Abstract
We present a new approach to the identification of degrees of freedom which comprise a reaction coordinate in a complex system. The method begins with the generation of an ensemble of reactive trajectories. Each trajectory is analyzed for its equicommittor position or transition state; then the transition state ensemble is identified as the stochastic separatrix. Numerical analysis of the points along the separatrix for variability of coordinate location correctly identifies the components of the reaction coordinate in a test system of a double well coupled to a promoting vibration and a bath of linearly coupled oscillators.
INTRODUCTION
Given a potential energy surface for a complex system and a method of dynamic propagation on that system, one of the central challenges of theoretical chemistry is the identification of mechanism. In a chemical reaction, this task corresponds to identification of the reaction mechanism and in microscopic variables, a reaction coordinate. Many challenges are present in such a definition. First, in complex systems, there are often many ways to move from one stable state to another. In a complex system, it is often hard to have appropriate intuition as to the dominant mode of transformation, even if there happens to be a dominant mode. A classic example of this is the recognition that charged particle transfer in polar solution is more appropriately followed as a reaction when solvent polarization is used as a reaction coordinate than the “obvious” choice of position of the transferred particle.1 Notwithstanding this difficulty, in biological simulation, where systems are extraordinarily complex, the standard method of approach is to guess at a reaction coordinate and impose this guess using methods such as umbrella sampling.2 It is worth noting that the recently developed potential energy and free energy string methods3 allow the identification of what is expected to be the dominant path through configuration space for very low temperature in the case of the potential energy string method, and a tube of reactive events in the case of the finite temperature version. In complex systems, however, it is possible that important but subtle motions that are part of the reaction coordinate will be missed.
The difficulty of finding the multiple paths that may connect one stable state to another has been addressed through the extraordinarily powerful technique of transition path sampling4 (TPS.) This computational approach allows a directed Monte Carlo walk through trajectory space, and thus the efficient generation of an ensemble of paths that connect stable states or transition paths. The trajectories themselves however, are of little use without a method to analyze their information. Reaction coordinate identification is possible through statistical analysis of the reactive trajectories. This short note describes a new approach to bypass the complexities of reaction coordinate identification simply by using information available from the reactive trajectories.
THE STOCHASTIC SEPARATRIX AND THE TRANSITION STATE ENSEMBLE
The first step in any identification of a reaction coordinate is the identification of the ensemble of transition states. In TPS, the definition of the transition state is most naturally given by a probabilistic assignment rather than any feature of the potential or free energy surface. For a single trajectory, the transition state is defined as that position (or positions as we will describe below) for which assignment of random momenta to all degrees of freedom chosen from a Boltzmann distribution results in a 0.5 probability of reaction. Because each trajectory may follow a different path through phase space, each trajectory may have a different transition state, and so gathering all transition states defines a lower dimensional hypersurface, each point of which has the property of an equicommittor, or 0.5 probability to go to either one stable state or another on random assignment of momenta. This hypersurface is known in the dynamics literature as the stochastic separatrix, and for example is useful in defining confinement bottles in plasma tokomacs.5 This hypersurface is a level surface of reaction probability, and so the reaction coordinate is logically defined as being orthogonal to it. This definition has resulted in one algorithm for identification of the reaction coordinate; one guesses at a reaction coordinate, holds these degrees of freedom fixed, and performs constrained molecular dynamics. After random periods of evolution, the constrained dynamics is halted and unconstrained dynamics is initiated with random momenta assigned to all degrees of freedom, again chosen from a Boltzmann distribution. If one has chosen the reaction coordinate correctly, then the distribution of reaction probabilities obtained should be peaked about 0.5. If it is not, then a new choice of reaction coordinate needs to be made. Bolhuis and Chandler6 described this algorithm in detail.
This method is formally well founded and has been used to identify the conformational transformation coordinates in alanine dipeptide.7 It is even possible to apply this approach to reactions as complex as those catalyzed by enzymes, and our group has done so for both the reactions catalyzed by lactate dehydrogenase8 and purine nucleoside phosphorylase.14 The difficulty with application is that the computational demand is very high. First, reactive trajectories must be generated. In each trajectory, the equicommittor point must be located to generate the equicommittor surface. A guess must be made (it should of course be an informed guess) of the reaction coordinate, and constrained dynamics on this surface must be run. Then from many points along that constrained trajectory, unconstrained trajectories must be shot with momenta chosen from a Boltzmann distribution. The result is the need to compute thousands of trajectories. In a complex system where the potential may be at least partially computed by way of quantum mechanical methods, this is a highly challenging computation. While the guess at the reaction coordinate is guided by intuition, it may not be correct, and the entire process needs to be repeated until a committor distribution can be obtained, which shows the diagnostic peak about 0.5.
Two approaches have been proposed to avoid the generation of many hundreds of trajectories to verify the correctness of a reaction coordinate. They are both methods, which can be viewed as numerical interpolation of properties of the trajectories, and so one collects a library of trajectories, and then this library is all that is needed to approximately check the correctness of a reaction coordinate. Ma and Dinner9 use a neural network algorithm to generate the needed distributions (along with a genetic algorithm to propose possible reaction coordinates, and the Trout group10 applies a maximum likelihood approach assuming Bayesian statistics. Similarly, Best and Hummer11 use a Bayesian relation between equilibrium and transition path ensembles to rank reaction coordinates. In the method described in this communication we take a completely different approach.
Here we recognize that the reaction coordinate test is really not a test of the reaction coordinate; it is a test of the stochastic separatrix. The separatrix may be defined as the hypersurface in which there is no progress toward or away from reaction. Of course we really know the separatrix if we have done a TPS computation along with a calculation of the transition state ensemble because this ensemble, the equicommittor ensemble, is the level surface of reaction probability,12 and so is the stochastic separatrix. What we do not have is a parametrization of this surface in terms of the standard configuration space descriptors of chemical reactions such as atomic coordinates. We can however locate which coordinates define the separatrix simply by recognition of the defining quality of this surface: these coordinates do not change or at least do not change nearly as much as all other degrees of freedom on the separatrix. As such, the algorithm is trivial to define: plot distributions of coordinates for all degrees of freedom in the problem. Examine the width of the distribution for all degrees of freedom. Coordinates which are components of the reaction coordinate should have a significantly smaller variation along the separatrix.
It is worth asking what advantages this method has over those previously published in Refs. 9, 10, 11. In those references we are told that the “postprocessing” of the committor probabilities does not involve too significant an expenditure of computational effort. The difficulty is that even if this effort of application of a neural net or Bayesian statistics is simple, it may not be right. Neural networks are highly nonlinear fitting functions that can easily either return only trained data or stray into unwanted areas of probability space. On the other hand, the use of Bayesian statistics may simply not be warranted. Overall we find our procedure more conceptually transparent.
TEST
To test the viability of this seemingly trivial approach to what is a highly complex numerical problem we apply the idea to a test system we have studied previously using TPS. This is a double well potential coupled symmetrically to a single oscillator (a promoting vibration) and also coupled to a bath of 100 linearly coupled oscillators. Transferred particle has mass equal to a proton mass. The potential barrier is a symmetric double well with barrier height 0.010 au (6.3 kcal∕mol), and transfer distance 1 Å. The barrier curvature is 1090 cm−1. The promoting oscillator Q has mass equal to 4 C mass, frequency=110 cm−1 and is symmetrically coupled to s with coupling strength k=0.0166 au, with coupling form: . With these parameters, the saddle point is at Q=0.67 au and the barrier height reduction at the saddle point is 50%. The bath consists of 100 oscillators, each one with mass equal to 4 C mass, linearly coupled to s (i.e., with coupling of the form ) with coupling strength 0.000 25 au. The frequencies of the bath oscillators are distributed like an Ohmic with exponential cutoff 110 cm−1. The frequencies are chosen randomly, and the specific realization that generated the data shown below is provided in supplementary information.15 The classical equations of motion were integrated with a velocity Verlet integrator.
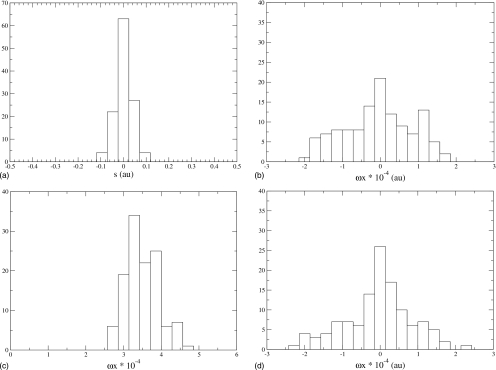
A TPS simulation was applied as follows. The molecular dynamics (MD) time step is 0.1 fs and the temperature is 300 K. We generated trajectories that were 40 000 MD steps long. The order parameter was a position of s, larger than 0.1 times the location of the well bottom. A trajectory was assumed to be reactive, if it stayed within the order parameter for the whole duration of the last 1500 MD steps. We found the isocommittor surface (121 transition states13) and plotted histograms of s, Q, and all bath oscillator positions on this surface.
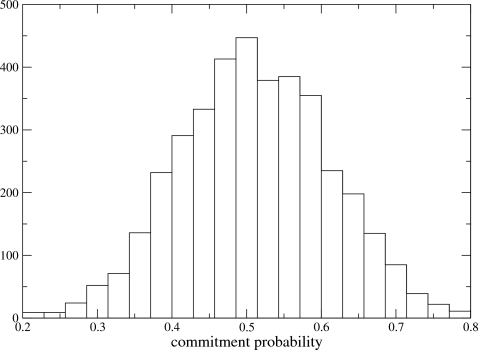
We now know from long experience that the reaction coordinate in this system is a mixture of the transferred particle (s) and the promoting vibration (Q). We examined all histograms of positions on the stochastic separatrix. In Fig. 1a, we show the distribution for the s degree of freedom, 1b is that for the promoting vibration, 1c is a randomly chosen bath oscillator, and finally 1d is the bath oscillator that happens to have the least variability (half-width.) Note that all oscillator location distributions are normalized by frequency, so the natural distribution of the motion is accounted for. In order to assess the actual widths of the distributions, we nonlinearly fit the data to Gaussians and extracted the half-width of the Gaussians. Figure 1b is the distribution for the promoting vibration on the separatrix, and the half-width of this distribution is 4.3×10−5. Figure 1c represents an exemplary chosen linearly coupled oscillator, and it has a half-width of 9.9×10−5. Finally Fig. 1d is the narrowest of all antisymmetrically coupled oscillators, and it has a width of 6.5×10−5. Thus analysis of the data along with the rather narrow double well coordinate shows one other degree of freedom with distribution on the separatrix less than 0.5, and all others with distributions of half-width 6.5×10−5 to about 1.0×10−4. If one knew nothing about this problem, it is easy to imagine at least segregating the double well coordinate, the promoting vibration, and potentially the oscillator shown in Fig. 1d. One now applies the standard test to confirm that we have chosen correctly. Inclusion of oscillator 1d in the reaction coordinate makes no noticeable difference in the committor distribution. A reaction coordinate chosen to be composed of the double well motion plus the promoting vibration results in a committor distribution of Fig. 2—clearly diagnostic of a correct choice for the reaction coordinate. It is to be admitted that this is not an especially complicated test system, but it does clearly show the reaction coordinate to be more complex than the coordinate of the transferring particle. It is also effective at eliminating almost all other degrees of freedom, and even in a highly conservative reading of the separatrix distributions, results in only two possible reactions coordinates; both contain the double well and the promoting vibration and differ only by inclusion of a single antisymmetrically coupled oscillator.
Figure 1.
The distributions of coordinate positions of points along the stochastic separatrix. Panel a shows for the double well potential, panel b for the symmetrically coupled vibration (promoting vibration), panel c for a representative antisymmetrically coupled vibration, and panel d for the antisymmetrically coupled vibration with narrowest width. The widths of b, c, and d are 4.3×10−5, 9.9×10−5, and 6.5×10−5, respectively.
Figure 2.
The committor distribution found when the double well and promoting vibration are held fixed while other degrees of freedom evolve. This demonstrates these two coordinates form the reaction coordinate. Inclusion of the vibration in panel d does not change this distribution indicating it is not part of the reaction coordinate.
CONCLUSIONS AND FUTURE WORK
This simple approach to analysis of the separatrix to isolate degrees of freedom, which are part of the reaction coordinate, shows potential for application to truly complex systems, in particular biological systems. We are currently applying these ideas to lactate dehydrogenase, where we have already isolated protein and substrate motions that are part of the reaction coordinate.8 One addition is needed in this application. In the model system we know the natural width of the oscillator degrees of freedom, which allow for appropriate normalization of the separatrix position distributions. We have no such a priori knowledge in a biological system, but we do have the maximum variability across all trajectories. This provides the needed normalization.
We point out that the system studied in this letter is not quite as trivial as it might appear. In fact until we employed a fairly fine grained binning for the position distribution of coordinates on the separatrix, we found a fairly large number of degrees of freedom that seemed to be part of the reaction coordinate, but upon testing did not improve on the sharpness of the committor distribution about 0.5. This indicated that these degrees of freedom were simply not part of the reaction coordinate, and our assumption from the computation was in error. Upon finer binning these spurious degrees of freedom disappeared. Another caveat is that fairly extensive statistics are needed to generate a reasonable picture of the separatrix. This is of course dependent on the spread in phase space of the generated TPS trajectories.
It is to be noted that the ideas contained in this communication will only work when the transition state for a specific trajectory is sharp in position space. This is certainly the case for lactate dehydrogenase, but equally not the case for other reactions. For example, our studies of purine nucleoside phosphorylase14 show a far more complex reaction, which has rather than a single equicommittor position, a relatively flat area extending over at least 10 fs of trajectory. This corresponds in turn to a two step chemical transformation in this reaction, and two peaks in the free energy surface. In such a case a single simple reaction coordinate is not meaningful, and the reaction must be analyzed separately for each part of the mechanism. Other cases of long barriers, more diffusive in character are found in such problems as protein folding, but it is worth noting that one would expect, for example, in all cases of enzymatic chemistry involving hydrogen transfer (a large percentage of all biological chemistry) committor distributions to be sharp. It is also worth noting that to obtain reasonable statistics, it is necessary to gather about 100 points along the committor surface—this for this problem ensured adequate coverage of configuration space. While not a trivial calculation, this is easily within the realm of possibility for reaction in an enzyme.
ACKNOWLEDGMENTS
We acknowledge the Chemistry Division of the National Science Foundation for support of this work through Award No. CHE-0714118 and the National Institutes of Health through Award No. GM068036. S.D.S. also acknowledges the Institut Des Hautes Études Scientifiques for their hospitality and creative environment where he was in residence when this work was begun.
References
- van der Zwan G. and Hynes J. T., J. Chem. Phys. 78, 4174 (1983). 10.1063/1.445094 [DOI] [Google Scholar]
- Dimelow R. J., Bryce R. A., Masters A. J., Hillier I. H., Burton N. A., J. Chem. Phys. 124, 114113 (2006). 10.1063/1.2172604 [DOI] [PubMed] [Google Scholar]
- Ren W. E. W., and Vanden-Eijnden E., Phys. Rev. B 66, 052301/1 (2002) 10.1103/PhysRevB.66.052301 [DOI] [Google Scholar]; Ren W. E. W., and Vanden-Eijnden E., J. Phys. Chem. B 109, 6688 (2005) 10.1021/jp0455430 [DOI] [PubMed] [Google Scholar]; Proceedings of the ICM 2002 (Higher Education Press, Beijing, 2002), Vol. I, pp. 621–630.
- Bolhuis P. G., Chandler D., Dellago C., and Geissler P. L., Annu. Rev. Phys. Chem. 53, 291 (2002) 10.1146/annurev.physchem.53.082301.113146; [DOI] [PubMed] [Google Scholar]; Dellago C., Bolhuis P. G., and Chandler D., J. Chem. Phys. 108, 9236 (1998) 10.1063/1.476378; [DOI] [Google Scholar]; Bolhuis P. G., Dellago C., and Chandler D., Faraday Discuss. 110, 421 (1998). 10.1039/a801266k [DOI] [Google Scholar]
- Punjabi A., Verma A., and Boozer A., Phys. Rev. Lett. 69, 3322 (1992). 10.1103/PhysRevLett.69.3322 [DOI] [PubMed] [Google Scholar]
- Bolhuis P. G. and Chandler D., J. Chem. Phys. 113, 8154 (2000). 10.1063/1.1315997 [DOI] [Google Scholar]
- Bolhuis P. G., Dellago C., and Chandler D., Proc. Natl. Acad. Sci. U.S.A. 97, 5877 (2000). 10.1073/pnas.100127697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaytman S. and Schwartz S. D., Proc. Natl. Acad. Sci. U.S.A. 104, 12253 (2007). 10.1073/pnas.0704304104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A. and Dinner A. R., J. Phys. Chem. B 109, 6769 (2005) 10.1021/jp045546c [DOI] [PubMed] [Google Scholar]; Hu J., Ma A., and Dinner A. R., Proc. Natl. Acad. Sci. U.S.A. 105, 4615 (2008). 10.1073/pnas.0708058105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. and Trout B. L., J. Chem. Phys. 10.1063/1.2234477, 125, 054108 (2006); [DOI] [PubMed] [Google Scholar]; Peters B., Beckham G. T., and Trout B. L., J. Chem. Phys., 127, 034109 (2007). [DOI] [PubMed] [Google Scholar]
- Best R. and Hummer G., Proc. Natl. Acad. Sci. U.S.A. 102, 6732 (2005). 10.1073/pnas.0408098102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragliano L., Fischer A. R., Vanden-Eijnden E., and Ciccotti G., J. Chem. Phys. 125, 024106 (2006). 10.1063/1.2212942 [DOI] [PubMed] [Google Scholar]
- Because this is a not fully dissipative systems, trajectories recross the transition state a number of times. Starting from an initial reactive trajectory, we did a total of 5000 shooting moves to obtain 40 reactive trajectories (we wanted to fully explore phase space.) We choose every five trajectories, and from recrossings obtained 121 distinct transition states. The entire analysis took minutes on a single linux cluster node.
- Saen-Oon S., Quaytman S., Schramm V. L., and Schwartz S. D., Proc. Natl. Acad. Sci. U.S.A. 105, 16543 (2008). 10.1073/pnas.0808413105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- See EPAPS Document No. E-JCPSA6-130-039917 for the values of the frequencies of the 100 bath oscillators used in the similation. For more information on EPAPS, see http://www.aip.org/pubservs/epaps.html.