Abstract
Methylation of Lys residues on histone proteins is a well known and extensively characterized epigenetic mark. The recent discovery of lysine-specific demethylase 1 (LSD1) demonstrated that lysine methylation can be dynamically controlled. Among the histone demethylases so far identified, LSD1 has the unique feature of functioning through a flavin-dependent amine oxidation reaction. Data base analysis reveals that mammalian genomes contain a gene (AOF1, for amine-oxidase flavin-containing domain 1) that is homologous to the LSD1-coding gene. Here, we demonstrate that the protein encoded by AOF1 represents a second mammalian flavin-dependent histone demethylase, named LSD2. The new demethylase is strictly specific for mono- and dimethylated Lys4 of histone H3, recognizes a long stretch of the H3 N-terminal tail, senses the presence of additional epigenetic marks on the histone substrate, and is covalently inhibited by tranylcypromine. As opposed to LSD1, LSD2 does not form a biochemically stable complex with the C-terminal domain of the corepressor protein CoREST. Furthermore, LSD2 contains a CW-type zinc finger motif with potential zinc-binding sites that are not present in LSD1. We conclude that mammalian LSD2 represents a new flavin-dependent H3-Lys4 demethylase that features substrate specificity properties highly similar to those of LSD1 but is very likely to be part of chromatin-remodeling complexes that are distinct from those involving LSD1.
Introduction
Histones pack the eukaryotic DNA into the nucleosome, the basic unit of chromatin (1). These highly conserved proteins are not merely spools to wind DNA, but they rather regulate gene expression by modulating the activity of the transcriptional machinery. This is achieved through recognition of histone post-translational modifications by specific transcription factors, according to a scheme dictated by the so-called “histone code” (2, 3). Methylation of Lys residues on the histone N-terminal tails is one of the most extensively characterized epigenetic marks, being involved in the regulation of a plethora of fundamental processes such as heterochromatin formation, X-chromosome inactivation, and DNA repair (4, 5). The recent discovery of lysine-specific demethylase 1 (LSD1)3 demonstrated that lysine methylation can be dynamically controlled and is not a static epigenetic mark as thought in the past (6, 7). LSD1 specifically acts on mono- and dimethylated Lys4 of histone H3 through an oxidative process that requires FAD as essential redox cofactor (Fig. 1a). More recently, several other histone demethylases have been uncovered; they feature the property of containing a JmjC catalytic domain that carries out the reaction through an iron-dependent mechanism (8). The JmjC enzymes are able to act on trimethylated lysines, which is mechanistically impossible for flavin-catalyzed oxidative demethylation reactions (7). Histone demethylases have been found in association with a number of chromatin-remodeling complexes and are involved in many diverse transcriptional programs (9, 10). Several recent articles indicate that histone demethylases may represent potential drug targets, in particular for the treatment of certain solid tumors such as prostate cancer (11, 12).
FIGURE 1.
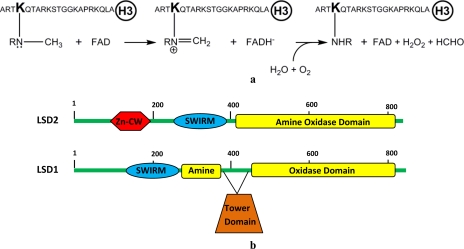
Comparative analysis of the mammalian flavin-dependent histone demethylases LSD1 and LSD2. a, reaction catalyzed by LSD1 and LSD2. The sequence of the N-terminal 21-residue histone H3 peptide used as substrate is shown. R is a hydrogen atom and a methyl group for mono- and dimethylated Lys4, respectively. b, domain organization of LSD1 and LSD2. Both proteins contain a SWIRM domain and the catalytic amine oxidase domain. LSD2 contains a N-terminal zinc finger domain (Zn-CW) that is not present in LSD1. Furthermore, the amine oxidase domain of LSD2 does not include the insertion (tower domain) that in LSD1 provides the binding site for the corepressor protein CoREST (19, 32).
Analysis of sequence data bases reveals that most mammalian genomes contain a gene (denoted as AOF1, for amine-oxidase flavin-containing domain 1) that is homologous to the gene encoding for LSD1 (Fig. 1b). Alignment of the corresponding protein sequences scores an overall 33% identity in the C-terminal amine oxidase domain (13) and in the preceding SWIRM domain (14) (Fig. 2). Indeed, phylogenetic analyses indicate that the two proteins are evolutionary related (15). Here we report on the expression, purification, and biochemical characterization of the enzyme encoded by the mouse AOF1 gene, and we demonstrate that the protein, which we name LSD2,4 is a flavin-dependent histone demethylase acting on mono- and dimethylated Lys4 of histone H3. LSD2 enzymatic activity is comparatively analyzed in the light of the homologous LSD1 and of the implications for its biological role in chromatin processes.
FIGURE 2.
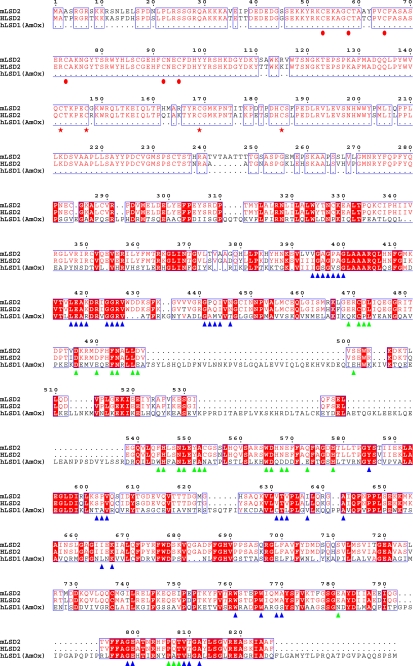
Sequence alignment of LSD2 from M. musculus (mLSD2; NCBI gene identifier 26986558), LSD2 from Homo sapiens (hLSD2; gene identifier 122889312), and LSD1 from H. sapiens (hLSD1; gene identifier 58761545). With regard to human LSD1, only the region that has been used for crystallographic studies (residues 171–852) (19, 20) has been included in the alignment because the N-terminal segment of LSD1 is not homologous to that of LSD2 (see Fig. 1b). Residues conserved in mLSD2 and hLSD2 are indicated by the red font, whereas amino acids that are conserved among all three aligned proteins are highlighted in red. The triangles outline the amino acids involved in FAD (blue) and substrate binding (green), as gathered from the analysis of the human LSD1·CoREST crystal structures (19, 20). The red circles indicate the conserved N-terminal Cys residues, which might represent potential zinc-binding sites. The red stars outline the conserved cysteines of the predicted CW-type zinc finger domain (30). Residue numbering refers to mLSD2. Identity values for the aligned sequences are: 92% between mLSD2 and hLSD2; 32% between mLSD2 and hLSD1; and 33% between hLSD1 and hLSD2.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Reagents were purchased from Sigma-Aldrich unless otherwise stated. The cDNA of LSD2 from Mus musculus was amplified by PCR to generate a full-length and two N-terminally truncated (Δ25 and Δ257) constructs. Amplified fragments were cloned between HindIII and XhoI restriction sites (enzymes from New England Biolabs) in a modified version of pET28b vector, and the His6-tagged fusion proteins were expressed in Escherichia coli BL21-DE3 RPplus cells (Stratagene). The construct LSD2 Δ25 proved to produce the protein with the highest degree of solubility and stability, and therefore, it was selected for biochemical studies (throughout the text we will refer to the Δ25 construct as LSD2). The cells were grown in a Bioflo 3000 fermentor (New Brunswick Scientific, NJ) at 37 °C up to a A600 of 2.0, and protein production was induced at 25 °C by the addition of 0.25 mm isopropyl β-d-thiogalactopyranoside. Harvested cells (30 g) were resuspended in 150 ml of lysis buffer containing 50 mm sodium phosphate, pH 8.0, 5% (w/v) glycerol, 300 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 0.3 mg deoxyribonuclease I, and 3 tablets of EDTA-free protease inhibitors mixture (Roche Applied Science). Cell disruption was accomplished using an Emulsiflex 3C (Avestin Inc.), and cell debris was removed by centrifuging at 70,000 × g for 45 min at 4 °C. Cell extract was loaded on a nickel affinity column (HisTrap FF 1 ml; GE Healthcare). The resin was washed with 50 mm sodium phosphate, pH 8.0, 300 mm NaCl, 5% (w/v) glycerol, and protein was eluted with 100 mm imidazole in the same buffer. Fractions enriched in LSD2 were pooled and loaded onto a HiScreen CaptoQ anion exchange column (GE Healthcare) equilibrated in 50 mm Hepes, pH 8.5, 10% (w/v) glycerol. Elution was performed by a linear gradient of NaCl (0–1 m). Fractions containing LSD2 were concentrated using an Amicon30 (Millipore) ultrafiltration device, and then gel filtered in 50 mm Hepes/NaOH, pH 8.0, 5% (w/v) glycerol on a Superdex200 HR 10/30 column (GE Healthcare). Sample purity was checked by SDS-PAGE, and protein concentration was estimated by the flavin absorption peak at 458 nm (Fig. 3). Extinction coefficient was determined to be 10.4 mm−1 cm−1 (16).
FIGURE 3.
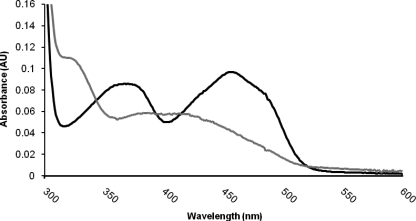
Absorption spectra of LSD2 (10 μm protein in 50 mm Hepes/NaOH, pH 8.0, 5% (w/v) glycerol) in the native oxidized state (black line) and after incubation with an excess of the inhibitor tranylcypromine (gray line). The oxidized enzyme exhibits the characteristic spectrum of a flavoprotein with maxima at 375 and 458 nm. The spectrum of the tranylcypromine-bound protein is fully consistent with the formation of a covalent adduct between the flavin cofactor and the inhibitor, which leads to the bleaching of the protein. This spectrum is very similar to that observed for the tranylcypromine-inhibited human LSD1 (27).
Activity and Inhibition Assays
Time course measurements were performed under aerobic conditions by using a Cary 100 UV-visible spectrophotometer (Varian Inc.) equipped with a thermostatted cell holder (T = 25 °C). Enzymatic activities were evaluated using both horseradish peroxidase-coupled (17) and formaldehyde dehydrogenase-coupled (18) assays with histone H3 peptides as substrates (Thermo Electron Corp., Ulm, Germany; Fig. 1a). In both cases, the reaction was started by adding 3 μl of the LSD2 enzyme (50 μm protein in 50 mm Hepes/NaOH buffer, pH 8.0, and 5% (w/v) glycerol) to 150-μl reaction mixtures. In the peroxidase-coupled assay, the reaction mixture consisted of 45 mm Hepes/NaOH, pH 8.5, 0.1 mm 4-aminoantipyrine, 1 mm 3,5-dichloro-2-hydroxybenzenesulfonic acid, 0.35 μm horseradish peroxidase, and variable concentrations (3–120 μm) of methylated peptides. Absorbance changes were monitored at 515 nm, and an extinction coefficient of 26 mm−1 cm−1 was used to calculate the initial velocities values (7). In the formaldehyde dehydrogenase-coupled assay, the mixture contained 45 mm Hepes/NaOH, pH 8.5, 0.2 unit/ml formaldehyde dehydrogenase, and 2 mm NAD+. The reaction was followed by monitoring NADH formation at 340 nm (ϵ340 = 6.22 mm−1 cm−1). The initial velocity values were fitted to the Michaelis-Menten equation using Grafit (Erithacus software), which provides the kcat and Km parameters along with their associated errors (see Table 1). Inhibition assays were performed by using the peroxidase-coupled assay in the presence of varied concentrations (1–120 μm) of dimethylated H3-K4 peptide and of the inhibitor under analysis. The initial velocity values were fitted to equations describing competitive, uncompetitive, and noncompetitive inhibition.
TABLE 1.
Comparative analysis of steady-state kinetic parameters and inhibition of LSD2 and LSD1 towards peptide substrate or inhibitors
| na | Modification/mutation | LSD2b |
LSD1b,c |
||||
|---|---|---|---|---|---|---|---|
| km | kcat | ki | km | kcat | Ki | ||
| μm | min−1 | μm | μm | min−1 | μm | ||
| Histone H3 peptides | |||||||
| 21 | Monomethyl-Lys4 | 9.2 ± 0.9 | 0.28 ± 0.01 | 3.4 ± 0.2 | 3.40 ± 0.10 | ||
| 21 | Dimethyl-Lys4 | 11.3 ± 1.3 | 2.00 ± 0.60 | 4.2 ± 0.5 | 8.10 ± 0.20 | ||
| 21 | Trimethyl-Lys4 | No activity | No activity | 58.0 ± 6.6 | No activity | No activity | 19.5 ± 3.2 |
| 8 | Monomethyl-Lys4 | No activity | No activity | No activity | No activity | ||
| 8 | Dimethyl-Lys4 | No activity | No activity | No activity | No activity | ||
| 16 | Monomethyl-Lys4 | No activity | No activity | No activity | No activity | ||
| 30 | Monomethyl-Lys4 | 5.1 ± 0.5 | 0.41 ± 0.01 | 3.4 ± 0.5 | 2.90 ± 0.10 | ||
| 21 | Monomethyl-Lys4, Monomethyl-Lys9 | 9.0 ± 1.5 | 0.24 ± 0.01 | 3.9 ± 0.5 | 5.60 ± 0.20 | ||
| 21 | Dimethyl-Lys4, dimethyl-Lys9 | 6.6 ± 0.9 | 1.33 ± 0.04 | 3.7 ± 0.4 | 4.20 ± 0.10 | ||
| 21 | Monomethyl-Lys4, acetyl-Lys9 | 70.5 ± 10.2 | 0.35 ± 0.01 | 17.5 ± 4.0 | 4.10 ± 0.30 | ||
| 21 | Monomethyl-Lys4, acetyl(Lys9, Lys14, Lys18) | No activity | No activity | No activity | No activity | ||
| 21 | Monomethyl-Lys9 | No activity | No activity | 6.6 ± 1.2 | No activity | No activity | 2.8 ± 0.3 |
| 21 | Monomethyl-Arg2, monomethyl-Lys4 | No activity | No activity | 5.5 ± 1.4 | 0.63 ± 0.03 | ||
| 21 | Monomethyl-Lys4, monomethyl-Arg8 | No activity | No activity | No activity | No activity | ||
| 21 | Monomethyl-Lys4, monomethyl-Arg17 | 8.1 ± 1.00 | 0.3 ± 0.02 | 5.7 ± 1.70 | 2.50 ± 0.20 | ||
| 21 | Monomethyl-Lys4, phospho-Ser10 | No activity | No activity | No activity | No activity | ||
| 21 | Phospho-Thr3, dimethyl-Lys4 | No activity | No activity | No activity | No activity | ||
| 21 | R2A, dimethyl-Lys4 | No activity | No activity | No activity | No activity | ||
| 21 | Monomethyl-Lys4, P16A | 12.5 ± 0.9 | 0.26 ± 0.07 | 4.9 ± 1.2 | 3.0 ± 0.2 | ||
| 21 | Monomethyl-Lys4, G12A, G13A | No activity | No activity | No activity | No activity | ||
| 21 | None (demethylated product) | No activity | No activity | 33.4 ± 4.3 | No activity | No activity | 1.8 ± 0.6 |
| 21 | K4M | No activity | No activity | 0.15 ± 0.1 | No activity | No activity | 0.05 ± 0.02 |
| 21 | K4Q | No activity | No activity | 2.0 ± 0.3 | No activity | No activity | 1.1 ± 0.1 |
| 21 | K4R | No activity | No activity | 46.0 ± 9 | No activity | No activity | 0.41 ± 0.05 |
| Non-histone substrated | |||||||
| 26 | Dimethyl-Lys370 p53 (residues 363–388) | No activity | No activity | No activity | No activity | ||
| 26 | Dimethyl-Lys370 p53 (residues 353–378) | No activity | No activity | No activity | No activity | ||
a The length (number of amino acids) of the histone H3 N-terminal peptide is indicated. The sequence of the H3 N-terminal tail is shown in Fig. 1a. All of the assays were carried out with a peroxidase-coupled method.
b Apparent steady-state kinetic parameters determined as described under “Experimental Procedures.”
c The kinetic parameters for human LSD1 are taken from Refs. 22 and 23 with the exception of the data on the two p53 peptides and the H3 peptides dimethyl-Lys4-dimethyl-Lys9, monomethyl-Lys9, phospho-Thr3, dimethyl-Lys4, K4Q. For these peptides, the parameters were determined following the assays described in Ref. 23.
d Peptides corresponding to residues 363–388 and 353–378 of human p53, comprising methylated Lys370. They have been tested following the data reported in Ref. 24. Both human LSD1 (unpublished) and mouse LSD2 do not show any activity on these peptides.
Effect of Ionic Strength and pH on LSD2 Activity
To evaluate the effects of ionic strength on LSD2 activity, the assays were performed by the peroxidase-coupled assay using increasing amounts of NaCl and KCl in the reaction mixture, starting from 1 mm up to 500 mm. In the determination of the pH dependence of the enzyme activity, the pH range was covered using Mes/NaOH (pH 6.0 or 6.5), Hepes/NaOH (pH 7.0, 7.5, 8.0, or 8.5), or Ches/NaOH (pH 9.0, 9.5, or 10.0) buffers at a final concentration of 45 mm. The ionic strength was kept constant at 40 mm by the addition of appropriate amounts of NaCl. The final enzyme and substrate concentrations were 1.0 and 30 μm, respectively.
Evaluation of Zinc Content
The amount of zinc present in protein solutions was measured using inductively coupled plasma-mass spectrometry on a PerkinElmer Life Sciences ELAN DRC-e instrument, following the standard procedures suggested by the manufacturer. Field blanks and blanks of reagents used throughout the procedure were evaluated and taken into account. Gel filtered protein solutions consisted of 60 μm LSD2 in 50 mm sodium phosphate, pH 7.0, and 10% (w/v) glycerol. The experiment was repeated on three different protein preparations without observing any significant variations. As a control, we evaluated whether the protein contained other cations such as manganese, nickel, and calcium. None of them was detected in significant amounts. Furthermore, we verified that other flavoproteins (a bacterial flavin-dependent monooxygenase and the human LSD1·CoREST complex) that do not specifically bind zinc did not exhibit any significant zinc content in the mass spectrometry analysis.
Binding Assays on CoREST
The LSD1-binding domain of human CoREST (residues 305–482; the C-terminal region of the protein) (19) was used to evaluate a possible interaction of LSD2 to this corepressor protein. Cell extracts containing glutathione S-transferase-tagged CoREST and His6-tagged LSD2 were subsequently passed through GSTrap and HisTrap columns (GE Healthcare) in a tandem affinity pulldown procedure, following the protocols for the in vitro reconstitution of the LSD1-CoREST complex (20). The eluted fractions were analyzed by SDS-PAGE. To rule out the possibility that the glutathione S-transferase and His6 tags may hamper the interaction, untagged CoREST and LSD2 were obtained by Prescission and TEV protease cleavage and incubated overnight in 1.5:1 molar ratio (CoREST:LSD2) at 4 °C. The sample was then loaded on a Superdex200 HR 10/30 gel filtration column (GE Healthcare) in 50 mm Hepes/NaOH, pH 8.0, 5% (w/v) glycerol, and the fractions were analyzed by SDS-PAGE. In vitro reconstitution and enzymatic assays with the human LSD1·CoREST complex were carried out as described (20).
RESULTS
LSD2 Acts on Mono- and Dimethylated Lys4 of Histone H3
We expressed the LSD2 protein encoded by AOF1 gene from M. musculus (Fig. 1b) as a recombinant enzyme lacking the first 25 amino acids, which are predicted to be disordered on the basis of bioinformatic sequence analysis (21). We purified the enzyme to homogeneity, and we found that it tightly binds a flavin cofactor as demonstrated by the characteristic UV-visible spectrum (Fig. 3). Protein denaturation (16) further showed that the cofactor is a noncovalently bound FAD molecule.
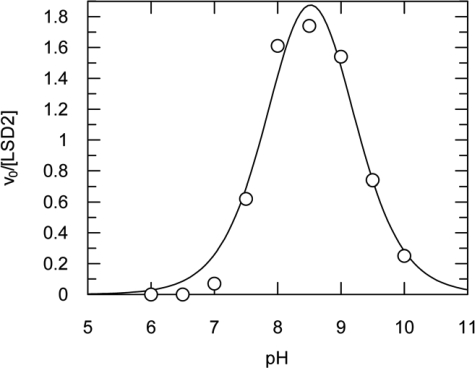
To investigate the catalytic activity of LSD2, we probed several histone peptides following procedures similar to those employed for human LSD1 (22, 23) (Table 1). We used two different enzymatic assays that detect hydrogen peroxide and formaldehyde production, respectively (Fig. 1a). These experiments clearly demonstrated that LSD2 is active on peptides corresponding to the N-terminal 21 amino acids of histone H3 that are mono- or dimethylated on Lys4 (Fig. 4). The enzyme exhibited maximal activity at pH 8.5 (Fig. 5), and turned out to be highly sensitive to ionic strength with a substantial drop in activity at ionic concentrations above 100 mm. The turnover numbers at pH 8.5 for the mono- and dimethylated H3-Lys4 substrates resulted to be 0.28 and 2.0 min−1, respectively (Table 1). Various peptides corresponding to different N-terminal segments of H3 were assayed to identify the minimal length required for productive binding and catalysis. Peptides of 8 and 16 amino acids were not substrates for LSD2, whereas the catalytic activity on a 30-residue peptide was very similar to that measured on the 21-amino acid substrate (Table 1 and Fig. 4b). Peptides with trimethylated Lys4 were not demethylated, consistently with the flavin-dependent oxidative mechanism, which requires a lone electron pair on the substrate nitrogen atom (9) (Fig. 1a). Moreover, like LSD1, LSD2 was inactive on a H3 peptide methylated on Lys9 (Fig. 4b) and on nonhistone substrates such as p53 peptides methylated on Lys370, which was suggested to represent a target site for LSD1 (24). Taken together, these data demonstrated that LSD2 is able to specifically catalyze the demethylation of Lys4 of histone H3 through an oxidative mechanism that (i) requires a redox cofactor for substrate oxidation, (ii) can use oxygen as electron acceptor generating hydrogen peroxide, and (iii) produces formaldehyde as a result of the hydrolysis of the imine group generated by the oxidation of the methylated lysine side chain (Fig. 1a).
FIGURE 4.
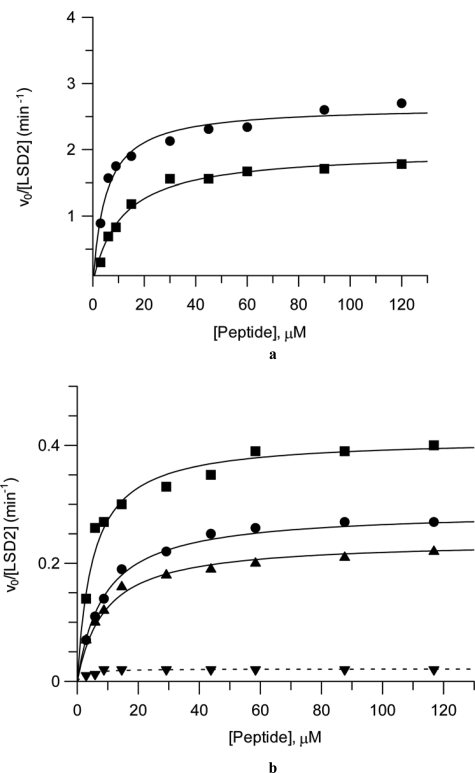
Demonstration of H3-Lys4 demethylase activity of LSD2. a, steady-state kinetic parameters were investigated by two assays that detect formation of two different reaction products (Fig. 1a), hydrogen peroxide and formaldehyde, respectively. The assays were carried out using 3–120 μm H3-K4 dimethylated peptide at 1.0 μm protein concentration and pH 8.5 (see “Experimental Procedures” and Table 1). Under these conditions, the kcat and Km values were 2.0 min−1 and 11.3 μm, respectively, in the peroxidase assay (squares) and 2.6 min−1 and 6.2 μm, respectively, in the formaldehyde dehydrogenase assay (circles). The curves are the best fit of the data to the Michaelis-Menten equation (v = Vmax[S]/Km + [S]). b, LSD2 specificity toward Lys4 of histone H3. The picture plots the dependence of the initial velocities of LSD2 reaction toward different peptides measured with the peroxidase-coupled assay: 30-amino acid peptide monomethylated on Lys4 (■), 21-amino acid peptide monomethylated on Lys4 (●), 21-amino acid peptide bearing monomethylated Lys4 and monomethylated Lys9 (▲), and 21-amino acid peptide monomethylated on Lys9 (▼).
FIGURE 5.
pH dependence of the LSD2 activity. The figure plots the dependence of the initial velocities measured using a 21-amino acid dimethyl-Lys4 peptide (30 μm) with the peroxidase-coupled assay. The highest activity is measured in the pH range 7.5–9, whereas it exhibits a sharp drop at lower and higher pH values. The curve shows the best fit of the data to a double pKa model (pKa1 = 7.9 ± 0.1 and pKa2 = 9.0 ± 0.1).
Substrate Specificity and Effect of Epigenetic Marks
To evaluate the effect on enzyme catalysis by additional epigenetic marks on the H3 tail, we used peptides carrying methylated Lys4 together with additional post-translational modifications (25). We found that acetylation of Lys9 makes substrate binding less efficient as shown by the 8-fold increase in the Km value (Table 1). Furthermore, hyperacetylation (i.e. simultaneous acetylation of Lys9, Lys14, and Lys18), which is a hallmark of gene activation, makes the peptide unable to function as LSD2 substrate (Fig. 4b). A similar effect is exerted by phosphorylation of Thr3 and Ser10 as well as by methylation of Arg2 and Arg8. The only two epigenetic modifications that did not exhibit any significant effect on catalysis were methylation of Lys9 and Arg17. As summarized in Table 1, these substrate recognition features are very similar to those exhibited by LSD1.
Next, to further explore the specificity of substrate binding, we used peptides mutated on the residues surrounding the site of demethylation (Table 1). Mutation of the only glycine residues of the peptide substrate (Gly12-Gly13) impairs LSD2 activity, whereas replacement of Pro16 with Ala has little effect. Moreover, on the basis of the crucial role of Arg2 highlighted by the crystal structure of LSD1 in complex with a histone peptide (20), we also tested a monomethylated H3-Lys4 peptide with Arg2 mutated to Ala. No demethylation activity was detected, confirming that Arg2 is central to substrate recognition also in LSD2. These observations, together with the finding that the complete stretch of the N-terminal 21 amino acids of H3 is necessary for catalytic activity, suggest that substrate binding does not rely only on methyl-Lys4 recognition, but it also involves extensive interactions with many residues of the H3 tail. Indeed, inhibition studies showed that the demethylated peptide (the product of the LSD2 reaction; Fig. 1a) competitively inhibits LSD2 with a Ki of 33 μm. Likewise, we found that the trimethylated Lys4 peptide and peptides in which Lys4 is mutated to Met, Gln, and Arg, are competitive inhibitors, with the K4M peptide exhibiting the highest affinity (Ki = 0.15 μm) as observed for human LSD1 (Table 1) (20).
In the framework of this biochemical analysis, we also investigated the inhibitory activity of tranylcypromine. This compound was originally developed as inhibitor of human monoamine oxidases and has been widely used as antidepressant (26). More recently, tranylcypromine has been shown to inhibit also LSD1 (Ki of 242 μm), although its potency is 100 times lower compared with that against monoamine oxidases (27–29). However, the mechanism of inhibition was found to be similar for all of these enzymes; tranylcypromine reacts with the flavin and forms a covalent adduct with the cofactor. Now, we find that this compound inhibits also LSD2 (Ki = 180 μm) and produces a modification of the flavin absorption spectrum that is fully consistent with formation of a covalent adduct (Fig. 3). As found for LSD1, other monoamine oxidase covalent inhibitors such as pargyline, deprenyl, and rasagiline (26) did not exert any inhibitory effect on LSD2 and did not perturb the enzyme absorption spectrum. These observations are relevant from two points of view. First, they further emphasize the similarity between the catalytic and substrate binding properties of LSD1 and LSD2. Second, they confirm that tranylcypromine is a nonspecific inhibitor of flavin-dependent amine oxidases, a feature that must be considered for a correct interpretation of the cell biology and pharmacology studies that are based on this compound.
Making the Difference: Zinc Finger Domain and Interactions with the Corepressor CoREST
The amino acid sequences of LSD1 and LSD2 are clearly homologous in their SWIRM and amino oxidase domains, but their N-terminal portions are unrelated (15). The first 150 amino acids of LSD1 are predicted to be disordered, whereas bioinformatic analyses on LSD2 identifies an N-terminal CW-type zinc finger domain (residues 130–200), which does not have a counterpart in LSD1 (Figs. 1b and 2) (15, 30). Zinc finger domains typically mediate interactions with either DNA or proteins, with the CW-type being characterized by conserved cysteine and tryptophan residues (30, 31). To obtain insight into this specific feature of LSD2, we performed an inductively coupled plasma mass spectrometry analysis, which indeed indicated that LSD2 contains zinc with 3:1 molar ratio (zinc:protein). Conversely, the same analysis on the LSD1·CoREST complex did not reveal any significant amount of this metal (molar ratio, <0.02). Although these data do not allow us to firmly establish the residues directly involved in zinc binding, they are fully consistent with the notion that the N-terminal region of LSD2 contains a CW-type zinc finger domain (30). In this regard, an interesting point is that the mass spectrometry indicates that there might be more than one zinc atom per enzyme molecule. The inspection of the amino acid sequence reveals that in addition to the Cys residues of the predicted zinc finger domain (Cys142, Cys147, Cys169, and Cys185), there are six other Cys residues (Cys53, Cys58, Cys65, Cys73, Cys92, and Cys95) that are conserved among mammalian LSD2 orthologues (Fig. 2). Although sequence similarity searches and phylogenetic analyses (15) do not identify the N-terminal 100 amino acids as a potential zinc finger domain, it is tempting to speculate that these conserved cysteine residues might represent a mono- or di-nuclear zinc-binding region or domain, in addition to the more clearly identified CW-type domain formed by residues 130–200. The discovery of the role of the zinc finger domains in LSD2 function, possibly in mediating the interactions with other chromatin proteins or nucleosomal DNA, will be a crucial line of investigation in future studies.
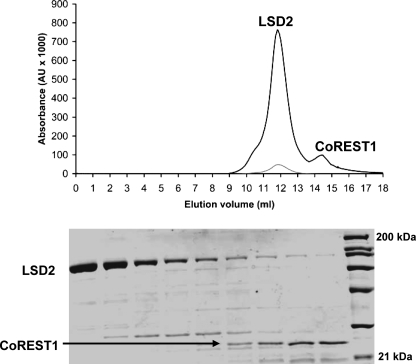
Another difference between LSD1 and LSD2 emerging from the comparison of their sequences is that LSD2 lacks the 100-amino acid insertion that in LSD1 forms the so-called “tower domain,” responsible for the binding to the corepressor protein CoREST (Figs. 1b and 2) (19). CoREST and LSD1 form a very tight heterodimeric complex that does not dissociate even at very high ionic strength. Moreover, CoREST has been shown to increase the stability and the activity of LSD1 both in vivo and in vitro (20, 32, 33). In particular, this effect is exerted by the C-terminal region of CoREST, which has been shown to bind to LSD1 and has been used for the crystallographic investigation of the LSD1·CoREST complex (19, 20). To probe the ability of LSD2 to associate to CoREST, we employed two chromatographic approaches using recombinant LSD2 and the C-terminal portion of human CoREST (residues 305–482; Fig. 6).5 First, we took advantage of the different purification tags present on the two recombinant proteins (His6 on LSD1 and glutathione S-transferase on CoREST) to attempt a tandem affinity purification of the complex. This experiment did not provide any evidence for the formation of a stable complex between the two proteins. Next, to rule out any interference on complex formation by the purification tags, we performed size exclusion chromatography experiments using untagged proteins. LSD2 and CoREST were incubated in a 1:1.5 molar ratio and subsequently loaded on a gel filtration column. The two proteins eluted in well separated peaks with no evidence in support of a significant association between them (Fig. 6). We remark that both these chromatographic methods are very effective in the reconstitution and purification of the LSD1·CoREST complex (19, 20, 32). As additional check, we measured the effect of CoREST on the enzymatic activity of LSD2. This experiment originated from the fact that the LSD1·CoREST complex exhibits a catalytic efficiency that is three times higher than that exhibited by isolated LSD1 (20, 32, 33). However, both kcat and Km values measured on LSD2 incubated with an excess of CoREST were undistinguishable from those exhibited by LSD2 alone (Table 1). Thus, all of these experiments consistently and clearly indicate that, unlike LSD1, LSD2 does not tightly associate to CoREST.
FIGURE 6.
Binding assay on LSD2 and CoREST. Recombinant untagged CoREST (residues 305–482) and LSD2 were incubated overnight in 1.5:1 molar ratio (CoREST:LSD2) at 4 °C. The sample was then loaded on a Superdex200 HR 10/30 gel filtration column (GE Healthcare) in 50 mm Hepes/NaOH, pH 8.0, 5% glycerol (w/v). The top panel shows the gel filtration chromatogram. Elution was followed by monitoring the absorbance at 280 nm (black line) and 458 nm (gray line; the flavin absorption peak). The 280-nm extinction coefficients for LSD2 and CoREST are 123 and 17 mm−1 cm−1, respectively. The 458-nm extinction coefficient for LSD2 is 10.4 mm−1 cm−1. The bottom panel shows the SDS-PAGE (12% v/v acrylamide-bisacrylamide) analysis of the 0.4-ml fractions eluted in the range 11.6–14.8 ml. The molecular mass markers are in lane 10 (21, 31, 45, 66, 97, 116, and 200 kDa). The molecular masses of LSD2 and CoREST are 90 and 22 kDa, respectively. The elution volumes of CoREST and LSD2 are identical to those observed with the isolated proteins. The weak LSD2 bands present in lanes 7–9 are due to the tail of the broad LSD2 elution peak, observed also in the chromatogram of the isolated protein.
DISCUSSION
Histone demethylases are responsible for the dynamic regulation of histone Lys methylation, an epigenetic mark crucial for the gene expression regulation and chromatin function. These enzymes are grouped in two classes according to the coenzyme employed to catalyze the demethylation reaction: the flavin-dependent and the JmjC iron-dependent histone demethylases, respectively. Despite the number of JmjC demethylases reported in the literature, the only mammalian flavin-dependent enzyme so far characterized is LSD1 (6, 7). Our biochemical studies show that the AOF1 gene, the only clearly identified paralogue of LSD1 present in human and other mammalian genomes, encodes for a second mammalian flavin-dependent histone demethylase, which, like LSD1, is strictly specific for mono- and dimethylated Lys4 of histone H3. Both LSD1 and LSD2 enzymes have a low turnover rate (Table 1) in line with the catalytic efficiency typically exhibited by histone-modifying enzymes (9). Their reactions produce hydrogen peroxide and formaldehyde and are similarly affected by high ionic strength and acidic pH. They also appear to be strictly specific for their substrates as indicated by the strong effect exerted by mutations in the side chains of the H3 N-terminal tail (Table 1 and Ref (34)). More important, LSD1 and LSD2 both feature the ability to sense the presence of additional epigenetic modifications on the H3 tail (6, 22, 32). In particular, epigenetic marks associated with gene activation such as Ser10 phosphorylation and Lys acetylation impair their catalytic activities by drastically affecting substrate binding and affinity. Altogether, these data indicate that LSD1 and LSD2 catalyze the same enzymatic reaction (H3-Lys4 demethylation) and have very similar, if not identical, mechanisms of recognition of the H3 N-terminal tail, which must be deprived of most epigenetic post-translational modifications for optimal binding. This biochemical similarity is in agreement with the high degree of homology displayed by the amino acid sequences of their catalytic domains (Fig. 2).
In contrast, LSD1 and LSD2 differ in the domains that are involved in protein-protein interactions. LSD2 lacks the “tower domain,” which forms the CoREST-binding site and is instrumental to the tight LSD1-CoREST association (Fig. 1b). Consistently, CoREST does not form a biochemically stable complex with LSD2 (Fig. 6). Moreover, LSD2 contains a CW-type zinc finger motif and other potential zinc-binding sites that are not present in LSD1. We conclude that LSD2 represents a new flavin-dependent histone demethylase, which features substrate specificity properties highly similar to those of LSD1 but is very likely to be part of chromatin-remodeling complexes and transcription programs that are distinct from those involving LSD1. A fundamental issue will be to advance our knowledge on the cross-talk with other histone-modifying enzymes. In this context, the strict specificity for deacetylated and dephosphorylated peptides raises the hypothesis that LSD2 might function in combination with deacetylases and/or phosphatases. The investigation of these problems will obviously be connected to the understanding of the function of the LSD2 N-terminal region and its role in protein-protein interactions.
This work was supported by the Italian Ministry of Science PRIN06 and FIRB programs, Fondazione Cariplo, and the Italian Association for Cancer Research.
4 Scientists in the chromatin field recently proposed to establish a rational nomenclature that envisions the acronym KDM (K-demethylases) for histone-Lys demethylases (10). Being the forerunner of this class of enzymes, LSD1 was named KDM1, whereas the JmjC proteins were sequentially denoted as KDM2 and so forth. Moreover, in a recent phylogenetic analysis (15), LSD1 and the AOF1-encoded protein have been named KDM1A and KDM1B, respectively. Given the widespread usage of the “LSD1” acronym in current literature, we have chosen the name “LSD2” for this new member of the flavin-dependent lysine demethylase family.
5 Human and mouse CoREST are highly similar, sharing 96% overall sequence identity. Moreover, their LSD1-binding regions (residues 305–482 of the human protein) differ for only six side chains that are all located in the C-terminal segment (residues 442–482), which is disordered in the human LSD1·CoREST crystal structure (19, 20).
- LSD
- lysine-specific demethylase
- Ches
- 2-(cyclohexylamino)ethanesulfonic acid
- Mes
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Zlatanova J., Bishop T. C., Victor J. M., Jackson V., van Holde K. ( 2009) Structure 17, 160– 171 [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T., Allis C. D. ( 2001) Science 293, 1074– 1080 [DOI] [PubMed] [Google Scholar]
- 3.Ruthenburg A. J., Li H., Patel D. J., Allis C. D. ( 2007) Nat. Rev. Mol. Cell Biol. 8, 983– 994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sims R. J., 3rd, Nishioka K., Reinberg D. ( 2003) Trends Genet. 19, 629– 639 [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. ( 2007) Cell 128, 693– 705 [DOI] [PubMed] [Google Scholar]
- 6.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. ( 2004) Cell 119, 941– 953 [DOI] [PubMed] [Google Scholar]
- 7.Forneris F., Binda C., Vanoni M. A., Mattevi A., Battaglioli E. ( 2005) FEBS Lett. 579, 2203– 2207 [DOI] [PubMed] [Google Scholar]
- 8.Anand R., Marmorstein R. ( 2007) J. Biol. Chem. 282, 35425– 35429 [DOI] [PubMed] [Google Scholar]
- 9.Smith B. C., Denu J. M. ( 2009) Biochim. Biophys. Acta 1789, 45– 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allis C. D., Berger S. L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., Shilatifard A., Workman J., Zhang Y. ( 2007) Cell 131, 633– 636 [DOI] [PubMed] [Google Scholar]
- 11.Lan F., Nottke A. C., Shi Y. ( 2008) Curr. Opin. Cell Biol. 20, 316– 325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marmorstein R., Trievel R. C. ( 2009) Biochim. Biophys. Acta 1789, 58– 68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binda C., Mattevi A., Edmondson D. E. ( 2002) J. Biol. Chem. 277, 23973– 23976 [DOI] [PubMed] [Google Scholar]
- 14.Da G., Lenkart J., Zhao K., Shiekhattar R., Cairns B. R., Marmorstein R. ( 2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2057– 2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X., Ma H. ( 2008) BMC Evol. Biol. 8, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aliverti A., Curti B., Vanoni M. A. ( 1999) Methods Mol. Biol. 131, 9– 23 [DOI] [PubMed] [Google Scholar]
- 17.Smith T. A., Barker J. H. ( 1988) Progress in Polyamine Research: Novel Biochemical, Pharmacological and Clinical Aspects ( Zappia V., Pegg A.E., eds) Plenum Press, New York [Google Scholar]
- 18.Lizcano J. M., Unzeta M., Tipton K. F. ( 2000) Anal. Biochem. 286, 75– 79 [DOI] [PubMed] [Google Scholar]
- 19.Yang M., Gocke C. B., Luo X., Borek D., Tomchick D. R., Machius M., Otwinowski Z., Yu H. ( 2006) Mol. Cell 23, 377– 387 [DOI] [PubMed] [Google Scholar]
- 20.Forneris F., Binda C., Adamo A., Battaglioli E., Mattevi A. ( 2007) J. Biol. Chem. 282, 20070– 20074 [DOI] [PubMed] [Google Scholar]
- 21.Rost B., Yachdav G., Liu J. ( 2004) Nucleic Acids Res. 32, W321– W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forneris F., Binda C., Vanoni M. A., Battaglioli E., Mattevi A. ( 2005) J. Biol. Chem. 280, 41360– 41365 [DOI] [PubMed] [Google Scholar]
- 23.Forneris F., Binda C., Dall'Aglio A., Fraaije M. W., Battaglioli E., Mattevi A. ( 2006) J. Biol. Chem. 281, 35289– 35295 [DOI] [PubMed] [Google Scholar]
- 24.Huang J., Sengupta R., Espejo A. B., Lee M. G., Dorsey J. A., Richter M., Opravil S., Shiekhattar R., Bedford M. T., Jenuwein T., Berger S. L. ( 2007) Nature 449, 105– 108 [DOI] [PubMed] [Google Scholar]
- 25.Lall S. ( 2007) Nat. Struct. Mol. Biol. 14, 1110– 1115 [DOI] [PubMed] [Google Scholar]
- 26.Edmondson D. E., Mattevi A., Binda C., Li M., Hubálek F. ( 2004) Curr. Med. Chem. 11, 1983– 1993 [DOI] [PubMed] [Google Scholar]
- 27.Schmidt D. M., McCafferty D. G. ( 2007) Biochemistry 46, 4408– 4416 [DOI] [PubMed] [Google Scholar]
- 28.Mimasu S., Sengoku T., Fukuzawa S., Umehara T., Yokoyama S. ( 2008) Biochem. Biophys. Res. Commun. 366, 15– 22 [DOI] [PubMed] [Google Scholar]
- 29.Binda C., Li M., Hubalek F., Restelli N., Edmondson D. E., Mattevi A. ( 2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9750– 9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry J., Zhao Y. ( 2003) Trends Biochem. Sci. 28, 576– 580 [DOI] [PubMed] [Google Scholar]
- 31.Gamsjaeger R., Liew C. K., Loughlin F. E., Crossley M., Mackay J. P. ( 2007) Trends Biochem. Sci. 32, 63– 70 [DOI] [PubMed] [Google Scholar]
- 32.Shi Y., Matson C., Lan F., Iwase S., Baba T., Shi Y. ( 2005) Mol. Cell 19, 857– 864 [DOI] [PubMed] [Google Scholar]
- 33.Lee M. G., Wynder C., Cooch N., Shiekhattar R. ( 2005) Nature 437, 432– 435 [DOI] [PubMed] [Google Scholar]
- 34.Forneris F., Binda C., Battaglioli E., Mattevi A. ( 2008) Trends Biochem. Sci. 33, 181– 189 [DOI] [PubMed] [Google Scholar]