Abstract
The integrity of the retinal pigment epithelial (RPE) cell is essential for the survival of rod and cone photoreceptor cells. Several stressors, including reactive oxygen species, trigger apoptotic damage in RPE cells preceded by an anti-inflammatory, pro-survival response, the formation of neuroprotectin D1 (NPD1), an oxygenation product derived from the essential omega-3 fatty acid family member docosahexaenoic acid. To define the ability of NPD1 and other endogenous novel lipid mediators in cell survival, we generated a stable knockdown human RPE (ARPE-19) cell line using short hairpin RNA to target 15-lipoxygenase-1. The 15-lipoxygenase-1-deficient cells exhibited 30% of the protein expression, and 15-lipoxygenase-2 remained unchanged, as compared with an ARPE-19 cell line control established using nonspecific short hairpin RNA transfected cells. NPD1 synthesis was stimulated by tumor necrosis factor α/H2O2-mediated oxidative stress in nonspecific cells (controls), whereas in silenced cells, negligible amounts of NPD1, 12(S)- and 15(S)-hydroxyeicosatetraenoic acid, and lipoxin A4 were found under these conditions. Neither control nor the deficient cells showed an increase in 15-lipoxygenase-1 protein content after 16 h of oxidative stress, suggesting that the increased activity of 15-lipoxygenase-1 is due to activation of pre-existing proteins. 15-Lipoxygenase-silenced cells also displayed an exacerbated sensitivity to oxidative stress-induced apoptosis when compared with the control cells. NPD1 selectively and potently rescued 15-lipoxygenase-silenced cells from oxidative stress-induced apoptosis. These results demonstrate that 15-lipoxygenase-1 is activated by oxidative stress in ARPE-19 cells and that NPD1 is part of an early survival signaling in RPE cells.
Introduction
Retinal pigment epithelial (RPE)2 cells are essential for the survival of rod and cone photoreceptors. RPE cells mediate the renewal of photoreceptor outer segments (1, 2); synthesis and secretion of neurotrophins (3); recycling of bleached visual pigments; and transport of vitamin A (4), docosahexaenoic acid (DHA), and other nutrients, and ions and fluids between photoreceptors and the choriocapillaries (5). Failure of RPE cells to accomplish their functions leads to photoreceptor damage or death and, as a consequence, decreased vision and eventually blindness. Apoptotic cell death of RPE cells takes place in retinal degenerative diseases, including retinitis pigmentosa and age-related macular degeneration; as a result, photoreceptors degenerate (6, 7).
Cell fate decisions made at the ectoderm yield either neuronal progeny or RPE cells. Thus RPE cells display similarities to neuronal lineages, even when differentiated (8), that make them suitable for conversion into neurons for therapeutic purposes (9). For example, recent studies revealed mechanisms for the transdifferentiation of RPE cells that depend on the expression status of certain genes (10). These studies have created a new interest in RPE cells caused by the potential applications not only in the treatment of retinal degenerative diseases (11, 12) but also in neurodegenerative diseases such as Parkinson disease (13). Moreover, a recent study has successfully reprogrammed chick embryonic RPE cells into photoreceptor-like cells that express red opsin and functionally emulate Ca2+ visual cycle recovery (11, 12). The RPE photoreceptor outer segment layer is located in an environment highly susceptible to oxidative stress because of the continuous flux of polyunsaturated fatty acids (omega-3 and omega-6), high oxygen consumption of the retina, and intermittent exposure to light (14). RPE cells, when confronted with oxidative stress in vitro, enhance an endogenous anti-inflammatory, pro-survival pathway that results in the production of neuroprotectin D1 (NPD1, 10R,17S-dihydroxy-docosa-4Z,7Z,11E,13E,15Z,19Z-hexaenoic acid), a DHA-derived mediator (15). NPD1 is a potent neuroprotective lipid mediator that inhibits the expression of pro-inflammatory genes such as cyclooxygenase-2 and, conversely, enhances the expression of anti-apoptotic proteins of the Bcl-2 family (15, 16), thus promoting cell survival. In addition, NPD1 signaling is involved in neurotrophin-mediated RPE cell survival (17).
The biosynthetic pathway leading to oxygenation of DHA into NPD1 in RPE cells is presently unknown. 15-Lipoxygenase-1 (15-LOX-1) catalyzes the formation of NPD1 in T-helper lymphocytes, where the silencing of 15-LOX-1 leads to reduced production of NPD1; however, no rescue bioactivity by this lipid mediator was studied (18). 15-LOX-1 is an enzyme that belongs to the family of nonheme iron-containing dioxygenases and stereospecifically inserts oxygen into free and/or esterified polyunsaturated fatty acids. 15-LOX-1 oxygenates arachidonic acid, dually forming 15(S)-HETE as a major product and 12(S)-HETE as a minor product. This enzyme also has the capability to oxygenate linoleic acid into 13-hydroxyoctadecadienoic acid (19). To address the role of 15-LOX-1 in the synthesis of NPD1 in RPE cells, we developed an ARPE-19 stable cell line that expresses a small interfering RNA (siRNA) targeting 15-LOX-1. In this cell line, we defined the involvement of 15-LOX-1 in NPD1 synthesis in the protective pathways elicited by tumor necrosis factor α (TNFα)/H2O2-mediated oxidative stress and demonstrated that the exacerbated response of these cells to oxidative stress-induced apoptosis was selectively and potently attenuated by the addition of NPD1, suggesting 15-LOX-1 as a key enzyme in NPD1 synthesis. This further highlights the significance of NPD1 bioactivity in RPE survival signaling.
EXPERIMENTAL PROCEDURES
Establishment of a Stable Cell Line Expressing shRNA Targeting 15-LOX-1
An ARPE-19 cell line was transfected using FuGENE 6 (Roche Applied Science) and SureSilencing vector system (Superarray Bioscience Corporation, Frederick, MD). Using Western blot analysis, four different predesigned shRNAs were tested for their ability to silence 15-LOX-1 in ARPE-19 cells. Cells transfected with vectors containing the selected shRNA sequence were (5′-CCAGCATGAGGAGGAGTATTT-3′) targeting either 15-LOX-1 or a nonspecific targeting sequence (scramble). After 72 h, the cells were trypsinized and plated in serial dilutions in Geneticin (1 mg/ml; Invitrogen) selective medium. After 10 days of incubation, the surviving colonies were replated separately in 5-ml culture flasks and allowed to grow to confluency. The cells were then passed to a 75-ml flask in Geneticin maintenance medium consisting of Dulbecco's modified Eagle's medium/F-12 (1:1), penicillin/streptomycin, 0.5 mg/ml Geneticin, and 10% fetal bovine serum.
Western Blot and Immunocytochemistry
For immunostaining analysis of 15-LOX-1 and 15-LOX-2, 25,000 cells/chamber were plated in a Nunc Lab-Tek two-chamber slide system on Permanox (Nalge Nunc International, Rochester, NY), incubated for 24 h at 37 °C, 5% CO2, and then fixed with 4% paraformaldehyde. For immunoblotting, 0.5 million cells were plated in each well of six well plates and incubated for 48 h. Three of those wells were lysed with radioimmune precipitation assay buffer containing anti-protease mixture (Sigma) and pooled together. The samples were assayed for protein content, and 30 μg of each was prepared following the Laemmli method (20). The samples were boiled for 3 min with 2× Laemmli buffer containing 100 mm of dithiothreitol and run in Novex Tris-glycine SDS-PAGE gel (Invitrogen) at 120 constant volts for 2 h. The transference was performed using the iBLOT dry transferring system (Invitrogen). The immunoblotting and immunostaining was performed using monoclonal antibodies against human 15-LOX-1 (Abnova, Taiwan), human 12-lipoxygenase (h12-LOX) (Santa Cruz Biotechnology, Santa Cruz, CA), and 15-LOX-2 (Cayman, Ann Arbor, MI). For the standardization of the immunoblotting, anti-glyceraldehyde-3-phosphate dehydrogenase monoclonal antibody was used (Chemicon, Temecula, CA), and the membranes were developed with ECL (Amersham Biosciences). For counterstaining ARPE-19 cells, anti-actin antibody produced either in rabbit or in mouse (Sigma) was used.
Oxidative Stress Treatment and Rescue Experiments
For oxidative stress experiments, 0.5 million cells/well were plated in six-well plates and incubated 72 h at 37 °C, 5% CO2, 99% relative humidity. Then they were serum-starved for 8 h in Dulbecco's modified Eagle's medium/F-12 (1:1), penicillin/streptomycin, 0.5 mg/ml Geneticin, and 0.5% fetal bovine serum. Oxidative stress was performed by adding 30% H2O2 in each case to reach the indicated final concentration and TNFα 10 ng/ml. The incubation time varied depending on the experiment. Western blot analysis and Hoechst staining were performed with samples treated for 16 h, whereas lipidomic analysis was done with samples incubated for 4 h. In the rescue experiments, the cells were treated simultaneously with H2O2 600 μm, and TNFα 10 ng/ml, plus either 50 nm of 15(S)-HETE, 12(S)-HETE, lipoxin A4, DHA, or NPD1. DHA was added along with PEDF (10 ng/ml).
Hoechst Staining and Apoptotic Cell Analysis
15-LOX-1 knockdown and nonspecific shRNA transfected cells were fixed with methanol for 15 min, washed with 1× phosphate-buffered saline, and then loaded with 2 μm of Hoechst in 1× phosphate-buffered saline for 15 min at room temperature before imaging using a Nikon DIAPHOT 200 microscope under UV fluorescence. Images were recorded by a Hamamatsu Color Chilled 3CCD camera and Photoshop 5.0 software (Adobe Systems, Mountain View, CA).
Mediator Lipidomic Analysis
Liquid chromatography-photodiode array-electrospray ionization-tandem MS-based lipidomic analysis was performed as previously described (15, 17). Briefly, 15-LOX-1 and NS cells, lipid extracts, and 1 ml of incubation medium were labeled with deuterated internal standards, purified by solid phase extraction, and loaded onto a C18 Pursuit column with 100-mm length × 2.1-mm OD loaded with 5-μm stationary phase (Varian), starting with solvent A (40:60:0.01 methanol:water:acetic acid, pH 4.5; 300 μl/min); the gradient reached 100% solvent B (99.99:0.01 methanol:acetic acid) in 30 min and was run isocratically for 5 min. We used a TSQ Quantum triple quadrupole mass spectrometer (Thermo-Finnigan, San Jose, CA); electrospray ionization spray voltage was 3 kV, and sheath gas was N2 (35 cm3/min, 275 °C). Calibration curves were obtained running in parallel synthetic NPD1, 15(S)-HETE, 12(S)-HETE, lipoxin A4, and DHA (Cayman Chemical, Ann Arbor, MI).
Statistical Analysis
Two-way analysis of variance was used in those experiments where the responses of two cell types (e.g. silenced and nonsilenced 15-LOX-1 cells) were compared against two or more different treatments, and the t test was used to compare any two treatments or samples to obtain error probability p.
RESULTS
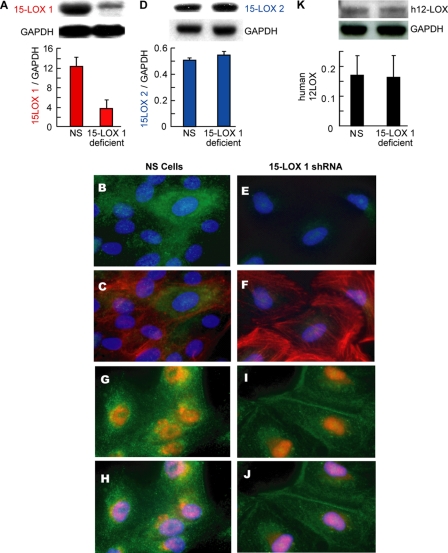
To test the hypothesis that 15-LOX-1 is involved in the biosynthetic pathway of NPD1 in RPE cells, we developed a stably silenced ARPE-19 cell line using shRNA technology. These knockdown cells showed more than a 70% decrease in expression of 15-LOX-1 when compared with the nonspecific (NS) shRNA stable transfected cells (Fig. 1, A–C, E, and F). Despite the fact that human 15-LOX-1 shares more than 70% homology with h12-LOX, the latter remained unaffected in silenced cells (Fig. 1K). Furthermore, the specific silencing was corroborated by immunocytochemistry, which revealed a cytoplasmic localization for 15-LOX-1 (Fig. 1, B, C, E, and F). On the other hand, 15-LOX-2 did show a nuclear localization (Fig. 1, G–J) but showed no modification in its expression by the knockdown (Fig. 1D). These results confirm the specificity of the siRNA expressed in the silenced cell line.
FIGURE 1.
15-Lipoxygenase-1 and -2 and h 12-LOX protein expression in ARPE-19 silenced cells. Both the stable cell line expressing shRNA targeting 15-LOX-1 as well as cells expressing a nonspecific scrambled targeting sequence (NS) were analyzed. A, D, and K show representative immunoblot of at least three experiments along the different cell passages. 15-LOX-1, h12-LOX, and 15-LOX-2 expressed in the knockdown cells were plotted as ratios between enzyme and the housekeeping control (glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) in relation with the control expressing nonspecific shRNA. These two cell types were immunostained against 15-LOX-1 in green (B and E) and actin in red (C and F). 15-LOX-2 signal (red) and actin (green) are shown in G–J. The nuclei were stained with 4′,6-diamidino-2-phenylindole, blue (B, C, and E–J).
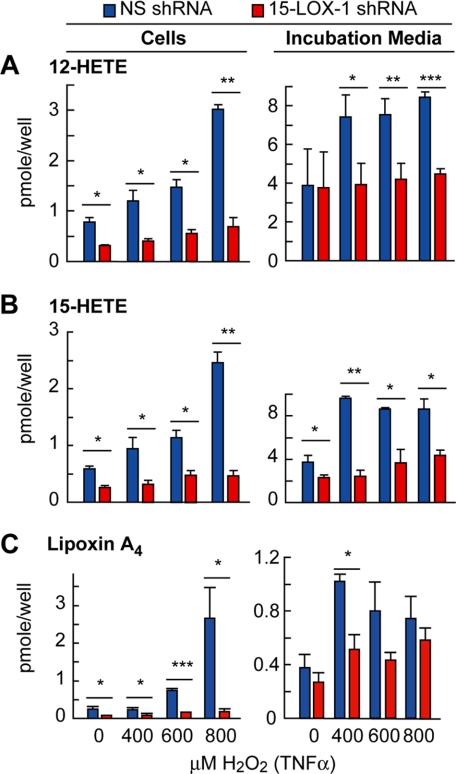
Human 15-LOX-1 catalyzes the oxygenation of arachidonic acid to 15(S)-HETE, 12(S)-HETE, and lipoxin A4, a product of its joint activity with 5-lipoxygenase (21). We examined the products of 15-LOX-1 by liquid chromatography-photodiode array-electrospray ionization-tandem MS-based lipidomic analysis and found an increase in 12-HETE, 15-HETE, and lipoxin A4 in NS cells and incubation media during TNFα/H2O2-induced oxidative stress, whereas these lipid mediators were at much lower levels in deficient cells (Fig. 2).
FIGURE 2.
Activity of 15-lipoxygenase-1 shRNA transfected cells. The cells were serum-starved, 72 h after plating, for 8 h and treated with 0, 400, 600, and 800 μm H2O2 plus 10 ng/ml TNFα for 4 h, after which both incubation medium and cells were collected for lipid extraction and analyzed by liquid chromatography tandem MS. 12-HETE (A), 15-HETE (B), and lipoxin A4 (C) were measured in cell extracts and medium. The bars represent the means ± S.E. of three different samples (*, p < 0.05; **, p < 0.005).
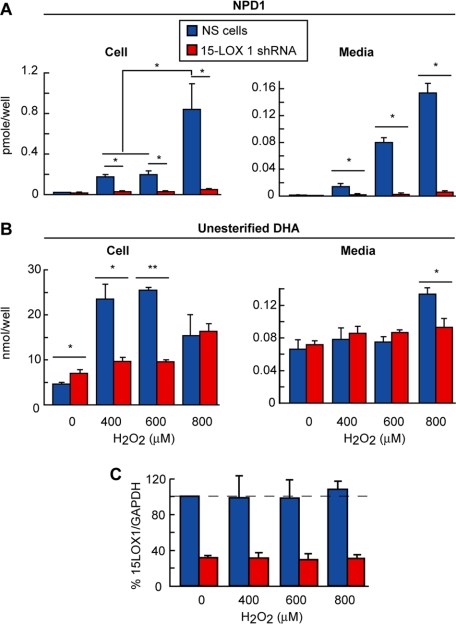
We then found that NPD1 content was negligible in silenced cells during oxidative stress, whereas in the ARPE-19 cells expressing nonspecific shRNA or NS cells, this lipid mediator was increased (Fig. 3A). These results suggest that 15-LOX-1 is the enzyme that mediates the synthesis of NPD1 in human ARPE-19 cells. We found that the content of unesterified DHA (micromolar range), the precursor of NPD1 (picomolar range), did not follow the same pattern of change in either type of cells when they were exposed to oxidative stress (Fig. 3B). This suggests that only a small pool of unesterified DHA is converted to NPD1, and hence DHA hydrolysis from the phospholipid pool is not a limiting process for the formation of this lipid mediator. Alternatively, TNFα/H2O2 may induce more than one phospholipase A2, and one of these may release a DHA pool that, in turn, is channeled through 15-LOX-1. These mechanisms remain to be explored.
FIGURE 3.
NPD1 synthesis in 15-lipoxygenase-1 silenced cells. NPD1 and its precursor DHA were measured by liquid chromatography tandem MS in 15-LOX-1 silenced cells and its incubation media after 8 h of serum starvation and 4 h of oxidative stress treatment using increasing amount of H2O2 (A and B). Under the same conditions as in A, the cells were lysed, and the supernatant was run in SDS-PAGE and immunoblotted against 15-LOX-1 (C). No significant differences in enzyme protein were found (*, p < 0.005; **, p < 0.0005).
The increase of NPD1 pool size was observed 4 h after application of the oxidative stress inducer. Activation of 15-lipoxygenase activity to enhance the production of NPD1 may be due either to the increase in the 15-lipoxygenase content or to the activation of an enzyme pool that, in resting conditions, remains inactive. We performed Western blot analysis to address this issue. Fig. 3C depicts the unchanged enzyme protein content during 16 h of oxidative stress; this implies that pre-existing 15-LOX-1 is activated to generate NPD1 under these conditions.
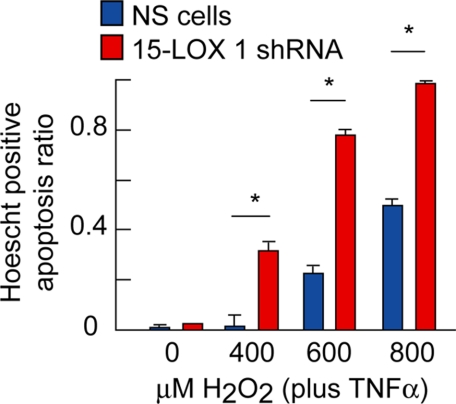
Because NPD1 protects RPE cells during oxidative stress (17), we tested the prediction that 15-LOX-1-deficient cells are more vulnerable to damage caused by oxidative stress damage by exposing them to TNFα/H2O2. We found that 15-LOX-1-deficient cells were more susceptible to perturbations that lead to apoptosis. The magnitude of the response in the 15-LOX-1-deficient cells, in which the formation of NPD1 remained at low levels, was higher at all H2O2 concentrations, especially at 800 μm, where it reached almost 100% (Fig. 4). These results, along with the unchanged content of 15-LOX-1 (Fig. 3C), suggest that the diminished 15-LOX-1 activity, which forms lesser amounts of NPD1, contributes to increased apoptosis in knockdown cells.
FIGURE 4.
15-Lipoxygenase-1 deficiency displays enhanced susceptibility of ARPE-19 cells to oxidative stress-induced apoptosis. Silenced and nonsilenced cells were fixed with paraformaldehyde and Hoechst stained after 8 h of serum starvation and 16 h of oxidative stress treatment using 0, 400, 600, and 800 μm of H2O2 plus 10 ng/ml TNFα. The apoptotic cells were counted, and the ratios of death cells/total cells were plotted for each stably transfected cell line and treatment applied. The bars represent the means ± S.E. of three samples or nine fields obtained from three wells in A (*, p < 0.005).
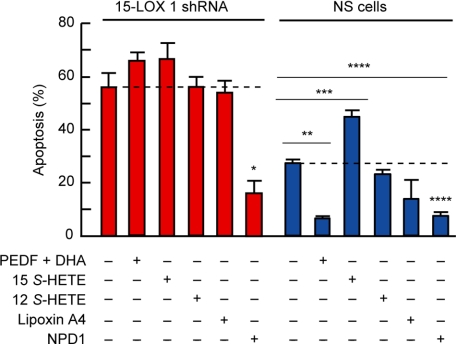
The increased sensitivity of ARPE-19 cells to oxidative stress-induced apoptosis may be due to a decreased availability either of NPD1 or of other 15-LOX-1 products. DHA and PEDF protect ARPE-19 cells synergistically from oxidative stress-induced apoptosis. NPD1 synthesis is enhanced under these conditions (15, 17). To corroborate that NPD1 was in fact the 15-LOX-1 product responsible for protecting ARPE-19 knockdown cells, we induced apoptosis in silenced cells using 600 μm H2O2 and 10 ng/ml TNFα and tested the protective bioactivity of PEDF/DHA, 15(S)-HETE, 12(S)-HETE, lipoxin A4, or NPD1. We found that only NPD1 rescued 15-LOX-1-deficient cells from oxidative stress-induced apoptosis. PEDF/DHA treatment was protective in NS cells but failed to prevent apoptosis in the knockdown cells (Fig. 5). Added DHA conversion to NPD1 is stimulated by PEDF; however, this conversion cannot take place in silenced cells. None of the arachidonic acid oxygenation products prevented cell death induced by oxidative stress.
FIGURE 5.
NPD1 rescues stably transfected 15-lipoxygenase-1 deficient cells from oxidative stress-induced apoptosis. 15-Lipoxygenase-1 silenced cells were serum-starved and treated with either PEDF-DHA, 15(S)-HETE, 12(S)-HETE, lipoxin A4, or NPD1 and 600 μm H2O2 plus 10 ng/ml TNFα for 16 h. 50 nm of each lipid was added. The bars represent the means of the ratio between the cells that were stained positive with Hoechst and the total cell count + S.E. (*, p < 0.05; **, p < 0.005; ***, p < 0.0005).
DISCUSSION
DHA, a membrane phospholipid acyl chain, highly enriched in brain and retina plays critical roles in membrane organization and function (22). The cleavage of DHA by phospholipases leads to the synthesis of bioactive docosanoids (22). Neuroprotectin D1, the first identified docosanoid, exerts homeostatic bioactivity of cell integrity, inhibition of neuroinflammation, down-regulation of apoptosis, and neuroprotection (15–17, 23, 24). In RPE cells, NPD1 synthesis is activated by oxidative stress (15), cytokines (15), neurotrophins (17), and A2E, a lipofusin component that accumulates in the RPE during aging and in certain forms of macular degeneration (17, 23), and after photoreceptor outer segments phagocytosis in the presence of oxidative stress (23). Outside the nervous system NPD1 has been shown to actively participate in the resolution of inflammation (25). Human 15-LOX-1 was proposed to catalyze the conversion of DHA into NPD1 in ARPE-19 (7, 17). In the present work we silenced 15-LOX-1 protein in ARPE-19 cells and tested the response to oxidative stress. We found that 15-LOX-1 deficiency remarkably increases the susceptibility of ARPE-19 cells to oxidative stress-induced apoptosis. Moreover, oxidative stress-induced NPD1 synthesis was negligible in the knockdown cells. The pool size of other products of the silenced enzyme, 15-HETE and 12-HETE, derivatives from arachidonic acid, did not show a decrease as marked as NPD1. Intriguingly, lipoxin A4, a derivative of 15-p-HETE through the action of 5-lipoxgenase, was decreased. This suggests that the formation of lipoxin A4 may be coupled with the production of 15-p-HETE by the activity of 15-LOX-1 but not 15-LOX-2, which remained unchanged. This observation is in agreement with previous reports (21).
Human 15-LOX-1 and human 12-LOX are highly homologous proteins (identity 65%), and although they are encoded by different genes their messenger RNAs are highly similar (about 70% identity). On the other hand, 15-LOX-2, a third lipoxygenase member, only shares 39% identity with human 15-LOX-1 (26). Because shRNA targets mRNA based on sequence complementarity, there is some chance for the siRNA to bind h12-LOX. Western blot analysis showed that neither 15-LOX-2 nor h12-LOX are affected by the 15-LOX-1 knockdown. Moreover, the cellular localization of 15-LOX-1 and 15-LOX-2 also differed (Fig. 1D, H--K, respectively), and the immunostaining analysis confirmed the Western blot analysis. The 15-LOX-1 showed a cytoplasmic localization with a perinuclear-increased signal by immunostaining, whereas the second isoform was found predominantly localized in the nucleus. This is in partial agreement with previous observations showing that the 15-LOX-2 green fluorescent protein localized in nuclei and cytoplasm of the HCE cell line and 15-LOX-1 was found only in cytoplasm (27), although in that report both enzymes were exogenously overexpressed and tagged with green fluorescent protein.
The arachidonic acid oxygenation activity of 15-LOX-1 leads to synthesis of 15(S)-HETE and 12(S)-HETE in a 9:1 ratio (28), whereas 15-LOX-2 produces only 15(S)-HETE (26). Although lipoxygenase function is not completely understood, other contributors to the formation of HETEs, CYP450 isozymes, may oxidase arachidonic acid to 12-HETE stereoisomer, thus contributing to the 12-HETE pool. Because CYP450s shares no similarity in their nucleotide sequence with 15-LOX-1, it is not likely that the siRNA may have had an effect in its protein expression. Hence, the differences in the 12-HETE measured during increasing oxidative stress in 15-LOX-1-silenced cells are most likely due to the 15-LOX-1 protein decrease. The same is applicable to the 15-HETE decrease (Fig. 2, A and B). The induction of HETE synthesis by oxidative stress and the role of 15-LOX-1 and other enzymes in this process still remain unclear.
In rabbit immune cells, translocation of reticulocyte 15-LOX-1 from the cytoplasm to cellular membranes (except nuclear ones) was shown to be promoted by calcium interactions that facilitate protein binding (29). The translocation results in increased activity in free and membrane-bound fatty acid oxygenation in that system (30).
In the present report, we investigated whether the oxidative stress-induced increase in activity was due to an augmented protein content. We found that the levels of 15-LOX-1 remained unchanged 16 h after oxidative stress treatment (Fig. 3C), even though its activity was induced only 4 h after the onset of oxidative stress and followed a dose-dependent response in ARPE-19 cells (Figs. 2 and 3). These results suggest that the induction of activity was not accompanied by a change in the steady state levels of protein. These observations are in agreement with interleukin-4 induction of 15-LOX-1 expression in fibroblasts that starts 16 h after cytokine treatment (31). Although it is unclear what mechanisms involved in oxidative stress induce changes 15-LOX-1 activity, the polycystin/lipoxygenase/α-toxin domain present in lipoxygenases may play a role in this induction (32) and in a possible translocation to the membrane (29).
Decreased 15-LOX-1 expression was accompanied by diminished production of NPD1 in a T-Helper cell culture by the means of siRNA; however, the involvement of the other isoform in this system could not be assessed because of the lack of 15-LOX-2 expression (18). We demonstrated that a stably silenced 15-LOX-1 ARPE-19 cell line produces diminished amounts of NPD1 despite the unaltered expression of 15-LOX-2, suggesting that the second isoform is not involved in the NPD1 synthesis triggered by oxidative stress. On the other hand, we found that the difference between the 15-HETE and 12-HETE produced in response to oxidative stress by control and deficient cells was not as steep as that observed in the production of NPD1. This could be due to the activity of other contributors to the mediator pool, such as 15-LOX-2 and h12-LOX. In addition, the levels of unesterified DHA, the NPD1 precursor, was found to be in excess in each case (Fig. 3B), revealing that the decrease in the product was not limited by the precursor, even though there were some differences in the cellular content of unesterified DHA in deficient and control cells when treated with 400 and 600 μm H2O2/10 ng/ml TNFα and in the released DHA when cells were exposed to 800 μm H2O2/10 ng/ml TNFα. These differences suggest a regulation in the sizes of the esterified and released DHA pools that did not follow a pattern complementing the decrease in NPD1.
In 12/15 lipoxygenase null (ALOX15−/−) mice, impairments in the re-epithelialization and recruitment of polymorphonuclear leukocytes to the cornea were found upon epithelial cell removal, especially when lipopolysaccharide was applied (33). These impairments were attributed to the exacerbation of the inflammatory process because of the lack of 15-LOX-1 products. They also performed partial rescue of this impairment by topical application of lipoxin A4. We found that in ARPE-19 cells, lipoxin A4 did not rescue ARPE-19 15-LOX-1-deficient cells from the damage caused by oxidative stress; however, NPD1 selectively exerted pro-survival rescue bioactivity. Finally, the initiation of oxidative stress-mediated pro-inflammatory/pro-apoptotic signaling allows the cell to compete with the ongoing pro-apoptotic signaling; the outcome decides the fate of the cell. In this sense, the cells that are deficient in NPD1 as a result of a decreased content of 15-LOX-1 tend to be more prone to apoptosis than the ones that can compensate for the pro-apoptotic fate with NPD1 action. Using this line of reasoning, we observed that ARPE-19 15-LOX-1-silenced cells are more susceptible to apoptosis than the control cells (Fig. 4), suggesting that the imbalance produced by the decreased production of NPD1 caused by the knockdown of its biosynthetic enzyme drove the cells into a pro-apoptotic state. This notion is supported by the rescue of the ARPE-19-deficient cells by exogenous NPD1. Together, these results show that the formation of NPD1 is dependent on 15-LOX-1 activity in RPE cells and that NPD1 successfully rescues RPE cells deficient in 15-LOX-1 from oxidative stress-induced apoptosis.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 EY005121 and P20 RR016816.
- RPE
- retinal pigment epithelial
- NPD1
- neuroprotectin D1
- DHA
- docosahexaenoic acid
- h12-LOX
- human 12-lipoxygenase
- 15-LOX-1
- 15-lipoxygenase-1
- PEDF
- pigment epithelium-derived factor
- shRNA
- short hairpin RNA
- TNF
- tumor necrosis factor
- HETE
- hydroxyeicosatetraenoic acid
- siRNA
- small interfering RNA
- MS
- mass spectrometry
- NS
- nonspecific.
REFERENCES
- 1.Bok D. ( 1993) J. Cell Sci. Suppl. 17, 189– 195 [DOI] [PubMed] [Google Scholar]
- 2.LaVail M. M. ( 1980) Invest. Ophthalmol. Vis. Sci. 19, 407– 411 [PubMed] [Google Scholar]
- 3.Tombran-Tink J., Barnstable C. J. ( 2003) Nat. Rev. Neurosci. 4, 628– 636 [DOI] [PubMed] [Google Scholar]
- 4.Scott B. L., Bazan N. G. ( 1989) Proc. Natl. Acad. Sci. U. S. A. 86, 2903– 2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strauss O. ( 2005) Physiol. Rev. 85, 845– 881 [DOI] [PubMed] [Google Scholar]
- 6.Rattner A., Nathans J. ( 2006) Nat. Rev. Neurosci. 7, 860– 872 [DOI] [PubMed] [Google Scholar]
- 7.Bazan N. G. ( 2007) Invest. Ophthalmol. Vis. Sci. 48, 4866– 4881 [DOI] [PubMed] [Google Scholar]
- 8.Engelhardt M., Bogdahn U., Aigner L. ( 2005) Brain Res. 1040, 98– 111 [DOI] [PubMed] [Google Scholar]
- 9.Bharti K., Arnheiter H. ( 2005) J. Invest. Dermatol. 125, x– xi [DOI] [PubMed] [Google Scholar]
- 10.Lanning J. L., Wallace J. S., Zhang D., Diwakar G., Jiao Z., Hornyak T. J. ( 2005) J. Invest. Dermatol. 125, 805– 817 [DOI] [PubMed] [Google Scholar]
- 11.Liang L., Yan R. T., Li X., Chimento M., Wang S. Z. ( 2008) Invest. Ophthalmol. Vis. Sci. 49, 4145– 4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osakada F., Ikeda H., Mandai M., Wataya T., Watanabe K., Yoshimura N., Akaike A., Akaike A., Sasai Y., Takahashi M. ( 2008) Nat. Biotechnol. 26, 215– 224 [DOI] [PubMed] [Google Scholar]
- 13.McKay B. S., Goodman B., Falk T., Sherman S. J. ( 2006) Exp. Neurol. 201, 234– 243 [DOI] [PubMed] [Google Scholar]
- 14.Organisciak D. T., Darrow R. M., Jiang Y. L., Blanks J. C. ( 1996) Invest. Ophthalmol. Vis. Sci. 37, 2243– 2257 [PubMed] [Google Scholar]
- 15.Mukherjee P. K., Marcheselli V. L., Serhan C. N., Bazan N. G. ( 2004) Proc. Natl. Acad. Sci. U. S. A. 101, 8491– 8496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukiw W. J., Cui J. G., Marcheselli V. L., Bodker M., Botkjaer A., Gotlinger K., Serhan C. N., Bazan N. G. ( 2005) J. Clin. Invest. 115, 2774– 2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee P. K., Marcheselli V. L., Barreiro S., Hu J., Bok D., Bazan N. G. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 13152– 13157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ariel A., Li P. L., Wang W., Tang W. X., Fredman G., Hong S., Gotlinger K. H., Serhan C. N. ( 2005) J. Biol. Chem. 280, 43079– 43086 [DOI] [PubMed] [Google Scholar]
- 19.Kühn H., Thiele B. J., Ostareck-Lederer A., Stender H., Suzuki H., Yoshimoto T., Yamamoto S. ( 1993) Biochim. Biophys. Acta 1168, 73– 78 [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U. K. ( 1970) Nature 227, 680– 685 [DOI] [PubMed] [Google Scholar]
- 21.Serhan C. N. ( 2007) Annu. Rev. Immunol. 25, 101– 137 [DOI] [PubMed] [Google Scholar]
- 22.Bazan N. G. ( 2006) Trends Neurosci. 29, 263– 271 [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee P. K., Marcheselli V. L., de Rivero Vaccari J. C., Gordon W. C., Jackson F. E., Bazan N. G. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 13158– 13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcheselli V. L., Hong S., Lukiw W. J., Tian X. H., Gronert K., Musto A., Hardy M., Gimenez J. M., Chiang N., Serhan C. N., Bazan N. G. ( 2003) J. Biol. Chem. 278, 43807– 43817 [DOI] [PubMed] [Google Scholar]
- 25.Schwab J. M., Chiang N., Arita M., Serhan C. N. ( 2007) Nature 447, 869– 874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brash A. R., Boeglin W. E., Chang M. S. ( 1997) Proc. Natl. Acad. Sci. U. S. A. 94, 6148– 6152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang M. S., Schneider C., Roberts R. L., Shappell S. B., Haselton F. R., Boeglin W. E., Brash A. R. ( 2005) Invest. Ophthalmol. Vis. Sci. 46, 849– 856 [DOI] [PubMed] [Google Scholar]
- 28.Gan Q. F., Browner M. F., Sloane D. L., Sigal E. ( 1996) J. Biol. Chem. 271, 25412– 25418 [DOI] [PubMed] [Google Scholar]
- 29.Walther M., Wiesner R., Kuhn H. ( 2004) J. Biol. Chem. 279, 3717– 3725 [DOI] [PubMed] [Google Scholar]
- 30.Brinckmann R., Schnurr K., Heydeck D., Rosenbach T., Kolde G., Kühn H. ( 1998) Blood 91, 64– 74 [PubMed] [Google Scholar]
- 31.Chen B., Tsui S., Boeglin W. E., Douglas R. S., Brash A. R., Smith T. J. ( 2006) J. Biol. Chem. 281, 18296– 18306 [DOI] [PubMed] [Google Scholar]
- 32.Aleem A. M., Jankun J., Dignam J. D., Walther M., Kühn H., Svergun D. I., Skrzypczak-Jankun E. ( 2008) J. Mol. Biol. 376, 193– 209 [DOI] [PubMed] [Google Scholar]
- 33.Gronert K., Maheshwari N., Khan N., Hassan I. R., Dunn M., Laniado Schwartzman M. ( 2005) J. Biol. Chem. 280, 15267– 15278 [DOI] [PubMed] [Google Scholar]