Abstract
The combination of reflectance, fluorescence, and Raman spectroscopy—termed multimodal spectroscopy (MMS)—provides complementary and depth-sensitive information about tissue composition. As such, MMS is a promising tool for disease diagnosis, particularly in atherosclerosis and breast cancer. We have developed an integrated MMS instrument and optical fiber spectral probe for simultaneous collection of all three modalities in a clinical setting. The MMS instrument multiplexes three excitation sources, a xenon flash lamp (370–740 nm), a nitrogen laser (337 nm), and a diode laser (830 nm), through the MMS probe to excite tissue and collect the spectra. The spectra are recorded on two spectrograph∕charge-coupled device modules, one optimized for visible wavelengths (reflectance and fluorescence) and the other for the near-infrared (Raman), and processed to provide diagnostic parameters. We also describe the design and calibration of a unitary MMS optical fiber probe 2 mm in outer diameter, containing a single appropriately filtered excitation fiber and a ring of 15 collection fibers, with separate groups of appropriately filtered fibers for efficiently collecting reflectance, fluorescence, and Raman spectra from the same tissue location. A probe with this excitation∕collection geometry has not been used previously to collect reflectance and fluorescence spectra, and thus physical tissue models (“phantoms”) are used to characterize the probe’s spectroscopic response. This calibration provides probe-specific modeling parameters that enable accurate extraction of spectral parameters. This clinical MMS system has been used recently to analyze artery and breast tissue in vivo and ex vivo.
INTRODUCTION
Spectroscopy is a promising means of extracting biochemical and morphological information about tissue that is relevant to disease progression and diagnosis.1, 2 Raman spectroscopy, which measures the molecular vibrations of a sample, has been utilized previously to study atherosclerosis,3 measure blood analytes,4 and detect dysplasia and cancer in various tissues including the breast,5, 6, 7 cervix,8, 9, 10 prostate,11, 12 and skin.13, 14, 15 By measuring changes in scattering and absorption properties, diffuse reflectance spectroscopy (DRS) has been used to track the progression of disease in tissues of the colon,16, 17 esophagus,18 cervix,19 and bladder.20, 21 Similarly, intrinsic fluorescence spectroscopy (IFS), free of distortions due to scattering and absorption, which measures the emission of endogenous fluorophores found in tissue, has been used to detect dysplasia in the uterine cervix,19, 22, 23 adenomatous colon polyps,24, 25 the esophagus,18, 26, 27 and the oral cavity.28, 29 Since these three spectroscopic modalities provide complementary information about tissue composition, their combination should provide useful information about disease status and its treatment.
Several groups have developed their own spectroscopic instrumentation and probes for tissue investigation.30, 31 Our laboratory has previously developed two separate clinical spectroscopic instruments, briefly described in the next two subsections, each with its own optical fiber probe for collecting different spectroscopic modalities. The clinical Raman instrument32 and probe33 have been developed for portable real-time study of diseases such as atherosclerosis and breast cancer. The fast excitation emission matrix (FastEEM) instrument and probe34, 35 have been developed to extract DRS and IFS spectra excited at multiple wavelengths, useful for studying cancer in organs such as the cervix and oral cavity.
Clinical Raman system
Our laboratory has previously developed a compact clinical Raman instrument for real-time tissue spectroscopy.32 The instrument uses an 830 nm diode laser to excite Raman scattering. The laser light is filtered and coupled into an optical fiber Raman probe, which delivers to and collects light from tissue. The Raman probe33 contains a single excitation fiber surrounded by a ring of 15 collection fibers, specialized filters on both the excitation and collection fibers at the distal tip of the probe, and a ball lens at the tip to optimize signal collection. The return light is passed through a spectrograph and dispersed onto a charge-coupled device (CCD) detector. The entire operation is fully automated and computer-controlled.
FastEEM system
Our laboratory has also developed the FastEEM instrument for real-time reflectance and fluorescence tissue spectroscopy.34, 35, 36 A fluorescence EEM is produced by means of a rotating wheel containing laser dyes, pumped by a 308 nm XeCl excimer laser, which generates nine laser pulses at excitation wavelengths ranging from 340 to 500 nm which, including the 308 nm pulses, provides ten laser excitation wavelengths in all. In addition, a xenon flash lamp produces white light pulses for reflectance. An unfiltered optical fiber probe delivers light to and collects it from the tissue. This probe contains a single central excitation fiber surrounded by a ring of six collection fibers, and an angled quartz “optical shield” at the probe tip37 to provide a fixed light delivery-collection geometry. Return light from the tissue is transmitted by the collection fibers to a spectrograph∕CCD detector for analysis. The operation of the instrument is automated via LABVIEW software.
Multimodal spectroscopy (MMS)
Previous studies demonstrated the diagnostic potential of Raman, DRS, and IFS as separate spectroscopic modalities. Recently, we have developed Raman, DRS, and IFS—or MMS—as a combined technique to yield complementary spectral and depth-sensitive information about tissue. DRS provides information about tissue absorption and scattering, IFS about tissue fluorophores, and Raman spectroscopy about molecular tissue composition. In addition, the three techniques utilize different portions of the wavelength range covered, 300–1000 nm. Light penetration in tissue varies substantially over this range, so the three modalities sample different depths, thus providing depth-sensitive capability. The depth-sensitive capability enables the measurement of the sizes of structures in layered tissue, which can be critical to disease diagnosis. For example, our laboratory envisions using MMS to measure the thickness of the top layer fibrous cap in diseased arteries relative to the size of the underlying plaque core, which is critical to being able to assess the propensity of the plaque to rupture and cause a heart attack or stroke.38
We have obtained promising results using MMS for disease diagnosis, specifically in atherosclerosis39 and breast cancer, with other organs also under investigation, and have developed a portable instrument for delivery, collecting, and processing the MMS data. In parallel, we have developed an efficient unitary MMS probe for simultaneously collecting spectra of all three modalities, thus eliminating sampling error. This paper describes the MMS system and the modeling for accurately extracting disease-related spectral parameters.
SYSTEM DESCRIPTION
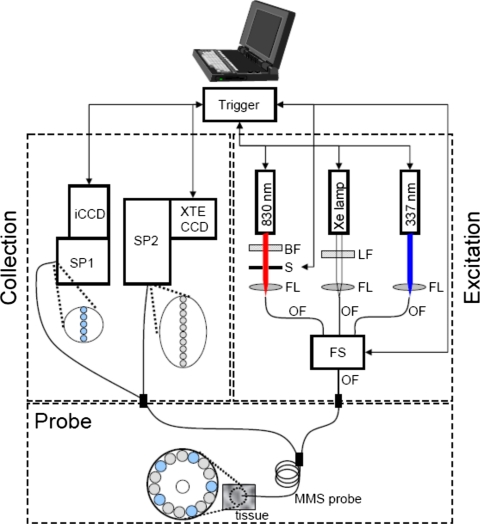
The clinical portable MMS instrument merges the designs of the previously developed clinical Raman and FastEEM systems. The design is modular. The MMS instrument contains all the critical functionality of both systems, with several improvements based on newer technology. A schematic diagram is provided in Fig. 1.
Figure 1.
Schematic diagram of the MMS instrument. BF=bandpass filter, LF=longpass filter, S=shutter, FL=focusing lens, OF=optical fiber, FS=fiber switch, SP1=visible spectrograph, SP2=NIR spectrograph, iCCD=intensified CCD, and XTE CCD=thermoelectrically cooled CCD.
Sources
The MMS instrument contains three excitation sources: a Xe flash lamp (L7684, Hamamatsu Corp., Bridgewater, NJ), a white light source used to obtain diffuse reflectance spectra (DRS); a 337 nm N2 laser used to excite fluorescence (NL100, Stanford Research Systems, Sunnyvale, CA); and an 830 nm diode laser used to excite Raman spectra (Process Instruments, Salt Lake City, UT). All are configured to be triggerable externally through automated software. The Xe lamp provides a 2.9 μs full width half maximum (FWHM) white light pulse, 1 J∕pulse max, externally triggered by the software. The light from the Xe lamp passes through a ∼370 nm long pass filter (LP02–364RS-25, Semrock, Rochester, NY) and is then focused into a fiber; this filter guarantees that the reflectance collected over the range 370–740 nm is free of second order diffraction from the spectrograph grating. The N2 laser provides externally triggered 3.5 ns FWHM pulses of 337 nm light, 170 μJ per pulse, with a 10 Hz repetition rate. The light is focused into an optical fiber by means of an UV-coated lens. The diode laser can be configured to output a range of up to 500 mW of continuous-wave 830 nm light driven by a user-specified analog signal. The 830 nm laser light is filtered by an 830 nm bandpass filter (LL01–830–12.5, Semrock, Rochester, NY) and gated by a mechanical shutter (LS6ZM2, Vincent Associates, Rochester, NY) before being focused into an optical fiber.
Coupling to the MMS probe
The excitation sources are coupled sequentially to the MMS probe (see Sec. 3A for details) using an optical fiber switch (FSM14, Piezo Jena, Germany). This is a microelectromechanical systems (MEMS) device that provides superior coupling of the individual sources and interfaces simply with the control software by means of a user-configurable binary code. The switch contains a four optical fiber inputs and a single optical fiber output with each fiber terminated by a SubMiniature version A (SMA) connector. The outputs of the Xe lamp, N2 laser, and diode laser are directly channeled into three input fibers of the switch. The remaining input channel is left vacant for a future light source. The output fiber of the switch is connected to the MMS probe by means of a SMA-to-SMA connector. The overall transmission efficiency of the switch is ∼60%, resulting from an 80% switching efficiency and an 80% SMA-to-SMA coupling efficiency.
Detection
The MMS system uses two spectrograph∕CCD modules, one optimized for near infrared (NIR) detection (Raman) and the other for visible wavelengths (reflectance and fluorescence). The operation of both cameras is controlled by modular LABVIEW drivers (R3 Software, Princeton, NJ).
The distal tips of the ten collection fibers carrying the Raman light are aligned in a linear array, effectively serving as an entrance slit to the spectrograph (Holospec f∕1.8i, Kaiser Optical Systems), resulting in an ∼9 cm−1 spectral resolution. The fibers are encased in a modified male fixed connection (FC) connector to ensure highly reproducible alignment with the connector plate of the spectrograph. Any spectral changes incurred by disconnecting and reconnecting the linear array are found to be below the system’s spectral resolution. The numerical aperture (NA) of the collection fibers is matched to the spectrograph (f/#=1.8) to conserve throughput. The spectrograph contains an 830 nm notch filter that further suppresses the elastically scattered Rayleigh light and a holographic grating to disperse the Raman scattered light onto a back-illuminated, deep-depletion CCD detector with a 1340×400 array of pixels (Spec10 XTE, Princeton Instruments∕Acton, Acton, MA). The CCD detector has a 16 bit dynamic range and can be thermoelectrically cooled within minutes to −90 °C, a significant practical improvement over the older Raman system, which had a liquid nitrogen cooled CCD. Spectra are obtained by vertically binning the signal from the ten collection fibers. Since the Raman signals are relatively weak compared to ambient background, and owing to an absence of gated detectors in the NIR region, all spectra are collected with the room lights off.
The five collection fibers carrying the reflectance or fluorescence light are arranged vertically and coupled to a diffraction grating spectrometer (Spectra Pro 150, Princeton Instruments∕Acton, Acton, MA), resulting in an ∼6 nm spectral resolution. The PIMAX camera has a 16 bit dynamic range and is thermoelectrically cooled to −20 °C. The collection fibers have individual male SMA adapters that connect to corresponding female SMA adapters mounted to a plate adjacent to the spectrograph; this insures easy reproducibility when the MMS probe is disconnected from and reconnected to the system. The collected light is then dispersed onto an intensified CCD detector (PIMAX, Princeton Instruments∕Acton, Acton, MA). The CCD is operated in a gated mode, where the intensifier is biased on for only a short period of time (∼30 μs) extending from immediately prior to immediately after each laser∕white light pulse. The short duration of this gate ensures that the background from ambient light is negligible, although this is less of a concern with the MMS instrument since the room lights are routinely turned off, as is required for Raman signal collection.
Real-time software
The entire MMS system is controlled by a single user-friendly interface developed in LABVIEW (National Instruments, Austin, TX). The control flow of the software for a single MMS acquisition is presented in Fig. 2, describing the timing by which the various sources and detectors are triggered to collect the MMS spectra. The software platform allows for calibration spectra to be taken and stored, provides easy operation of the experimental data collection and storage, and provides instant feedback about the collected spectra, fits, and extracted diagnostic spectral parameters. The sources and cameras are triggered by two data acquisition (DAQ) cards (6062 and 6035E, National Instruments, Austin, TX).
Figure 2.
Control flow of the MMS instrument software in which the Raman, reflectance, and fluorescence signals are collected sequentially through the optical fiber switch. Collection of the Raman signals is facilitated by opening the laser shutter and triggering the camera to collect a spectrum for a specified amount of time. Collection of the reflectance signal is facilitated by appropriately triggering the Xe lamp and the camera gate. The fluorescence signal is collected by triggering the nitrogen laser and the camera gate.
System calibration
In order to provide real-time data analysis and spectral diagnosis, spectra for calibration and background subtraction must be acquired prior to data collection. The Raman shift axis is calibrated by collecting a spectrum from a known Raman scatterer, 4-acetamidophenol (Tylenol). The Raman fiber background signal from the MMS probe is measured through acquisition of a spectrum from a roughened aluminum block. The spectral response of the Raman collection system is obtained by recording the spectrum of a calibrated tungsten white light source diffusely scattered by a reflectance standard (BaSO4). The wavelength of the reflectance and fluorescence spectra is calibrated by recording a spectrum from a mercury lamp. Background calibration for reflectance and fluorescence is used to remove effects of CCD dark current and stray light and is typically acquired by collecting a spectrum from an opaque cup of de-ionized water. The reflectance amplitude is calibrated by collecting a spectrum from a reflectance standard (20% reflectance standard, Labsphere Inc., North Sutton, NH). To normalize the fluorescence spectra appropriately, the output from the nitrogen laser through the MMS probe is noted with an energy meter. The system is designed to be stable from day to day; thus disconnecting and reconnecting the probe to the system do not change the calibration parameters. However, different MMS probes can have significantly different calibration parameters.
Data analysis
This section describes the data analysis steps for each modality.
Raman
A sequence of steps is executed to extract quantitative Raman information from the raw spectrum acquired at the CCD. First, the data are corrected for the system spectral response by dividing it by the white light spectrum. The resulting data are then normalized to maximum peak height and truncated to the biological Raman region of interest appropriate to the models used in this study (686 to 1788 cm−1). Next, the fiber background from the tissue is removed by subtracting the spectrum acquired from roughened aluminum. The relationship between the intensity of spectra from the aluminum and the tissue spectrum depends on the tissue’s optical properties, which are unknown a priori. Therefore, we iteratively subtract the same aluminum spectrum scaled by a range of different intensities to determine the optimal ratio for background removal. The spectrum that results in the lowest standard deviation of the residual between the data and the model fit is used for analysis. After fiber background removal, the remaining broadband tissue luminescence is removed by subtracting a slowly varying polynomial (sixth order) that is fit to the data.33 This final correction results in the calibrated Raman spectrum of the examination site. This spectrum is numerically arranged into a standard set of wavenumber bins. It is analyzed with the appropriate disease model, atherosclerosis40 or breast cancer,41 using ordinary least-squares, which provides the relative fit coefficients of the various chemical∕morphological components of the tissue. The user can toggle between the two different models, as mandated by the clinical situation.
Reflectance and fluorescence
A similar sequence of steps is executed to extract reflectance and fluorescence information. The DRS spectrum is obtained from the raw reflectance spectrum by subtracting the background spectrum and dividing by the background-subtracted reflectance standard response, as specified by the manufacturer. This ensures that the excitation∕collection geometry of the probe is properly accounted for. The DRS spectrum is fit to the model of Farrell et al.42 to extract the DRS spectral parameters that characterize the scattering and absorption coefficients (Sec. 3B1). The IFS spectrum is obtained from the raw fluorescence spectrum by subtracting the background spectrum and applying the IFS extraction formula from Zhang et al.,43 enabling extraction of IFS spectral parameters that characterize the tissue fluorophores (Sec. 3B2).
Safety
The MMS system must adhere to strict safety guidelines. For breast or artery studies, in the clinical environment, any part of the system that enters the sterile field (touches the patient directly or indirectly) must be kept sterile. Sterilization of the MMS probe was performed using cold-gas ethylene oxide. Furthermore, the excitation laser energies cannot exceed predetermined maximum values, which are determined and safeguarded prior to entering the operating room. The Xe flash lamp and the nitrogen laser are pulsed sources and thus internally shuttered. The continuous-wave diode laser beam is gated by means of a high-speed shutter also controlled by LABVIEW. The shutter opens automatically just before DAQ begins and closes immediately after acquisition is complete. The shutter then remains closed until the system receives a signal through LABVIEW for the next acquisition.
Clinical layout
In order for the MMS instrument to be operated in a clinical environment, it needs to have a suitable portable layout. The various components of the system described in Fig. 1 were optimally positioned on optical breadboards with the help of SOLIDWORKS software. This modular placement was used to confine the components within a metal enclosure (dimensions of 27×27×58 in.3) that can be wheeled into the operating room for clinical data collection. The layout schematic and a photo of the finished instrument are provided in Fig. 3.
Figure 3.
Modular layout of the MMS instrument (left) and a photo of the actual system (right). The various components are indicated.
PROBE DESCRIPTION
The MMS probe design is a modification of the Raman probe that enables efficient delivery and collection of DRS and IFS in the UV∕visible wavelength region, as well as Raman in the near IR. Because the excitation∕collection geometry is altered from that of the FastEEM probe, the modeling of DRS and IFS spectra collected with this probe must be adjusted to enable correct extraction of spectral parameters.
MMS probe design
The MMS probe is similar to the Raman probe described in detail elsewhere.33 A schematic is shown in Fig. 4a. The MMS probe differs from the Raman probe in several ways: (1) the optical fibers that are used for delivery and collection, (2) the filter characteristics, and (3) the type of adhesives used to glue the components in the probe tip.
Figure 4.
(a) Schematic of the MMS probe with a side view on the left and a cross sectional view on the right. A central excitation fiber (200 μm core diameter, 0.22 NA) is optically isolated from the surrounding 15 collection fibers, ten of which (200 μm core diameter, 0.26 NA) collect Raman spectra and five of which (200 μm core diameter, 0.22 NA) collect DRS and IFS return light. The probe tip contains a module to filter the excitation and collection light and a sapphire ball lens to optimize collection. (b) Filter characteristics of the MMS probe. The excitation fiber light is filtered by a short-pass filter that transmits 300–830 nm light and blocks light >850 nm. The collection fiber tube contains a notch filter that transmits 300–800 and 850–1000 nm while blocking light around the Raman laser line (830 nm).
Optical fibers
Owing to the broad range of wavelengths of light used, the MMS probe fibers need to be optimized for their particular modalities. The central excitation fiber must efficiently transmit excitation light in the range of 300–830 nm, so a Superguide fiber (200 μm core diameter, 0.22 NA, Fiberguide Industries, Stirling, NJ) is used. The collection fibers need to be partitioned in a manner so as to provide sufficient Raman return light at NIR wavelengths and reflectance∕fluorescence return light in the visible. The collection fibers can thus be optimized based on modalities, so Superguide fibers are used for the visible and Anhydroguide fibers (200 μm core diameter, 0.26 NA, Fiberguide Industries, Stirling, NJ) are used for the NIR. Therefore, out of the 15 collection fibers, five fibers are employed for DRS∕IFS, while the remaining ten are used for the much weaker Raman light. This allows almost equivalent signal collection with the single MMS probe as with the individual FastEEM and Raman probes used individually in previous studies.
Filter characteristics
The tiny filter module of the MMS probe must process light properly in order that DRS, IFS, and NIR Raman spectroscopies can all be implemented. As shown in Fig. 4a, the filter module is in the form of a fused silica rod (0.5 mm diameter, 1 mm length, polished and dielectrically coated for optimal light delivery) surrounded by a snug-fitting fused silica tube of 0.7 mm inner diameter, 1.8 mm outer diameter, 1 mm length, polished and dielectrically coated for optimal light collection. The excitation fiber rod contains a short-pass filter that efficiently transmits 300–830 nm wavelength light and blocks light at wavelengths of >850 nm. The collection fiber tube contains a notch filter that blocks light around Raman excitation wavelength (800–850 nm) while efficiently transmitting 300–800 and 850–1000 nm wavelength. These filter characteristics are shown in Fig. 4b. The filters are custom manufactured (Barr Associates, Westford, MA). One should note that owing to the significant amount of quartz Raman background generated in the fibers themselves and the inherently weak Raman signals from tissue, the described filters must be placed at the probe tip to enable the extraction of Raman spectra; more details about the filters can be found in Ref. 33.
Adhesives
Sodium silicate or epoxy was used in the Raman probe to glue the rods and tubes to the fibers. Ultimately, epoxy was deemed more effective as a result of its durability and optical properties. However, ordinary epoxy is not optically clear in the UV wavelength range. As a result, a special epoxy (Epo-tek 305, Epoxy Technology, Billerica, MA) is used for the MMS probe. Note that a sapphire ball lens is still optimal for the MMS probe, as sapphire is optically clear in the range of 300–1000 nm and its Raman features occur at low wavenumbers, well below the Raman features in the fingerprint of diagnostic interest. Also, in the MMS probe the ball lens is not glued in the probe but instead held by means of a stainless steel retaining sleeve.
The ball lens has an antireflection coating at 830 nm to substantially reduce reflection losses at the boundary between the fused silica fiber and the ball lens for Raman spectra, while reflection losses at 337 nm (∼6%) are acceptable as fluorescence has much higher signal strength.
Probe calibration
In the past, reflectance and fluorescence spectra were collected with an unfiltered FastEEM probe, consisting of six collection fibers surrounding one excitation fiber, with a 1.5 mm long quartz shield at the tip. The probe geometry was such that the excitation and collection spots on the sample were in the form of partially overlapping circles. The diffuse reflectance spectral model developed by Zonios et al.16 is dependent on this probe geometry. Now, with the ball lens of the MMS probe, the excitation∕collection geometry has changed specifically by increasing the spatial overlap between the excitation and collection regions and the range of angles collected, as compared to the FastEEM probe. The IFS modeling can, in principle, stay the same as that presented by Zhang et al.,43 with modifications of several probe-specific parameters. Therefore, the modeling in this new probe geometry for both DRS and IFS spectra was revisited so that each MMS probe could be calibrated appropriately to ensure accurate extraction of tissue parameters. Note that the spectral modeling of the Raman signals collected with the MMS probe is the same as for the previous Raman probes since the excitation∕collection geometry has not been changed for this modality.
DRS modeling
Farrell et al.42 calculated the diffuse reflectance from a narrow beam of light incident on the surface of a semi-infinite turbid medium in the diffusion approximation. The Green’s function for reflectance density, R(λ,r) (in units mm−2), at a distance r from the point of incidence, depends on tissue parameters: the absorption coefficient μa(λ), the reduced scattering coefficient , and A, a diffusion model parameter that depends on the refractive index of the tissue, which is assumed to be A≡3.2.16 In order to calculate the calibrated reflectance collected by a probe, Rp(λ), the Green’s function must be integrated over the delivery and collection areas characterized by the radius of delivery (rd) and radius of collection (rc) and normalized by the delivery area to obtain the unitless quantity
| (1) |
where ∣r−r′∣ specifies the distance between a point on the delivery area and a point on the collection area. Equation 1 can be evaluated numerically, but it can further simplified by making the approximation of point source delivery (rd⪡1). With this approximation, Zonios et al.16 obtained a closed form solution for the reflectance collected by a FastEEM probe, which was used for previous DRS modeling of the FastEEM probe. As discussed below, this approach does not hold for the MMS probe.
Our objective is to accurately extract and μa(λ) from the observed reflectance spectrum of tissue. To do this, rd and rc must be evaluated for a given probe, which can be accomplished by means of the following calibration technique. Five physical tissue models (“phantoms”) were prepared to mimic artery tissue reflectance properties. Polystyrene beads with 1 μm diameter and 1% solids by volume (64030 Polysciences, Warrington, PA) were used to simulate scattering. The values were calculated using a standard MIE code that evaluates the total scattering cross section σs(λ), which translated into for the specified bead solution.44 For biological tissue in the visible region, μa(λ) is dominated by hemoglobin absorption. Therefore, the phantoms were mixed with a solution of dry hemoglobin (H0267, Sigma-Aldrich, St. Louis, MO) dissolved in de-ionized H2O. The μa(λ) parameter depends on the known extinction spectrum of hemoglobin, the concentration of hemoglobin cHb, and the oxygen saturation of hemoglobin α, which was taken to be 100% for the phantoms, as the hemoglobin is completely oxygenated. The five phantoms included variations in scattering amplitude and hemoglobin concentration similar to that found in human artery tissue.45
The phantoms were mixed in small glass vials and thoroughly agitated to prevent clumping of the beads. Diffuse reflectance spectra were then collected using the MMS probe and instrument. The DRS spectra were obtained using the following expression:
| (2) |
where reflectance(λ) is the recorded reflectance spectrum from the phantom, background(λ) is the background spectrum collected from an opaque cup of de-ionized water, spectralon(λ) is the reflectance spectrum recorded from a calibrated standard (20% reflectance standard, Labsphere Inc., North Sutton, NH), and calibStd(λ) is the manufacturer-specified response of the standard. In this way, the Rp,observed(λ) is expressed in calibrated reflectance units.
The probe-specific rd and rc values were calculated through an optimization that minimized the residual [Rp,observed(λ)−Rp(λ)]2, where Rp,observed(λ) was obtained from Eq. 2 and Rp(λ) was calculated by numerically evaluating Eq. 1 with the known and specified values of and μa(λ). Although the optimal values of rd and rc varied slightly across the five phantoms, we found for the particular probe used that the intermediate values (rd=0.45 mm and rc=0.30 mm) can model well all five phantoms. (The values obtained were slightly different for a second probe.) These average parameters also make physical sense, given the MMS probe ball lens that specifies both delivery spot and also a range of reflected angles that can couple into the collection fibers. In contrast, poor agreement was observed between Rp,observed(λ) and Rp(λ) calculated using the simplified closed form solution of Zonios et al.16 Thus, the Farrell model of Eq. 1 rather than the Zonios model should be used to model DRS spectra collected with the MMS probe.
Once the calibrated values of rd and rc were obtained, we were able to prospectively extract accurate values of and μa(λ) from an unknown tissue sample. For this purpose we used a lookup table in which DRS spectra are generated by numerically integrating Eq. 1 over a range of values of and μa(λ) while keeping the calibrated probe-specific parameters rd and rc fixed. In this case, was calculated by a power law, Aλ−B, with two free parameters A and B. In general, the choice of μa(λ) depends on the concentrations of the particular absorbers present in the tissue and their extinction coefficients, but in the present case hemoglobin is the dominant absorber, so μa(λ) is then specified by quantities cHb and α defined earlier.
An unknown tissue spectrum is then compared to the entries in the lookup table to obtain the optimal values of A, B, cHb, and α that in turn specify and μa(λ). To check its accuracy, the lookup table was applied to the phantom spectra, and the extracted spectral quantities and μa(λ) were compared to the known (measured) quantities in the phantom mix. The deviation between the two was on average only a few percent, which demonstrates the accuracy of the predictive fitting. Another way to prospectively extract the absorption and scattering coefficients from an unknown tissue sample is to find the optimal and μa(λ) that minimize the residual [Rp,observed(λ)−Rp(λ)]2 for the fixed calibrated values of rd and rc. However, the optimization method is computationally more intensive, as it involves multiple numerical integrations of Eq. 1, which is why the lookup table is preferred for real-time fitting applications.
IFS modeling
In addition to containing the emission spectrum of fluorophores, raw tissue fluorescence is also distorted by absorption and scattering. Intrinsic fluorescence, on the other hand, is defined as the signal free from this interference and is thus a linear combination of the concentrations of the fluorophores. Zhang et al.43 presented a method based on photon migration for extracting IFS spectra from turbid media, which built on the previously developed model of Wu et al.46 The IFS spectrum fxm(λ) is related to the experimentally measured fluorescence spectrum Fxm(λ) and DRS spectrum Rm(λ) by means of the following expression:43
| (3) |
where Rx is a constant obtained from Rp,observed(λ) at the fluorescence excitation wavelength, R0x is a constant similar to Rx but computed using Eq. 1 by setting μa=0, R0m(λ) is a spectrum evaluated from Rp,observed(λ) but for all needed fluorescence emission wavelengths, and μsx is evaluated at the fluorescence excitation wavelength and is extracted from using the known value of anisotropy g. Finally, εx and εm(λ) are evaluated at the excitation and all emission wavelengths, respectively, using ε=eβ−1, with β=S(1−g), for a fixed parameter S and anisotropy g, and l is related to the optical thickness of the sample. Once the IFS spectrum fxm(λ) is extracted, it can be related to tissue fluorophores that are relevant to disease diagnosis.
Of all these parameters, S and l are probe-specific parameters that need to be calibrated for a particular probe. This was done in a similar fashion as described in the case of DRS to calibrate rd and rc. First, a set of known phantoms using polystyrene beads, hemoglobin, and furan for fluorescence was used to calibrate the values of S and l. In this process, we determined that the optimal values of these parameters for one particular MMS probe were S=0.7 and l=40 μm. These probe-specific parameters were then held fixed for the particular probe and used with Eq. 3 to extract fxm(λ) prospectively from unknown samples. Once the correct IFS spectrum is extracted, it can be fit using least-squares to a specific linear model of constituent spectra.
Raman modeling
The modeling of the Raman spectra is the same for the MMS probe as it is for the previously developed Raman probe, as nothing has been changed in the excitation∕collection geometry for this modality. The modeling remains as described previously.33
SYSTEM PERFORMANCE
We have used the clinical MMS instrument and probe for in vivo and ex vivo studies to detect vulnerable plaques in arteries and also to diagnose breast cancer. MMS spectra were collected in approximately 5 s total: 2.5 s for Raman, ∼1 s for DRS, and ∼1 s for IFS, with a fraction of a second consumed by switching between the three excitation sources. Typical excitation powers utilized were 100 mW at 830 nm continuous-wave for Raman, seven white light pulses of 2 μJ∕pulse for DRS (energy meter PD10, Ophir Optronics, City), and seven pulses of 4 μJ∕pulse at 337 nm for IFS. Representative MMS spectra from artery tissue in vivo during a carotid endarterectomy are shown in Fig. 5. The signal-to-noise ratios of all three spectral modalities collected by the MMS instrument and probe are equivalent to those collected with the previous instruments. Physical parameters accurately extracted from this spectroscopy data contain important information about disease status of tissue, specifically in this case about plaque vulnerability. The detailed clinical results from both of these studies will be published elsewhere.
Figure 5.
Representative spectra of (a) DRS, (b) IFS, and (c) Raman collected ex vivo from artery tissue. The data are in blue, the appropriate fits are in red, and the residual between the data and fit is given in black.
One should note that the light sources and camera detectors used in the MMS system are identical to (or newer versions of) the ones used in the previous FastEEM and clinical Raman systems. The main difference in data collection compared to previous systems is the single MMS probe that is used for all spectral measurements. However, since each probe is calibrated, for each modality, using standards and∕or phantoms, any probe-specific effects on the signal are thus neutralized. Consistent with proper calibration, we were able to model and fit data collected with the MMS system equally well as data collected with the previous systems.
CONCLUSION
We have developed an integrated MMS instrument and unitary probe for simultaneously collecting reflectance, fluorescence, and Raman spectra. The MMS instrument is designed to be compact and portable and thus appropriately suited for clinical applications. All the critical diagnostic functionality of previous stand-alone Raman and FastEEM instruments has been preserved, taking advantage of technological improvements to reduce the size and cost of the system. It is easy to use and backward compatible with previously developed probes for Raman and reflectance∕fluorescence, contains a software platform that automates instrument calibration and data collection, and provides real-time analysis capability.
The MMS probe has a ball lens tip and appropriate filters at the tip to enable efficient collection of Raman signals, as well as fluorescence and reflectance, over the entire 300–1000 nm spectral range. The ability to collect reflectance, fluorescence, and Raman signals at the same time from the same location in tissue eliminates sampling error and enables realistic clinical use. A probe with this excitation∕collection geometry has not been used previously to collect reflectance and fluorescence spectra, and careful calibration is required to enable the accurate extraction of spectral parameters.
This MMS system has been used recently to analyze tissue in vivo, and results from those studies will be published separately. Additional improvements to the system, which include a spectrograph with two gratings that can be used with a single CCD camera as well as a smaller diameter MMS probe, will make the instrument more compact and clinic-friendly.
ACKNOWLEDGMENTS
The authors thank Jonathan Nazemi for his contribution to instrumentation and software development. This research was conducted at the MIT Laser Biomedical Research Center under NIH Grant No. P41-RR-02594.
References
- Hanlon E. B., Manoharan R., Koo T. W., Shafer K. E., Motz J. T., Fitzmaurice M., Kramer J. R., Itzkan I., Dasari R. R., and Feld M. S., Phys. Med. Biol. 45, R1 (2000). 10.1088/0031-9155/45/2/201 [DOI] [PubMed] [Google Scholar]
- Wagnieres G. A., Star W. M., and Wilson B. C., Photochem. Photobiol. 68, 603 (1998). [PubMed] [Google Scholar]
- Motz J. T., Fitzmaurice M., Miller A., Gandhi S. J., Haka A. S., Galindo L. H., Dasari R. R., Kramer J. R., and Feld M. S., J. Biomed. Opt. 11, 021003 (2006). 10.1117/1.2190967 [DOI] [PubMed] [Google Scholar]
- Enejder A. M. K., Koo T. W., Oh J., Hunter M., Sasic S., Feld M. S., and Horowitz G. L., Opt. Lett. 27, 2004 (2002). 10.1364/OL.27.002004 [DOI] [PubMed] [Google Scholar]
- Alfano R. R., Liu C. H., Sha W. L., Zhu H. R., Akins D. L., Cleary J., Prudente R., and Cellmer E., Lasers Life Sci. 4, 23 (1991). [Google Scholar]
- Frank C. J., McCreery R. L., and Redd D. C. B., Anal. Chem. 67, 777 (1995). 10.1021/ac00101a001 [DOI] [PubMed] [Google Scholar]
- Haka A. S., Shafer-Peltier K. E., Fitzmaurice M., Crowe J., Dasari R. R., and Feld M. S., Proc. Natl. Acad. Sci. U.S.A. 102, 12371 (2005). 10.1073/pnas.0501390102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan-Jansen A. and Richards-Kortum R., J. Biomed. Opt. 1, 31 (1996). 10.1117/12.227815 [DOI] [PubMed] [Google Scholar]
- Utzinger U., Heintzelman D. L., Mahadevan-Jansen A., Malpica A., Follen M., and Richards-Kortum R., Appl. Spectrosc. 55, 955 (2001). 10.1366/0003702011953018 [DOI] [Google Scholar]
- Jess P. R. T., Smith D. D. W., Mazilu M., Dholakia K., Riches A. C., and Herrington C. S., Int. J. Cancer 121, 2723 (2007). 10.1002/ijc.23046 [DOI] [PubMed] [Google Scholar]
- Crow P., Stone N., Kendall C. A., Uff J. S., Farmer J. A. M., Barr H., and Wright M. P. J., Br. J. Cancer 89, 106 (2003). 10.1038/sj.bjc.6601059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow P., Barrass B., Kendall C., Hart-Prieto M., Wright M., Persad R., and Stone N., Br. J. Cancer 92, 2166 (2005). 10.1038/sj.bjc.6602638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers P. J., Lucassen G. W., and Puppels G. J., Biophys. J. 85, 572 (2003). 10.1016/S0006-3495(03)74501-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers P. J., Lucassen G. W., Bruining H. A., and Puppels G. J., J. Raman Spectrosc. 31, 813 (2000). [DOI] [Google Scholar]
- Hata T. R., Scholz T. A., Ermakov I. V., McClane R. W., Khachik F., Gellermann W., and Pershing L. K., J. Invest. Dermatol. 115, 441 (2000). 10.1046/j.1523-1747.2000.00060.x [DOI] [PubMed] [Google Scholar]
- Zonios G., Perelman L. T., Backman V. M., Manoharan R., Fitzmaurice M., Van Dam J., and Feld M. S., Appl. Opt. 38, 6628 (1999). 10.1364/AO.38.006628 [DOI] [PubMed] [Google Scholar]
- Hidovic-Rowe D. and Claridge E., Phys. Med. Biol. 50, 1071 (2005). 10.1088/0031-9155/50/6/003 [DOI] [PubMed] [Google Scholar]
- Georgakoudi I., Jacobson B. C., Van Dam J., Backman V., Wallace M. B., Muller M. G., Zhang Q., Badizadegan K., Sun D., Thomas G. A., Perelman L. T., and Feld M. S., Gastroenterology 120, 1620 (2001). 10.1053/gast.2001.24842 [DOI] [PubMed] [Google Scholar]
- Georgakoudi I., Sheets E. E., Muller M. G., Backman V., Crum C. P., Badizadegan K., Dasari R. R., and Feld M. S., Am. J. Obstet. Gynecol. 186, 374 (2002). 10.1067/mob.2002.121075 [DOI] [PubMed] [Google Scholar]
- Mourant J. R., Bigio I. J., Boyer J., Conn R. L., Johnson T., and Shimada T., Lasers Surg. Med. 17, 350 (1995). 10.1002/lsm.1900170403 [DOI] [PubMed] [Google Scholar]
- Koenig F., Larne R., Enquist H., McGovern F. J., Schomacker K. T., Kollias N., and Deutsch T. F., Urology 51, 342 (1998). 10.1016/S0090-4295(97)00612-2 [DOI] [PubMed] [Google Scholar]
- Mitchell M. F., Cantor S. B., Ramanujam N., Tortolero-Luna G., and Richards-Kortum R., Obstet. Gynecol. (N.Y., NY, U. S.) 93, 462 (1999). 10.1016/S0029-7844(98)00385-8 [DOI] [PubMed] [Google Scholar]
- Wu Y. C., Xi P., Qu J. N. Y., Cheung T. H., and Yu M. Y., Opt. Express 13, 382 (2005). 10.1364/OPEX.13.000382 [DOI] [PubMed] [Google Scholar]
- Fiarman G. S., Nathanson M. H., West A. B., Deckelbaum L. I., Kelly L., and Kapadia C. R., Dig. Dis. Sci. 40, 1261 (1995). 10.1007/BF02065535 [DOI] [PubMed] [Google Scholar]
- DaCosta R. S., Andersson H., Cirocco M., Marcon N. E., and Wilson B. C., J. Clin. Pathol. 58, 766 (2005). 10.1136/jcp.2004.023804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo-Dinh T., Panjehpour M., and Overholt B. F., Ann. N.Y. Acad. Sci. 838, 116 (1998). 10.1111/j.1749-6632.1998.tb08192.x [DOI] [PubMed] [Google Scholar]
- Kara M. A., DaCosta R. S., Streutker C. J., Marcon N. E., Bergman J. J. G. H. M., and Wilson B. C., Dis. Esophagus 20, 141 (2007). 10.1111/j.1442-2050.2007.00660.x [DOI] [PubMed] [Google Scholar]
- Muller M. G., Valdez T. A., Georgakoudi I., Backman V., Fuentes C., Kabani S., Laver N., Wang Z., Boone C. W., Dasari R. R., Shapshay S. M., and Feld M. S., Cancer 97, 1681 (2003). 10.1002/cncr.11255 [DOI] [PubMed] [Google Scholar]
- Schantz S. P., Kolli V., Savage H. E., Yu G. P., Shah J. P., Harris D. E., Katz A., Alfano R. R., and Huvos A. G., Clin. Cancer Res. 4, 1177 (1998). [PubMed] [Google Scholar]
- Lane P. M., Gilhuly T., Whitehead P., Zeng H. S., Poh C. F., Ng S., Williams P. M., Zhang L. W., Rosin M. P., and MacAulay C. E., J. Biomed. Opt. 11, 024006 (2006). 10.1117/1.2193157 [DOI] [PubMed] [Google Scholar]
- Lieber C. A. and Mahadevan-Jansen A., Opt. Express 15, 11874 (2007). 10.1364/OE.15.011874 [DOI] [PubMed] [Google Scholar]
- Motz J. T., Gandhi S. J., Scepanovic O. R., Haka A. S., Kramer J. R., Dasari R. R., and Feld M. S., J. Biomed. Opt. 10, 031113 (2005). 10.1117/1.1920247 [DOI] [PubMed] [Google Scholar]
- Motz J. T., Hunter M., Galindo L. H., Gardecki J. A., Kramer J. R., Dasari R. R., and Feld M. S., Appl. Opt. 43, 542 (2004). 10.1364/AO.43.000542 [DOI] [PubMed] [Google Scholar]
- Tunnell J. W., Desjardins A. E., Galindo L., Georgakoudi I., McGee S. A., Mirkovic J., Mueller M. G., Nazemi J., Nguyen F. T., Wax A., Zhang Q. G., Dasari R. R., and Feld M. S., Technol. Cancer Res. Treat. 2, 505 (2003). [DOI] [PubMed] [Google Scholar]
- Muller M. G., Wax A., Georgakoudi I., Dasari R. R., and Feld M. S., Rev. Sci. Instrum. 73, 3933 (2002). 10.1063/1.1511795 [DOI] [Google Scholar]
- Zangaro R. A., Silveira L., Manoharan R., Zonios G., Itzkan I., Dasari R. R., VanDam J., and Feld M. S., Appl. Opt. 35, 5211 (1996). 10.1364/AO.35.005211 [DOI] [PubMed] [Google Scholar]
- Cothren R., Hayes G., Kramer J. R., Sacks B., Kittrell C., and Feld M. S., Lasers Life Sci. 1, 1 (1986). [Google Scholar]
- Virmani R., Burke A. P., Farb A., and Kolodgie F. D., Prog. Cardiovasc. Dis. 44, 349 (2002). 10.1053/pcad.2002.122475 [DOI] [PubMed] [Google Scholar]
- Scepanovic O. R., Fitzmaurice M., Gardecki J. A., Angheloiu G. O., Awasthi S., Motz J. T., Kramer J. R., Dasari R. R., and Feld M. S., J. Biomed. Opt. 11, 021007 (2006). 10.1117/1.2187943 [DOI] [PubMed] [Google Scholar]
- Buschman H. P., Deinum G., Motz J. T., Fitzmaurice M., Kramer J. R., van der Laarse A., Bruschke A. V., and Feld M. S., Cardiovasc. Pathol. 10, 69 (2001). 10.1016/S1054-8807(01)00064-3 [DOI] [PubMed] [Google Scholar]
- Shafer-Peltier K. E., Haka A. S., Fitzmaurice M., Crowe J., Myles J., Dasari R. R., and Feld M. S., J. Raman Spectrosc. 33, 552 (2002). 10.1002/jrs.877 [DOI] [Google Scholar]
- Farrell T. J., Patterson M. S., and Wilson B., Med. Phys. 19, 879 (1992). 10.1118/1.596777 [DOI] [PubMed] [Google Scholar]
- Zhang Q. G., Muller M. G., Wu J., and Feld M. S., Opt. Lett. 25, 1451 (2000). 10.1364/OL.25.001451 [DOI] [PubMed] [Google Scholar]
- Welch A. J. and van Gemert M. J. C., in Lasers, Photonics, and Electro-optics, edited by Kogelnik H. (Plenum, New York, 1995). [Google Scholar]
- Keijzer M., Richards-Kortum R. R., Jacques S. L., and Feld M. S., Appl. Opt. 28, 4286 (1989). 10.1364/AO.28.004286 [DOI] [PubMed] [Google Scholar]
- Wu J., Feld M. S., and Rava R. P., Appl. Opt. 32, 3585 (1993). 10.1364/AO.32.003585 [DOI] [PubMed] [Google Scholar]