Abstract
Palmitoylation of the yeast vacuolar protein Vac8 is important for its role in membrane-mediated events such as vacuole fusion. It has been established both in vivo and in vitro that Vac8 is palmitoylated by the Asp-His-His-Cys (DHHC) protein Pfa3. However, the determinants of Vac8 critical for recognition by Pfa3 have yet to be elucidated. This is of particular importance because of the lack of a consensus sequence for palmitoylation. Here we show that Pfa3 was capable of palmitoylating each of the three N-terminal cysteines of Vac8 and that this reaction was most efficient when Vac8 is N-myristoylated. Additionally, when we analyzed the Src homology 4 (SH4) domain of Vac8 independent of the rest of the protein, palmitoylation by Pfa3 still occurred. However, the specificity of palmitoylation seen for the full-length protein was lost, and the SH4 domain was palmitoylated by all five of the yeast DHHC proteins tested. These data suggested that a region of the protein C-terminal to the SH4 domain was important for conferring specificity of palmitoylation. This was confirmed by use of a chimeric protein in which the SH4 domain of Vac8 was swapped for that of Meh1, another palmitoylated and N-myristoylated protein in yeast. In this case we saw specificity mimic that of wild type Vac8. Competition experiments revealed that the 11th armadillo repeat of Vac8 is an important element for recognition by Pfa3. This demonstrates that regions distant from the palmitoylated cysteines are important for recognition by DHHC proteins.
Introduction
S-Palmitoylation (hereafter referred to as palmitoylation) is a widespread post-translational modification in which the fatty acid palmitate (16:0) is attached to a cysteine residue via a thioester linkage. Palmitate can be released from a cysteine by hydrolysis of the thioester linkage; thus palmitoylation is a reversible modification. Numerous eukaryotic proteins are palmitoylated, and substrates include both peripheral and transmembrane domain proteins. The functional consequences of palmitoylation are diverse. Membrane association, protein stability, and protein trafficking are among the properties that can be modulated by the palmitoylation status of a protein (reviewed in 1, 2).
The enzymes responsible for palmitoylation have only recently been identified. The first protein acyltransferases (PATs)3 discovered were Erf2 and Akr1 in yeast, which palmitoylate Ras2 and Yck2, respectively (3, 4). Both Erf2 and Akr1 are transmembrane proteins with an Asp-His-(His/Tyr)-Cys (DHHC) motif embedded in a cysteine-rich domain (CRD). Homology with this domain has identified five additional DHHC proteins in Saccharomyces cerevisiae. These seven proteins account for the bulk of palmitoylating activity in the cell (5).
To date 23 DHHC proteins have been identified in mammals (6). As the field has progressed DHHC proteins have been linked to the regulation of neurotransmission (7–9), nitric oxide release (10), and cell-cell contacts (11). Palmitoylation of Huntingtin by HIP14 (DHHC17) plays a protective role in inhibiting aggregation of the Huntingtin protein (12). Mutations in DHHC9 and DHHC15 are associated with forms of X-linked mental retardation (13, 14). Accordingly, understanding the structure, function, and mechanism of this large family of enzymes is of obvious interest.
An important goal is to understand what features of substrates DHHC proteins recognize. A subclass of palmitoylated proteins consists of peripheral proteins that are modified at cysteines near a site of N-myristoylation. N-Myristoylation is the addition of a myristate fatty acid (14:0) to an N-terminal glycine residue via an irreversible amide linkage (15, 16). N-Myristoylated proteins that are palmitoylated include members of the Gαi family of G proteins and most nonreceptor tyrosine kinases (17). The S. cerevisiae protein Vac8 is also palmitoylated at cysteines that are just downstream of its N-myristoylated glycine (18). Vac8 is a scaffolding protein consisting primarily of 11 armadillo repeats involved in protein-protein interactions (19, 20). Located at the vacuolar limiting membrane, Vac8 is involved in several membrane-mediated events, including vacuolar fusion, vacuolar inheritance, cytoplasm-to-vacuole targeting, and nuclear autophagy (18, 21–24). The N-terminal cysteines, and presumably the palmitoylation that occurs there, are required for the function of Vac8 in vacuolar fusion and vacuolar inheritance (18, 24, 25). We recently demonstrated both genetically and biochemically that the yeast DHHC protein Pfa3 is a Vac8 PAT (26).
Palmitoylated proteins are often broadly grouped into subclasses defined by the sequence context in which the palmitoylated cysteine occurs, such as the N-myristoylated, palmitoylated subclass described above. However, unlike the well characterized consensus motifs of N-myristoylation and prenylation, there is no single sequence requirement for palmitoylation outside of the presence of a cysteine residue. This has made the identification of palmitoylation candidates difficult.
The lack of a universal palmitoylation consensus motif led us to analyze the region(s) of Vac8 that are specifically recognized by its PAT Pfa3. It is currently unknown how DHHC PATs recognize their substrates for palmitoylation. Here we used Pfa3 palmitoylation of Vac8 as a model to understand what elements in substrates with Src homology 4 (SH4) domains determine specific palmitoylation by DHHC PATs. SH4 domains are N-terminal membrane-anchoring regions that consist of an N-myristoylation motif followed by a second membrane-targeting signal (17). In Src that second signal is a polybasic region that interacts at membranes with acidic phospholipid head groups (27). In Vac8 that second signal is palmitoylation (18). Gaining an understanding of how Vac8 is palmitoylated by Pfa3 will give us insight into how other SH4 domain proteins are palmitoylated, including several proteins that play roles in important mammalian signaling pathways.
EXPERIMENTAL PROCEDURES
Plasmids
Standard molecular biology techniques were used to manipulate DNA. Unless otherwise noted, all PCR products were ligated into pCR2.1-TOPO (Invitrogen) prior to subcloning into expression vectors and sequenced. Plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Name | Description |
|---|---|
| pML311 | pQE60, Gαi1-6xHIS |
| pML938 | NpT7Q |
| pML964 | NpT7Q, VAC8(C4S)-MYC |
| pML965 | NpT7Q, VAC8(C5S)-MYC |
| pML966 | NpT7Q, VAC8(C7S)-MYC |
| pML967 | NpT7Q, VAC8(C4,5S)-MYC |
| pML1024 | NpT7Q, VAC8-MYC |
| pML1025 | NpT7Q, VAC8(C4,7S)-MYC |
| pML1026 | NpT7Q, VAC8(C4,5,7S)-MYC |
| pML1027 | NpT7Q, VAC8(C5,7S)-MYC |
| pML1064 | pQE60, GFP-6xHIS |
| pML1065 | pQE60, VAC8[SH4]-GFP-6xHIS |
| pML1133 | pQE60, YGL108c-MYC-6xHIS |
| pML1135 | pQE60, MEH1-MYC-6xHIS |
| pML1164 | pQE60, PSR2-MYC-6xHIS |
| pML1226 | pQE60, MEH1[SH4]-GFP-6xHIS |
| pML1228 | pQE60, MEH1[SH4]-VAC8-MYC-6xHIS |
| pML1234 | pQE60, VAC8(C4,5S)-MYC |
| pML1241 | pQE60, VAC8(C4,5,7S)-MYC |
| pML1245 | pQE60, VAC8[SH4](C4,5,7S)-GFP-6xHIS |
| pML1249 | pQE60, MEH1[SH4](C4,5,7S)-GFP-6xHIS |
| pML1374 | pQE60, VAC8[ARM1–11](C4,5,7S)-MYC-6xHIS |
| pML1393 | pQE60, VAC8[ARM1–10](C4,5,7S)-MYC-6xHIS |
| pML1417 | pQE60, VAC8[ARMΔ11](C4,5,7S)-MYC-6xHIS |
| Vac8-10 | pRS416, VAC8Δarm11 (18) |
| pBB131 | NMT1 (53) |
| pQE60 | Qiagen |
| NpT7-5 | Ref. (54) |
| pML658 | pQE60, VAC8-MYC-6xHIS (26) |
| pML125 | pEG(KG), GST-ERF4 (3) |
| pML124 | pESC-TRP, FLAG-ERF2 (3) |
| pML393 | pESC-TRP, PFA4-FLAG (26) |
| pML394 | pESC-TRP, PFA5-FLAG (26) |
| pML395 | pESC-TRP, PFA3-FLAG (26) |
| pML477 | pESC-TRP, AKR1-FLAG (26) |
| pML851 | pESC-TRP, PFA3-FLAG-6xHIS |
The bacterial expression vector NpT7Q (pML938) was constructed by inserting a double-stranded oligonucleotide containing the ribosomal binding site, multiple cloning site, and hexahistadine tag of pQE60 into the EcoRI and HindIII sites of NpT7-5. VAC8 coding sequence was amplified from pML658. The mutations in pML964, pML965, pML966, and pML967 were incorporated into the 5′ primer of a standard PCR and subcloned into NpT7Q as NcoI/HindIII fragments. The mutations in pML1024, pML1025, pML1026, and pML1027 were incorporated using the QuikChange site-directed mutagenesis kit (Stratagene). To obtain higher expression levels, the C7 and CΔ mutants were subcloned from NpT7Q into pQE60 as an NcoI/HindIII and EcoRI/HindIII fragment, respectively. Vac8[Arm1–11](CΔ) (pML1374) was generated using a 3′ primer that annealed to the 3′ end of armadillo repeat 11 (amino acid 480) and a 5′ primer that incorporated the cysteine mutations. The resulting PCR fragment was subcloned into pQE60 as an NcoI/BglII fragment. Vac8[Arm1–10](CΔ) (pML1393) was generated using a 3′ primer that annealed to the 3′ end of armadillo repeat 10 (amino acid 440), amplifying from pML1374 to incorporate the cysteine mutations, and subcloned the PCR fragment into pQE60 as an NcoI/BglII fragment.
To generate Vac8[ArmΔ11]CΔ, the cysteine mutations in pML1417 were incorporated into the 5′ primer of a standard PCR that amplified VAC8 from vac8–10 (18). The resulting PCR fragment was subcloned into pQE60 as an NcoI/BglII fragment. MEH1 and PSR2 coding sequences were amplified from genomic DNA and inserted into pQE60 as NcoI/BglII fragments. YGL108c coding sequence was amplified from genomic DNA and inserted into pQE60 as an NcoI/BamHI fragment.
To produce a C-terminally hexahistadine-tagged GFP (pML1064), the coding region of monomerized GFP was amplified and ligated into pQE60 as an NcoI/BglII fragment. SH4-GFP-6xHIS constructs were generated by ligating double-stranded oligos containing a ribosomal binding site and the SH4 sequence of interest into pML1064 at the EcoRI and NcoI sites. The SH4 chimera (pML1228) was generated using overlap extension PCR as described previously (28). The chimera was then subcloned into pQE60 as NcoI/BglII fragments.
Gαi1-6xHIS (pML311) was generated by amplifying the coding sequence of rat Gαi1 and ligating it into pQE60 as an NcoI/BamHI fragment. PFA3–6xHIS-FLAG (pML851) was generated by ligating a double-stranded oligonucleotide containing a hexahistadine sequence into PFA3-FLAG (pML395) at the PacI and BglII sites.
Isolation of Yeast Membranes
yPH499 (Stratagene) was transformed with the appropriate pESC expression construct(s). Cells were grown, and membranes were isolated as described previously (29). Protein concentration was determined by Bradford assay.
In Vitro PAT Assay
[3H]Palmitoyl-CoA was synthesized using [3H]palmitate (45 Ci/mmol; PerkinElmer Life Sciences), coenzyme A (Sigma), and acyl-CoA synthase (Sigma) as described (30) with the following modification. Following synthesis, [3H]palmitoyl-CoA was separated from palmitate by chloroform/methanol extraction (31) and subsequently purified on a C8 reversed phase cartridge (30). In vitro PAT assays and fluorography were performed essentially as described (29). Palmitoyl-CoA concentrations varied from 0.8 to 1.5 μm, and all reactions were allowed to proceed for 10 min. For the quantitation presented in Fig. 1, filter binding was performed essentially as described (32) except protein was precipitated in the presence of 0.04 mg/ml bovine brain membranes at room temperature for 15 min, and precipitated protein was filtered on a 25-mm APFA glass filter (Millipore) using a Millipore filtration unit. For the quantitation presented in Fig. 2C, soluene extraction of the protein of interest was performed as described previously (26, 29). For the quantitation presented in Figs. 4–, densitometry was performed by pre-flashing hyperfilm MP (GE Healthcare) prior to exposure. The films were scanned and quantitated by ImageJ software (National Institutes of Health). To ensure that the signal of interest was in the linear range of the film, a standard curve was included with the exposures.
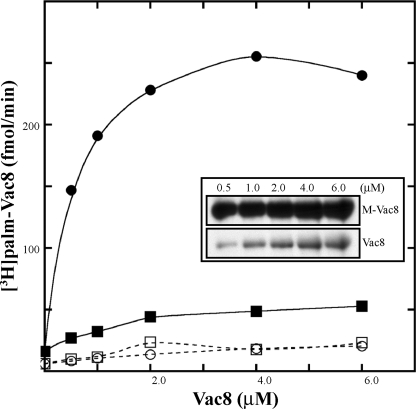
FIGURE 1.
Palmitoylation of nonmyristoylated Vac8 by Pfa3. Vac8 proteins were expressed in bacteria with or without Nmt1 and purified using conventional chromatography. The amount of Vac8 protein was determined as detailed under “Experimental Procedures.” Increasing amounts of Vac8 were incubated with [3H]palmitoyl-CoA and with partially purified Pfa3–6xHis-FLAG (●, myr-Vac8; ■, Vac8) or without partially purified Pfa3–6xHis-FLAG (○, myr-Vac8; □, Vac8) for 10 min. For each experiment three replicates were assayed. Two were quantitated by scintillation spectroscopy as described under “Experimental Procedures” (averaged in graph), and the third was analyzed by fluorography (inset) (3-day exposure). Data shown are from a single experiment, which is representative of three independent experiments.
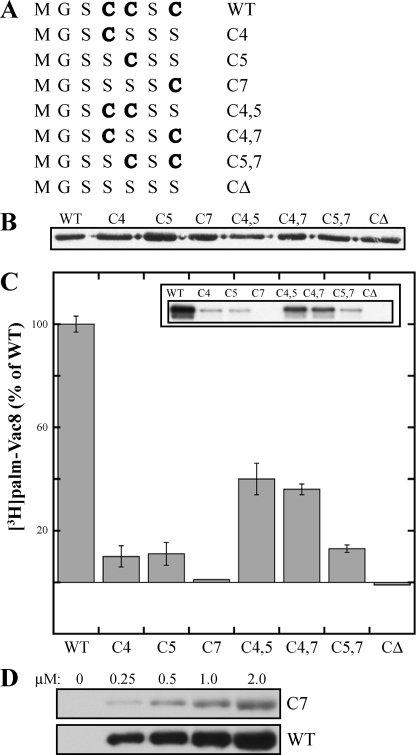
FIGURE 2.
Palmitoylation of Vac8 cysteine mutants by Pfa3. A, Vac8 N-terminal cysteine residues at positions 4, 5, and 7 were mutated in various combinations to serine residues. The remaining cysteine(s) is denoted in boldface and determines the name given to each mutant. B, WT or mutant Vac8 proteins were expressed in bacteria with Nmt1 and partially purified. The amount of myr-Vac8 protein in each preparation was determined by quantitative Western blot. Samples of 250 ng myr-Vac8 were analyzed by an anti-Myc Western blot. C, partially purified Pfa3–6xHis-FLAG was incubated with 80 nm myr-Vac8 mutants and [3H]palmitoyl-CoA for 10 min. Half of each reaction (20 μl) was quantitated by scintillation spectroscopy as described under “Experimental Procedures” (graph), and the other half was analyzed by fluorography (inset) (5-day exposure). Data are the average of three independent experiments each performed in duplicate. The error bars represent the standard deviation. The 100% values are as follows: experiment 1, 18.1 fmol/min; experiment 2, 34.6 fmol/min; experiment 3, 41.2 fmol/min. D, increasing amounts of WT or C7 Vac8 were incubated with partially purified Pfa3–6xHis-FLAG and [3H]palmitoyl-CoA for 10 min. The reactions were analyzed by fluorography (top panel, 7-day exposure; bottom panel, 1-day exposure).
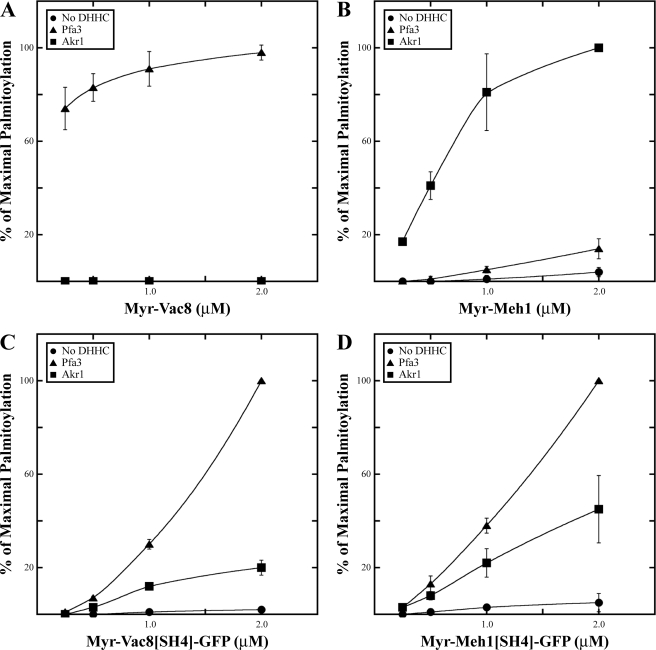
FIGURE 4.
Specificity of palmitoylation by Pfa3 and Akr1. Increasing amounts of myr-Vac8-myc-6xHIS (A), myr-Meh1-myc-6xHIS (B), myr-Vac8[SH4]-GFP-6xHIS (C), or myr-Meh1[SH4]-GFP-6xHIS (D) were incubated with [3H]palmitoyl-CoA and either no enzyme (●), 10 nm Pfa3-FLAG (▲), or 10 nm Akr1-FLAG (■). Reactions were analyzed by fluorography and quantitated by densitometry with exposure lengths ranging from 8 h to 2 days. Data are the average of three independent experiments. The error bars represent the standard deviation. Representative films are shown in supplemental Fig. S3A, and 100% values are shown in supplemental Table 1.
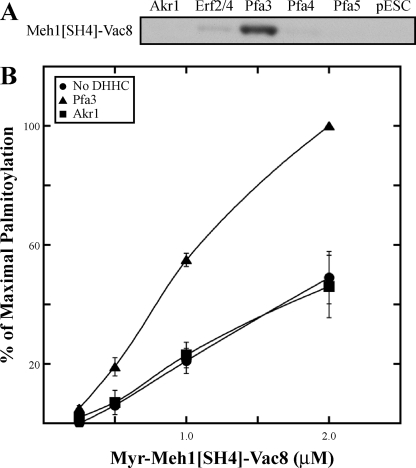
FIGURE 5.
Palmitoylation of Meh1[SH4]-Vac8. A, membranes expressing Akr1-FLAG, FLAG-Erf2/GST-Erf4, Pfa3-FLAG, Pfa4-FLAG, Pfa5-FLAG, or empty vector (pESC) were incubated with [3H]palmitoyl-CoA and 1 μm myr-Meh1[SH4]-Vac8-myc-6xHIS. Reactions were analyzed by fluorography with an exposure length of 2 days. B, increasing amounts of myr-Meh1[SH4]-Vac8-myc-6xHIS was incubated with [3H]palmitoyl-CoA and either no enzyme (●), 10 nm Pfa3-FLAG (▲), or 10 nm Akr1-FLAG (■). Reactions were analyzed by fluorography and quantitated by densitometry with an exposure length of 2 days. Data are the average of three independent experiments. The error bars represent the standard deviation. Representative films are shown in supplemental figure S3A, and 100% values are shown in supplemental Table 1.
FIGURE 6.
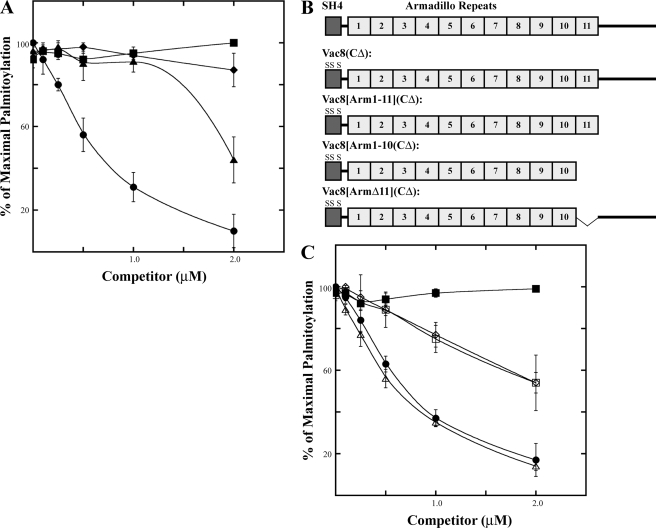
Pfa3 recognizes Vac8 armadillo repeat 11. A, increasing amounts of myr-Vac8(CΔ)-myc (●), myr-Vac8[SH4]-GFP-6xHIS (▲), myr-Vac8[SH4](CΔ)-GFP-6xHIS (■), or myr-Gαi1-6xHIS (♦) were incubated with 0.1 μm myr-Vac8-myc-6xHis, [3H]palmitoyl-CoA, and partially purified Pfa3–6xHis-FLAG. B, diagram representing the proteins analyzed in C. C, increasing amounts of myr-Vac8(CΔ)-6xHIS (●), myr-Vac8[SH4](CΔ)-GFP-6xHIS (■), myr-Vac8[Arm1–11](CΔ)-myc-6xHIS ( ), myr-Vac8[Arm1–10](CΔ)-myc-6xHIS (♢), or myr-Vac8[ArmΔ11](CΔ)-myc-6xHIS (□) were incubated with 0.1 μm myr-Vac8-myc-6xHis, [3H]palmitoyl-CoA and partially purified Pfa3–6xHis-FLAG. A and C, reactions were analyzed by fluorography and quantitated by densitometry with an exposure length of 4 h. Data are the average of three independent experiments. The error bars represent the standard deviation. Representative films are shown in supplemental Fig. S3, B and C, and 100% values are shown in supplemental Tables 2 and 3.
), myr-Vac8[Arm1–10](CΔ)-myc-6xHIS (♢), or myr-Vac8[ArmΔ11](CΔ)-myc-6xHIS (□) were incubated with 0.1 μm myr-Vac8-myc-6xHis, [3H]palmitoyl-CoA and partially purified Pfa3–6xHis-FLAG. A and C, reactions were analyzed by fluorography and quantitated by densitometry with an exposure length of 4 h. Data are the average of three independent experiments. The error bars represent the standard deviation. Representative films are shown in supplemental Fig. S3, B and C, and 100% values are shown in supplemental Tables 2 and 3.
Purification of N-Myristoylated Proteins
Escherichia coli JM109 cells were transformed with the vector carrying the substrate to be N-myristoylated and pBB131 (NMT1), grown at 37 °C to A600 = 0.4, induced with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside in the presence of 1 μg/ml chloramphenicol, and incubated for 21 h at 30 °C. Cells were harvested, washed, and suspended in lysis buffer (50 mm Tris, 1 mm EDTA, 1 mm DTT, pH 7.4). Complete protease inhibitor tablets (Roche Applied Science) were included only in the lysis step. Cells were lysed using a French press at 1000 p.s.i. for three passages and cleared by centrifugation at 25,400 × g for 30 min. To purify nonmyristoylated Vac8-myc-6xHis, the protein was expressed in E. coli in the absence of pBB131. Exceptions to this protocol are appropriately noted.
TX114 Partitioning
French press lysis supernatants were adjusted to 1% Triton X-114 (Sigma) and subjected to partitioning first described by Bordier (33). Briefly, the supernatant/Triton mixture was incubated at 4 °C for 5 min, layered onto a 6% sucrose cushion, incubated at 4 °C for 5 min, incubated at 37 °C for 5 min, and centrifuged at a low speed for 10 min in a swinging bucket rotor. The aqueous phase was recovered; the detergent phase was set aside, and the aqueous phase was subjected to another round of partitioning. The two detergent phases were then pooled and further purified.
Purification of N-Myristoylated and Nonmyristoylated Vac8-myc-6xHis
Cells were processed as described above except the lysis buffer was pH 8. The supernatant was applied to a HiLoad 26/10 fast flow 64-ml Q-Sepharose column using a fast protein liquid chromatography system (Amersham Biosciences) equilibrated with lysis buffer. The column was washed with 80 ml of lysis buffer, and protein was eluted with 500 ml of an ascending NaCl gradient (0–500 mm) in lysis buffer. Fractions containing Vac8-myc-6xHis were identified by Western blot, pooled, and applied to a 20-ml hydroxylapatite column (Bio-Rad) that was equilibrated with 200 ml of HAP buffer (20 mm Tris, 1 mm DTT, 100 mm NaCl, pH 8.0). The column was washed with 200 ml of HAP buffer, and protein was eluted with 300 ml of an ascending KPi gradient (0–150 mm) in HAP buffer. Fractions containing Vac8-myc-6xHis were identified by Western blot and pooled. The pool was buffer-exchanged into final buffer (20 mm Tris, 1 mm EDTA, 1 mm DTT, 100 mm NaCl, pH 8) and concentrated using an Amicon ultrafiltration device. An E. coli culture of 2 liters yielded 2.4–3.8 mg of full-length myr-Vac8-myc-6xHis as determined by comparison with standards on Coomassie Blue-stained gels. An E. coli culture of 1 liter yielded 4 mg of full-length nonmyristoylated Vac8-myc-6xHis.
Partial Purification of Myristoylated Vac8-myc Cysteine Mutants
Cells were processed as described above except these mutants were expressed in E. coli BL21(DE3), cultured to A600 = 0.6, and induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. The supernatant was diluted 1:1 with lysis buffer and applied to a 10-ml Q-Sepharose column (Amersham Biosciences) equilibrated with 50 ml of buffer A (50 mm Tris, 1 mm EDTA, 1 mm DTT, pH 7.4). The column was washed with 100 ml of buffer A, and protein was eluted with 100 ml of an ascending NaCl gradient (0–500 mm) in buffer A. Fractions containing myr-Vac8 were identified by Western blot and pooled. Only the C7 mutant was concentrated in a Centricon-30 (Amicon). Each partially purified protein maintained its N-myristoylation as determined by electrophoretic mobility shift (supplemental Fig. S1). The concentrations of full-length wild type and cysteine mutants were determined by quantitative Western blot in which extrapolation to a linear curve of known amounts of wild type myr-Vac8-myc-6xHis was performed using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Purification of N-Myristoylated C7 and CΔ Vac8-myc Cysteine Mutants
To improve protein yields, the C7 (pML1234) and CΔ (pML1241) mutants were expressed from pQE60 plasmids in JM109 E. coli. Cells were processed as described above. To enrich for N-myristoylated protein, the supernatant was subjected to Triton X-114 partitioning. The detergent fractions were pooled and diluted to 0.5% Triton X-114. Q-Sepharose (10 ml) was equilibrated with 50 ml of buffer A (50 mm Tris, 1 mm EDTA, 1 mm DTT, 5 mm NaCl, pH 7.4) and incubated with the pooled, diluted detergent fractions at 4 °C for 30 min. The resin was reconstituted into a column and was washed with 100 ml of buffer A. Protein was batch-eluted three times with buffer B (10 ml) (50 mm Tris, 1 mm EDTA, 1 mm DTT, 500 mm NaCl, pH 7.4). The fraction containing the most Vac8 was diluted 1:4 in buffer A and concentrated using an Amicon ultrafiltration device. E. coli cultures of 500 ml yielded 2.5 mg of full-length C7 and 1.5 mg of full-length CΔ.
Purification of N-Myristoylated Meh1-myc-6xHis
Cells were processed as described above except the culture was harvested after 4 h at 30 °C, and the lysis buffer (50 mm Tris, 10 mm β-mercaptoethanol, pH 7.4) contained EDTA-free complete protease inhibitor tablets (Roche Applied Science). To enrich for N-myristoylated protein, the supernatant was subjected to Triton X-114 partitioning. The detergent fractions were pooled, diluted to 0.5% Triton X-114, and applied to a 4- ml Ni-NTA column (Qiagen) that was equilibrated with 40 ml of buffer A (50 mm Tris, 5 mm β-mercaptoethanol, 10% glycerol, 100 mm NaCl, pH 7.4). The column was washed with 100 ml of buffer A, and protein was batch-eluted five times with 200 mm imidazole (4 ml) in buffer A. Fractions containing myr-Meh1-myc-6xHis were identified by Western blot and pooled. The pool was buffer-exchanged into buffer B (50 mm Tris, 1 mm EDTA, 1 mm DTT, 100 mm NaCl, pH 7.4) and concentrated using an Amicon ultrafiltration device. An E. coli culture of 1 liter yielded 0.9 mg of myr-Meh1-myc-6xHis.
Purification of N-Myristoylated Ygl108-myc-6xHis
Cells were processed as described above except the culture was harvested after 4 h at 30 °C, and the lysis buffer (50 mm Tris, 10 mm β-mercaptoethanol, 300 mm NaCl, pH 7.2) contained EDTA-free complete protease inhibitor tablets (Roche Applied Science). The supernatant was diluted 1:1 with lysis buffer and incubated with 4 ml of Ni-NTA, equilibrated with 40 ml of buffer A (50 mm Tris, 10 mm β-mercaptoethanol, 300 mm NaCl, pH 7.2), at 4 °C for 60 min. The resin was reconstituted into a column and washed with 80 ml of buffer B (50 mm Tris, 10 mm β-mercaptoethanol, 300 mm NaCl, 10% glycerol, 10 mm imidazole, pH 7.2) followed by 20 ml of buffer C (50 mm Tris, 10 mm β-mercaptoethanol, 100 mm NaCl, 10% glycerol, 10 mm imidazole, pH 7.2). Protein was batch-eluted five times with 300 mm imidazole (4 ml) in buffer C. Fractions containing myr-Ygl108-myc-6xHis were determined by Western blot analysis.
Purification of N-Myristoylated SH4-GFP-6xHis Proteins
myr-Vac8[SH4]-GFP-6xHis, myr-Vac8[SH4](C4,5,7S)-GFP-6xHis, myr-Meh1[SH4]-GFP-6xHis, and myr-Meh1[SH4](C7,8S)-GFP-6xHis were purified as follows. It was confirmed that each SH4-GFP fusion could be N-myristoylated by [3H]myristate incorporation (supplemental Fig. S2C). Cells were processed, and protein was purified as described for myr-Meh1-myc-6xHis except the culture was harvested after 21 h. E. coli cultures of 1 liter yielded 5 mg of myr-Vac8[SH4]-GFP-6xHis, 5 mg of myr-Vac8[SH4](C4,5,7S)-GFP-6xHis, 2.9 mg of myr-Meh1[SH4]-GFP-6xHis, and 1.6 mg of myr-Meh1[SH4](C7,8S)-GFP-6xHis.
Purification of N-Myristoylated Meh1[SH4]-Vac8-myc-6xHis
Cells were processed as described above for N-myristoylated proteins. myr-Meh1[SH4]-Vac8-myc-6xHis was purified as described in the partial purification of N-myristoylated myr-Vac8-myc cysteine mutants above. An E. coli culture of 500 ml yielded 38 mg of full-length myr-Meh1[SH4]-Vac8-myc-6xHis.
Purification of N-Myristoylated Vac8[Arm1–11](C4,5,7S)- myc-6xHis
Cells were processed as described above for N-myristoylated proteins. myr-Vac8[Arm1–11](C4,5,7S)-myc-6xHis was purified as described in the purification of N-myristoylated C7 and CΔ myr-Vac8-myc cysteine mutants above except a Q-Sepharose column was gravity loaded, and protein was eluted with 100 ml of an ascending NaCl gradient (0–500 mm) in buffer A (50 mm Tris, 1 mm EDTA, 1 mm DTT, pH 7.4). Fractions containing Myr-Vac8[Arm1–11](C4,5,7S)-myc-6xHis were pooled, diluted 1:4 in buffer A, and concentrated using an Amicon ultrafiltration device. An E. coli culture of 1 liter yielded 1.8 mg of full-length myr-Vac8[Arm1–11](C4,5,7S)-myc-6xHis.
Purification of N-Myristoylated Vac8[Arm1–10](C4,5,7S)-myc-6xHis and Vac8[ArmΔ11](C4,5,7S)-myc-6xHis
Cells were processed as described above for N-myristoylated proteins. Cells were processed, and protein was purified as described for myr-Meh1-myc-6xHis except the culture was harvested after 21 h and the Ni-NTA column was gravity loaded. E. coli cultures of 1 liter yielded 2 mg of myr-Vac8[Arm1–10](C4,5,7S)-myc-6xHis and 0.6 mg of myr-Vac8[ArmΔ11](C4,5,7S)-myc-6xHis.
Purification of N-Myristoylated Gαi1-6xHis
Gαi1-6xHis was expressed in JM101 cells with pBB131 as described above except protein was induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. The supernatant was applied to a 5-ml Ni-NTA column that was equilibrated with 25 ml of buffer A (50 mm Tris, 20 mm β-mercaptoethanol, 100 mm NaCl, 0.1 mm phenylmethylsulfonyl fluoride, pH 8). The column was washed with 50 ml of buffer A, and protein was eluted with ascending imidazole gradient (10–200 mm) in buffer A supplemented with 10% glycerol. Fractions containing myr-Gαi1-6xHis were identified by Western blot and pooled. The pool was buffer-exchanged by dialysis into buffer B (20 mm Hepes, 1 mm EDTA, 1 mm DTT, 10% glycerol, pH 8) and concentrated using an Amicon ultrafiltration device. An E. coli culture of 1.2 liter yielded 4.5 mg of myr-Gαi1-6xHis.
Partial Purification of Pfa3–6xHis-FLAG
Membranes from cells transformed with pML851 were extracted with 1% Triton X-100 at 5 mg/ml total protein in extraction buffer (50 mm Tris, 100 mm NaCl, 10% glycerol, 5 mm β-mercaptoethanol, 10 μg/ml pepstatin A, 1.6 μg/ml leupeptin, 1.6 μg/ml lima bean trypsin inhibitor, 0.7 μg/ml aprotinin, 0.1 mm phenylmethylsulfonyl fluoride, pH 8) rotating for 1 h at 4 °C. Insoluble material was pelleted at 200,000 × g for 20 min. The extract (7.5 mg of total protein) was diluted 1:1 in extraction buffer and allowed to bind to 2 ml of Ni-NTA resin for 1 h at 4 °C. The resin was reconstituted in a column and washed with 20 ml of wash buffer (50 mm Tris, 100 mm NaCl, 10% glycerol, 5 mm β-mercaptoethanol, 0.1% Triton X-100, 0.5 mg/ml bovine liver lipids (Avanti Polar Lipids), 1 μg/ml pepstatin A, 1.6 μg/ml leupeptin, 1.6 μg/ml lima bean trypsin inhibitor, 0.7 μg/ml aprotinin, 0.1 mm phenylmethylsulfonyl fluoride, pH 8). Bound proteins were batch-eluted four times with 2 ml of elution buffer (50 mm Tris, 100 mm NaCl, 10% glycerol, 200 mm imidazole, 1 mm β-mercaptoethanol, 0.1% Triton X-100, 0.5 mg/ml bovine liver lipids, 1 μg/ml pepstatin A, 1.6 μg/ml leupeptin, 1.6 μg/ml lima bean trypsin inhibitor, 0.7 μg/ml aprotinin, pH 8). Fractions containing Pfa3 were identified by in vitro PAT assays using myr-Vac8-myc-6xHis as substrate, and the two fractions with the most activity were pooled.
Purification of Pfa3-FLAG and Akr1-FLAG
Membranes from cells expressing pML395 or pML477 were extracted as described above, and the proteins were purified as described (26) with the following exceptions: 1 mm β-mercaptoethanol was present in all buffers; all buffers containing 0.1 mg/ml bovine liver lipids were increased to 0.5 mg/ml; and Akr1-FLAG was eluted with 3xFLAG peptide (Sigma). The peak fractions were identified by Western blot and pooled. DHHC-FLAG concentrations were determined by extrapolation to a linear curve of known bovine serum albumin concentrations using SYPRO Ruby protein gel stain (Invitrogen).
RESULTS AND DISCUSSION
N-Myristoylation Is Required for Efficient Palmitoylation of Vac8 by Pfa3
To date there has been inconsistency in whether palmitoylation of N-myristoylated substrates requires prior N-myristoylation. On one hand, members of the Src family of protein-tyrosine kinases need to be N-myristoylated before palmitoylation. In vivo, mutation of the N-myristoylation site (G2A) in Lck or Fyn yields protein that is neither N-myristoylated nor palmitoylated (34). Fyn G2A mutants cannot be palmitoylated in vitro, and in competition assays only an N-myristoylated Fyn peptide could inhibit palmitoylation of the wild type (WT) protein (35). On the other hand, palmitoylation of Gαi subunits of heterotrimeric G proteins does not require prior N-myristoylation. Mutation of the N-myristoylated glycine of Gαi1 does not abolish in vivo palmitoylation of Gαi1 if it is co-expressed with Gβγ subunits (36). It is postulated that by localizing Gαi1 to the membrane, Gβγ is bringing Gαi1 into the proximity of a membrane-bound PAT. Thus, it is membrane association and not N-myristoylation that confers the ability of Gαi1 to be palmitoylated. Dunphy et al. (30) reported similar results with reconstitution of Gαi1 PAT activity in vitro.
In yeast, a Vac8 mutant that is not N-myristoylated (G2A) is able to incorporate [3H]palmitate, although at a much reduced level. This small amount of palmitoylation is able to maintain vacuolar inheritance at a WT level (18). It is thought that transient membrane interactions, potentially via protein-protein interactions, are sufficient to allow for palmitoylation of the nonmyristoylated Vac8. This would suggest that it is membrane association, and not N-myristoylation, that promotes Vac8 palmitoylation in yeast.
To investigate how N-myristoylation of Vac8 affects in vitro palmitoylation by Pfa3, we prepared both N-myristoylated and nonmyristoylated Vac8 and assayed them as substrates for Pfa3. Consistent with in vivo data, palmitoylation of N-myristoylated Vac8 was considerably greater than that of nonmyristoylated Vac8 (Fig. 1). However, nonmyristoylated Vac8 was palmitoylated by Pfa3 to a detectable level and in a manner that was dependent on substrate concentration. Hence, N-myristoylation significantly enhances palmitoylation but is not an absolute requirement for substrate recognition. These data suggest that the primary role of N-myristoylation is to increase Vac8 hydrophobicity, thereby allowing it to sample the membrane (or detergent micelle) where Pfa3 resides.
Vac8 Is Palmitoylated by Pfa3 at All Three N-terminal Cysteine Residues
The N-terminal N-myristoylated glycine of Vac8 is followed by three cysteine residues at positions 4, 5, and 7. Mutation of all three cysteine residues to serine produces a Vac8 protein that is no longer palmitoylated in vivo (18) or in vitro by Pfa3 (26). The mutation also produces several phenotypes, including defective vacuolar inheritance (18), defective homotypic vacuole fusion in vitro, and fragmentation of the vacuole in vivo (24).
Evidence suggests that Pfa3 is responsible for the majority of Vac8 palmitoylation (26, 37). However, deletion of PFA3 does not eliminate palmitoylation of overexpressed Vac8 (26). It is possible that Pfa3 is only capable of palmitoylating a subset of three cysteines of Vac8 and that another enzyme is responsible for the palmitoylation of the remaining cysteine(s), accounting for the residual Vac8 palmitoylation seen in pfa3Δ cells. To determine whether Pfa3 is capable of palmitoylating Vac8 at each of the three N-terminal cysteine residues, we generated Vac8 cysteine mutants (Fig. 2A) and assessed their ability to be palmitoylated by Pfa3 in vitro. The mutants were N-myristoylated by co-expression in bacteria with Nmt1 and partially purified. The amount of myr-Vac8 protein for each mutant was normalized by immunoblot (Fig. 2B) and assayed.
myr-Vac8 C4 or C5 was palmitoylated by Pfa3 to 10% of WT (Fig. 2C). Palmitoylation of myr-Vac8 C7 was low but detectable at 1% of WT. To confirm C7 palmitoylation, we further purified this mutant and used it as a substrate in a PAT assay. C7 was palmitoylated by Pfa3 in a concentration-dependent manner (Fig. 2D). Thus, Pfa3 could palmitoylate myr-Vac8 at each of the three N-terminal cysteine residues, although not all equally.
A common phenomenon in palmitoylation is the dependence of palmitoylation at one cysteine on the presence of an additional cysteine(s) (38). C4,5 and C4,7 were the best mutant substrates for Pfa3. Both were palmitoylated to significantly higher levels than C5,7. One possibility is that the proximity of Cys-4 to the N-myristoylated glycine enhanced palmitoylation at the second site. Additionally, palmitoylation of the C4,7 mutant was greater than the additive palmitoylation of the proteins containing one cysteine at either position. This suggests that the addition of a cysteine at position 4 improved palmitoylation at position 7. Interestingly, palmitoylation of the C5,7 mutant was not much greater than the additive palmitoylation of C5 and C7. Unlike Cys-4, it appears that the absence or presence of Cys-5 has little effect on palmitoylation of Cys-7. Of note, palmitoylation of myr-Vac8 was not additive in that the cumulative palmitoylation of C4, C5, and C7, or of any double mutant with the remaining single mutant, was at most 50% of WT.
Other groups provide support for our findings in vivo. Vacuolar binding of the C4 and C5 mutants is decreased by 60%, and that of the C7 mutant is decreased by 90% (39). These data are consistent with the decreased palmitoylation we observed for C4 and C5 and almost absent palmitoylation of C7. Analysis of Vac8 function showed that the C7 mutant could not rescue Vac8 function in vacuole fusion or vacuole inheritance better than Vac8 lacking all three cysteines. Interestingly, the C4 and C5 mutants can rescue function despite their decreased vacuolar localization. Subramanian et al. (39) argued that the cysteine at position 7 is capable of being palmitoylated because addition of Cys-7 to the C4 or C5 mutants restores vacuole localization to WT levels and suggested that palmitoylation of Cys-7 occurs only if the proximal cysteine at position 4 or 5 has already been modified. Peng et al. (40) found similar results. We provide the first direct evidence both that Cys-7 is palmitoylated and that a DHHC PAT can palmitoylate a substrate at more than one site. An important question for future investigations is to determine whether the enzyme palmitoylates a single site, followed by release of the substrate, or whether the reaction is processive. Mechanistic insight into the action of DHHC PATs will be key to developing and characterizing inhibitors of these enzymes.
Pfa3 Does Not Palmitoylate All SH4 Domain Proteins
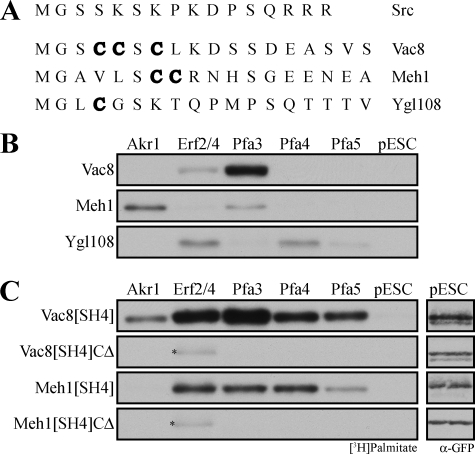
We sought to test the role of SH4 domains in recognition by DHHC PATs. Yeast has five proteins with palmitoylated SH4 domains as follows: Vac8, Ygl108, Meh1, Gpa1, and Gpa2. It was suggested that the redundant phosphatases Psr1 and Psr2 each contain a N-myristoylated and palmitoylated SH4 domain (5). However, neither was identified as a substrate for N-myristoylation using an algorithm to identify N-myristoylation motifs. To determine whether Psr1 and Psr2 should be considered, we cloned PSR2 and co-expressed it with Nmt1 in bacteria. No incorporation of [3H]myristate into Psr2 was detected (supplemental Fig. S2A), and therefore Psr1 and Psr2 were not included in our analysis of palmitoylated SH4 domains. Gpa1 and Gpa2, the yeast G protein α subunits, were not studied because they were not amenable to purification in our system. Hence, we analyzed Ygl108 and Meh1, in addition to Vac8 (Fig. 3A), to determine whether Pfa3 universally recognizes SH4 domain proteins as substrates for palmitoylation.
FIGURE 3.
Palmitoylation of SH4 domains. A, alignment of palmitoylated yeast SH4 domains analyzed in this study. The sites, or presumed sites, of palmitoylation are denoted in boldface. B and C, membranes expressing Akr1-FLAG, FLAG-Erf2/GST-Erf4, Pfa3-FLAG, Pfa4-FLAG, Pfa5-FLAG, or empty vector (pESC) were incubated with [3H]palmitoyl-CoA and 1 μm (B) myr-Vac8-myc-6xHIS, myr-Meh1-myc-6xHIS, or myr-Ygl108-myc-6xHIS (C) myr-Vac8[SH4]-GFP-6xHIS, myr-Vac8[SH4](CΔ)-GFP-6xHIS, myr-Meh1[SH4]-GFP-6xHIS, or myr-Meh1[SH4](CΔ)-GFP-6xHIS. Reactions were analyzed by fluorography with exposure lengths ranging from 1 to 3 days. The vector-incubated (pESC) reactions from C were also analyzed by immunoblot with GFP antibodies to ensure that the substrate was not proteolyzed during the reaction. The amount analyzed by Western blot represents 40% of that analyzed by fluorography. Asterisk denotes Erf2 autoacylation.
Ygl108 (open reading frame name YGL108c) is a protein of unknown function localized at the cell periphery (41). Palmitoylation of Ygl108 was detected in a large scale analysis of yeast palmitoylated proteins (5). Ygl108 contains a predicted N-myristoylation site (5), and when we expressed it in bacteria with Nmt1, Ygl108 incorporated [3H]myristate (supplemental Fig. S2B). Collectively, these data support the presence of a bona fide SH4 domain in Ygl108. Meh1/Ego1/Gse2 (hereafter referred to as Meh1) is part of the EGO/GSE protein complex involved in regulating rapamycin-induced microautophagy (42) and intracellular sorting of the amino acid permease Gap1 (43). Meh1 distributes to the vacuole membrane (42, 44) and endosomes (43). Palmitoylation of Meh1 was demonstrated by mass spectrometry and confirmed by acyl-biotin exchange (5). Meh1 is predicted to be N-myristoylated (44), and when we expressed Meh1 in bacteria with Nmt1, it incorporated [3H]myristate (supplemental Fig. S2B).
We have previously established an in vitro assay to analyze the specificity of palmitoylation by yeast DHHC proteins (29). Five (Akr1, Erf2/Erf4, Pfa3, Pfa4, and Pfa5) of the seven yeast DHHC proteins are amenable to overexpression in yeast cells, and membranes derived from these cells can be used as a source of enzyme. Previous studies using this assay showed that Pfa3 is the primary DHHC protein to palmitoylate myr-Vac8, with very weak palmitoylation by Erf2/4 (26, 29) (Fig. 3B), whereas Erf2/Erf4 displayed specificity for Ras2 (29). Here we used this assay to analyze the specificity of palmitoylation of myr-Ygl108 and myr-Meh1. myr-Ygl108 was not palmitoylated by Pfa3. Instead it was palmitoylated by Erf2, Pfa4, and slightly by Pfa5. myr-Meh1 was most robustly palmitoylated by Akr1. This confirmed data demonstrating that in akr1Δ cells Meh1 palmitoylation is reduced (5). However, a small amount of palmitoylation by Pfa3 was detectable. Thus, there may be some DHHC redundancy in regard to Meh1 palmitoylation. This would explain results observed by Hou et al. (37) showing that membrane association of Meh1 was unchanged in akr1Δ cells, whereas mutation of cysteines 7 and 8 produced a soluble protein (43). Neither myr-Ygl108 nor myr-Meh1 was a good substrate for Pfa3, suggesting that Pfa3 does not universally recognize SH4 domain proteins as substrates.
An SH4 Domain Is Not Sufficient to Confer Specificity for a DHHC Protein
To analyze more directly the role of the SH4 domain in substrate palmitoylation, we fused the first 18 amino acids of Vac8 or Meh1 to GFP. N-Myristoylated fusion proteins were expressed and purified from bacteria. In a membrane PAT assay, the SH4 domain of either myr-Vac8 or myr-Meh1 was sufficient to support palmitoylation at the appropriate cysteines of the fusion protein (Fig. 3C). However, the fusion proteins lost the substrate specificity of the full-length proteins (Fig. 3B). Whereas full-length myr-Vac8 was palmitoylated almost exclusively by Pfa3, the myr-Vac8 SH4 domain fused to GFP was palmitoylated by each of the five DHHC proteins tested. This is consistent with in vivo studies demonstrating that Vac8[SH4]-GFP is palmitoylated to similar levels in WT and pfa3Δ cells (37). Additionally, WT Vac8-GFP localized exclusively to the limiting membrane of the vacuole, but Vac8[SH4]-GFP was found on both the plasma membrane and internal membranes (37). This is accordant with Vac8[SH4]-GFP being palmitoylated by multiple DHHC proteins, potentially resulting in its expanded subcellular distribution.
myr-Meh1 was palmitoylated best by Akr1 and Pfa3 (Fig. 3B), but the Meh1 SH4 domain fused to GFP was palmitoylated by each of the DHHC proteins tested except Akr1 (Fig. 3B). Collectively the data show that both SH4-GFP fusion proteins are promiscuous substrates. Thus, despite harboring the palmitoylated cysteines and surrounding amino acids, the SH4 domain does not account solely for the specificity of palmitoylation.
The failure of Akr1 to palmitoylate Meh1[SH4]-GFP palmitoylation in the membrane assay was surprising. To reconcile this result, we tested the ability of purified Pfa3 and Akr1 to palmitoylate various substrates (Fig. 4). First, we tested the specificity of myr-Vac8 and myr-Meh1 palmitoylation. The specificity observed in the membrane assays was recapitulated in assays with purified components. myr-Vac8 was palmitoylated specifically by Pfa3 (Fig. 4A). myr-Meh1 was palmitoylated efficiently by Akr1 and poorly by Pfa3 (Fig. 4B). Both myr-Vac8[SH4]-GFP and myr-Meh1[SH4]-GFP were capable of being palmitoylated in this assay by Akr1 (Fig. 4, C and D) but were significantly better substrates for Pfa3. These data suggest that although Pfa3 is capable of palmitoylating isolated SH4 domains, elements in the rest of the substrate restrict specificity of palmitoylation to a particular enzyme(s).
The SH4-GFP data point to the importance of regions downstream of the SH4 domain in directing palmitoylation. If this is correct, this region of Vac8 might confer Pfa3 specificity to a non-Vac8 SH4 domain. To address this question we generated a chimeric protein in which the SH4 domain of Vac8 was swapped with that of Meh1. The N-myristoylated chimeric protein was purified from bacteria. In a membrane PAT assay, myr-Meh1[SH4]-Vac8 was palmitoylated specifically by Pfa3 (Fig. 5A). This pattern of palmitoylation mimicked that of full-length Vac8, not full-length Meh1 (Fig. 3B). When assayed using purified DHHC proteins, the chimera was palmitoylated best by Pfa3 (Fig. 5B), and palmitoylation by Akr1 was no higher than the spontaneous acylation rate by the substrate alone. These data support the importance of elements distant from the palmitoylated cysteines in regulating palmitoylation. Additionally, any elements that may be present in the Meh1 SH4 domain that confer specificity to Akr1 were not potent enough to achieve robust palmitoylation by Akr1 in the context of the chimera, similar to the results for the myr-Meh1[SH4]-GFP fusion.
Vac8 Armadillo Repeat 11 Is Partially Responsible for Recognition by Pfa3
Manipulation of the SH4 domain described above suggests a role for a Vac8 region(s) outside of the SH4 domain in substrate recognition. To address this question directly, we performed competition experiments (Fig. 6). Initially we determined if the CΔ mutant could compete for palmitoylation of the WT protein. CΔ cannot be palmitoylated by Pfa3, but it was able to compete with WT myr-Vac8 (Fig. 6A, circles). Modest competition was seen at equimolar concentrations of WT and CΔ (0.1 μm), and at 20-fold excess CΔ (2 μm) palmitoylation of WT was almost undetectable. This suggests that Pfa3 recognizes regions of Vac8 downstream of the SH4 domain. To exclude the possibility that the sequence surrounding the palmitoylated cysteines was responsible for the competition seen with CΔ, we next determined if myr-Vac8[SH4]CΔ-GFP could compete for palmitoylation of WT myr-Vac8 (Fig. 6A, squares). myr-Vac8[SH4]CΔ-GFP cannot be palmitoylated by Pfa3 nor does it contain the C-terminal elements of Vac8 we propose are recognized by Pfa3. myr-Vac8[SH4]CΔ-GFP was unable to compete with WT, even at 20-fold excess concentrations. myr-Vac8[SH4]-GFP was a substrate and therefore expected to compete. However, because it lacked the non-SH4 elements of Vac8, it was only competitive at the highest concentration tested (Fig. 6A, triangles). As another test of specificity, we assayed the ability of myr-Gαi1 to compete with WT myr-Vac8. myr-Gαi1 is not a substrate for Pfa3 (data not shown), but it is N-myristoylated. Accordingly, it can partition into detergent micelles with Pfa3. As expected, myr-Gαi1 did not compete (Fig. 6A, diamonds), suggesting that the competition seen by the other proteins was specific.
To further define the region(s) of Vac8 that is recognized by Pfa3, we generated Vac8 truncation proteins with the cysteines mutated (Fig. 6B). The N-myristoylated proteins were purified from bacteria. Mutation of the cysteines allowed us to assess the ability of the proteins to compete with WT myr-Vac8 independent of their ability to be palmitoylated by Pfa3. When Vac8 was truncated after armadillo repeat (Arm) 11 (Vac8[Arm1–11]CΔ), the protein was able to compete with WT myr-Vac8 (Fig. 6C, open triangles) in a manner similar to full-length CΔ (Fig. 6C, circles). This suggests that the region downstream of Arm 11 is not important for recognition of Vac8 by Pfa3. Vac8 truncated after Arm 10 (Vac8[Arm1–10]CΔ) (Fig. 6C, open diamond) competed less well than full-length CΔ but better than myr-Vac8[SH4]CΔ-GFP (Fig. 6C, squares). To determine whether this partial reduction was due to the loss of Arm 11 and not the combined loss of the entire region downstream of Arm 10, we tested a Vac8 protein lacking just Arm 11 (Vac8[ArmΔ11]CΔ) (Fig. 6B). myr-Vac8[ArmΔ11]CΔ competed with WT myr-Vac8 similarly to truncation after Arm 10 (Fig. 6C, open squares), suggesting that Pfa3 is recognizing sequences in Arm 11. Because Myr-Vac8[ArmΔ11]CΔ demonstrated only a partial loss of competition, there are probably additional sites between the SH4 domain and Arm 11 that contribute to the interaction between Pfa3 and Vac8.
Our findings suggest that DHHC proteins recognize multiple determinants on their substrates. The isolated SH4 domain of Vac8 is sufficient for palmitoylation by Pfa3. However, the presence of additional contacts between Pfa3 and Vac8 is strongly supported by the competition experiments described above. We have mapped one of these elements to Arm 11. Arms are imperfect ∼42-amino acid repeats with a conserved structure consisting of three α-helices. Tandem Arms fold over each other and form a superhelix, yielding a crescent-shaped tertiary structure (45). We speculate that the myristoyl group on Vac8 inserts into the membrane, bringing the SH4 domain into proximity of the presumed catalytic domain of Pfa3, the DHHC-CRD. Membrane topology predictions suggest that the DHHC-CRD is located at the membrane interface, coinciding with the end of a cytoplasmic loop and the beginning of the third transmembrane domain. If the Vac8 structure is similar to that of other Arm proteins, then Arm 11 is at the end of the crescent and is likely to contact Pfa3 at a site distant from the DHHC-CRD. The large cytoplasmic tail of Pfa3 is a candidate for this interaction. It is possible that DHHC PATs are similar to mitogen-activated protein kinases that recognize their substrates through interactions at the active site and a docking site distant from the active site (46). Multiple contact sites between the enzyme and substrate would permit greater specificity. Our data show that myr-SH4-GFP fusions clearly can access the active site of multiple DHHC PATs. In the context of the full-length protein, however, we speculate that the substrate is not able to bind appropriately to other regions of the enzyme to permit proper orientation of the SH4 domain at the active site.
While this manuscript was in revision, two studies of mammalian DHHC proteins were published that support our findings. In a study documenting substrate specificity of neuronal DHHC proteins, Huang et al. (47) reported that regions outside the DHHC-CRD contribute to specificity. DHHC3 and Huntingtin interacting protein (HIP)14/DHHC17 are neuronal palmitoyltransferases that have overlapping substrate specificity. Huntingtin is a substrate for HIP14 but not for DHHC3. HIP14 has an N-terminal cytoplasmic domain composed of ankyrin repeats. Appendage of the ankyrin repeat domain to the N terminus of DHHC3 enables it to palmitoylate Huntingtin, suggesting that binding of Huntingtin to the ankyrin repeats positions it for interaction with the catalytic domain of DHHC3. In a separate study, Greaves et al. (48) provide support for our contention that residues distant from the palmitoylated cysteines are important for enzyme recognition. SNAP-25 is a soluble NSF attachment protein receptor protein palmitoylated at a cluster of cysteine residues near the N terminus of the interhelical domain that connects two coiled coil domains. A previous study demonstrated the importance of a glutamine-proline-alanine-arginine-valine motif at the end of the interhelical domain for palmitoylation and membrane association of SNAP-25 (49). Greaves et al. (48) showed that mutation of the proline within that motif resulted in the loss of palmitoylation by DHHC3.
When presented with SH4 domains as GFP fusions, the specificity of the DHHC proteins studied here was lost. Our findings suggest that DHHC proteins broadly recognize short peptide sequences. This has implications for the use of fluorescent acylated peptides as reporters for acylation-dependent trafficking in cells (50, 51). These peptides may be promiscuous substrates for DHHC proteins in vivo, whereas full-length proteins might be palmitoylated by a restricted set of DHHC enzymes.
The discovery that a region(s) of Vac8 distant from the SH4 domain is key to specific recognition by Pfa3 also has important implications for the development of inhibitors of DHHC proteins. If our evidence for specific Pfa3 and Vac8 interactions extends to other DHHC:substrate pairs, then it should be possible to identify small molecules that specifically disrupt the interaction between an individual DHHC protein and its substrate. Although there is evidence for functional redundancy among the DHHC proteins (5, 26, 52), the existence and conservation of a large family of DHHC proteins suggest that they have specific and important roles in physiology.
Supplementary Material
Acknowledgments
We thank Wendy Greentree and Andrea Nichols for providing technical support and advice and Ben Jennings for helpful discussions. We thank Dr. Lois Weisman for providing the Vac8-10 plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant GM51466.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables SI–SIII.
- PAT
- protein acyltransferase
- WT
- wild type
- DHHC
- aspartic acid, histidine, histidine, cysteine
- CRD
- cysteine-rich domain
- myr
- N-myristoylated
- SH4
- Src homology 4
- Arm
- armadillo repeat
- DTT
- dithiothreitol
- Ni-NTA
- nickel-nitrilotriacetic acid
- GFP
- green fluorescent protein.
REFERENCES
- 1.Linder M. E., Deschenes R. J. ( 2007) Nat. Rev. Mol. Cell Biol. 8, 74– 84 [DOI] [PubMed] [Google Scholar]
- 2.Resh M. D. ( 2006) Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 3.Lobo S., Greentree W. K., Linder M. E., Deschenes R. J. ( 2002) J. Biol. Chem. 277, 41268– 41273 [DOI] [PubMed] [Google Scholar]
- 4.Roth A. F., Feng Y., Chen L., Davis N. G. ( 2002) J. Cell Biol. 159, 23– 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. ( 2006) Cell 125, 1003– 1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell D. A., Vasudevan A., Linder M. E., Deschenes R. J. ( 2006) J. Lipid Res. 47, 1118– 1127 [DOI] [PubMed] [Google Scholar]
- 7.Stowers R. S., Isacoff E. Y. ( 2007) J. Neurosci. 27, 12874– 12883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohyama T., Verstreken P., Ly C. V., Rosenmund T., Rajan A., Tien A. C., Haueter C., Schulze K. L., Bellen H. J. ( 2007) J. Cell Biol. 179, 1481– 1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. ( 2004) Neuron 44, 987– 996 [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Hernando C., Fukata M., Bernatchez P. N., Fukata Y., Lin M. I., Bredt D. S., Sessa W. C. ( 2006) J. Cell Biol. 174, 369– 377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma C., Yang X. H., Hemler M. E. ( 2008) Mol. Biol. Cell 19, 3415– 3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanai A., Huang K., Kang R., Singaraja R. R., Arstikaitis P., Gan L., Orban P. C., Mullard A., Cowan C. M., Raymond L. A., Drisdel R. C., Green W. N., Ravikumar B., Rubinsztein D. C., El-Husseini A., Hayden M. R. ( 2006) Nat. Neurosci. 9, 824– 831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raymond F. L., Tarpey P. S., Edkins S., Tofts C., O'Meara S., Teague J., Butler A., Stevens C., Barthorpe S., Buck G., Cole J., Dicks E., Gray K., Halliday K., Hills K., Hinton J., Jones D., Menzies A., Perry J., Raine K., Shepherd R., Small A., Varian J., Widaa S., Mallya U., Moon J., Luo Y., Shaw M., Boyle J., Kerr B., Turner G., Quarrell O., Cole T., Easton D. F., Wooster R., Bobrow M., Schwartz C. E., Gecz J., Stratton M. R., Futreal P. A. ( 2007) Am. J. Hum. Genet. 80, 982– 987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansouri M. R., Marklund L., Gustavsson P., Davey E., Carlsson B., Larsson C., White I., Gustavson K. H., Dahl N. ( 2005) Eur. J. Hum. Genet 13, 970– 977 [DOI] [PubMed] [Google Scholar]
- 15.Magee A. I., Courtneidge S. A. ( 1985) EMBO J. 4, 1137– 1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Towler D. A., Eubanks S. R., Towery D. S., Adams S. P., Glaser L. ( 1987) J. Biol. Chem. 262, 1030– 1036 [PubMed] [Google Scholar]
- 17.Resh M. D. ( 1994) Cell 76, 411– 413 [DOI] [PubMed] [Google Scholar]
- 18.Wang Y. X., Catlett N. L., Weisman L. S. ( 1998) J. Cell Biol. 140, 1063– 1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott S. V., Nice D. C., 3rd, Nau J. J., Weisman L. S., Kamada Y., Keizer-Gunnink I., Funakoshi T., Veenhuis M., Ohsumi Y., Klionsky D. J. ( 2000) J. Biol. Chem. 275, 25840– 25849 [DOI] [PubMed] [Google Scholar]
- 20.Tang F., Peng Y., Nau J. J., Kauffman E. J., Weisman L. S. ( 2006) Traffic 7, 1368– 1377 [DOI] [PubMed] [Google Scholar]
- 21.Fleckenstein D., Rohde M., Klionsky D. J., Rüdiger M. ( 1998) J. Cell Sci. 111, 3109– 3118 [DOI] [PubMed] [Google Scholar]
- 22.Pan X., Goldfarb D. S. ( 1998) J. Cell Sci. 111, 2137– 2147 [DOI] [PubMed] [Google Scholar]
- 23.Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., Goldfarb D. S. ( 2000) Mol. Biol. Cell 11, 2445– 2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y. X., Kauffman E. J., Duex J. E., Weisman L. S. ( 2001) J. Biol. Chem. 276, 35133– 35140 [DOI] [PubMed] [Google Scholar]
- 25.Veit M., Laage R., Dietrich L., Wang L., Ungermann C. ( 2001) EMBO J. 20, 3145– 3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smotrys J. E., Schoenfish M. J., Stutz M. A., Linder M. E. ( 2005) J. Cell Biol. 170, 1091– 1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigal C. T., Zhou W., Buser C. A., McLaughlin S., Resh M. D. ( 1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12253– 12257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. ( 1989) Gene 77, 61– 68 [DOI] [PubMed] [Google Scholar]
- 29.Budde C., Schoenfish M. J., Linder M. E., Deschenes R. J. ( 2006) Methods 40, 143– 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunphy J. T., Greentree W. K., Manahan C. L., Linder M. E. ( 1996) J. Biol. Chem. 271, 7154– 7159 [DOI] [PubMed] [Google Scholar]
- 31.Duncan J. A., Gilman A. G. ( 1996) J. Biol. Chem. 271, 23594– 23600 [DOI] [PubMed] [Google Scholar]
- 32.Reiss Y., Goldstein J. L., Seabra M. C., Casey P. J., Brown M. S. ( 1990) Cell 62, 81– 88 [DOI] [PubMed] [Google Scholar]
- 33.Bordier C. ( 1981) J. Biol. Chem. 256, 1604– 1607 [PubMed] [Google Scholar]
- 34.Koegl M., Zlatkine P., Ley S. C., Courtneidge S. A., Magee A. I. ( 1994) Biochem. J. 303, 749– 753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berthiaume L., Resh M. D. ( 1995) J. Biol. Chem. 270, 22399– 22405 [DOI] [PubMed] [Google Scholar]
- 36.Degtyarev M. Y., Spiegel A. M., Jones T. L. ( 1994) J. Biol. Chem. 269, 30898– 30903 [PubMed] [Google Scholar]
- 37.Hou H., Subramanian K., LaGrassa T. J., Markgraf D., Dietrich L. E., Urban J., Decker N., Ungermann C. ( 2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17366– 17371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srinivasa S. P., Bernstein L. S., Blumer K. J., Linder M. E. ( 1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5584– 5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian K., Dietrich L. E., Hou H., LaGrassa T. J., Meiringer C. T., Ungermann C. ( 2006) J. Cell Sci. 119, 2477– 2485 [DOI] [PubMed] [Google Scholar]
- 40.Peng Y., Tang F., Weisman L. S. ( 2006) Traffic 7, 1378– 1387 [DOI] [PubMed] [Google Scholar]
- 41.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. ( 2003) Nature 425, 686– 691 [DOI] [PubMed] [Google Scholar]
- 42.Dubouloz F., Deloche O., Wanke V., Cameroni E., De Virgilio C. ( 2005) Mol. Cell 19, 15– 26 [DOI] [PubMed] [Google Scholar]
- 43.Gao M., Kaiser C. A. ( 2006) Nat. Cell Biol. 8, 657– 667 [DOI] [PubMed] [Google Scholar]
- 44.Gao X. D., Wang J., Keppler-Ross S., Dean N. ( 2005) FEBS J. 272, 2497– 2511 [DOI] [PubMed] [Google Scholar]
- 45.Huber A. H., Nelson W. J., Weis W. I. ( 1997) Cell 90, 871– 882 [DOI] [PubMed] [Google Scholar]
- 46.Reményi A., Good M. C., Lim W. A. ( 2006) Curr. Opin. Struct. Biol. 16, 676– 685 [DOI] [PubMed] [Google Scholar]
- 47.Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M. R., El-Husseini A. ( 2009) FASEB J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greaves J., Prescott G. R., Fukata Y., Fukata M., Salaun C., Chamberlain L. H. ( 2009) Mol. Biol. Cell 20, 1845– 1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalo S., Greentree W. K., Linder M. E. ( 1999) J. Biol. Chem. 274, 21313– 21318 [DOI] [PubMed] [Google Scholar]
- 50.Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., Bastiaens P. I. ( 2005) Science 307, 1746– 1752 [DOI] [PubMed] [Google Scholar]
- 51.Draper J. M., Xia Z., Smith C. D. ( 2007) J. Lipid Res. 48, 1873– 1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartels D. J., Mitchell D. A., Dong X., Deschenes R. J. ( 1999) Mol. Cell. Biol. 19, 6775– 6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duronio R. J., Jackson-Machelski E., Heuckeroth R. O., Olins P. O., Devine C. S., Yonemoto W., Slice L. W., Taylor S. S., Gordon J. I. ( 1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1506– 1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graziano M. P., Freissmuth M., Gilman A. G. ( 1989) J. Biol. Chem. 264, 409– 418 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.