Abstract
Upon massive DNA damage, hyperactivation of the nuclear enzyme poly(ADP-ribose) polymerase (PARP)-1 causes severe depletion of intracellular NAD and ATP pools as well as mitochondrial dysfunction. Thus far, the molecular mechanisms contributing to PARP-1-dependent impairment of mitochondrial functioning have not been identified. We found that degradation of the PARP-1 product poly(ADP-ribose) through the concerted actions of poly(ADP-ribose) glycohydrolase and NUDIX (nucleoside diphosphate-X) hydrolases leads to accumulation of AMP. The latter, in turn, inhibits the ADP/ATP translocator, prompting mitochondrial energy failure. For the first time, our findings identify NUDIX hydrolases as key enzymes involved in energy derangement during PARP-1 hyperactivity. Also, these data disclose unanticipated AMP-dependent impairment of mitochondrial exchange of adenine nucleotides, which can be of relevance to organelle functioning and disease pathogenesis.
Introduction
Poly(ADP-ribosylation) is a post-translational modification of proteins operated by poly(ADP-ribose) polymerases (PARPs),2 a family of NAD-consuming enzymes whose best characterized member is nuclear PARP-1 (1–4). PARP-1 activation upon DNA damage had been originally identified as a key event in signaling genotoxic stress. Intense investigation on poly(ADP-ribosylation) confirmed its role in DNA repair and also disclosed a fundamental role in regulation of chromatin dynamics (5) and cell death (3). In keeping with an involvement of PARP-1 in disease pathogenesis, inhibitors of its enzymatic activity afford cytoprotection in disparate experimental models of human disorders and are currently being evaluated in several clinical trials (6, 7).
As for the involvement of PARP-1 in cell demise, a great deal of attention has been directed at investigating the underpinning molecular mechanisms. In this regard, it has been repeatedly demonstrated that hyperactivation of PARP-1 following massive DNA damage leads to severe impairment of energy metabolism with almost complete depletion of NAD and ATP pools. Concomitant loss of the nucleotide pools allowed the formulation of the so-called “suicide hypothesis.” The latter posits that derangement of energy metabolism is due to excessive consumption of ATP to resynthesize NAD, which is continuously consumed by PARP-1 (8, 9). Indeed, eukaryotic cells, being unable to convert nicotinamide into nicotinic acid (which is transformed into NAD through the Preiss-Handler pathway), have developed an ATP-consuming “NAD rescue pathway” converting nicotinamide into NAD thanks to the enzymatic activities of nicotinamide phosphoribosyl transferase (NaPRT) and nicotinamide mononucleotide adenylyl transferase (NMNAT) (see Fig. 1A) (10, 11). The suicide metabolic pathway prompted by excessive PARP-1 activation has been conserved throughout evolution of multicellular organisms, probably because of its ability to eliminate cells that have undergone excessive genotoxic stress and are therefore at risk of neoplastic transformation. However, despite its apparent biochemical soundness and evolutionary logic, it remains to be established whether PARP-1-dependent derangement of energy metabolism is causally and entirely linked to NAD resynthesis.
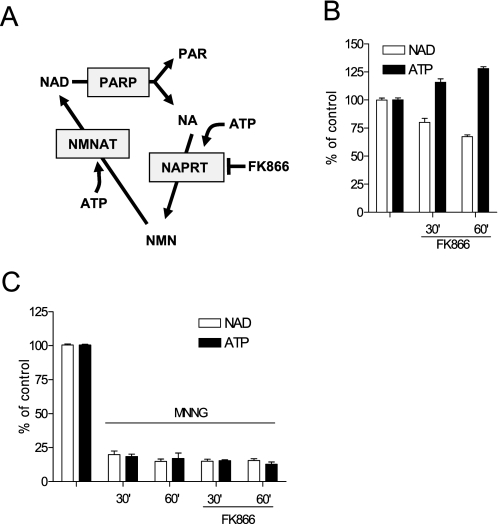
FIGURE 1.
NAD resynthesis is not causative to ATP loss during PARP-1 hyperactivation. A, the ATP-dependent NAD rescue pathway showing the site of action of FK866. B and C, effect of 100 μm FK866 on the contents of NAD and ATP in resting (B) or MNNG (100 μm)-challenged (C) cells. Bars represent the mean ± S.E. of at least three experiments conducted in duplicate.
We previously reported that HeLa cells exposed to the genotoxic agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) undergo massive NAD and ATP loss, which is dependent on PARP-1 hyperactivity (12). In that study, we reported the striking observation that upon PARP-1 activation, ATP contents decrease earlier in mitochondria than in cytosol and that the organelle respiration shifts from state 3 to state 4 (12). These findings are at apparent odds with the suicide hypothesis and, along with other contributions (13–15), point to mitochondria as a direct target of nuclear PARP-1 activity.
In the present study, we sought to elucidate the metabolic pathways and molecular mechanisms responsible for energy derangement during PARP-1 hyperactivation. We have identified a metabolic route through which the PARP-1 product poly(ADP-ribose) (PAR) is catabolized into AMP thanks to the concerted enzymatic activities of poly(ADP-ribose) glycohydrolase (PARG) and NUDIX (nucleoside diphosphate-X) hydrolases. AMP, in turn, abrogates mitochondrial ATP production by competing with ADP for its binding to the adenine nucleotide transporter (ANT). Our findings disclose an unprecedented metabolic link between PAR degradation and derangement of mitochondrial energy production.
EXPERIMENTAL PROCEDURES
Cells and Culture Conditions
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 2 mm glutamine, 10% fetal bovine serum, and antibiotics. Cultures were brought to 50–70% confluence and exposed to MNNG and other drugs directly dissolved in the culture media. Unless otherwise specified, MNNG and N-bis-(3-phenyl-propyl)9-oxo-fluorene-2,7-diamide (GPI-16552) were 100 μm, gallotannin was at 1 mm, and atractyloside (ATR) as at 10 μm.
Western Blotting and Immunocytochemistry
Western blotting and immunocytochemistry were performed as described previously (16). The anti-PAR antibodies (10H) were from ALEXIS Biochemicals (Vinci, Italy). Imaging was performed using a Nikon fluorescence microscope equipped with a CCD camera.
Measurement of Mitochondrial ATP Production
Mitochondrial ATP production was evaluated by means of mitochondria-targeted luciferase as described (12). mit-luciferase-transfected cells were exposed to MNNG and then to 0.001% Triton X-100 in the presence or absence of ADP, AMP, or atractyloside. Later on, luciferin was added, and light emission was evaluated with the TopCount-NXT luminometer (PerkinElmer Life Sciences). Recordings did not exceed 10 min because longer measurements are biased by factors other than intramitochondrial ATP (17, 18).
Nucleotide Measurement
Methods adopted for ATP and NAD quantitation have been previously described (12). AMP, ADP, and ADP-ribose (ADPR) were quantified in HCl cell extracts by high pressure liquid chromatography using a Supelco 25-cm column (5 μm), 0.1 m mobile phase KH2PO4, 1% acetonitrile, 10 mm tetrabutylammonium bromide, pH 6.9, and UV detection at 260 nm. NUDIX activity was measured in whole cell homogenates obtained with a Teflon Potter homogenizer and buffer A (50 mm Tris-HCl, pH 7.4, 225 mm mannitol, 75 mm saccharose, 1 mm phenylmethylsulfonyl fluoride, 10 μl of protease inhibitor mixture). Reactions were carried out in 100 μl of buffer containing 50 mm KH2PO4, pH 8.5, 1 mm dithiothreitol, 50 mm MgCl2, 100 μm ADPR, and an aliquot of cell extract. Samples were incubated at 37 °C for 30 min, and the reaction was stopped with an equal volume of HCl 0.5 m for AMP quantitation.
siRNA and RT-PCR
siRNA and RT-PCR were conducted as described (19, 20). siRNA was purchased from Dharmacon (Chicago, IL). The following primers were used for RT-PCR: NUDT-5, 5′-ACCGAAGTGTCGAGAGAGGA-3′ (sense) and 5′-CCAAGGTGTAGGGGTGAAGA-3′ (antisense); NUDT-9, 5′-TGGAAAAGGGATAGCAGTGG-3′ (sense) and 5′-TCTCTCCTGGATCCACCATC-3′ (antisense).
Mitochondrial Isolation and Handling
Mitochondria were isolated from cells using a glass/glass homogenizer in 500 μl of buffer A and centrifuged at 600 × g. Supernatants were centrifuged at 7000 × g to obtain the mitochondrial pellet, which was resuspended in respiration buffer (10 mm NaCl, 140 mm KCl, 2 mm MgSO4, 1 mm KH2PO4, 100 nm CaCl2, and 20 mm HEPES, pH 7, 2 mm succinate, 1 mm pyruvate) in the presence or absence of different compounds. Mitochondrial suspension was centrifuged to measure ADP uptake in the pellet (after two washes) and ATP output in the supernatant. ATP was measured by the ATPlight kit (PerkinElmer Life Sciences, Milan, Italy). [14C]ADP binding was performed as described (21).
Computational Methods
As for computational methods, molecules were drawn with Cerius-2 and optimized using Universal force-field version 1.2 (22) and the Smart Minimizer protocol of the Open Force Field module. ADP and AMP were considered with a formal charge of −3 and −2. Atomic charges were calculated using the semiempirical method AM1 as implemented in the MOPAC interface of Cerius-2. NUDT-5 in complex with magnesium ion and ADPR (23), and ANT in complex with ATR (24) were from the Protein Data Bank (PDB number 1OKC). Water and heteroatoms were removed with the exception of magnesium ion in NUDT-5. Hydrogen atoms were added, and the resulting hydrogen bond network was optimized using MolProbity (25). Docking calculations were carried out using the program Surflex-dock 2.1 (26). Conformational gap energies between bioactive conformations and the most close local minima of ADP and AMP were calculated using Universal force-field version 1.2 and the Smart Minimizer protocol of the Open Force Field module. During the calculations, the dielectric constant was set to a value of 80 to implicitly consider the water environment.
Statistical Analysis
Evaluation of significant differences among groups was performed using the Student's t test or analysis of variance followed by Tukey's w test.
RESULTS
NaPRT Inhibition Does Not Prevent PARP-1-dependent ATP Loss
Unequivocal evidence that excessive ATP consumption for NAD resynthesis is the mechanism responsible for energy failure during PARP-1 hyperactivation is lacking. Similarly, the amount of ATP consumed by the NAD rescue pathway under control conditions or when nicotinamide availability increases because of PARP-1-dependent NAD hydrolysis is unknown. Lack of information mainly depends on the fact that the NAD rescue pathway has never been pharmacologically or genetically targeted in cells undergoing PARP-1 activation.
To gather information of the energetic cost of NAD resynthesis from nicotinamide, we took advantage of (E)-N-[4-(1-benzoylpiperidin-4-yl) butyl]-3-(pyridin-3-yl) acrylamide) (FK866), a recently identified inhibitor of NaPRT (27). We reasoned that pharmacological inhibition of the first step of the NAD rescue pathway should prevent ATP consumption by NaPRT and, indirectly, by NMNAT (Fig. 1A). Under resting conditions, contents of NAD and ATP in HeLa cells were 8.2 ± 0.9 and 16 ± 2.3 nmol/mg of protein, respectively. HeLa cells exposed to FK866 underwent time-dependent depletion of NAD contents and, remarkably, concomitant accumulation of ATP (Fig. 1B). These findings on the one hand provide the biochemical proof that FK866 does inhibit the NAD rescue pathway, and on the other hand, they demonstrate that significant amounts of cellular NAD are constitutively consumed, in keeping with the increasing numbers of enzymes able to irreversibly hydrolyze NAD into different metabolites (10, 28). Of note, these data also allow us to unmask and quantitate constitutive utilization of ATP for NAD resynthesis. We then reasoned that if the suicide hypothesis holds true, inhibition of NAD resynthesis by FK866 should prevent, or at least reduce, PARP-1 hyperactivation-dependent ATP depletion (Fig. 1A). At variance with our hypothesis, however, FK866 did not affect ATP loss in cells undergoing PARP-1 hyperactivity (Fig. 1C). These data suggest that mechanisms in addition to or different from NAD resynthesis caused energy failure.
PARG Activity Contributes to PARP-1-dependent ATP Depletion
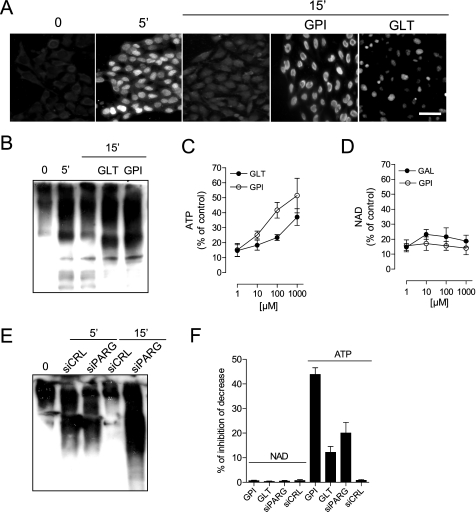
We next sought to determine whether PAR catabolism plays any role in energy failure during PARP-1 hyperactivity. It is well appreciated that, upon its formation, PAR is rapidly degraded by PARG into free monomers of ADPR (29). To understand the kinetics of PAR formation and degradation under our experimental conditions, we analyzed cellular PAR contents by immunohistochemistry and Western blotting at different times after PARP-1 activation. We found rapid synthesis and degradation of PAR in the cell nucleus of HeLa cells exposed to MNNG, consistent with nuclear localization of PARP-1 and prompt cytoplasm-to-nucleus shuttling of PARG (29). That the latter was responsible for PAR degradation was confirmed by evidence that polymer contents during the degradation phase were higher in cells exposed to PARG inhibitors such GPI-16552 or gallotannin (30) (Fig. 2, A and B). Next, to understand whether polymer degradation contributes to PARP-1-dependent energy derangement, we checked the effect of the two PARG inhibitors on NAD and ATP loss in cells undergoing MNNG-dependent PARP-1 activation. The two compounds did not affect NAD depletion but, strikingly, counteracted ATP loss (Fig. 2, C and D). To corroborate pharmacological evidence that PARG activity contributes to energy depletion, we next silenced PARG (20). In keeping with data obtained with PARG inhibitors, PARG siRNA impaired PAR degradation (Fig. 2E) and reduced ATP but not NAD depletion (Fig. 2F) upon PARP-1 activation. Altogether, these findings indicate a causal role of PAR degradation by PARG in PARP-1-dependent energy failure.
FIGURE 2.
PARG activity contributes to ATP but not NAD depletion during PARP-1 hyperactivation. A and B, immunocytochemical detection (A) or Western blotting (B) of PAR by means of an antibody directed against the polymer. The effect of MNNG on PAR formation at 5 min and of the two PARG inhibitors GPI-16552 (GPI) and gallotannin (GLT) on PAR degradation at 15 min is shown. Note that poly(ADP-ribosyl)ated proteins appear as a smear due to their highly increased molecular weight. In A, bar = 20 μm. C and D, GPI-16552 and gallotannin (GAL) inhibit ATP (C) but not NAD (D) loss in cells exposed 60 min to MNNG. E, effect of control siRNA (siCRL) and PARG siRNA (siPARG) on PAR contents in cells exposed 5 and 15 min to MNNG. F, effects of PARG inhibitors GPI-16552 and gallotannin as well as PARG siRNA on NAD and ATP contents in cells exposed 30 min to MNNG. Bars/points represent the mean ± S.E. of at least three experiments conducted in duplicate.
Transformation of ADPR into AMP by NUDIX Hydrolases Underlies PARP-1-dependent ATP Loss
Despite evidence for PARG activity, however, the intracellular contents of ADPR did not increase in HeLa cells exposed to MNNG (not shown). To reconcile these findings, we hypothesized that upon its formation, ADPR is rapidly catabolized. ADPR undergoes transformation into ATP by ADPR pyrophosphorylase (31, 32) and into AMP by the two ADPR pyrophosphatases NUDT-5 and -9 (members of the NUDIX hydrolase superfamily) (33). To unmask the possible ATP production by ADPR catabolism, we abrogated glycolytic and mitochondrial ATP synthesis by culturing HeLa cells in the absence of glucose and in the presence of oligomycin, rotenone, and antimycin. Of note, a PARP-1-dependent ATP increase occurred in these cells upon MNNG exposure (supplemental Fig. S1), revealing pyrophosphorylase-dependent ADPR transformation into ATP during PARP-1 activation. The latter also caused a 6-fold increase of cellular AMP contents (Fig. 3A). Given that AMP accumulation was reduced by PARP-1 and PARG inhibitors as well as by PARG siRNA (Fig. 3A), these findings suggest that once ADPR is formed through the concerted action of PARP-1 and PARG, it is also promptly converted into AMP. Prior work by Ivana Scovassi and co-workers (34) is in keeping with these findings.
FIGURE 3.
Origin and effect of AMP accumulation during PARP-1 activation. A, time-dependent AMP accumulation in cells exposed to MNNG and effect of pharmacological inhibition of PARP-1 (phenanthridinone (PHE) 30 μm and N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-N,N-dimethylacetamide (PJ34) 10 μm), PARG (GPI-16552, 100 μm), or PARG siRNA. B and C, effect of LY294002, chelerythrine, cumaric acid, and flavone on cellular NUDIX activity (B) and MNNG-induced ATP depletion (C). D, the capacity of a set of compounds to prevent MNNG-induced ATP depletion correlates with their ability to inhibit NUDIX activity or prevent AMP increase. Bars/points represent the mean ± S.E. of at least three experiments conducted in duplicate.
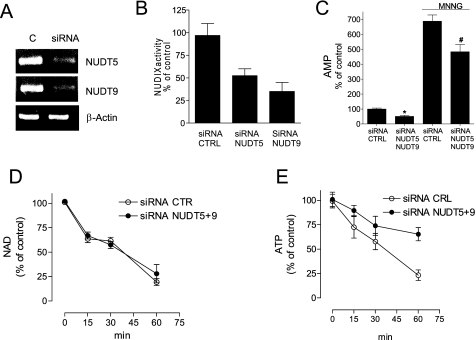
To understand whether NUDIX hydrolases have a causative role in PARP-1-depenent energy depletion, we next screened an in-house library of compounds and identified LY294002, chelerythrine, p-cumaric acid, and flavone as inhibitors of NUDIX activity in HeLa extracts (Fig. 3B and supplemental Fig. S2). Remarkably, the four compounds inhibited AMP increase as well as ATP but not NAD depletion in cells undergoing PARP-1 activation (Fig. 3C and supplemental Fig. S2). The ATP rescue effect correlated with the ability of the compounds to suppress NUDIX activity and AMP accumulation (Fig. 3D). To rule out the possibility that LY294002 prevented ATP depletion through its well known ability to inhibit phosphoinositide-3 kinase, we tested the effect of a more potent kinase inhibitor, wortmannin, on PARP-1-dependent energy failure. Evidence that wortmannin did not affect PARP-1-induced ATP depletion (supplemental Fig. S3) indicates that the effects of LY294002 were not due to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Accordingly, LY294002 nicely fits the catalytic site of NUDT-5 (supplemental Fig. S3). We next silenced NUDT-5 and -9 by means of siRNA. Silencing of the two NUDIX hydrolases (Fig. 4A) reduced cellular ADPR pyrophosphatase activity (Fig. 4B) and AMP contents in resting and MNNG-challenged cells (Fig. 4C). In keeping with pharmacological inhibition of NUDIX activity, NUDT-5 and -9 silencing reduced PARP-1-dependent ATP but not NAD loss (Fig. 4, D and E). Overall, these data indicate that NUDIX hydrolase-dependent ADPR conversion into AMP contributes to energy derangement during hyperpoly(ADP-ribosylation).
FIGURE 4.
Silencing of NUDT-5 and -9 reduces AMP accumulation as well as ATP but not NAD depletion during PARP-1 activation. A–C, effect of NUDT-5 and -9 siRNA on their transcript levels (A), cellular NUDIX activity (B), and AMP content in cells under resting conditions or exposed 30 min to MNNG (C). CTRL, control. D and E, impact of NUDT-5 and -9 siRNA on NAD (D) or ATP (E) depletion in cells exposed to MNNG. Bars/points represent the mean ± S.E. of at least three experiments conducted in duplicate. CTR, control. In A, a representative RT-PCR is shown. *, p < 0.05 versus siRNA control; #, p < 0.05 versus siRNA control + MNNG, analysis of variance plus Tukey's post hoc test.
AMP Binds Mitochondrial ANT and Impairs Its Functioning
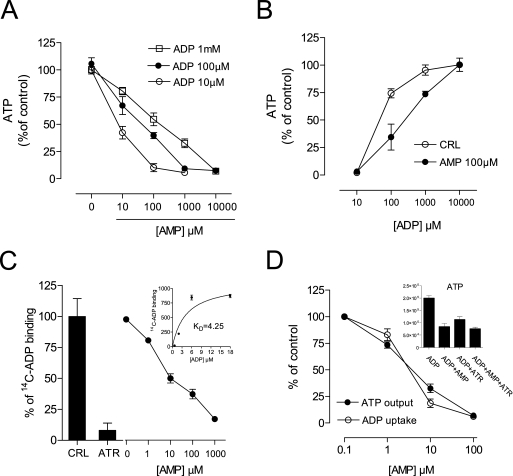
In cells undergoing PARP-1 hyperactivation, NAD depletion abrogates glycolytic ATP production at the level of the NAD-dependent enzyme glyceraldehyde dehydrogenase (35). Hence, we reasoned that the detrimental effects of AMP on energy dynamics should be ascribed to an effect of the nucleotide on mitochondria. It is worth noting that early mitochondrial dysfunction has been repeatedly reported during PARP-1 activation (12–15, 36). To our knowledge, the impact of PAR degradation products on mitochondria has not been investigated. The impact of AMP on ATP production by isolated mitochondria was therefore investigated. Somehow unexpectedly, AMP competitively inhibited ADP-dependent ATP production by isolated state 3 mitochondria (Fig. 5, A and B). Given that cytosolic ADP is exchanged 1:1 with intramitochondrial ATP by the ANT (37, 38), we next focused on ANT as a possible target of AMP. We found that AMP inhibited binding of [14C]ADP to ANT (Fig. 5C), as well as mitochondrial ADP uptake and ATP extrusion (Fig. 5D). However, AMP content increased neither in the matrix of isolated mitochondria exposed to the nucleotide (up to 10 mm) nor in the mitochondria of cells undergoing PARP-1 activation (not shown). These findings, along with evidence that suppression of mitochondrial ATP production by the prototypical ANT inhibitor atractyloside was not additive to that caused by AMP (Fig. 5D, inset), suggest that AMP impaired mitochondrial ATP production by competing with the binding of ADP to ANT.
FIGURE 5.
AMP impairs ANT functioning by competing with the ADP binding site. A, effect of AMP on ADP-dependent ATP production by isolated mitochondria. B, effect of increasing ADP concentrations on AMP-dependent inhibition of mitochondrial ATP production. CRL, control. C, effect of ATR (100 μm) or AMP on binding of [14C]ADP to mitochondria (inset, saturation curve). D, effect of AMP on mitochondrial ADP uptake and ATP output; inset, the effect of AMP is not additive to that of ATR. Bars/points represent the mean ± S.E. of at least three experiments conducted in duplicate.
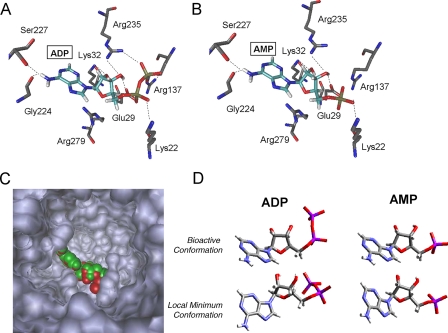
Crystal structure analysis of the translocator reveals that the protein is composed of six helices forming a barrel with a deep conic-shaped cavity (24). By means of molecular mechanic modeling, we evaluated the docking properties of AMP to ANT and found that AMP reaches the bottom of the gorge and docks almost identically to ADP (Fig. 6, A–C). ADP binding is stabilized by interactions among the phosphate moieties and the positively charged residues Lys-22, Lys-32, Arg-137, and Arg-235. Residues stabilizing AMP binding were identical, with the exception of bonds with Arg-137 and Arg-235 involved in the binding of the β-phosphate of ADP. In keeping with this, the docking scores (which are an index of the binding affinity) of ADP and AMP were 7.99 and 7.73 (pKd units), respectively. The question then arises as to why AMP was not transported inside mitochondria. According to the “transition fit theory” (38, 39), the catalytic energy for ADP uptake stems from ANT conformational changes. During these changes, ANT undergoes a transition state in which the nucleotide binding site assumes the best fit to the substrate. Conversely, the fit is poor both in the external (c-state) and in the internal (m-state) ground states. The maximum binding energy released in the transition state provides enough catalytic energy to enable large conformational changes of the carrier protein associated with the transport. During the journey of ADP down through the ANT cavity, rigid interactions with specific residues induce high conformational strain to the bioactive conformation of the nucleotide. We calculated the conformational energy acquired by ADP and AMP once inside the ANT cavity and found that bioactive conformation of ANT-bound AMP (ΔE = +3.25 kcal/mol) was energetically favored when compared with docked ADP (ΔE = +7.60 kcal/mol) (Fig. 6D). Hence, in the case of ADP, ANT undergoes a transition state in which the carrier optimizes the fit to the substrate, thereby lowering the strain energy of the nucleotide and acquiring the internal ground state (m-state). Conversely, in the case of AMP, the energy strain induced by the carrier to the nucleotide is less than the half of that of ADP. We speculate, therefore, that AMP binding to the external grown state (c-state) of ANT is sufficiently stable and does not prompt conformational rearrangements. Our data provide the biophysical basis as to why AMP is not transported inside mitochondria.
FIGURE 6.
Molecular mechanic analysis of AMP and ADP binding to ANT. A and B, docking pose of ADP (A) or AMP (B) bound to ANT. Interactions with different amino acidic residues are shown. C, visualization of ADP (red) and AMP (green) bound at the bottom of the ANT cavity. D, comparison of the bioactive (i.e. bound to ANT) and local minima conformations of ADP and AMP. ADP and not AMP is twisted and energetically strained when bound to ATN.
Competition between AMP and ADP for binding to ANT can therefore explain AMP-dependent inhibition of ATP production in both isolated mitochondria as well as cells undergoing PARP-1 hyperactivation. Further strengthening the concept of nucleotide competition, intracellular ADP contents are reduced during PARP-1 hyperactivity (12), a condition clearly promoting AMP-dependent impairment of ANT functioning. Given the competitive nature of ADP and AMP binding to ANT, the ratio between the intracellular contents of the two nucleotides should be the key determinant for ADP uptake and ATP production. We therefore measured the ADP/AMP ratio in cells under control conditions or undergoing PARP-1 hyperactivation and found that in cells exposed for 30 min to MNNG, the ADP/AMP ratio dramatically reduced to 0.7 ± 0.25 from its constitutive value of 9.9 ± 2.2. This is a more than 14-fold reduction of the ADP/AMP ratio, which, according to our findings, is expected to severely impair mitochondrial adenine nucleotide exchange and energy production.
Inverted ADP/AMP Ratio Is Causative to ANT Inhibition and Impairment of Mitochondrial ATP Production during PARP-1 Hyperactivity
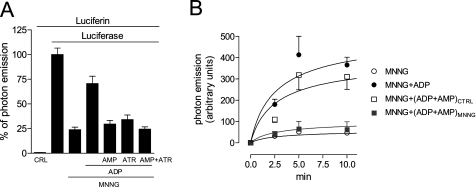
To confirm our hypothesis, we next evaluated the effects of externally added ADP and AMP on mitochondrial ATP production by permeabilized cells transfected with a mitochondrially targeted luciferase, a tool adopted to quantify real-time, intramitochondrial ATP (17, 18). In conditions in which permeabilization did not affect mitochondrial membrane potential and respiration (supplemental Fig. S4), PARP-1 activation by MNNG severely reduced photon emission. Of note, light emission readily recovered when 100 μm ADP was added to the culture medium but not when 100 μm AMP or 10 μm atractyloside was also present. The AMP and atractyloside effects were not additive (Fig. 7A), again indicating that AMP and the ANT blocker were acting on the same binding site. Most importantly, in cells challenged with MNNG, light emission recovered when ADP and AMP were concomitantly added at concentrations measured in control cells but not at those found in cells undergoing PARP-1 activation (Fig. 7B). Overall, these novel findings indicate that mitochondrial energy failure triggered by PARP-1 can be readily recovered by ADP and that AMP abrogate this rescue by inhibiting ANT activity.
FIGURE 7.
Effect of ADP and AMP on ATP production by in situ mitochondria. A, effect of externally added ADP or AMP (both at 100 μm) or ATR (10 μm) on light emission by permeabilized mit-luciferase-transfected cells previously exposed to MNNG. MNNG was added at time 0, compounds were added together with permeabilization at time 15 min, luciferin was added at time 45 min, and light emission was recorded at time 50 min. CRL, control. B, effect of ADP and AMP concomitantly added at concentrations found in cells under control conditions (ADP+AMPCTRL) or challenged for 30 min with MNNG (ADP+AMPMNNG). MNNG was added at time 0, compounds were added together with permeabilization at time 30 min, luciferin was added at time 35 min, and light emission was recorded 2.5, 5, and 10 min after luciferin addition. Bars/points represent the mean ± S.E. of at least three experiments conducted in duplicate. In B, an experiment representative of three is shown.
DISCUSSION
A great deal of effort has been directed at understanding the molecular mechanisms contributing to NAD and ATP loss once PARP-1 gets hyperactivated. Such knowledge could shed light on still obscure metabolic pathways in eukaryotic cells and also clarify how this energetic catastrophe contributes to mitochondrial release of proapoptotic factors of relevance to cell death and disease pathogenesis (36). According to the suicide hypothesis, energy failure is caused by the utilization of ATP for the resynthesis of NAD through the rescue pathway originating from nicotinamide (8). In principle, this metabolic network could certainly represent an energy sink of relevance to energy homeostasis. It should be stressed, however, that when additional metabolic parameters are taken into account, the strength of the theory significantly weakens. Indeed, if one considers the metabolic potential of mitochondrial ATP production, it is hard to believe that a single enzyme, even if hyperactivated, can consume all the ATP made available by the organelles. One should also consider that, according to the suicide hypothesis, energy failure is not directly caused by PARP-1, but putatively due to increased flux through the ATP-consuming enzymes NaPRT and NMNAT, the activation of which is still to be demonstrated. To our knowledge, there is no enzyme or single metabolic pathway able to prompt complete cellular energy failure by simple ATP consumption. The application of metabolic control analysis to mitochondria allowed the identification of several regulatory steps at the level of respiration and ATP production endowed with a significant degree of elasticity and thereby warranting increased energy production on demand (39). On this basis, we speculate that the possibility that PARP-1-dependent energy dysfunction is not due to excessive ATP utilization but rather to impairment of ATP production is more conceivable. In keeping with this, we report that ongoing NAD resynthesis is indeed responsible for considerable ATP consumption under resting conditions, but it is not the primary cause of ATP loss when PARP-1 is hyperactivated. By showing that pharmacological or genetic approaches can prevent PARP-1-dependent ATP but not NAD loss, we provide evidence that NAD depletion is not invariantly linked to that of ATP, further weakening the soundness of the suicide hypothesis.
Our data point to PAR degradation into ADPR through PARG, and ensuing degradation of ADPR into AMP by NUDIX hydrolases, as a key metabolic pathway responsible for PARP-1-induced mitochondrial energy failure. The present study is in keeping with prior work demonstrating that alteration of PAR metabolism causes mitochondrial dysfunction (12–15, 36). That PARG contributes to energy failure is in good agreement with studies showing that inhibition of PARG activity affords cytoprotection in different models of PARP-1-dependent cell death (40–43). In apparent contrast with this, however, PARG deletion prompts cytotoxicity (44–47). These findings might be reconciled considering that cell death is triggered each time homeostasis of PAR metabolism is deranged, either because of PAR degradation block and ensuing accumulation or because of excessive polymer production and massive formation of degradation products.
We identify NUDIX hydrolases as new players in the mechanisms responsible for PARP-1-induced energy failure. In eukaryotic cells, members of the NUDIX family are ancestral enzymes of bacterial origin that afford cytoprotection by degrading xenobiotics or toxic compounds originating from cell metabolism. However, our study provides evidence that specific members of the NUDIX hydrolase superfamily are also causally involved in a cell death pathway originating from PAR. We also identify pharmacological inhibitors of NUDIX hydrolases, which can be harnessed to understand the role of the enzymes in cell metabolism and disease pathogenesis. It is likely that NUDT-5 and -9 constitutively prevent non-enzymatic ADPR-dependent direct protein glycation (48). In conditions of massive PAR formation, however, metabolism of ADPR into AMP by NUDT-5 and -9 triggers mitochondrial impairment and energetic catastrophe. Indeed, when we counteracted AMP accumulation by pharmacological inhibition of NUDIX activity or genetic suppression of NUDT-5 and -9, PARP-1-dependent ATP loss was reduced. Correlation of the ATP-sparing effect with the degree of inhibition of both NUDIX activity and AMP accumulation underscores the causal role of NUDIX-dependent AMP formation in PARP-1-induced energy failure.
ANT is a well characterized protein present in the internal mitochondrial membrane delegated to the import of ADP and extrusion of ATP, and therefore, critical for energetic metabolism. Prior work demonstrates that ANT has a strict selectivity for ADP and ATP, being unable to transport other endogenous nucleotides (37). Adopting different methodological approaches, we report that ANT also binds AMP, which is not transported but still able to prevent mitochondrial ADP uptake and ATP production. Comparison of AMP and ADP binding to ANT by means of molecular mechanic allowed us to demonstrate that energetic conformation of ANT-bound AMP is significantly lower than that of ANT-bound ADP. These data disclose the biophysical mechanism by which ANT does not transport AMP and, along with those showing a good binding affinity of AMP to ANT, clarify why AMP impairs ANT activity. Thus, transport selectivity of ANT does not correlate with its binding ability, a concept with remarkable metabolic consequences. Our findings are in good agreement with the transition fit theory of ANT functioning proposed by Klingenberg (49) and represent the first demonstration that accumulation of an endogenous compound triggers mitochondrial dysfunction by inhibiting ANT. This notion might have important implications in bioenergetics, cell metabolism, and pathophysiology.
Evidence that hyperactivation of PARP-1 triggers mitochondrial dysfunction through an AMP-dependent impairment of ANT functioning also has important implications when considered on an evolutionary perspective. According to the “endosymbiont theory” (50), eukaryotic cells evolved when fermentative cells acquired archaebacteria as a permanent source of intracellular ATP. It is conceivable that fundamental rearrangements of the metabolic pathways of the cell and the protomitochondrion occurred at that time as a sort of trade off. Likely, a high grade of binding selectivity of ANT able to protect the transporter functioning and energy homeostasis had been a critical term of the agreement. This homeostatic condition, however, collapses upon massive formation and degradation of PAR. Indeed, intracellular nucleotide imbalance brought about by hyperpoly(ADP-ribosylation) prompts a metabolic state incompatible with oxidative phosphorylation and mitochondrial energy production. This metabolic condition leads to a sort of mitochondrial uncoupling, and accordingly, increase of mitochondrial membrane potential during PARP-1 activation has been reported (12). Findings obtained by measuring in situ mitochondrial ATP production demonstrate that PARP-1-induced energy failure is not irreversible but readily rescued by increasing extramitochondrial ADP concentration. On the one hand, this corroborates the consistency of the “nucleotide imbalance hypothesis”; on the other hand, this suggests innovative cytoprotective strategies.
In conclusion, we report that upon PARP-1 hyperactivity, PAR degradation leads to impairment of ANT functioning. This, in turn, contributes to energy failure in cells facing glycolytic block because of NAD depletion (35). Overall, these data further our understanding of both poly(ADP-ribosylation) and mitochondrial functioning, pointing for the first time to NUDIX hydrolases as enzymes of relevance to the pathogenesis of disorders characterized by PARP-1 hyperactivation or bioenergetic derangements. This study also highlights the unexpected pathogenetic relevance of AMP metabolism to mitochondrial dysfunction and related human disorders.
Supplementary Material
Acknowledgments
We thank Prof. R. Rizzuto for providing the mit-luciferase plasmid.
This study was supported by grants from the Italian Ministry of University and Research PRIN 2007, Fondazione Italiana Sclerosi Multipla, and Ente Cassa di Risparmio di Firenze.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- PAR
- poly(ADP-ribose)
- PARP
- poly(ADP-ribose) polymerase
- PARG
- poly(ADP-ribose) glycohydrolase
- ADPR
- ADP-ribose
- ANT
- adenine nucleotide transporter
- ATR
- atractyloside
- FK866
- (E)-N-[4-(1-benzoylpiperidin-4-yl) butyl]-3-(pyridin-3-yl) acrylamide)
- GPI-16552
- N-bis-(3-phenyl-propyl)9-oxo-fluorene-2,7-diamide
- MNNG
- N-methyl-N′-nitro-N-nitrosoguanidine
- NaPRT
- nicotinamide phosphoribosyl transferase
- NMNAT
- nicotinamide mononucleotide adenylyl transferase
- NUDIX
- nucleoside diposphate-X
- RT-PCR
- reverse transcription-PCR
- siRNA
- small interfering RNA.
REFERENCES
- 1.D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. ( 1999) Biochem. J. 342, 249– 268 [PMC free article] [PubMed] [Google Scholar]
- 2.Amé J. C., Spenlehauer C., de Murcia G. ( 2004) BioEssays 26, 882– 893 [DOI] [PubMed] [Google Scholar]
- 3.Schreiber V., Dantzer F., Ame J. C., de Murcia G. ( 2006) Nat. Rev. Mol. Cell. Biol. 7, 517– 528 [DOI] [PubMed] [Google Scholar]
- 4.Hassa P. O., Haenni S. S., Elser M., Hottiger M. O. ( 2006) Microbiol. Mol. Biol. Rev. 70, 789– 829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraus W. L. ( 2008) Curr. Opin. Cell Biol. 20, 294– 302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Virág L., Szabó C. ( 2002) Pharmacol. Rev. 54, 375– 429 [DOI] [PubMed] [Google Scholar]
- 7.Jagtap P., Szabó C. ( 2005) Nat. Rev. Drug Discov. 4, 421– 440 [DOI] [PubMed] [Google Scholar]
- 8.Berger N. A. ( 1985) Radiat. Res. 101, 4– 15 [PubMed] [Google Scholar]
- 9.Chiarugi A. ( 2002) Trends Pharmacol. Sci 23, 122– 129 [DOI] [PubMed] [Google Scholar]
- 10.Berger F., Ramírez-Hernández M. H., Ziegler M. ( 2004) Trends Biochem. Sci. 29, 111– 118 [DOI] [PubMed] [Google Scholar]
- 11.Bogan K. L., Brenner C. ( 2008) Annu. Rev. Nutr. 28, 115– 130 [DOI] [PubMed] [Google Scholar]
- 12.Cipriani G., Rapizzi E., Vannacci A., Rizzuto R., Moroni F., Chiarugi A. ( 2005) J. Biol. Chem. 280, 17227– 17234 [DOI] [PubMed] [Google Scholar]
- 13.Yu S. W., Wang H., Poitras M. F., Coombs C., Bowers W. J., Federoff H. J., Poirier G. G., Dawson T. M., Dawson V. L. ( 2002) Science 297, 259– 263 [DOI] [PubMed] [Google Scholar]
- 14.Alano C. C., Ying W., Swanson R. A. ( 2004) J. Biol. Chem. 279, 18895– 18902 [DOI] [PubMed] [Google Scholar]
- 15.Yu S. W., Andrabi S. A., Wang H., Kim N. S., Poirier G. G., Dawson T. M., Dawson V. L. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18314– 18319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rapizzi E., Pinton P., Szabadkai G., Wieckowski M. R., Vandecasteele G., Baird G., Tuft R. A., Fogarty K. E., Rizzuto R. ( 2002) J. Cell Biol. 159, 613– 624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy H. J., Rafiq I., Pouli A. E., Rutter G. A. ( 1999) Biochem. J. 342, 275– 280 [PMC free article] [PubMed] [Google Scholar]
- 18.Jouaville L. S., Pinton P., Bastianutto C., Rutter G. A., Rizzuto R. ( 1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13807– 13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rapizzi E., Fossati S., Moroni F., Chiarugi A. ( 2004) Mol. Pharmacol. 66, 890– 898 [DOI] [PubMed] [Google Scholar]
- 20.Cohausz O., Blenn C., Malanga M., Althaus F. R. ( 2008) Cell Mol. Life Sci. 65, 644– 655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weidemann M. J., Erdelt H., Klingenberg M. ( 1970) Eur. J. Biochem. 16, 313– 335 [DOI] [PubMed] [Google Scholar]
- 22.Rappe A. K., Casewit C. J., Colwell K. S., Goddard W. A., Skiff W. M. ( 1992) J. Am. Chem. Soc. 114, 10024– 10035 [Google Scholar]
- 23.Zha M., Zhong C., Peng Y., Hu H., Ding J. ( 2006) J. Mol. Biol. 364, 1021– 1033 [DOI] [PubMed] [Google Scholar]
- 24.Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trézéguet V., Lauquin G. J., Brandolin G. ( 2003) Nature 426, 39– 44 [DOI] [PubMed] [Google Scholar]
- 25.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. ( 2007) Nucleic Acids Res. 35, W375– W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain A. N. ( 2007) J. Comput. Aided Mol. Des 21, 281– 306 [DOI] [PubMed] [Google Scholar]
- 27.Hasmann M., Schemainda I. ( 2003) Cancer Res. 63, 7436– 7442 [PubMed] [Google Scholar]
- 28.Sauve A. A. ( 2008) J. Pharmacol. Exp. Ther. 324, 883– 893 [DOI] [PubMed] [Google Scholar]
- 29.Davidovic L., Vodenicharov M., Affar E. B., Poirier G. G. ( 2001) Exp. Cell Res. 268, 7– 13 [DOI] [PubMed] [Google Scholar]
- 30.Zhang J., Li J. H. ( 2002) Drugs Future 27, 371– 383 [Google Scholar]
- 31.Tanuma S. ( 1989) Biochem. Biophys. Res. Commun. 163, 1047– 1055 [DOI] [PubMed] [Google Scholar]
- 32.Oei S. L., Ziegler M. ( 2000) J. Biol. Chem. 275, 23234– 23239 [DOI] [PubMed] [Google Scholar]
- 33.McLennan A. G. ( 2006) Cell Mol. Life Sci. 63, 123– 143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi L., Denegri M., Torti M., Poirier G. G., Ivana Scovassi A. ( 2002) Biochimie 84, 1229– 1235 [DOI] [PubMed] [Google Scholar]
- 35.Ying W., Garnier P., Swanson R. A. ( 2003) Biochem. Biophys. Res. Commun. 308, 809– 813 [DOI] [PubMed] [Google Scholar]
- 36.Hong S. J., Dawson T. M., Dawson V. L. ( 2004) Trends Pharmacol. Sci. 25, 259– 264 [DOI] [PubMed] [Google Scholar]
- 37.Nury H., Dahout-Gonzalez C., Trezeguet V., Lauquin G. J., Brandolin G., Pebay-Peyroula E. ( 2006) Annu. Rev. Biochem. 75, 713– 741 [DOI] [PubMed] [Google Scholar]
- 38.Klingenberg M. ( 2008) Biochim. Biophys. Acta 1778, 1978– 2021 [DOI] [PubMed] [Google Scholar]
- 39.Murphy M. P. ( 2001) Biochim. Biophys. Acta 1504, 1– 11 [DOI] [PubMed] [Google Scholar]
- 40.Ying W., Sevigny M. B., Chen Y., Swanson R. A. ( 2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12227– 12232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu X. C., Massuda E., Lin Q., Li W., Li J. H., Zhang J. ( 2003) Brain Res. 978, 99– 103 [DOI] [PubMed] [Google Scholar]
- 42.Cuzzocrea S., Di Paola R., Mazzon E., Cortes U., Genovese T., Muià C., Li W., Xu W., Li J. H., Zhang J., Wang Z. Q. ( 2005) FASEB J. 19, 558– 566 [DOI] [PubMed] [Google Scholar]
- 43.Formentini L., Arapistas P., Pittelli M., Jacomelli M., Pitozzi V., Menichetti S., Romani A., Giovannelli L., Moroni F., Chiarugi A. ( 2008) Br. J. Pharmacol. 155, 1235– 1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cortes U., Tong W. M., Coyle D. L., Meyer-Ficca M. L., Meyer R. G., Petrilli V., Herceg Z., Jacobson E. L., Jacobson M. K., Wang Z. Q. ( 2004) Mol. Cell. Biol. 24, 7163– 7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cozzi A., Cipriani G., Fossati S., Faraco G., Formentini L., Min W., Cortes U., Wang Z. Q., Moroni F., Chiarugi A. ( 2006) J. Cereb. Blood Flow Metab. 26, 684– 695 [DOI] [PubMed] [Google Scholar]
- 46.Koh D. W., Lawler A. M., Poitras M. F., Sasaki M., Wattler S., Nehls M. C., Stöger T., Poirier G. G., Dawson V. L., Dawson T. M. ( 2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17699– 17704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrabi S. A., Kim N. S., Yu S. W., Wang H., Koh D. W., Sasaki M., Klaus J. A., Otsuka T., Zhang Z., Koehler R. C., Hurn P. D., Poirier G. G., Dawson V. L., Dawson T. M. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18308– 18313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobson E. L., Cervantes-Laurean D., Jacobson M. K. ( 1994) Mol. Cell Biochem. 138, 207– 212 [DOI] [PubMed] [Google Scholar]
- 49.Klingenberg M. ( 2007) Biochimie 89, 1042– 1048 [DOI] [PubMed] [Google Scholar]
- 50.Kroemer G. ( 1997) Cell Death Differ. 4, 443– 456 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.