Abstract
Accurate pre-mRNA splicing is critical for gene expression. The 5' splice site (5' ss) — the highly diverse element at the 5' end of introns — is initially recognized via base-pairing to the 5' end of U1 small nuclear RNA (snRNA). However, many natural 5' ss have a very poor match to the consensus sequence, and are predicted to be very weak. Using genetic suppression experiments in human cells, we demonstrate that some atypical 5' ss are actually efficiently recognized by U1, in an alternative base-pairing register that is shifted by one nucleotide. These atypical 5' ss are phylogenetically widespread, and many of them are conserved. Moreover, shifted base-pairing provides an explanation for the effect of a 5' ss mutation associated with pontocerebellar hypoplasia. The unexpected flexibility in 5' ss/U1 base-pairing challenges an established paradigm, and has broad implications for splice-site prediction algorithms and gene-annotation efforts in genome projects.
Accurate pre-mRNA splicing is critical for the correct transmission of information from gene to protein1. Splicing is catalyzed by the spliceosome, a large and dynamic complex composed of five small nuclear ribonucleoprotein particles (snRNPs) made up of snRNAs and associated polypeptides, as well as many other protein factors2. Conserved sequences that match degenerate consensus motifs at both ends of introns are essential for splicing1. As first proposed in 19803,4, and definitively demonstrated in 19865, the 5' ss is initially recognized via base-pairing to the 5' end of the U1 snRNA. The 5' ss consensus sequence for the major, or U2-type GT-AG introns in mammals, which comprise >98% of all introns6, has perfect complementarity to the 5' end of the U1 snRNA3–5,7,8, establishing up to eleven base pairs in a defined register, here referred to as the 'canonical' register (Fig. 1a; see Methods). However, the major spliceosome can accurately recognize a highly diverse set of 5' ss sequences: using SpliceRack6, a comprehensive database of splice sites, we find 2,503 unique human 5' ss sequences – considering only the classical 9-nt motif (see Methods) – that are used at least three times in the transcribed genome, in 186,630 introns.
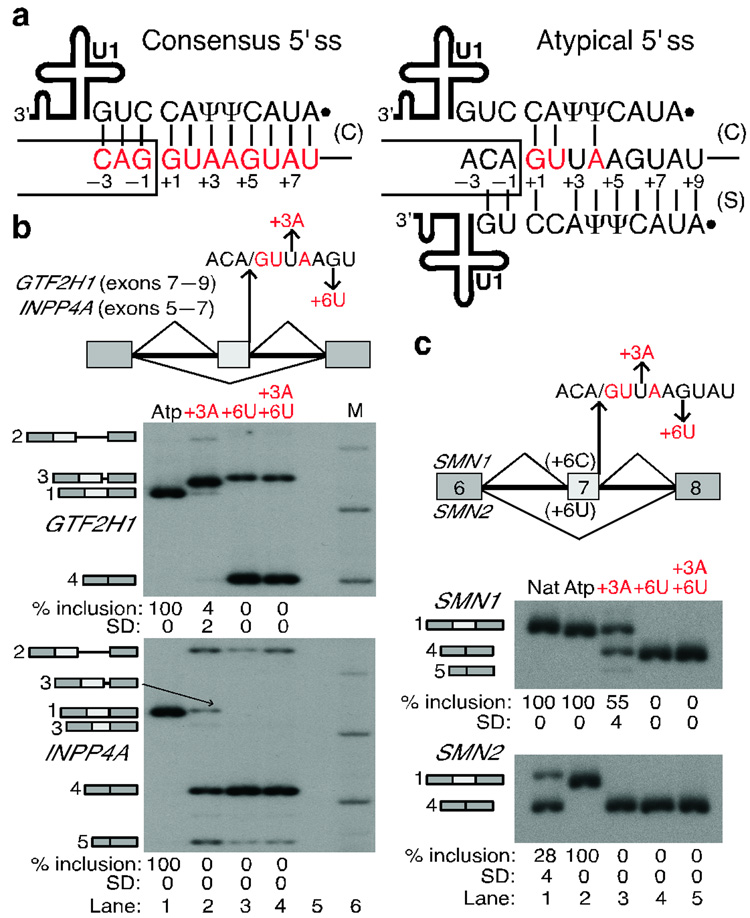
Figure 1. Shifted base-pairing between atypical 5' ss and the 5' end of U1 snRNA.
a, Diagram of the two base-pairing registers between the 5' ss (positions are numbered) and U1. Consensus nucleotides are shown in red in all figures (see Methods). Ψ, pseudouridine. Solid dot, 2,2,7-trimethylguanosine cap at the 5' end of U1. Box, upstream exon; line, intron. Base pairs in the canonical (C) or shifted (S) register are indicated by vertical lines. Note that the atypical 5' ss can form seven more base pairs to U1 in the shifted arrangement. b, Mutations at atypical (Atp) 5' ss that disrupt shifted but enhance canonical base-pairing abolish correct splicing. The human GTF2H1 and INPP4A minigenes are schematically represented at the top, indicating the mutations introduced at the atypical 5' ss. M, Molecular weight markers. The identity of the various spliced mRNAs, detected by radioactive RT-PCR, is schematically shown on the left of the gels and corresponds to: #1, correctly spliced mRNA; #2, retention of the downstream intron; #3, use of cryptic 5' ss in the middle exon; #4, skipping of the middle exon; #5, activation of a cryptic 5' ss in the first exon. The percentage of correct splicing is shown at the bottom. See Supplementary Fig. 1 online for details about the aberrantly spliced mRNAs. c, RT-PCR analysis of the atypical 5' ss in the SMN1/2 context (schematic at the top). Nat, natural SMN1/2 exon 7 5' ss. Numbers below the panels show the percentage and Standard Deviation (SD) of exon 7 inclusion.
Many of these bona-fide 5' ss have very few predicted base pairs to U16,9,10, and selection of these atypical 5' ss cannot be explained by other known mechanisms, such as splicing via the minor, U12-type spliceosome6,11. We noticed that a subset of atypical 5' ss have a sequence (ACA/GUUAAGUAU, where / marks the exon-intron boundary) that is reminiscent of the consensus motif (Fig. 1a). This sequence can form only three potential base pairs with the 5' end of U1 in the canonical scheme (+1G of the 5' ss base-pairing with C8 of U1); however, this can be increased to 10 base pairs by shifting the 5' end of U1 snRNA one position downstream of the 5' ss (+1G of the 5' ss base-pairing with C9 of U1). We refer to this alternative base-pairing arrangement as the 'shifted' register. Thus, we hypothesized that these 5' ss are recognized via shifted base-pairing to the 5' end of U1, and we present here experimental evidence to support this model.
RESULTS
Some 5' ss do not base-pair to U1 by the canonical register
To test the shifted base-pairing hypothesis experimentally, we first analyzed the atypical 5' ss associated with exons 6 and 8 of the human INPP4A and GTF2H1 genes, respectively. We transiently transfected three-exon, two-intron minigene constructs into HeLa cells, and analyzed the inclusion or skipping of the middle exon carrying the atypical 5' ss by RT-PCR. We found in both cases that the atypical 5' ss was efficiently used for splicing of the minigene transcripts (Fig. 1b, lane 1) as well as of the endogenous transcripts in HeLa cells (Supplementary Fig. 1 online), with slight retention of the second intron in the case of GTF2H1.
We also mutated the atypical 5' ss in both minigenes, so as to restore the consensus nucleotides at positions +3 and/or +6 of the 5' ss. Paradoxically, these mutant minigenes with improved base-pairing potential to U1 in the canonical register (four or five base pairs) and decreased base-pairing in the shifted register (8 or 9 base pairs) expressed many aberrantly spliced mRNAs, generated by skipping of the internal exon, retention of the second intron, or cryptic 5' ss activation (Fig. 1b, lanes 2–4). This observation indicates that these 5' ss are not recognized via the classical base-pairing register with U1 snRNA.
Next, we used SMN1/2 minigenes12 to test the atypical 5' ss in a heterologous context. The SMN1 and 2 paralog pre-mRNAs give different extents of exon 7 inclusion, providing two distinct contexts to analyze the efficiency of the test 5' ss. This difference is mainly attributable to a single-nucleotide divergence at the sixth position of this exon. Whereas a C in SMN1 results in virtually complete exon 7 inclusion, a U in SMN2 results predominantly in exon skipping13, because of the sequence change in a cis-acting element(s) recognized by a splicing activator in SMN112 and/or a repressor in SMN214.
We substituted the natural 5' ss of SMN1/2 exon 7 (GGA/GUAAGUCU; eight base pairs with U1) with different versions of the atypical 5' ss. In SMN2, the atypical 5' ss was three-fold more efficient than the natural one (Fig. 1c, lanes 1 and 2). This finding is remarkable, because all available computational methods15–22 predict the natural SMN1/2 exon 7 5' ss to be much stronger than the atypical 5' ss (Table 1). The splicing compatibility of the atypical 5' ss with the canonical 3' splice site of SMN1/2 exon 8 also indicates that splicing via this 5' ss is catalyzed by the major spliceosome1.
Table 1.
Scores of the SMN1/2 exon 7 5' ss (upper sequence) and of the atypical 5' ss (lower sequence).
| S&Si | ΔGii | H-Bondiii | NNiv | MAXENTv | MDDvi | MMvii | |
|---|---|---|---|---|---|---|---|
| 77.48 | −8.70 | 14.50 | 0.99 | 8.57 | 12.28 | 6.36 | |
| 51.65 | −2.20 | 1.90 | 0.00 | −12.18 | −2.72 | −4.30 |
SMN1/2 minigenes with mutations at the atypical 5' ss positions +3 and +6 that restore the consensus nucleotide (but disrupt shifted base-pairing) showed increased exon 7 skipping (Fig. 1c, lanes 3–6), consistent with the above results with the GTF2H1 and INPP4A substrates, and suggesting that the shifted base-pairing register is being used. The simpler mRNA patterns obtained with the SMN1/2 minigenes made them more amenable to further mutational analyses.
Suppressor U1 analysis demonstrates shifted base-pairing
We next sought to determine whether the atypical 5' ss is indeed recognized by shifted base-pairing to U1 (Fig. 1a). To this end, we transfected a series of SMN1/2 minigenes carrying mutations at the atypical 5' ss, along with U1 snRNA expression plasmids with compensatory mutations that restore base-pairing. This type of informational suppression analysis is known as suppressor- or shift-U1 experiments5,7–9,23–25.
First, we tested a series of mutations that introduced a consensus nucleotide at different positions of the atypical 5' ss (Fig. 2a,b; Supplementary Fig. 2a online). All mutations, with the exception of −2A and −1G in the SMN1 context, resulted in partial or complete loss of exon 7 inclusion, further indicating that canonical base-pairing with U1 does not occur at the atypical 5' ss (Fig. 2c). The corresponding suppressor U1 snRNAs in the shifted base-pairing register partially restored exon 7 inclusion for some of these mutants: +5G and +7A in SMN1 (Fig. 2c, upper panel, lanes 8–9 and 12–13), and −1G and +5G in SMN2 (Fig. 2c, lower panel, lanes 4–5 and 8–9). For one mutant 5' ss, +3A in the SMN1 context, the suppressor U1 snRNA decreased exon 7 inclusion (Fig. 2c, upper panel, lanes 6–7), perhaps reflecting a block in a subsequent step in the splicing reaction. The −2A and +6U mutations could not be rescued by suppressor U1s in either of the two contexts. The −2A mutation resulted in very slight exon 7 skipping. The +6U mutation (as well as +6C in Fig. 3, see below) was not rescued by suppressor U1, perhaps because this mutation eliminates a strong G-C base pair essential for efficient binding of U1. Nevertheless, the rescue of exon 7 inclusion by suppressor U1s for mutants −1G, +5G and +7A is consistent with the hypothesis that atypical 5' ss are recognized via shifted base-pairing to U1.
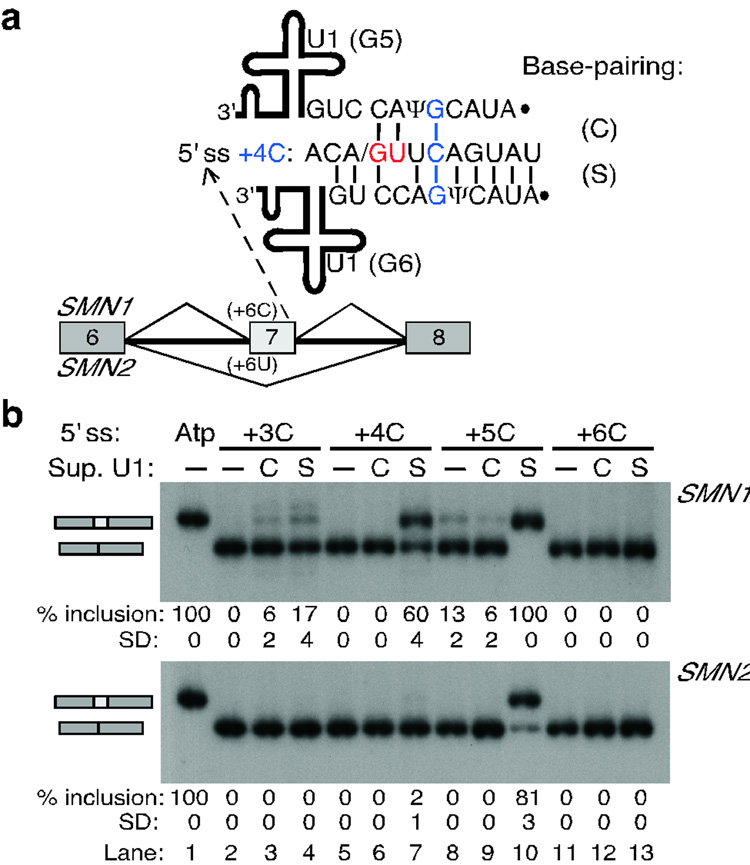
Figure 2. Suppressor U1 snRNAs in the shifted register can rescue splicing.
a, Schematic of the single mutations introduced at the atypical 5' ss in the SMN1/2 context. These mutations substitute a non-consensus nucleotide by a consensus nucleotide. b, Base-pairing of the mutant 5' ss with the corresponding suppressor U1 snRNA. As an example, we show the base-pairing of the +5G mutant 5' ss with the suppressor U1 snRNA carrying the corresponding compensatory mutation (C5) in the shifted register. The mutant nucleotide at the 5' end of U1 in each case is shown in red. See Supplementary Fig. 2 online for the base-pairing of all mutant 5' ss with their respective suppressor U1s. c, RT-PCR analysis of the SMN1/2 minigenes carrying the wild-type (lane 1) or mutant atypical 5' ss (lanes 2–13). The 5' ss mutation is indicated at the top, without (−) or with (+) the corresponding suppressor U1 snRNA. The mRNA products are schematically indicated on the left of each panel. The fastest migrating band in SMN1 corresponds to an mRNA that skipped exon 7 and used a cryptic 5' ss 50 nucleotides upstream of the exon 6 5' ss. The percentage and Standard Deviation (SD) of exon 7 inclusion is indicated below each autoradiogram.
Figure 3. Compensatory U1 mutations that restore shifted but not canonical base-pairing rescue splicing at atypical 5' ss.
a, Scheme of the experimental design. SMN1/2 minigenes carrying point mutations at a heterologous atypical 5' ss in exon 7 were co-transfected with suppressor U1 snRNAs. The 5' ss nucleotides at positions +3 to +6 were individually mutated to C (+3C to +6C). The 5' end of U1 was mutated so as to rescue base-pairing in the canonical or the shifted arrangement (suppressor U1 mutations G3 to G6). Mutant +4C is shown as a representative example, for which U1 mutations G5 or G6 restore base-pairing in the canonical (C) or shifted (S) register, respectively. For the other three mutations, see Supplementary Fig. 2 online. b, RT-PCR analysis of the +3C to +6C 5' ss mutations in SMN1/2 with suppressor U1. Top labels indicate the 5' ss mutant and the suppressor U1 in either register. Atp, wild-type atypical 5' ss. The percentage and Standard Deviation of exon 7 inclusion is shown below each autoradiogram.
Second, we analyzed a series of mutant 5' ss with a C at one intronic position (+3 to +6), with or without co-transfected suppressor U1 snRNAs carrying the compensatory mutation in either the classical or the shifted register (Fig. 3a, Supplementary Fig. 2b–d online). We chose the nucleotide C because it cannot form a base pair with wild-type U1 in either of the two arrangements. In all cases, each mutation resulted in predominant SMN1/2 exon 7 skipping (Fig. 3b). Splicing via the mutant 5' ss with C at positions +4 or +5 was rescued by suppressor U1s in the shifted, but not in the canonical base-pairing register (Fig. 3b, lanes 5–10). Splicing via the +3C mutant 5' ss in SMN1 was rescued by both suppressor U1s (Fig. 3b, lanes 2–4), but the suppressor in the shifted register showed substantially higher activity. We also tested for suppression of the +4C mutation in the INPP4A and GTF2H1 minigenes, and found that the suppressor U1 in the shifted but not in the canonical register restored recognition of the mutant 5' ss (Supplementary Fig. 3 online). Furthermore, analysis of 5' ss with two mutations in the context of SMN1/2 minigenes gave consistent results (Supplementary Fig. 4 online). Although not all suppressor U1s are effective in this type of experiment5,7,8, our data show that many of the suppressor U1 snRNAs in the shifted register can rescue mutations at atypical 5' ss. Together, our U1-suppressor experiments formally demonstrate that recognition of these 5"ss is mediated by base-pairing to U1 that is shifted by one nucleotide, relative to the canonical scheme.
Atypical 5' ss are recognized by U1 and not U1A7 snRNA
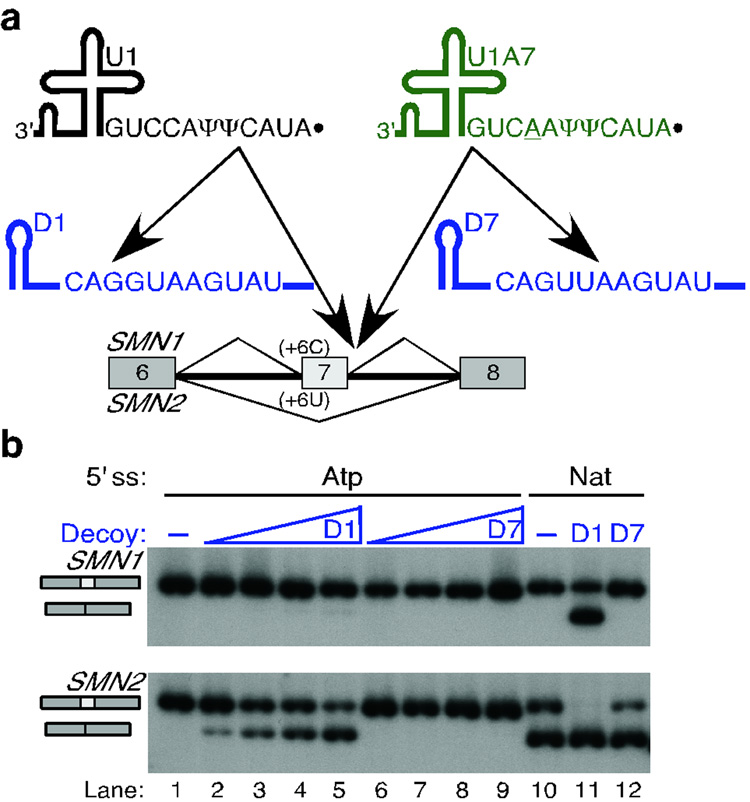
Recently, Kyriakopolou et al.26 reported the expression of three human U1 snRNA variants with 5' ends different from that of U1, and several nucleotide changes at other positions. Interestingly, the U1A7 snRNA 5' end has perfect complementarity to the atypical 5' ss, also in the shifted register. To test the role of the U1A7 snRNA in the recognition of atypical 5' ss, we performed a series of experiments with suppressor U1/U1A7 snRNAs or RNA decoys.
We used suppressor U1 and U1A7 snRNAs both in the canonical or shifted register to try to rescue the +4C mutation in the SMN1/2 contexts (Supplementary Fig. 5 online). In addition, the 5' ends and the snRNA bodies of U1 and U1A7 were swapped to make chimeric snRNAs. None of the suppressors with the U1A7 snRNA body rescued exon 7 inclusion. With the U1 body, both the U1 and U1A7 5' ends carrying the compensatory mutation in the shifted but not in the canonical register rescued splicing. As expected, the 5' end of U1A7 was more effective than that of U1, because it can form an extra base pair to the +4C 5' ss. However, due to the lack of activity of suppressors with the U1A7 body, the much greater abundance of U1, and the fact that an snRNA with the U1 body and the U1A7 5' end does not exist in human cells, we infer that U1A7 is not involved in the recognition of atypical 5' ss.
In addition, we used U1- and U1A7-specific RNA decoys to further test which of these two transacting factors is involved in the recognition of atypical 5' ss (Fig. 4). The D1 and D7 decoys are short RNAs that carry a sequence with perfect complementarity to the 5' end of U1 or U1A7 snRNAs, respectively (Fig. 4a). The D1 decoy has the consensus 5' ss sequence, and the D7 decoy has the atypical 5' ss sequence. We determined that RNA decoys bind to their cognate snRNAs only when they have perfect complementarity to them (Supplementary Fig. 6 online), thereby reducing the free levels of these snRNPs in the cell and affecting the splicing of certain introns (data not shown). The decoy RNAs were cotransfected with SMN1/2 minigenes with the natural exon 7 5' ss or the atypical 5' ss. The D1 decoy reduced recognition of both the natural (Fig. 4b, lane 11 vs. 10) and the atypical 5' ss in exon 7 (Fig. 4b, lanes 2–5 vs. 1) in a dose-dependent manner. The D7 decoy did not substantially affect recognition of the atypical 5' ss (Fig. 4b, lanes 6–9 vs. 1), and had only a subtle effect on the natural exon 7 5' ss (Fig. 4b, lane 12 vs. 10). The results obtained with the U1/U1A7 suppressors and the decoys demonstrate that both the atypical and the natural SMN1/2 exon 7 5' ss are recognized by the same transacting factor, U1 snRNA, and not by U1A7.
Figure 4. U1 but not U1A7 snRNA decoys reduce splicing via the atypical 5' ss.
a, Schematic of the U1 (black) and U1A7 (green) snRNA decoys. The D1 and D7 decoys are short RNAs expressed from the potent U6 promoter, and comprise the first 27 nucleotides of the U6 snRNA for stability, and a sequence with perfect complementarity to the 5' end of U1 (black) or U1A7 (green) snRNAs, respectively. These decoys reduce the free levels of their cognate snRNAs in the cell, affecting the splicing of certain introns. b, The D1 but not the D7 decoy reduced SMN1/2 exon 7 inclusion in minigenes carrying the natural or an atypical 5' ss. The top labels indicate the identity of the 5' ss in exon 7 and the decoy used. The triangle depicts an increasing amount of decoy plasmid transfected with the minigene.
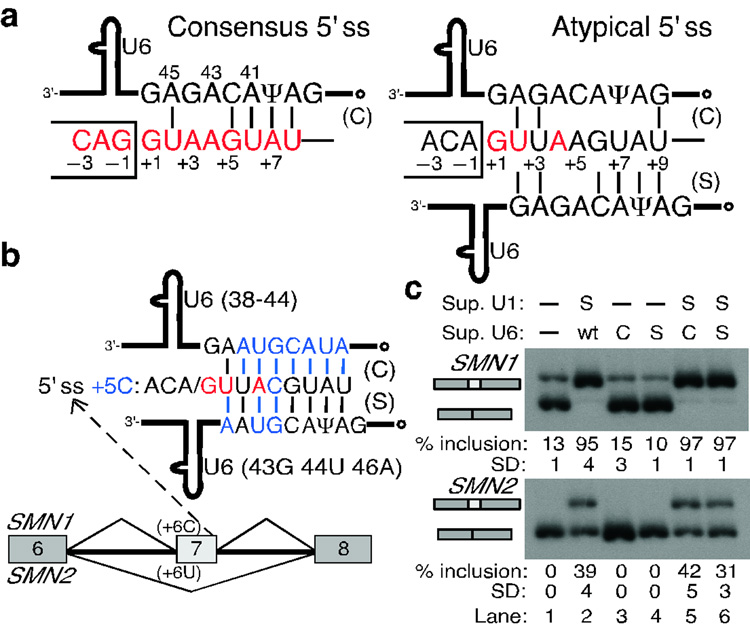
Atypical 5' ss do not base-pair to U6 in a shifted register
During spliceosome assembly, U1 is displaced from the 5' ss to allow base-pairing of U5 and U6 snRNAs to the exonic and intronic portions of the 5' ss, respectively27–31. This replacement is critical for spliceosome assembly and catalysis. The atypical 5' ss has an extended potential base-pairing to the phylogenetically invariant U6 ACAGAG box, when its position is shifted by one nucleotide (Fig. 5a; six vs. three base pairs). To test whether this shifted base-pairing to U6 can occur, we used suppressor U6 snRNAs30,32–34 in combination with suppressor U1 snRNAs to try to rescue atypical 5' ss mutations in the SMN1/2 context (Fig. 5b,c, and Supplementary Fig. 7 online). Suppressor U6s with only one compensatory mutation had no effect on exon 7 inclusion (Supplementary Fig. 7 online), but suppressor U6s with several mutations did (Fig. 5b,c). Suppressor U6 in the canonical register resulted in higher levels of exon 7 inclusion than suppressor U6 in the shifted register (Fig. 5c, lanes 5 and 6 in SMN2). These data suggest that shifted base-pairing between the atypical 5' ss and U6 does not occur. In other words, the same positions of the 5' ss base-pair to the same positions in U6 in both conventional and atypical 5' ss, such as U at 5' ss position +2 base-pairing to 45A in U6. This observation is consistent with the proposed prominent role of the 5' ss/U6 RNA helix in catalysis1,31 (see Discussion).
Figure 5. U6 snRNA does not base-pair to the atypical 5' ss in a shifted register.
a, Schematic of the base-pairing between consensus (left) or atypical (right) 5' ss and the conserved U6 ACAGAG box (positions are numbered). The open dot indicates the γ-monomethyl cap. The atypical 5' ss has an extended base-pairing potential to U6 in the shifted register. b, Schematic of the suppressor U6 snRNAs carrying compensatory mutations in either the canonical (C) or the shifted (S) register. These mutations (blue font) restore base-pairing for the +5C mutation at atypical 5' ss in the SMN1/2 context. c, RT-PCR analysis of the SMN1/2 minigenes cotransfected with suppressor U1 and U6 snRNAs. Top labels indicate the suppressor U1/U6 used. wt, wild-type U6 snRNA. Suppressor U6s alone had no effect (lanes 3, 4 vs. lane 1). In combination with suppressor U1, suppressor U6 in the canonical register resulted in more exon 7 inclusion than suppressor U6 in the shifted register (lanes 5 and 6 in SMN2). These results suggest that atypical 5' ss establish canonical base-pairing to U6 snRNA.
Estimated counts and conservation of atypical 5' ss
Atypical 5' ss that can be recognized by shifted base-pairing to the U1 snRNA 5' end are present in the five genomes in the SpliceRack database6: H. sapiens, M. musculus, D. melanogaster, C. elegans and A. thaliana (Table 2 and Supplementary Tables 1 and 2 online). Conservative estimates of the number of 5' ss recognized by this new mechanism, based on our current understanding of the shifted base-pairing requirements, range from 20 in D. melanogaster to 115 in A. thaliana. Remarkably, the C. elegans genome, which has lost all minor, U12-type introns6, has 63 5' ss predicted to be recognized by shifted base-pairing. Furthermore, a comparison of orthologous 5' ss pairs between human and mouse showed that the shifted base-pairing arrangement is partially conserved (~50%): we found 27 atypical 5' ss that have either no nucleotide change between the two species, or have changes that maintain shifted base-pairing to U1; in contrast, we found 21 orthologous 5' ss pairs recognized by shifted base-pairing in only one of the two species (Supplementary Table 2 online). These predictions strongly suggest that shifted base-pairing between 5' ss and U1 is a minor but phylogenetically widespread phenomenon, and that many of these atypical 5' ss are conserved.
Table 2.
Counts for the atypical 5' ss in five species, and for the conserved 5' ss between human and mouse
| Homo sapiens |
Mus musculus |
Drosophila melanogaster |
Caenorhabditis elegans |
Arabidopsis thaliana |
|
|---|---|---|---|---|---|
| Total | 59 | 59 | 20 | 63 | 115 |
| Conserved | 27 | 27 |
DISCUSSION
Here we have shown a phylogenetically conserved mechanism of 5' ss selection by shifted base-pairing to U1 snRNA, with important implications for genomics, evolution, and human disease. Shifted base-pairing provides a basis for the efficient recognition of a subset of 5' ss that are predicted to be very weak (Table 1). This unprecedented mechanism also reveals that the interaction between the 5' ss and U1 is not as rigid as previously believed, allowing for alternative base-pairing arrangements that result in efficient splicing. The plasticity of the interaction between the 5' ss and U1 is probably tolerated because the U1 snRNP defines the 5' ss early on, and is displaced from the spliceosome prior to catalysis1,31. Furthermore, the 5' ss and U6 snRNA do not appear to show such base-pairing flexibility. Shifted base-pairing between atypical 5' ss and U6 would imply that an extra nucleotide has to be inserted between the 5' ss/U6 helix and the scissile bond. Since the 5' ss/U6 helix is at the spliceosomal catalytic core35, subtle perturbations of the positioning of this helix could impair catalysis. Thus, whereas U1 has enough flexibility to recognize the atypical 5' ss in a shifted register, U6 probably needs to base-pair in the conventional register to allow the first trans-esterification step to occur at the correct position.
Early in splicing, 5' ss and neighboring sequences are also bound by proteins that influence base-pairing to U1 and hence 5' ss selection22. For instance, the U1-snRNP-specific polypeptide U1C binds to the 5' ss prior to base-pairing with U136,37. Shifted base-pairing between the 5' ss and U1 could also rely on proteins by mechanisms that might differ from those for canonical base-pairing. In addition, proteins involved in 5' ss selection perhaps account for the differences in splicing patterns seen for different mutations at atypical 5' ss, as well as for the differences in rescue by suppressor U1s (Fig. 2 and 3, Supplementary Fig. 3 and 4 online).
We ruled out the possibility that atypical 5' ss are recognized by the U1 snRNA variant U1A726 instead of U1. We have shown that suppressor U1A7 snRNAs did not rescue mutations at atypical 5' ss (Supplementary Fig. 5 online), and that the U1A7-specific decoy D7 did not compromise recognition of any 5' ss (Fig. 4). Since these atypical 5' ss were the most likely 5' ss to be recognized by U1A7, considering their perfect complementarity (11 bp), our data also suggest that U1A7 is unlikely to function in splicing. Nevertheless, it remains possible that U1A7 is involved in processes other than splicing, as is U118,38,39, or that other U1 variants26 play a role in 5' ss selection.
A mechanism distinct from shifted base-pairing was proposed for one unusual intron in the HOP2 gene in S. cerevisiae40. Mutational analysis of this non-canonical 5' ss suggested that it is recognized via an alternative base-pairing arrangement with U1, involving a bulged nucleotide at position +2 or +3 of the 5' ss. In the case of the human atypical 5' ss we analyzed here, our mutational analyses and suppressor U1 data for position −1 (Fig. 2c, lanes 4,5) rule out the possibility of a bulged nucleotide in the interaction between these atypical 5' ss and U1: the rescue of the −1G mutation in SMN2 by the U1 suppressor C10 indicates that the exonic positions of the atypical 5' ss base-pair to U1 in the shifted register. This observation rules out a base-pairing register between the atypical 5' ss and U1 that involves a bulged nucleotide at the 5' ss, as this arrangement implies that position −1 would not base-pair to position 10 of U1.
Our study leaves open the possibility that other subclasses of atypical 5' ss base-pair to U1 in other 'shifted' registers. We searched SpliceRack6 for other base-pairing arrangements between 5' ss and U1, by shifting the 5' end of U1 by two or three positions downstream, as well as by shifting it by one-three positions upstream (data not shown). We found very few (15 or less) 5' ss for each of these categories. Furthermore, most of these 5' ss can establish a similar number of base pairs to U1 in the canonical register, as opposed to the atypical 5' ss analyzed in this study (Supplementary Table 2 online). We conclude that if other shifted base-pairing arrangements between naturally-occurring 5' ss and U1 actually occur, the number of 5' ss recognized by these putative mechanisms should be far lower than the counts for atypical 5' ss presented here (Table 2). Finally, we did not find any obvious candidate 5' ss that could be recognized by shifted base-pairing to U11 snRNA or to the other two U1 variants26 (data not shown).
Interestingly, a +5 A to G mutation at the atypical 5' ss (AGA/GUUAAGUAU) in intron 2 of the human RARS2 gene results in exon 2 skipping and is associated with pontocerebellar hypoplasia41. The pathogenic effects of this mutation, which paradoxically changes a non-consensus to a consensus nucleotide, can now be explained by weakening of shifted base-pairing between this 5' ss and U1: an A-Ψ base pair at position +5 is substituted by a weaker wobble G-Ψ base pair in the shifted register. Indeed, we found that this transition at a similar atypical 5' ss tested in the SMN1/2 context compromised exon 7 inclusion, and exon 7 inclusion could be partially rescued by the U1 suppressor C5, which restored shifted base-pairing (Fig. 2c, lanes 8 and 9). Thus, shifted base-pairing can explain the effects at the molecular level of the +5 A to G mutation in intron 2 of the human RARS2 gene. These observations further strengthen the shifted base-pairing hypothesis, and highlight its implications for molecular diagnosis of 5' ss mutations10,41,42.
Atypical 5' ss that are recognized by shifted base-pairing to U1 snRNA are found in a wide range of eukaryotic genomes. Even though the estimated number of these atypical 5' ss in the genome is rather low at present, further experimental analysis of the tolerance of mutations at these 5' ss will very likely expand the set of predicted atypical 5' ss. Furthermore, experimental analysis of the numerous 5' ss sequences that can potentially base-pair to U1 with similar stability in both registers should allow a reassessment of their mechanism of recognition. In addition, characterization of this alternative mechanism of 5' ss selection should prompt a recalculation of the 5' ss motifs recognized in each base-pairing register, as these two categories of 5' ss should have different consensus motifs (Fig. 1a). This in turn could lead to improved splice-site prediction tools, considering that all current 5' ss scoring methods estimate these atypical 5' ss to be very weak (Table 1). Finally, this study should facilitate the development of improved algorithms to find genes and exons in sequenced genomes, as well as to predict the effects of disease-causing mutations and SNPs that map at these atypical 5' ss.
METHODS
In silico analyses
In addition to base-pairing to the classical 5' ss motif spanning from positions −3 to +6, we took into account positions +7 and +8, which can also base-pair to U1 and contribute to splicing18,43,44, even though they do not show appreciable conservation in 5' ss compilations6.
The SpliceRack database is a comprehensive collection of splice sites from five different genomes6: Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans and Arabidopsis thaliana. We used the built-in tool 'Locate splice site sequence patterns' to search for 5' ss that are presumably recognized via shifted base-pairing. We restricted the query to 'splice-site type GT_AG_U2, donor', and 'motif start position 5'. We used the following query sequences: NNHGTYRAGT, NYGGTYRAGT, NYAGTRRAGT, NYAGTYYAGT, NYAGTYRBGT, NYAGTYRAHT, and NYAGTYRAGV, where N = A, G, C, or T; Y = C or T; R = A or G; H = A, T, or C; B = G, C, or T; V = G, A, or C. We chose these patterns to single out 5' ss that base-pair to U1 much more efficiently in the shifted than in the canonical register. We selected the intronic positions (+3 to +7) to base-pair to U1 in the shifted but not in the canonical register, but also allowing for one nucleotide mismatch to the putative 'shifted' consensus (CA/GUUAAGU). The requirements for the exonic positions (−2 and −1) are less stringent, in that mutations at these positions have weaker effects (Fig. 2). We avoided sequences with U2-type consensus nucleotides at both positions −2 and −1 (−2A−1G) in the searches, because this combination substantially strengthens canonical base-pairing to U1.
We performed all searches for the five species, retrieved and manually curated hits using the ENSEMBL45 and UCSC46 genome browsers. We also derived human-mouse orthologous pairs of 5' ss. In many cases, the orthologous gene or intron could not be identified in the other species. Nevertheless, the comparison of 5' ss between human and mouse resulted in the addition of a few extra 5' ss to the lists from both species, because these genes were missing from the SpliceRack database in one of the species. We provide the total counts of 5' ss predicted to be recognized by shifted base-pairing to U1 snRNA in Table 2 and Supplementary Table 1 online, as well as the counts for conserved human-mouse orthologous pairs. We show the complete list of atypical 5' ss for the five species in Supplementary Table 2 online.
We calculated the 5' ss scores using several methods15–21. See Refs18,22 for detailed descriptions and comparisons between algorithms.
Cloning procedures
We amplified the three-exon and two-intron GTF2H1 and INPP4A fragments from human genomic DNA and subcloned them into the pcDNA3.1+ vector (Invitrogen). We internally deleted intron 5 of INPP4A to leave only 225 nt at each end. Likewise, we deleted intron 7 of GTF2H1 to leave only 200 nt at each end. The SMN1/2 mutant minigenes in the pCI vector were previously described12. The U1 and U6 expression plasmids, termed pN/S6 and pGemU6, respectively, were a generous gift from N. Hernandez. We derived the plasmid containing the decoy RNAs from the pU6/Hae/RA.2 plasmid47, also obtained from N. Hernandez. This plasmid includes a U6 RNA pol III promoter and 27 nucleotides of the U6 snRNA 5' stem-loop structure to stabilize the small RNA48. In addition, we added unique restriction sites to subclone the different decoy RNA sequences, as well as a pol III termination sequence.
We used PCR mutagenesis with Pfu I Turbo (Stratagene) and oligonucleotides carrying the various mutations to generate the different mutant constructs. The sequences of all the primers used in this study are available upon request. We digested the PCR products with Dpn I (New England Biolabs) prior to transformation of competent DH5α cells. We verified all mutants by DNA sequencing.
Minigene transfection into HeLa cells
We cultured HeLa cells in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) containing 10% (v/v) fetal bovine serum and antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin). We mixed the various GTF2H1, INPP4A, or SMN1/2 plasmid constructs with control or suppressor U1/U6, or decoy plasmids, and with the pEGFP-N1 plasmid (Clontech). For the suppressor snRNA experiments, we transfected 80 ng of the SMN1/2 minigene and EGFP-N1 plasmids, and 800 ng of control (pcDNA3.1+ or pUC19) or suppressor U1/U6 plasmid. For the decoy experiments, we transfected 55 ng of the SMN1/2 minigene and EGFP-N1 plasmids, and 890 ng of decoy plasmids. We transfected a total of 1 µg of plasmid mixture into ~50%-confluent HeLa cells in 6-well plates, using FuGENE 6 (Roche Diagnostics) at a 3:1 (plasmid:reagent) ratio.
RNA extraction, reverse transcription, and PCR
We harvested cells 48 hr after transfection, and extracted total RNA using TRIzol (Invitrogen). We eliminated residual DNA by RQ-DNase1 (Promega) digestion, and we phenol-extracted and ethanol-precipitated the RNA. We used a total of 1 µg of RNA for reverse transcription with Superscript II RT (Invitrogen) and oligo-dT as a primer.
We amplified cDNAs derived from expression of the pcDNA3.1+ constructs by PCR using primers located in the transcribed portion of the plasmid. We amplified cDNAs from endogenous GTF2H1 and INPP4A transcripts using primers in the exons flanking the exon with the atypical 5' ss. We amplified cDNAs from the SMN1/2 minigenes with pCI-Fwb and pCI-Rev primers12. In each case, we radiolabeled the 5' end of one of the PCR primers using T4 polynucleotide kinase (New England Biolabs) and γ-32P-ATP, and we purified the primers using MicroSpin G-25 columns (GE Healthcare). We performed 23 cycles of PCR, ensuring that amplification remained in the exponential phase (data not shown). We separated the PCR products by 6% native PAGE, followed by phosphorimage analysis to quantify the intensity of the bands. We performed three experimental replicas (RT-PCRs from three independent transfections) to derive the mean percentage of inclusion for each experiment. In all cases, the standard deviations were <5%, such that the exon-inclusion percentage values can be compared between experiments. We determined the identity of each PCR product by using the Original TA Cloning kit (Invitrogen) to subclone gel-purified bands, followed by sequencing on an ABI3730 automated sequencer.
Supplementary Material
Acknowledgements
We thank Michelle Hastings and David Horowitz for insightful comments on the manuscript, Ravi Sachidanandam for helpful suggestions, and Yimin Hua and Zuo Zhang for technical advice. X.R. and A.R.K. acknowledge support from NIH grant GM42699.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Brow DA. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 2.Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 3.Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. Are snRNPs involved in splicing? Nature. 1980;283:220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- 4.Rogers J, Wall R. A mechanism for RNA splicing. Proc. Natl. Acad. Sci. USA. 1980;77:1877–1879. doi: 10.1073/pnas.77.4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 6.Sheth N, et al. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Séraphin B, Kretzner L, Rosbash MH. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siliciano PG, Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 9.Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5' splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roca X, et al. Features of 5'-splice-site efficiency derived from disease-causing mutations and comparative genomics. Genome Res. 2008;18:77–87. doi: 10.1101/gr.6859308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Will CL, Lührmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 2005;386:713–724. doi: 10.1515/BC.2005.084. [DOI] [PubMed] [Google Scholar]
- 12.Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 15.Senapathy P, Shapiro MB, Harris NL. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 16.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serra MJ, Turner DH. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 18.Hartmann L, Theiss S, Niederacher D, Schaal H. Diagnostics of pathogenic splicing mutations: does bioinformatics cover all bases? Front. Biosci. 2008;13:3252–3272. doi: 10.2741/2924. [DOI] [PubMed] [Google Scholar]
- 19.Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 20.Burge C. Modeling dependencies in pre-mRNA splicing signals. Chapter 8. In: Salzberg SL, Searls DB, Kasif S, editors. Computational Methods in Molecular Biology, Elsevier Science. 1998. pp. 129–164. [Google Scholar]
- 21.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 22.Roca X, Sachidanandam R, Krainer AR. Determinants of the inherent strength of human 5' splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mount SM, Anderson P. Expanding the definition of informational suppression. Trends Genet. 2000;16:157. doi: 10.1016/s0168-9525(99)01964-2. [DOI] [PubMed] [Google Scholar]
- 24.Lo PC, Roy D, Mount SM. Suppressor U1 snRNAs in Drosophila. Genetics. 1994;138:365–378. doi: 10.1093/genetics/138.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen JB, Snow JE, Spencer SD, Levinson AD. Suppression of mammalian 5' splice-site defects by U1 small nuclear RNAs from a distance. Proc. Natl. Acad. Sci. U S A. 1994;91:10470–10474. doi: 10.1073/pnas.91.22.10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyriakopoulou C, et al. U1-like snRNAs lacking complementarity to canonical 5' splice sites. RNA. 2006;12:1603–1611. doi: 10.1261/rna.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 28.Wassarman DA, Steitz JA. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 29.Kandels-Lewis S, Séraphin B. Involvement of U6 snRNA in 5' splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 30.Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;6:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 31.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 32.Hwang DY, Cohen JB. U1 snRNA promotes the selection of nearby 5' splice sites by U6 snRNA in mammalian cells. Genes Dev. 1996;10:338–350. doi: 10.1101/gad.10.3.338. [DOI] [PubMed] [Google Scholar]
- 33.Brackenridge S, Wilkie AO, Screaton GR. Efficient use of a 'dead-end' GA 5' splice site in the human fibroblast growth factor receptor genes. EMBO J. 2003;22:1620–1631. doi: 10.1093/emboj/cdg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 35.Rhode BM, Harmuth K, Westhof E, Lührmann R. Proximity of conserved U6 and U2 snRNA elements to the 5' splice site region in activated spliceosomes. EMBO J. 2006;25:2475–2486. doi: 10.1038/sj.emboj.7601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du H, Rosbash M. Yeast U1 snRNP-pre-mRNA complex formation without U1snRNA-pre-mRNA base pairing. RNA. 2001;7:133–142. doi: 10.1017/s1355838201001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du H, Rosbash M. The U1 snRNP protein U1C recognizes the 5 splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 38.Lu XB, Heimer J, Rekosh D, Hammarskjold ML. U1 small nuclear RNA plays a direct role in the formation of a rev-regulated human immunodeficiency virus env mRNA that remains unspliced. Proc. Natl Acad. Sci. USA. 1990;87:7598–7602. doi: 10.1073/pnas.87.19.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boelens WC, et al. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993;72:881–892. doi: 10.1016/0092-8674(93)90577-d. [DOI] [PubMed] [Google Scholar]
- 40.Leu JY, Roeder GS. Splicing of the meiosis-specific HOP2 transcript utilizes a unique 5' splice site. Mol. Cell. Biol. 1999;19:7933–7943. doi: 10.1128/mcb.19.12.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edvarson S, et al. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am. J. Hum. Genet. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buratti E, et al. Aberrant 5' splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lund M, Kjems J. Defining a 5' splice site by functional selection in the presence and absence of U1 snRNA 5' end. RNA. 2002;8:166–179. doi: 10.1017/s1355838202010786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz SH, et al. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008;18:88–103. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stalker J, et al. The Ensembl Web Site: Mechanics of a Genome Browser. Genome Res. 2004;14:951–955. doi: 10.1101/gr.1863004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent WJ, et al. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lobo SM, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 48.Good PD, et al. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.