-5 exerts its fusion inhibition activity independently from its neutralizing and NOB activities. Our result therefore points to a crucial and specific role played by E2 in the fusion process. It is likely that steric hindrance caused by binding of antibodies to HCVcc-resident E2 proteins would prevent close apposition between HCV particles and liposomes, thus resulting in fusion inhibition. Similarly in vivo, binding of these antibodies may not only directly interfere with attachment to CD81 via direct occlusion of the CD81-binding region but also affect fusion by steric hindrance. Alternatively or in addition, both in our in vitro fusion assay and in vivo, bound antibodies may preclude crucial conformational changes required for fusion. Taken together, these results highlight the important role of the CBH-5 epitope in E2 for HCV fusion. This antibody is known to bind a conformational epitope within the immunogenic domain B of E2 (55), the latter comprising binding sites for various human monoclonal antibodies that all have NOB activity (68). This region includes four highly conserved residues, of which three are involved in CD81 binding to E2 (17).
The second set of experiments addressing the role of E2 in HCV infectivity and fusion was based upon the observation that single mutations in the HCVpp context on a glycine at position 418 led to a decrease in particle infectivity and a dramatic loss of fusogenicity of mutant HCVpp, despite a close-to-normal incorporation of E1-E2 onto the retroviral particles (25). Here we introduced the analogous mutations in the Jc1 context (Fig. 4). Intracellular glycoprotein expression and processing as well as coprecipitation of E1 and E2 were dramatically affected neither by the G418A nor the G418D mutation. However, in the case of the Jc1G418A mutant, we observed an ∼10-fold lower virus release using core protein quantities in the culture fluid as surrogate marker for the number of released virions. Strikingly, infectivity was reduced by ∼4 orders of magnitude, and fusion activity was almost completely abolished, indicating that the G418A mutation rendered the released particles almost completely noninfectious, possibly via inactivation of the fusion activity of the E1-E2 complex. Interestingly, when Gly-418 was replaced by an aspartic acid, virus release, infectivity, and fusion were only slightly reduced compared with the wild-type virus. Although in the absence of an envelope protein incorporation assay for HCVcc particles we cannot distinguish if these defects were because of a lower incorporation of E1-E2 or because of poor function of the mutant E1-E2 complex in virus entry, these results nevertheless highlight the importance of the conserved glycine at position 418 and the E2 protein in general in fusion and HCV infection. Interestingly Asp at position 418 is found in 0.4% of the 4822 nonidentical HCV sequences analyzed, which could explain the relative “tolerance” toward this mutation in our studies, although Gly is found in 99.2% and Ala is not reported (71). Examining the relative hydropathy properties of these amino acids and in the absence of tridimensional structural data, one could only speculate that increasing locally the hydrophobicity around this position with Ala might induce rearrangements in its immediate vicinity, moderately affecting virus assembly but detrimental to its fusion. Surprisingly, introduction of the comparatively large and charged Asp was tolerated much better than exchange of Gly to the supposedly very similar Ala. However, Ala unlike Asp is not found in any known natural HCV isolates, further corroborating that Ala at this position is detrimental to the function of the HCV glycoprotein complex. Availability of a crystal structure may help in the future to better understand the precise structural requirements at this particular position of E2. In conclusion our data confirm our recent proposal that this region of E2 could be a key fusion determinant of HCV (25) and firmly establish the crucial role played by E2 in the fusion process.
The influence of cholesterol (chol) and sphingomyelin (SM) on viral entry and fusion has been widely studied (reviewed in Ref. 72). In the Flaviviridae family of viruses, the cellular internalization and entry of the West Nile and Dengue viruses were shown to depend on chol-enriched microdomains (73, 74); along the same lines, chol depletion of the plasma membrane impaired cell entry of the Japanese encephalitis virus and the Dengue (75), although conflicting results have been reported (76). The fusion step per se of several flaviviruses is only facilitated by the presence of chol or chol/SM in the target membranes (66, 77, 78), whereas the presence of these lipids is a strict prerequisite for the fusion of the closely related alphaviruses. This facilitation is most likely not because of a direct binding of their envelope proteins to any of these lipids, in striking contrast to what was observed/suggested for the envelope glycoproteins of alphaviruses (76, 79, 80). Increasing evidence indicates that CD81-mediated HCV entry is dependent upon chol (43, 81) and ceramide levels at the plasma membrane (82). HCVpp fusion was facilitated by the presence of chol in target liposomes (33). Here we have shown that HCVcc fusion is not only facilitated by chol but further enhanced when a combination between SM and chol is present in the target membranes (Fig. 5). This major enhancement suggests that chol and SM could act in synergy to convey optimal HCV fusion.
Finally, we confirmed a strong difference with regard to infectivity of HCV structures containing core resolved by density gradient centrifugation (Fig. 6). Interestingly, these structures displayed varying levels of fusogenicity. There was a rough correlation that structures containing core associated with low density fractions (between 1.02 and 1.12 g/ml) were both more infectious and fusogenic than those with intermediate and high density. Initial studies of HCVcc separated by density gradients demonstrated a broad distribution of highly infectious material at low density with a peak at 1.09–1.10 g/ml similar to that previously observed in chimpanzees infected with natural HCV isolates (29, 44). Further analyses revealed that low density fractions of HCVcc contained apolipoproteins B and E (67), components of low and very low density lipoproteins (83). HCV particles circulating in blood were found associated with β-lipoproteins (38, 40, 44), and the notion that highly infectious HCV represents in fact a “lipo-viro-particle” (low density fractions containing HCV RNA) has recently emerged (58, 59, 61). Our novel finding that fusogenicity, in addition to infectivity, is highest in low density fractions of HCVcc further emphasizes the notion that virus composition has a pronounced impact on virus infectivity and suggests that this may in part be caused by differential fusion properties of the virions. In the context of the natural infection process, the aforementioned factors may play a role at the plasma membrane, through modulation of interactions with the low density lipoprotein or scavenger receptor BI receptors for instance, thus facilitating HCV internalization (41, 58, 84). In addition they may influence trafficking of the virus-receptor complex on the cell surface and into the cells via endocytosis and finally the actual fusion step, through subtle protein-lipid interactions with the endosomal membrane.
In conclusion, this study is the first molecular investigation of the membrane fusion features of cell-cultured grown HCV. It is also the first description that HCV particles of lower density display the highest fusogenicity, which is a strong indication that the lipids associated with HCV, whatever this association may be (61), play a key role in the process of HCV membrane fusion. Our novel assay should provide the opportunity for a more comprehensive analysis of the fusion characteristics of HCV and may contribute to the identification and development of small molecules targeting HCV through inhibition of the fusion process.
Acknowledgments
We thank Brett D. Lindenbach for thoughtful comments; Christophe Combet for help with the euHCVdb data base; François Penin for critical reading of the manuscript; Charles Rice for the kind gift of the Huh-7.5 cells and NS5A-specific monoclonal antibodies; Steven Foung for the kind gift of the CBH-5 antibody; and Fabienne Archer and Kathy Gallay for the expert management of the Level 3 security laboratory from the IFR128 BioSciences Lyon-Gerland.
This work was supported by the CNRS and the National Agency for AIDS and Viral Hepatitis Research (to E.-I. P.), by Deutsche Forschungsgemeinschaft Grant PI 734/1-1, and by Helmholtz Association Grant SO-024 (to T. P.). This work was presented in part at the 15th International Symposium on Hepatitis C Virus and Related Viruses, October 4–9, 2008, in San Antonio, TX.
- HCV
- hepatitis C virus
- HCVcc
- cell culture-produced HCV
- HCVpp
- HCV pseudotyped particles
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- PC
- phosphatidylcholine
- SM
- sphingomyelin
- chol
- cholesterol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- GFP
- green fluorescent protein
- ARB
- arbidol
- NOB
- neutralization-of-binding.
REFERENCES
- 1.Lindenbach B. D., Rice C. M. ( 2001) in Fields Virology ( Knipe D. M., Howley P. M. eds) Vol. 1, pp. 991– 1041, Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 2.Moradpour D., Penin F., Rice C. M. ( 2007) Nat. Rev. Microbiol. 5, 453– 463 [DOI] [PubMed] [Google Scholar]
- 3.Gottwein J. M., Scheel T. K., Jensen T. B., Lademann J. B., Prentoe J. C., Knudsen M. L., Hoegh A. M., Bukh J. ( 2009) Hepatology 49, 364– 377 [DOI] [PubMed] [Google Scholar]
- 4.Murphy D., Chamberland J., Dandavino R., Sablon E. ( 2007) Hepatology 46, Suppl. 1, 623A ( Abstr. 872) [Google Scholar]
- 5.Simmonds P., Bukh J., Combet C., Deléage G., Enomoto N., Feinstone S., Halfon P., Inchauspé G., Kuiken C., Maertens G., Mizokami M., Murphy D. G., Okamoto H., Pawlotsky J. M., Penin F., Sablon E., Shin-I T., Stuyver L. J., Thiel H. J., Viazov S., Weiner A. J., Widell A. ( 2005) Hepatology 42, 962– 973 [DOI] [PubMed] [Google Scholar]
- 6.Helle F., Goffard A., Morel V., Duverlie G., McKeating J., Keck Z. Y., Foung S., Penin F., Dubuisson J., Voisset C. ( 2007) J. Virol. 81, 8101– 8111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Op De Beeck A., Voisset C., Bartosch B., Ciczora Y., Cocquerel L., Keck Z., Foung S., Cosset F. L., Dubuisson J. ( 2004) J. Virol. 78, 2994– 3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barth H., Schafer C., Adah M. I., Zhang F., Linhardt R. J., Toyoda H., Kinoshita-Toyoda A., Toida T., Van Kuppevelt T. H., Depla E., Von Weizsacker F., Blum H. E., Baumert T. F. ( 2003) J. Biol. Chem. 278, 41003– 41012 [DOI] [PubMed] [Google Scholar]
- 9.Barth H., Schnober E. K., Zhang F., Linhardt R. J., Depla E., Boson B., Cosset F. L., Patel A. H., Blum H. E., Baumert T. F. ( 2006) J. Virol. 80, 10579– 10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint M., Maidens C., Loomis-Price L. D., Shotton C., Dubuisson J., Monk P., Higginbottom A., Levy S., McKeating J. A. ( 1999) J. Virol. 73, 6235– 6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A. J., Houghton M., Rosa D., Grandi G., Abrignani S. ( 1998) Science 282, 938– 941 [DOI] [PubMed] [Google Scholar]
- 12.Scarselli E., Ansuini H., Cerino R., Roccasecca R. M., Acali S., Filocamo G., Traboni C., Nicosia A., Cortese R., Vitelli A. ( 2002) EMBO J. 21, 5017– 5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartosch B., Cosset F. L. ( 2006) Virology 348, 1– 12 [DOI] [PubMed] [Google Scholar]
- 14.Helle F., Dubuisson J. ( 2008) Cell. Mol. Life Sci. 65, 100– 112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartosch B., Bukh J., Meunier J. C., Granier C., Engle R. E., Blackwelder W. C., Emerson S. U., Cosset F. L., Purcell R. H. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14199– 14204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keck Z. Y., Op De Beeck A., Hadlock K. G., Xia J., Li T. K., Dubuisson J., Foung S. K. ( 2004) J. Virol. 78, 9224– 9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owsianka A. M., Tarr A. W., Keck Z. Y., Li T. K., Witteveldt J., Adair R., Foung S. K., Ball J. K., Patel A. H. ( 2008) J. Gen. Virol. 89, 653– 659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perotti M., Mancini N., Diotti R. A., Tarr A. W., Ball J. K., Owsianka A., Adair R., Patel A. H., Clementi M., Burioni R. ( 2008) J. Virol. 82, 1047– 1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamataki Z., Grove J., Balfe P., McKeating J. A. ( 2008) Clin. Liver Dis. 12, 693– 712, x [DOI] [PubMed] [Google Scholar]
- 20.Shimizu Y. K., Hijikata M., Iwamoto A., Alter H. J., Purcell R. H., Yoshikura H. ( 1994) J. Virol. 68, 1494– 1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farci P., Shimoda A., Wong D., Cabezon T., De Gioannis D., Strazzera A., Shimizu Y., Shapiro M., Alter H. J., Purcell R. H. ( 1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15394– 15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penin F., Combet C., Germanidis G., Frainais P. O., Deléage G., Pawlotsky J. M. ( 2001) J. Virol. 75, 5703– 5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarr A. W., Owsianka A. M., Timms J. M., McClure C. P., Brown R. J., Hickling T. P., Pietschmann T., Bartenschlager R., Patel A. H., Ball J. K. ( 2006) Hepatology 43, 592– 601 [DOI] [PubMed] [Google Scholar]
- 24.Drummer H. E., Boo I., Poumbourios P. ( 2007) J. Gen. Virol. 88, 1144– 1148 [DOI] [PubMed] [Google Scholar]
- 25.Lavillette D., Pécheur E. I., Donot P., Fresquet J., Molle J., Corbau R., Dreux M., Penin F., Cosset F. L. ( 2007) J. Virol. 81, 8752– 8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drummer H. E., Maerz A., Poumbourios P. ( 2003) FEBS Lett. 546, 385– 390 [DOI] [PubMed] [Google Scholar]
- 27.Bartosch B., Dubuisson J., Cosset F. L. ( 2003) J. Exp. Med. 197, 633– 642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu M., Zhang J., Flint M., Logvinoff C., Cheng-Mayer C., Rice C. M., McKeating J. A. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7271– 7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindenbach B. D., Evans M. J., Syder A. J., Wölk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., Rice C. M. ( 2005) Science 309, 623– 626 [DOI] [PubMed] [Google Scholar]
- 30.Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., Mizokami M., Bartenschlager R., Liang T. J. ( 2005) Nat. Med. 11, 791– 796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D. R., Wieland S. F., Uprichard S. L., Wakita T., Chisari F. V. ( 2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9294– 9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartosch B., Vitelli A., Granier C., Goujon C., Dubuisson J., Pascale S., Scarselli E., Cortese R., Nicosia A., Cosset F. L. ( 2003) J. Biol. Chem. 278, 41624– 41630 [DOI] [PubMed] [Google Scholar]
- 33.Lavillette D., Bartosch B., Nourrisson D., Verney G., Cosset F. L., Penin F., Pécheur E. I. ( 2006) J. Biol. Chem. 281, 3909– 3917 [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi M., Bennett M. C., Bercot T., Singh I. R. ( 2006) J. Virol. 80, 1817– 1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tscherne D. M., Jones C. T., Evans M. J., Lindenbach B. D., McKeating J. A., Rice C. M. ( 2006) J. Virol. 80, 1734– 1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchard E., Belouzard S., Goueslain L., Wakita T., Dubuisson J., Wychowski C., Rouillé Y. ( 2006) J. Virol. 80, 6964– 6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koutsoudakis G., Kaul A., Steinmann E., Kallis S., Lohmann V., Pietschmann T., Bartenschlager R. ( 2006) J. Virol. 80, 5308– 5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomssen R., Bonk S., Propfe C., Heermann K. H., Köchel H. G., Uy A. ( 1992) Med. Microbiol. Immunol. 181, 293– 300 [DOI] [PubMed] [Google Scholar]
- 39.André P., Perlemuter G., Budkowska A., Bréchot C., Lotteau V. ( 2005) Semin. Liver Dis. 25, 93– 104 [DOI] [PubMed] [Google Scholar]
- 40.Nielsen S. U., Bassendine M. F., Burt A. D., Martin C., Pumeechockchai W., Toms G. L. ( 2006) J. Virol. 80, 2418– 2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agnello V., Abel G., Elfahal M., Knight G. B., Zhang Q. X. ( 1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12766– 12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aizaki H., Morikawa K., Fukasawa M., Hara H., Inoue Y., Tani H., Saito K., Nishijima M., Hanada K., Matsuura Y., Lai M. M., Miyamura T., Wakita T., Suzuki T. ( 2008) J. Virol. 82, 5715– 5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kapadia S. B., Barth H., Baumert T., McKeating J. A., Chisari F. V. ( 2007) J. Virol. 81, 374– 383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradley D., McCaustland K., Krawczynski K., Spelbring J., Humphrey C., Cook E. H. ( 1991) J. Med. Virol. 34, 206– 208 [DOI] [PubMed] [Google Scholar]
- 45.Pietschmann T., Kaul A., Koutsoudakis G., Shavinskaya A., Kallis S., Steinmann E., Abid K., Negro F., Dreux M., Cosset F. L., Bartenschlager R. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7408– 7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blight K. J., McKeating J. A., Rice C. M. ( 2002) J. Virol. 76, 13001– 13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Hoff M. J., Christoffels V. M., Labruyère W. T., Moorman A. F., Lamers W. H. ( 1995) Methods Mol. Biol. 48, 185– 197 [DOI] [PubMed] [Google Scholar]
- 48.Kärber G. ( 1951) Naunyn Schmiedebergs Arch. Exp. Pathol. Pharmakol. 212, 509– 521 [PubMed] [Google Scholar]
- 49.Spearman C. ( 1987) Am. J. Psychol. 100, 441– 471 [PubMed] [Google Scholar]
- 50.Lindenbach B. D., Meuleman P., Ploss A., Vanwolleghem T., Syder A. J., McKeating J. A., Lanford R. E., Feinstone S. M., Major M. E., Leroux-Roels G., Rice C. M. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3805– 3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooks M. J., Sasadeusz J. J., Tannock G. A. ( 2004) Curr. Opin. Pulm. Med. 10, 197– 203 [DOI] [PubMed] [Google Scholar]
- 52.Boriskin Y. S., Leneva I. A., Pécheur E. I., Polyak S. J. ( 2008) Curr. Med. Chem. 15, 997– 1005 [DOI] [PubMed] [Google Scholar]
- 53.Boriskin Y. S., Pécheur E. I., Polyak S. J. ( 2006) Virol. J. 3, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pécheur E. I., Lavillette D., Alcaras F., Molle J., Boriskin Y. S., Roberts M., Cosset F. L., Polyak S. J. ( 2007) Biochemistry 46, 6050– 6059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keck Z. Y., Li T. K., Xia J., Bartosch B., Cosset F. L., Dubuisson J., Foung S. K. ( 2005) J. Virol. 79, 13199– 13208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartosch B., Verney G., Dreux M., Donot P., Morice Y., Penin F., Pawlotsky J. M., Lavillette D., Cosset F. L. ( 2005) J. Virol. 79, 8217– 8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Hahn T., Lindenbach B. D., Boullier A., Quehenberger O., Paulson M., Rice C. M., McKeating J. A. ( 2006) Hepatology 43, 932– 942 [DOI] [PubMed] [Google Scholar]
- 58.André P., Komurian-Pradel F., Deforges S., Perret M., Berland J. L., Sodoyer M., Pol S., Bréchot C., Paranhos-Baccalà G., Lotteau V. ( 2002) J. Virol. 76, 6919– 6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diaz O., Delers F., Maynard M., Demignot S., Zoulim F., Chambaz J., Trépo C., Lotteau V., André P. ( 2006) J. Gen. Virol. 87, 2983– 2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maillard P., Huby T., Andréo U., Moreau M., Chapman J., Budkowska A. ( 2006) FASEB J. 20, 735– 737 [DOI] [PubMed] [Google Scholar]
- 61.Icard V., Diaz O., Scholtes C., Perrin-Cocon L., Ramière C., Bartenschlager R., Penin F., Lotteau V., André P. ( 2009) PLoS ONE 4, e4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meertens L., Bertaux C., Dragic T. ( 2006) J. Virol. 80, 11571– 11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kielian M., Rey F. A. ( 2006) Nat. Rev. Microbiol. 4, 67– 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stegmann T., Hoekstra D., Scherphof G., Wilschut J. ( 1986) J. Biol. Chem. 261, 10966– 10969 [PubMed] [Google Scholar]
- 65.Smit J. M., Bittman R., Wilschut J. ( 1999) J. Virol. 73, 8476– 8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gollins S. W., Porterfield J. S. ( 1986) J. Gen. Virol. 67, 157– 166 [DOI] [PubMed] [Google Scholar]
- 67.Chang K. S., Jiang J., Cai Z., Luo G. ( 2007) J. Virol. 81, 13783– 13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hadlock K. G., Lanford R. E., Perkins S., Rowe J., Yang Q., Levy S., Pileri P., Abrignani S., Foung S. K. ( 2000) J. Virol. 74, 10407– 10416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keck Z. Y., Xia J., Cai Z., Li T. K., Owsianka A. M., Patel A. H., Luo G., Foung S. K. ( 2007) J. Virol. 81, 1043– 1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keck Z. Y., Li T. K., Xia J., Gal-Tanamy M., Olson O., Li S. H., Patel A. H., Ball J. K., Lemon S. M., Foung S. K. ( 2008) J. Virol. 82, 6061– 6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Combet C., Garnier N., Charavay C., Grando D., Crisan D., Lopez J., Dehne-Garcia A., Geourjon C., Bettler E., Hulo C., Le Mercier P., Bartenschlager R., Diepolder H., Moradpour D., Pawlotsky J. M., Rice C. M., Trépo C., Penin F., Deléage G. ( 2007) Nucleic Acids Res. 35, D363– 366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teissier E., Pécheur E. I. ( 2007) Eur. Biophys. J. 36, 887– 899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Medigeshi G. R., Hirsch A. J., Streblow D. N., Nikolich-Zugich J., Nelson J. A. ( 2008) J. Virol. 82, 5212– 5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reyes-Del Valle J., Chávez-Salinas S., Medina F., Del Angel R. M. ( 2005) J. Virol. 79, 4557– 4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee C. J., Lin H. R., Liao C. L., Lin Y. L. ( 2008) J. Virol. 82, 6470– 6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Umashankar M., Sánchez-San Martín C., Liao M., Reilly B., Guo A., Taylor G., Kielian M. ( 2008) J. Virol. 82, 9245– 9253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stiasny K., Koessl C., Heinz F. X. ( 2003) J. Virol. 77, 7856– 7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Corver J., Ortiz A., Allison S. L., Schalich J., Heinz F. X., Wilschut J. ( 2000) Virology 269, 37– 46 [DOI] [PubMed] [Google Scholar]
- 79.Moesby L., Corver J., Erukulla R. K., Bittman R., Wilschut J. ( 1995) Biochemistry 34, 10319– 10324 [DOI] [PubMed] [Google Scholar]
- 80.Ahn A., Gibbons D. L., Kielian M. ( 2002) J. Virol. 76, 3267– 3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bertaux C., Dragic T. ( 2006) J. Virol. 80, 4940– 4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Voisset C., Lavie M., Helle F., Op De Beeck A., Bilheu A., Bertrand-Michel J., Tercé F., Cocquerel L., Wychowski C., Vu-Dac N., Dubuisson J. ( 2008) Cell. Microbiol. 10, 606– 617 [DOI] [PubMed] [Google Scholar]
- 83.Smith L. C., Pownall H. J., Gotto A. M., Jr. ( 1978) Annu. Rev. Biochem. 47, 751– 757 [DOI] [PubMed] [Google Scholar]
- 84.Molina S., Castet V., Fournier-Wirth C., Pichard-Garcia L., Avner R., Harats D., Roitelman J., Barbaras R., Graber P., Ghersa P., Smolarsky M., Funaro A., Malavasi F., Larrey D., Coste J., Fabre J. M., Sa-Cunha A., Maurel P. ( 2007) J. Hepatol. 46, 411– 419 [DOI] [PubMed] [Google Scholar]