Abstract
We report a synthetic biology approach to demonstrate substrate channeling in an unusual bifunctional flavoprotein dimethylglycine oxidase. The catabolism of dimethylglycine through methyl group oxidation can potentially liberate toxic formaldehyde, a problem common to many amine oxidases and dehydrogenases. Using a novel synthetic in vivo reporter system for cellular formaldehyde, we found that the oxidation of dimethylglycine is coupled to the synthesis of 5,10-methylenetetrahydrofolate through an unusual substrate channeling mechanism. We also showed that uncoupling of the active sites could be achieved by mutagenesis or deletion of the 5,10-methylenetetrahydrofolate synthase site and that this leads to accumulation of intracellular formaldehyde. Channeling occurs by nonbiased diffusion of the labile intermediate through a large solvent cavity connecting both active sites. This central “reaction chamber” is created by a modular protein architecture that appears primitive when compared with the sophisticated design of other paradigm substrate-channeling enzymes. The evolutionary origins of the latter were likely similar to dimethylglycine oxidase. This work demonstrates the utility of synthetic biology approaches to the study of enzyme mechanisms in vivo and points to novel channeling mechanisms that protect the cell milieu from potentially toxic reaction products.
Introduction
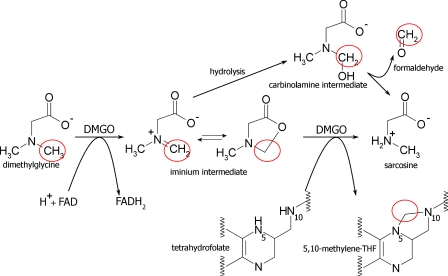
The catabolism of betaine (a breakdown product of choline) and other methylated amines liberates one-carbon (C1) units derived from substrate methyl groups (1). Whereas C1 units derived from the oxidation of betaine are coupled to methionine production, the subsequent oxidation of dimethylglycine, sarcosine, and glycine can potentially release formaldehyde, a toxic product (2), by hydrolysis of unstable imine intermediates (3–5), which is a problem common to many amine oxidases and dehydrogenases (6). Thus, rapid hydrolysis of these imines yields formaldehyde and the demethylated amine product (Fig. 1).
FIGURE 1.
Dimethylglycine oxidation by DMGO. Schematic overview of dimethylglycine oxidation by DMGO with the reactive C1 units circled in red for the respective molecular species. The iminium intermediate generated can interconvert between a linear iminium and cyclic lactone species prior to the enzymatic reaction with tetrahydrofolate. On the other hand, non-enzymatic rapid hydrolysis of the iminium intermediate leads to a transient carbinolamine that is converted into formaldehyde and sarcosine.
Many organisms can further metabolize formaldehyde (7–8), but for most the inherent reactivity of this compound makes it an undesirable byproduct of methyl group oxidation (9). In mitochondria and a significant proportion of bacteria, formaldehyde production is avoided by coupling amine oxidation to the synthesis of 5,10-methylenetetrahydrofolate (5,10-CH2-THF)3 (5), thereby enabling transfer to the folate C1 pool (1, 10). In the glycine cleavage system, comprising several monofunctional proteins (11), premature hydrolysis is avoided by covalent addition of the substrate to the carrier H-protein. This shields the lipoyl-bound intermediates until a productive complex with one of the other glycine cleavage system components is formed. Enzymes responsible for the oxidation of dimethylglycine or sarcosine contain both amine oxidase/dehydrogenase and 5,10-CH2-THF synthase activities on a single polypeptide chain. This suggests that channeling of labile intermediates between active sites prevents release of formaldehyde into the cell.
Channeling of substrates has been suggested in a number of metabolic pathways (12–17). Examples include purine and pyrimidine synthesis, DNA replication, glycolysis, the tricarboxylic acid cycle, lipid metabolism, and amino acid metabolism. That said, direct and compelling evidence for channeling is often lacking. Channeling has been established in only a few systems including tryptophan synthase (18) and carbamoyl-phosphate synthase (19), imidazoleglycerol-phosphate synthase, (20), and 4-hydroxy-2-ketovalerate aldolase/aldehyde dehydrogenase (21) for which both structural and biochemical data are available. These proteins employ “tunnels” to connect active sites (13, 14) where they protect reaction intermediates and steer them to a second active site. Allosteric mechanisms aid communication between active sites and thus ensure that production of a reaction intermediate is balanced with respect to its consumption.
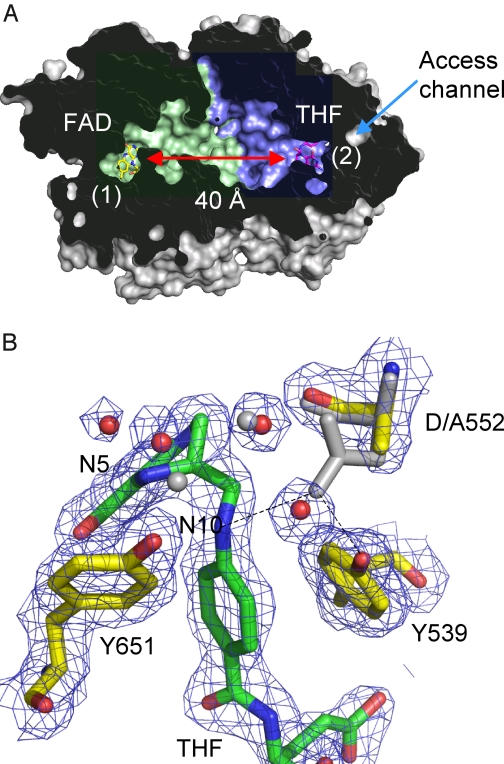
The potential for channeling in amine oxidation pathways has been recognized, based primarily on structural analysis of the component enzymes (22). We determined recently the crystal structure of a bifunctional enzyme (dimethylglycine oxidase (DMGO)) from Arthrobacter globiformis, revealing an unusual protein architecture that could support a channeling mechanism. The crystal structure reveals a fusion of two catalytic domains, separating two active sites by ∼40 Å (Fig. 2A). The amine oxidase site (site 1) is accessed only via the 5,10-CH2-THF synthase site (site 2), which in turn is separated from active site 1 by an irregular and water-filled internal cavity made by the tight packing of the two bowl-shaped catalytic domains. There are no clear access channels for substrate entry or product release except for the funnel that leads to the THF-binding site (active site 2). This funnel connects the internal cavity with bulk solvent, but in the presence of tetrahydrofolate the cavity is largely isolated from the external solvent.
FIGURE 2.
DMGO structure. A, cross-sectional view of a DMGO monomer solvent-accessible surface, with the internal cavity colored blue for the FAD domain and green for the THF-binding domain (Ref. 22; Protein Data Bank code 3GSI). FAD and THF are shown as sticks designating the respective active sites (labeled 1 and 2), which are separated by ∼40 Å. The only access channel to the internal cavity coincides with the THF-binding site. B, detailed view of the THF-binding site for the D552A mutant crystal structure (Protein Data Bank code 3GSI). 2 Fo− Fc sigmaA-weighted electron density is shown as a blue mesh contoured at 1 σ surrounding THF and key amino acids. These are shown in atom-colored sticks (THF, green carbons; amino acids, yellow carbons) in addition to selected water molecules shown as red spheres. The corresponding wt DMGO structure is shown as gray sticks and spheres. This figure was made using PyMOL (DeLano Scientific).
The unique architecture of DMGO suggests that efficient coupling of imine formation (active site 1) to 5,10-CH2-THF synthesis (active site 2) might prevent the accumulation of formaldehyde, formed by hydrolysis of the iminium intermediate product of active site 1. This is a seductive but unproven hypothesis. This raises key questions, such as the extent (if any) of coupling between the active sites and the degree of retention of the iminium (or hydrolysis products) in the DMGO internal reservoir. These questions are difficult to resolve through mechanistic analyses that rely on in vitro measurements owing to the reactivity of formaldehyde and 5,10-CH2-THF, the instability of THF substrate, and the lack of appropriate real-time assays. We report here the construction and use of a novel reporter system that enables in vivo analysis of formaldehyde formation using a synthetic biology approach. Our system employs components of the Escherichia coli formaldehyde detoxification system (the detector) in conjunction with an optimized green fluorescence protein (the reporter), which allowed us to analyze intracellular formaldehyde accumulation following dimethylglycine exposure of E. coli cells expressing DMGO and mutant forms. Together with computational simulations, our studies provide compelling evidence for a highly efficient channeling mechanism in DMGO. We conclude that a relatively simple design, comprising a central reaction chamber and a single entry/exit point, is sufficient to protect an organism from the toxic effects of formaldehyde by internal sequestration prior to the detoxification chemistry catalyzed by active site 2.
EXPERIMENTAL PROCEDURES
Plasmids and Cloning
Plasmids expressing different forms of DMGO are listed in supplemental Table 1. An in vitro reporter system for formaldehyde was created using fusions of the gfp gene and the E. coli frm operon (23, 24). A UV light-sensitive version of the gfp gene (25) optimized for prokaryotic expression was used (Clontech). The gene was amplified using PCR; the corresponding primers are listed in supplemental Table 2. Using the corresponding restriction endonucleases the gfp gene was fused to the amplified promoter region of regulator frmR (pETfrmRGfp). Plasmid pET-24(+) (Novagen) was used as a vector for most of the gfp reporter constructs. Experiments measuring the formaldehyde release of mutant forms DMGOY259F used a version of the reporter where the “frmRGfp” construct was inserted as a EcoRI-HindIII fragment into the first multiple cloning site (MCS1) of pColaDuet-1 (Novagen) and the corresponding dmg mutant gene as a PCR product into MCS2 (supplemental Table 2).
Detection and Quantification of gfp Expression
Microtiter plate assays were routinely used for the in vivo detection of formaldehyde using a 96-well plate reader (BioTek Synergy HT, Biotek) with optical microtiter plates and measuring GFPuv fluorescence (excitationmax = 395 nm, emissionmax = 509 nm) with a 360ex/528em nm filter set. Microtiter wells with a total volume of 300 μl were half-filled with 150 μl of minimal medium and inoculated with 4 μl of fresh starter culture each. The cultures were grown with vigorous shaking at 25 °C before measuring the fluorescence in the early stationary phase (≅22–24 h after inoculation). Fluorescence measurements were scaled to the highest values found on each plate and calibrated for the optical cell density (measured at 580 nm, typically at ∼0.5–0.6) before further analysis. E. coli W3GM with pDMGO and pETfrmRGfp ± DMG was included on each plate to allow for comparison of different experiments on different plates and to assess background fluorescence by means of uninduced cells (= fluorescence of pDMGO in absence of DMG). Data calibrated for differences in the optical density, background fluorescence, and individual plates were then adjusted for the different levels of DMGO enzyme activity found in the corresponding mutant. A typical example of values before enzymatic activity calibration is shown in supplemental Fig. 8. All assays presented were done with at least 6 biological by 10 technical replicates, and the results were found to be statistically significant within a confidence interval of 3%.
Bacteria and Growth Conditions
E. coli DH5α was routinely used as the containment strain in all clonings. Experiments detecting the in vivo formation of formaldehyde were done using a formaldehyde-sensitive strain, E. coli W3GM (24). Bacteria were either grown on Luria-Bertani medium (26) or on minimal medium (27) with 10 mm glucose as the sole source of carbon and energy. Antibiotics for plasmid selection (25 μg/ml kanamycin and/or 50 μg/ml ampicillin) were added as necessary. Dimethylglycine (5 mm), glycine (0.1 mm), and N-methyltryptophan (0.1 mm) were added from pH-adjusted (∼6.5–7.5) stocks to the experiments as appropriate.
Enzyme Assays
Cells were grown overnight in minimal medium in the presence of the corresponding substrates and additives (5 mm dimethylglycine (DMG), 0.1 mm Gly, or 0.2 mm N-methyltryptophan (N-MeTrp)). Equal amounts of cells were harvested, washed twice in 10 mm potassium phosphate, pH 7.2, and lysed using Novagen's primary amine-free BugBuster (700 μl for 14 ml of cells, OD580 ≅ 5–6) supplemented with 200 μm phenylmethylsulfonyl fluoride, 200 ng/ml DNase/RNase, and 100 μg/ml lysozyme (20 min, with shaking, at room temperature). After centrifugation (15 min, 16,000 × g, 4 °C) 40 μl of supernatant was used for 1 ml of assay in 10 mm potassium phosphate, pH 7.2, with 2 mm o-dianisidine and 7.5 units/ ml (defined as 2,2-azinobis(3-ethylbisthiazolinesulfonic acid) (ABTS) units) horseradish peroxidase (28). Control measurements were performed using boiled supernatant. Formation of the stable red bisazobiphenyl end product (Σ430 = 10800 m−1 × cm−1) was measured after a 20-min incubation at room temperature as A430. To ensure the A430 increase reflected all H2O2 production, competing catalase activity in the cell extract was assessed by monitoring A430 response to the addition of 3 μm H2O2. Following 10 min of incubation, substrates for DMGO and N-methyltryptophan oxidase (MTOX), as well as any other additives, were added. The reaction was allowed to proceed for another 20 min before the final reading was taken. Enzyme activities were calculated from the absorption difference before and after addition of the substrate and calibrated against the optical density of the original cell culture. All biochemical assays were done at least as a 5-fold replicate.
Computational Modeling
All simulations were performed using the structure of the folinic acid-DMGO complex (Protein Data Bank code 1PJ7 (22)). Brownian dynamics simulations were done using SDA software (29). Poisson-Boltzmann equations, solved using APBS software (30), were used as input files for the simulations. Up to 20,000 trajectories were simulated at 300 K for each experiment, and the coordinates of nitrogen and oxygen atoms of iminium (or the carbon and oxygen atoms in the case of formaldehyde) from 10,000 positions were randomly recorded to generate occupation maps (Fig. 5, A and B; supplemental Fig. 3). From each of 20,000 formaldehyde trajectories, the residence time was calculated as the time needed to reach a minimum distance between the formaldehyde carbon atom and residues surrounding the outside of the exit channels. For iminium intermediates, the same protocol was applied using the distance between the C1 atom of intermediates and residues surrounding the N-10 atom of folinic acid. Molecular dynamics simulations (MD) were performed with NAMD2 software (31) using AMBER force field parameters (32). Topology and parameter files for dimethyglycine derivatives and the FAD cofactor were obtained using the Antechamber program (33) with AM1-BCC charges (34). The DMGO peptidic backbone was kept fixed during the simulations. Water molecules outside the protein cavity and folinic acid were removed, and the protein was immersed in a periodic TIP3 water box (35) (dimension, 85 × 75 × 80 Å). The temperature and pressure were kept at 298 K and 1 bar, respectively using the Nosé-Hoover method with Langevin dynamics and Langevin piston pressure, respectively. The time step was set to 2 fs, with energies and trajectories stored every 5 ps. Local interactions, including bonded interactions and short-range van der Waals and electrostatic interactions, were calculated at every time step. The cutoff distance for the van der Waals interactions was set to 12 Å, with the pair list distance extended to 13.5 Å. The energy was minimized, and the system was then equilibrated during 1 ns. The iminium derivative was placed near the FAD molecule, according to the positioning observed in acetate-bound DMGO (Protein Data Bank code 1PJ5 (22)). Finally, energy minimization was followed by MD simulations for up to 10 ns.
FIGURE 5.
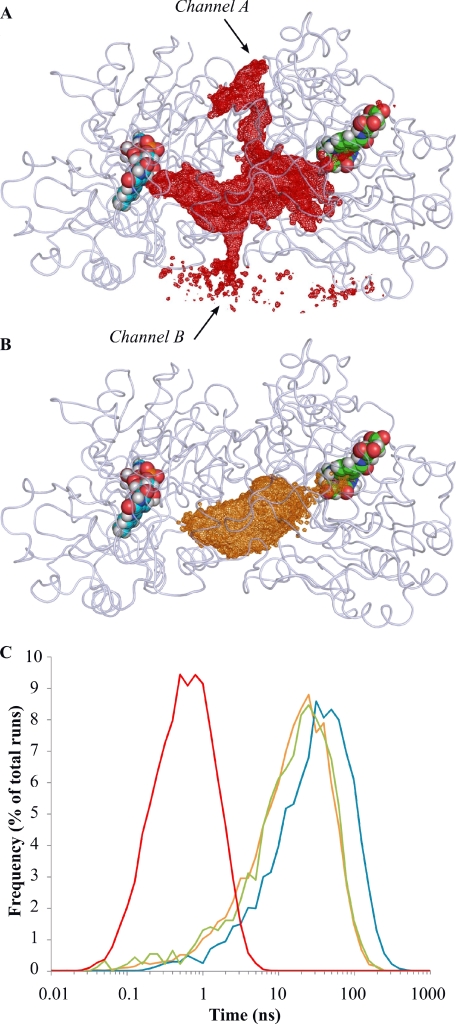
Brownian dynamics diffusion of intermediates in DMGO. A and B, positions of formaldehyde (A) and lactone intermediate (B) molecules during Brownian dynamics simulations. The volume explored by formaldehyde and the lactone are represented, respectively, as red and orange mesh (similar volumes were obtained for other intermediates species). Alternative escape channels for formaldehyde are indicated by arrows. Bound FAD and folinic acid molecules are rendered as cyan and green Van der Waals spheres, respectively. C, residence times for formaldehyde and iminium intermediates. Times were calculated from 20,000 Brownian dynamics trajectories using formaldehyde (red), lactone intermediate (orange), and two rotamers of iminium intermediate (blue and green) as moving molecules.
FIGURE 3.
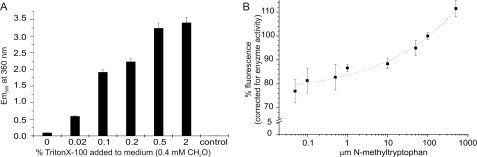
GFP fluorescence increase with formaldehyde (extra- and intracellular). A comparison of the fluorescent response of pETfrmRGfp to external (A) and internal (B) formaldehyde is shown. A, cells containing the reporter system were grown as batch cultures in the presence of 0.4 mm formaldehyde. Different amounts of Triton X-100 were added in the exponential phase to facilitate diffusion of formaldehyde into the cells. Fluorescence was measured using cell extract from equal amounts of cells (1.5 ml, A580 = 2.2). Cells grown in the presence of 0.5% Triton X-100 but without formaldehyde were used as control. B, cells were grown as batch cultures in the presence of different concentrations of N-methyltryptophan as indicated. Fluorescence calibrated against the internal MTOX enzyme activity was measured using a microtiter plate assay. The dotted line represents the result of a logistic dose-response fit against the data.
Crystallization and Structure Elucidation
Crystallization of D552A DMGO was achieved using the sitting-drop vapor diffusion method at 277 K with a well solution of 15% polyethylene glycol-monomethyl ether 5,000, 0.1 m Hepes, pH 7.5, and 0.2 m MgCl. A saturated THF solution was made anaerobically in mother liquor adjusted to contain 1 m Hepes, pH 7.5. Crystals were soaked in the THF solution for ∼60 s before being transferred to oil for cryoprotection and plunge-frozen in liquid nitrogen. Data to 2 Å resolution was collected from a single cryofrozen crystal at Diamond Light Source, Didcot, UK. The data were scaled and integrated using CrystalClear (Rigaku Corp., UK) and subsequently handled using the CCP4 suite (37). The structure was solved using difference Fourier methods. Refinement and model building were carried out using Refmac5 (38) and COOT (39). Structure validation was carried out using Molprobity (40). Data and final refinement statistics are given in supplemental Table 3. Structure factors and atomic coordinates have been deposited with the Protein Data Bank under code 3GSI.
RESULTS
An in Vivo Reporter System for Intracellular Formaldehyde
Determination of the relative levels of THF, 5,10-CH2-THF, and formaldehyde during DMGO catalysis is difficult because these compounds can easily interconvert non-enzymatically. Also, THF is sensitive to oxygen (a substrate) and hydrogen peroxide (a product). These problems prohibited the in vitro determination of kinetic parameters for active site 2. Therefore, the DMGO catalyzed reaction needs to be studied in a cellular context to ensure physiologically relevant levels of the various folate species are present.
The E. coli formaldehyde detoxification system contains a putative formaldehyde sensor, FrmR, which regulates expression of the glutathione-dependent formaldehyde dehydrogenase gene, frmA, and the putative S-formylglutathione hydrolase gene, frmB (23, 24). We fused a UV light-sensitive version of gfp optimized for prokaryotic expression (25) to the E. coli promoter region of frmR and cloned this into pET- or pCola-based plasmid vectors. This created a synthetic construct with the potential to monitor formaldehyde levels in vivo using the fluorescence properties of the gfp-encoded protein. To ensure maximum formaldehyde sensitivity, all experiments were carried out using an E. coli strain deficient in various glutathione-dependent formaldehyde dehydrogenases, E. coli W3GM (supplemental Fig. 1; (24)). Increases in fluorescence levels were observed for cells transformed with pETfrmRGfp grown in the presence of formaldehyde (Fig. 3A). Given that formaldehyde reacts with both the medium and cell wall and periplasmic components, the level of intracellular formaldehyde corresponding to the various extracellular concentrations is not clear, but the experiment demonstrated that the synthetic system is responsive to the presence of formaldehyde. Direct intracellular enzymatic formaldehyde release can be induced in E. coli by providing N-MeTrp in the growth medium. This serves as substrate for MTOX, a constitutively expressed E. coli enzyme homologous to the N-terminal amine oxidase domain of DMGO (41, 42). Unlike DMGO, MTOX does not contain (or is not associated with) 5,10-CH2-THF synthase activity, and oxygen-dependent N-MeTrp oxidation results in the formation of Trp and formaldehyde. We observed increases in fluorescence levels for cells containing pETfrmRGfp grown in the presence of N-MeTrp (Fig. 3B). The expression of MTOX is enhanced by the presence of N-MeTrp (42), and the fluorescence values we obtained were therefore normalized versus total MTOX enzyme activity in the corresponding cellular extracts. We demonstrated that a response to intracellular formaldehyde formation is obtained from levels as low as 0.1 μm N-MeTrp with a nonlinear increase in the fluorescence per unit of enzyme activity obtained within the range of N-MeTrp tested.
Dimethylglycine Catabolism by wt DMGO Does Not Increase Intracellular Formaldehyde Levels
We cultured cells containing both pETfrmRGfp and a plasmid containing the wt dmg gene (pDMGO) in the absence or presence of dimethylglycine. Compared with normal MTOX activity, significantly higher levels of DMGO oxidase activity were detected in the corresponding cellular extracts. However, only a minor fluorescence increase was observed in the presence of dimethylglycine compared with values obtained when N-MeTrp was added to the external medium (Fig. 4). Also, the fluorescence increase was reduced to background level when cells were grown in the presence of glycine. Glycine is a substrate for serine hydroxymethyltransferase, which catalyzes the interconversion of THF and serine to 5,10-CH2-THF and glycine (43). The presence of excess glycine ensures that DMGO is operating at sufficiently high ratios of cellular THF to 5,10-CH2-THF (supplemental Fig. 1).
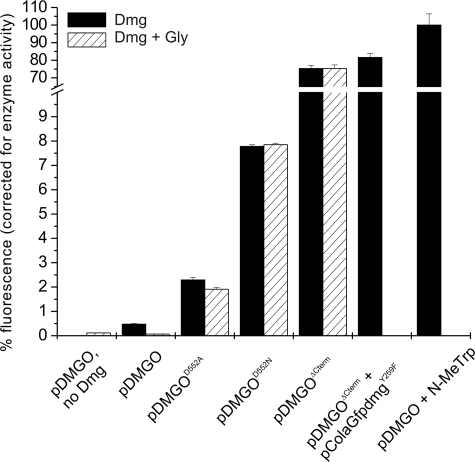
FIGURE 4.
GFP fluorescence increase with various DMGO forms. Efficient formaldehyde detoxification requires an intact DMGO structure as well as a sufficient supply of THF. Cells containing different mutant forms of DMGO were grown in the presence of DMG (black bars) or DMG + glycine (shaded bars). DMGO-specific production of formaldehyde was measured in vivo using pETfrmRGfp (or pColaGfpdmgY259F, as indicated) and calibrated against the corresponding DMGO enzyme activity. The results were scaled using cells containing pDMGO in the absence of DMG and 100 μm N-MeTrp-treated cells as reference points for 0 and 100%, respectively.
Asp-552 Mutations Lead to Increased Formaldehyde Release
All of the 5,10-CH2-THF synthases that are similar to the C-terminal domain of DMGO (e.g. the glycine cleavage system T-protein) contain an acidic residue (Asp-552 in DMGO) that binds to the THF N-10 amino group, thus increasing its nucleophilic character (44). This residue is most likely involved in the mechanism of 5,10-methylene-THF synthesis from the substrate iminium and THF, thereby preventing release of formaldehyde through iminium hydrolysis (22). We created two DMGO mutants (D552A and D552N) to test this hypothesis specifically. We solved the crystal structure of the DMGOD552A-THF complex to ensure that mutation of Asp-552 to Ala or Asn in DMGO does not significantly alter protein structure or the THF binding mode (Fig. 2B). This structure revealed little difference from the previously determined DMGO crystal structures (0.2 Å root mean square deviations for all Cα atoms with Protein Data Bank entry 1PJ6) with the exception of the presence of one water molecule instead of the Asp-552 side chain. We demonstrated that cells containing both pETfrmRGfp and either pDMGOD552A or pDMGOD552N give rise to elevated levels of fluorescence in the presence of dimethylglycine when compared with the cells containing the wild-type DMGO expression plasmid, pDMGO (Fig. 4). Furthermore, fluorescence levels remain sensitive to the presence of dimethylglycine even when cells are grown in the presence of glycine, although to a lesser extend for the pDMGOD552A mutant. When fluorescence levels were normalized to the DMGO activity measured in the corresponding cell extracts, we found that DMGOD552N released more formaldehyde than DMGOD552A. Despite these findings, neither of the mutant DMGO enzymes generated levels of intracellular formaldehyde comparable with those observed with N-MeTrp.
The DMGOΔCterm Deletion Mutant Generates High Levels of Intracellular Formaldehyde
Complete removal of the DMGO 5,10-CH2-THF synthase domain through expression of a truncated version effectively generated a DMGO form (pDMGOΔCterm) that is highly similar to MTOX. This mutant enzyme is unable to avoid hydrolysis of the imine species generated following amine oxidation. Cells containing both pETfrmRGfp and pDMGOΔCterm displayed very high levels of dimethylglycine-dependent and glycine-independent fluorescence (when normalized to account for the lower DMGOΔCterm oxidase activity in the cells (Fig. 4)). The normalized fluorescence observed was similar in magnitude to that obtained with MTOX-dependent N-MeTrp oxidation. We also provided a 5,10-CH2-THF synthase functional DMGO in trans. This was achieved by cloning the gfp-frm promoter construct together with DMGOY259F into pColaDuet-1, generating pColaDMGOY259F. The DMGOY259F mutant, which is severely impaired in DMGO amine oxidase activity (45), was used to ensure that the ratio of 5,10-CH2-THF synthase activity to amine oxidase activity for pColaDMGOY259F/pDMGOΔCterm cells was similar to that present in cells expressing wild-type DMGO. We observed no significant differences in fluorescence levels when comparing pETfrmRGfp/pDMGOΔCterm with pColaDMGOY259F/pDMGOΔCterm cells (Fig. 4; a small increase in fluorescence observed for the pColaDMGOY259F is likely due to the increased copy number of this plasmid (colA) when compared with pETfrmRGfp (colE1)).
Molecular Simulations Reveal Unbiased Diffusion between Active Sites
Brownian dynamics was used to simulate intermediate diffusion from the amine oxidase site (active site 1) to the THF-binding active site (active site 2) in wild-type DMGO. Little is known about the identity of the intermediate that is channeled between the active sites. Stopped flow analysis of the DMGO amine oxidase activity has identified a purple charge transfer complex involving the reduced FAD and product of amine oxidation (46). This complex decays at a rate of 16 s−1 and is the rate-limiting step for amine oxidation (e.g. turnover number for active site 1). The chemistry of charge transfer (CT) decay is unclear, with various possibilities existing. Hydrolysis of the product iminium is a possible reason for CT decay, but equally possible is direct attack of the substrate carboxylate on the oxidized C1 of the iminium to form a cyclic lactone (Fig. 1). Also, CT decay could simply correspond to product release, which could be followed by hydrolysis or cyclization within the internal cavity of DMGO. In addressing all possibilities, Brownian dynamics simulations were used to simulate diffusion of formaldehyde itself (assuming iminium hydrolysis precedes channeling) as well as for the various imine forms (i.e. linear and cyclic forms in various protonation states).
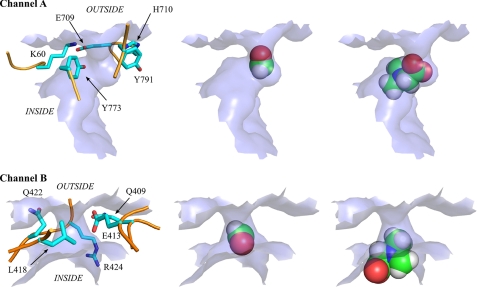
Formaldehyde (or hydrated formaldehyde) was seen to randomly explore the entire protein cavity and readily escape from the “reaction chamber” through several pores (Fig. 5A; supplemental Fig. 3). This is in contrast to the imine species that each explored the protein cavity but could only escape by passing through the THF-binding site funnel (Figs. 5B and 6). The average time needed for the imine C1 to reach the THF N-5 to N-10 atoms (15–30 ns depending on the imine form used) contrasts with the shorter time (0.5 ns) for the escape of a formaldehyde molecule from the cavity (Fig. 5C). As formaldehyde entry into the cavity should be equally facile, we concluded that formaldehyde is unlikely to be the channeled species given that the high levels of intracellular formaldehyde (either generated by pDMGOΔCterm or via addition of N-MeTrp) were unaffected by the presence of a functional C-terminal THF synthase domain of DMGO (Fig. 4; supplemental Fig. 4). In turn, this suggests that an iminium species is the substrate for active site 2 (5,10-CH2-THF synthase).
FIGURE 6.
Topological description of alternative exit channels and size exclusion of formaldehyde versus iminium intermediate. Channels A and B are annotated according to the legend for Fig. 5. In all panels, the molecular surface defining the exit channels is rendered as a light blue surface. The left panels depict the amino acid residues surrounding the exit channels. Protein backbone and residues side chains, respectively, are rendered as orange tube and sticks. Hydrogen atoms are not represented (for figure clarity). The middle panels show an example of positioning of the formaldehyde molecule in the exit channel, and the right panels show a similar view for the iminium intermediate. Atoms are rendered as Van der Waals spheres.
Brownian dynamics simulations do not account for the conformational freedom of molecules involved, nor do they explicitly model water molecules (water is treated as a continuum with same physical/chemical properties). These factors might impact on the diffusion mechanism. We therefore conducted more sophisticated MD simulations using explicit water molecules to model intermediate diffusion within the cavity. Individual MD trajectories spanning 7 ns revealed distinct trajectories associated with the individual components (supplemental Fig. 2). When we performed several 0.5-ns simulations for four distinct starting positions obtained along the path of the longer simulation trajectories, the average behavior was stochastic (including an increase in substrate-THF distance) and was not influenced by the chemical nature of the imine (supplemental Figs. 2, 4, 6, and 7). We concluded that there was no preferential path from FAD to THF that would indicate that diffusion was guided (e.g. via electrostatic interactions) for any of the putative channeled intermediates.
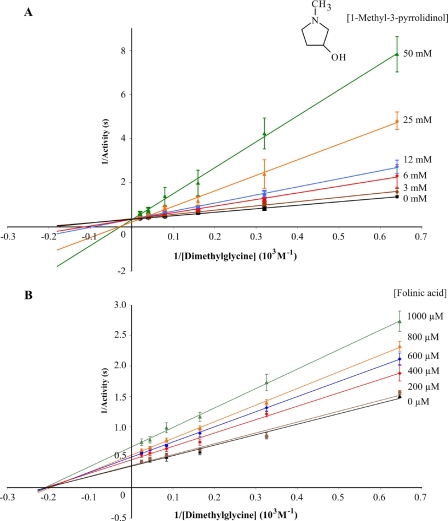
FIGURE 7.
Lineweaver-Burk plots of DMGO-catalyzed dimethylglycine oxidation inhibition by an analogue of the lactone intermediate and folinic acid. 1-Methyl-3-pyrrolidinol (A) and folinic acid (B) were used as inhibitors of DMGO oxidase activity. Results are means ± S.D. calculated from three independent experiments using the indicated inhibitor concentrations. The Ki for 1-methyl-3-pyrrolidinol was determined as 4 ± 0.2 mm.
Significant Inhibition of Active Site 1 by an Iminium Lactone Mimic
Although we concluded that the iminium product of active site 1 is the channeled species, we could not easily discriminate between the linear and cyclic forms. However, we demonstrated that significant competitive inhibition of active site 1 activity occurs with stable compounds that mimic the cyclic lactone form as opposed to those that resemble the linear iminium species (Fig. 7; linear iminium like compounds tested included N,N-dimethylglycinamide, N,N-dimethylacrylic acid, and N,N-dimethylglycine methyl ester). Folinic acid, on the other hand, inhibited the activity at active site 1 through a noncompetitive mechanism, reaffirming the notion that dimethylglycine needs to enter the internal cavity via active site 2. Our data suggest that a cyclic lactone is released by active site 1, and we noted that in DMGO (and related bifunctional amine oxidases) the amine oxidase site is structurally different from the related monofunctional amine oxidases (which do not channel the product to a second “detoxifying” active site) (6, 42). We postulated that this divergent evolution is a consequence of the requirement for release of the appropriate product (e.g. lactone) in the bifunctional amine oxidases.
DISCUSSION
The lifetimes of iminium ions in aqueous solution are short because of the rapid rates of hydrolysis (∼1.8 × 107 s−1 for aliphatic imines (47). The resulting carbinolamine products are in rapid equilibrium with their corresponding amines and aldehydes (48). The oxidation of dimethylglycine to the corresponding iminium ion will therefore lead to rapid formation of formaldehyde. This is a major biological problem, as liberated formaldehyde reacts quickly with susceptible cellular components through common functional groups (e.g. amino groups of biological macromolecules). Our studies suggest that DMGO (and related enzymes) avoid formaldehyde production, and in doing so they provide alternative chemistry that can compete with the very fast hydrolysis rates for iminium ions that are the product of active site 1.
The DMGO crystal structure suggests a central cavity that connects active sites might support a channeling mechanism that negates the production of intracellular formaldehyde (22). That said, this cavity bears little resemblance to the nanotunnel in carbamoyl-phosphate synthase that likewise channels an intermediate to prevent premature hydrolysis (13, 14, 19). The DMGO cavity is several times larger in diameter than needed to transfer intermediates between active sites. Moreover, it is lined with several hydrophilic residues that could either react with, or assist hydrolysis of, the intermediate. The cavity is filled with water molecules and appears to have a single entry-exit point at active site 2 for molecules that are approximately the size of dimethylglycine. These features appear to be at odds with the notion that this enzyme has evolved to avoid hydrolysis of a highly unstable iminium intermediate. Furthermore, oxidase activity appears independent of THF availability with the presence of high levels of THF leading to a reduction in turnover by blocking entry to the reaction chamber (22, 46) (Fig. 7). That the cavity appears readily permeable to (smaller) molecules suggests that significant leakage of formaldehyde can occur.
The puzzles presented by the DMGO structure prompted us to create a simple GFP-based reporter system for in vivo formaldehyde levels in E. coli to address the function of the unusual internal cavity. This reporter system was based on the transcriptional regulator frmR (23). Our synthetic reporter shows a logistic response for a wide range of exogenously added N-methyltryptophan (the formaldehyde-generating precursor through the action of MTOX (42)). GFP fluorescence can be normalized to enzyme activity by monitoring enzyme activity in corresponding cellular extracts (by following substrate-dependent production of hydrogen peroxide). We were surprised to find that very little formaldehyde is produced in vivo when cells expressing wt DMGO are grown in the presence of dimethylglycine; this is despite the high levels of enzyme activity in the cell. By feeding cells with glycine (a substrate for serine hydroxymethyltransferase (43)), a high cellular ratio of THF to 5,10-CH2-THF is maintained even when DMGO activity is elevated above that normally seen under physiological conditions. Under these conditions, glycine significantly reduces the dimethylglycine-dependent GFP fluorescence to background levels, indicating that DMGO turnover does not lead to significant leakage of formaldehyde under nonlimiting THF conditions.
We have shown that impairment of active site 2 (mutants DMGOD552A and DMGOD552N), or complete removal of this activity (mutant DMGOΔCterm), generates substantially elevated levels of intracellular formaldehyde. This is consistent with a coupling (through channeling) of the chemistry of active sites 1 and 2 in wt DMGO and formally establishes the operation of a channeling mechanism. Intracellular formaldehyde production is more elevated with DMGOD552N compared with DMGOD552A. The crystal structure of DMGOD552A reveals a water molecule at the position originally occupied by one of the Asp-552 carboxylate oxygen atoms in the wt structure. This water is likely to be a functional surrogate for the carboxyl oxygen of Asp-552 (wt DMGO) in the THF activation. The counterpart amide group in DMGOD552N is unlikely to be involved in this activity, thereby accounting for the higher levels of intracellular formaldehyde seen with this mutant enzyme. Formaldehyde levels normalized for enzyme activity for the DMGO Asp-552 mutants compared with the level observed for native MTOX indicate that the mutant DMGO enzymes retain some ability to protect the cell against formaldehyde (only 2.3 ± 0.1 and 7.8 ± 0.1% of the MTOX formaldehyde levels were observed for DMGOD552A and DMGOD552N, respectively). This contrasts with observations we have made on deleting the entire 5,10-CH2-THF synthase domain (DMGOΔCterm). Deletion resulted in a substantially less stable enzyme, but importantly significant amine oxidase activity could still be detected in cell extracts. In this case, formaldehyde levels normalized to amine oxidase activity are similar to the level observed for native MTOX, which clearly establishes the trade-off between active site coupling (through channeling) and formaldehyde generation in DMGO. This is further substantiated by the attempted trans complementation assay in which DMGOY259F (impaired at active site 1) is co-expressed with DMGOΔCterm. Our inability to reduce intracellular formaldehyde levels in this complementation assay establishes the need for internal channeling of the iminium product of active site 1 to the 5,10-CH2-THF site (active site 2) within the same enzyme molecule (a mechanism also supported by our simulations of the channeling mechanism).
In developing a channeling model for DMGO, one needs to consider the competing chemistries and their associated rate constants. The average time to reach active site 2 is diffusion-controlled, which imposes an absolute upper limit of ∼1 × 108 s−1 for the 5,10-CH2-THF synthase active site (in all likelihood the active site 2 turnover will be substantially slower). Also, DMGO turnover does not lead to formaldehyde production, and the competing intermediate hydrolysis rate must therefore be several orders of magnitude slower than the rate of 5,10-methylene-THF production at active site 2. At first glance, this appears at odds with the fast hydrolysis rates for aliphatic imines reported in the literature (47). However, this is resolved should DMGO specifically release the more stable lactone at active site 1 (i.e. through CT decay) rather than the more reactive aliphatic imine. The fact that competitive inhibition of active site 1 can be observed only by compounds resembling the lactone species supports this notion. Although the formation of the lactone is disfavored (kinetically) in solution for stereoelectronic reasons, the modeled substrate-enzyme complex reveals that the carboxylate moiety is held in the appropriate position for nucleophilic attack on the oxidized carbon atom (22).
CONCLUSIONS
The DMGO central reaction chamber allows the enzyme to maintain ownership of the labile intermediate created, with simple diffusion responsible for the transfer between both active sites. Compared with the more sophisticated substrate channeling systems in which a directive effect and/or allosteric change is involved, a term such as substrate “containment” is more descriptive of the DMGO process than substrate “channeling.” We suggest that the DMGO amino oxidase active site has evolved to release the most stable intermediate (e.g. the cyclic lactone form) to ensure a favorable kinetic ratio of intermediate hydrolysis versus 5,10-CH2-THF synthesis. Hence, no evolutionary pressure exists to remove water from the internal reaction chamber and/or reduce the volume. Furthermore, DMGO and related enzymes are likely to operate within a narrow range of THF concentrations, so that allosteric communication between active sites to regulate intermediate generation under limiting THF conditions is not needed. Both the reduction of volume of the internal reaction chamber and the development of allosteric regulation have occurred in more sophisticated substrate-channeling enzymes (18–21). The evolutionary origins of the latter can be assumed as similar to those for DMGO. Furthermore, the modular design of DMGO reveals how (transient) association of individual enzymes to create similar reaction chambers can quickly give rise to remarkably efficient substrate-channeling units. Association between enzymes leading to substrate channeling has been postulated to occur for fundamental pathways such as purine biosynthesis (15), glycolysis (16), and respiration (49).
Supplementary Material
Acknowledgment
We gratefully acknowledge access to Diamond Beamlines through a BAG allocation.
This work was supported by Biotechnology and Biological Sciences Research Council (BBSRC) Grant BBE0170101.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–8 and Tables 1–3.
The atomic coordinates and structure factors (code 3GSI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- 5,10-CH2-THF
- 5,10-methylenetetrahydrofolate
- CT
- charge transfer
- DMG
- dimethylglcyine
- DMGO
- dimethlylglycine oxidase
- GFP
- green fluorescent protein
- MD
- molecular dynamics
- MTOX
- N-methyltryptophan oxidase
- N-MeTrp
- N-methyltryptophan
- THF
- tetrahydrofolate.
REFERENCES
- 1.Barak A. J., Tuma D. J. ( 1983) Life Sci. 32, 771– 774 [DOI] [PubMed] [Google Scholar]
- 2.Bosetti C., McLaughlin J. K., Tarone R. E., Pira E., La Vecchia C. ( 2008) Ann. Oncol. 19, 29– 43 [DOI] [PubMed] [Google Scholar]
- 3.Mackenzie C. G., Frisell W. R. ( 1958) J. Biol. Chem. 232, 417– 427 [PubMed] [Google Scholar]
- 4.Wittwer A. J., Wagner C. ( 1981) J. Biol. Chem. 256, 4109– 4115 [PubMed] [Google Scholar]
- 5.Porter D. H., Cook R. J., Wagner C. ( 1985) Arch. Biochem. Biophys 243, 396– 407 [DOI] [PubMed] [Google Scholar]
- 6.Scrutton N. S. ( 2004) Nat. Prod. Rep. 21, 722– 730 [DOI] [PubMed] [Google Scholar]
- 7.Yurimoto H., Kato N., Sakai Y. ( 2005) Chem. Rec. 5, 367– 375 [DOI] [PubMed] [Google Scholar]
- 8.Duine J. A. ( 1999) Biofactors 10, 201– 206 [DOI] [PubMed] [Google Scholar]
- 9.Levovich I., Nudelman A., Berkovitch G., Swift L. P., Cutts S. M., Phillips D. R., Rephaeli A. ( 2008) Cancer Chemother. Pharmacol. 62, 471– 482 [DOI] [PubMed] [Google Scholar]
- 10.Maden B. E. ( 2000) Biochem. J. 350, 609– 629 [PMC free article] [PubMed] [Google Scholar]
- 11.Douce R., Bourguignon J., Neuburger M., Rébeillé F. ( 2001) Trends Plant Sci. 6, 167– 176 [DOI] [PubMed] [Google Scholar]
- 12.Huang X., Holden H. M., Raushel F. M. ( 2001) Annu. Rev. Biochem. 70, 149– 180 [DOI] [PubMed] [Google Scholar]
- 13.Raushel F. M., Thoden J. B., Holden H. M. ( 2003) Acc. Chem. Res. 36, 539– 548 [DOI] [PubMed] [Google Scholar]
- 14.Weeks A., Lund L., Raushel F. M. ( 2006) Curr. Opin. Chem. Biol. 10, 465– 472 [DOI] [PubMed] [Google Scholar]
- 15.An S., Kumar R., Sheets E. D., Benkovic S. J. ( 2008) Science 320, 103– 106 [DOI] [PubMed] [Google Scholar]
- 16.Graham J. W., Williams T. C., Morgan M., Fernie A. R., Ratcliffe R. G., Sweetlove L. J. ( 2007) Plant Cell 19, 3723– 3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mouilleron S., Golinelli-Pimpaneau B. ( 2007) Curr. Opin. Struct. Biol. 17, 653– 664 [DOI] [PubMed] [Google Scholar]
- 18.Dunn M. F., Niks D., Ngo H., Barends T. R., Schlichting I. ( 2008) Trends Biochem. Sci. 33, 254– 264 [DOI] [PubMed] [Google Scholar]
- 19.Holden H. M., Thoden J. B., Raushel F. M. ( 1999) Cell. Mol. Life Sci. 56, 507– 522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers R. S., Jensen J. R., Deras I. L., Smith J. L., Davisson V. J. ( 2003) Biochemistry 42, 7013– 7022 [DOI] [PubMed] [Google Scholar]
- 21.Manjasetty B. A., Powlowski J., Vrielink A. ( 2003) Proc. Natl. Acad. Sci. U. S. A. 100, 6992– 6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leys D., Basran J., Scrutton N. S. ( 2003) EMBO J. 22, 4038– 4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herring C. D., Blattner F. R. ( 2004) J. Bacteriol. 186, 6714– 6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez C. F., Proudfoot M., Brown G., Korniyenko Y., Mori H., Savchenko A. V., Yakunin A. F. ( 2006) J. Biol. Chem. 281, 14514– 14522 [DOI] [PubMed] [Google Scholar]
- 25.Crameri A., Whitehorn E. A., Tate E., Stemmer W. P. ( 1996) Nat. Biotechnol. 14, 315– 319 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch E. F., Maniatis T. ( 1989) Molecular Cloning: A Laboratory Manual, Vol. 3, A1#, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Thurnheer T., Köhler T., Cook A. M., Leisinger T. ( 1986) J. Gen. Microbiol. 132, 1215– 1220 [Google Scholar]
- 28.Meskys R., Harris R. J., Casaite V., Basran J., Scrutton N. S. ( 2001) Eur. J. Biochem. 268, 3390– 3398 [DOI] [PubMed] [Google Scholar]
- 29.Gabdoulline R. R., Wade R. C. ( 1997) Biophys. J. 72, 1917– 1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. ( 2001) Proc. Natl. Acad. Sci. U. S. A. 98, 10037– 10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. ( 2005) J. Comput. Chem. 26, 1781– 1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakalian A., Jack D. B., Bayly C. I. ( 2002) J. Comput. Chem. 23, 1623– 1641 [DOI] [PubMed] [Google Scholar]
- 33.Cornell W. D., Cieplak P., Bayly C. I., Gould I. R., Merz K. M., Ferguson D. M., Spellmeyer D. C., Fox T., Caldwell J. W., Kollman P. A. ( 1995) J. Am. Chem. Soc. 117, 5179– 5197 [Google Scholar]
- 34.Wang J., Wang W., Kollman P. A., Case D. A. ( 2006) J. Mol. Graph. Model. 25, 247– 260 [DOI] [PubMed] [Google Scholar]
- 35.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. ( 1983) J. Chem. Phys. 79, 926– 935 [Google Scholar]
- 36.Deleted in proof
- 37.Collaborative Computational Project, Number 4 ( 1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760– 76315299374 [Google Scholar]
- 38.Murshudov G. N., Vagin A. A., Dodson E. J. ( 1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240– 255 [DOI] [PubMed] [Google Scholar]
- 39.Emsley P., Cowtan K. ( 2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126– 2132 [DOI] [PubMed] [Google Scholar]
- 40.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. ( 2007) Nucleic Acids Res. 35, W375– 383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ilari A., Ilari A., Bonamore A., Franceschini S., Fiorillo A., Boffi A., Colotti G. ( 2008) Proteins 71, 2065– 2075 [DOI] [PubMed] [Google Scholar]
- 42.Khanna P., Schuman-Jorns M. ( 2001) Biochemistry 40, 1441– 1450 [DOI] [PubMed] [Google Scholar]
- 43.Schirch V., Szebenyi D. M. ( 2005) Curr. Opin. Chem. Biol. 9, 482– 487 [DOI] [PubMed] [Google Scholar]
- 44.Scrutton N. S., Leys D. ( 2005) Biochem. Soc. Trans. 33, 776– 779 [DOI] [PubMed] [Google Scholar]
- 45.Basran J., Fullerton S., Leys D., Scrutton N. S. ( 2006) Biochemistry 45, 11151– 11161 [DOI] [PubMed] [Google Scholar]
- 46.Basran J., Basran J., Bhanji N., Basran A., Nietlispach D., Mistry S., Meskys R., Scrutton N. S. ( 2002) Biochemistry 41, 4733– 4743 [DOI] [PubMed] [Google Scholar]
- 47.Eldin S., Digits J. A., Huang S. T., Jencks W. P. ( 1995) J. Am. Chem. Soc. 117, 6631– 6632 [Google Scholar]
- 48.Hall N. E., Smith B. J. ( 1998) J. Phys. Chem. A 102, 4930– 4938 [Google Scholar]
- 49.Schägger H., Pfeiffer K. ( 2000) EMBO J. 19, 1777– 1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.