Abstract
Spinocerebellar ataxia type 1 (SCA1) is one of a group of nine expanded CAG repeat diseases, in which polyglutamine (polyQ) expansion above a threshold is associated with increased disease risk and aggregation. SCA1 is unique in which the polyQ in the disease protein, ataxin1, often contains a few His residues that appear to block toxicity. Here, we ask how His insertions affect aggregation by comparing a Q30 peptide with and without a centrally inserted His-Gln-His sequence. We found that at pH 7.5–8.5, His interruptions decrease polyQ aggregation rates but do not change the spontaneous growth mechanism: nucleated growth polymerization with a critical nucleus of one without non-fibrillar intermediates. The decreased aggregation rates are because of reductions in nucleation equilibrium constants. At pH 6, however, the His-interrupted peptide aggregates by a different mechanism that involves a low ThT-binding intermediate and produces a polymorphic amyloid product. In aggregates grown at pH 7.5, the His residues are solvent-accessible. Aggregates of His-inserted polyQ are good seeds for Q30 elongation, suggesting the potential to recruit polyQ proteins in the cell. Our data are therefore most consistent with His insertions blocking toxicity by suppressing rates and/or altering pathways of spontaneous aggregation.
Keywords: amyloid, ataxin 1, kinetics, nucleated growth polymerization, polymorphism
Introduction
Spinocerebellar ataxia type 1 (SCA1; Chung et al., 1993; Klement et al., 1998) is one of nine expanded CAG repeat diseases, neurodegenerative disorders caused by the cytotoxicity associated with long polyglutamine (polyQ) sequences harbored within specific disease proteins (Bates and Benn, 2002). For eight of the nine diseases, there is a relatively sharp transition between benign and pathological polyQ repeat lengths in the same approximate range of 35–45 Gln residues. While SCA1 is one of the diseases with a pathological repeat length cutoff in this range, it is unusual in that the predicted sequence of the SCA1 protein, ataxin-1 (AT-1), sometimes contains one to several histidine-encoding CAT codons within the CAG repeat region (Zoghbi and Orr, 1995; Klement et al., 1998). These CAT insertions appear to play several critical roles in controlling SCA1 risk. First, the presence of one or more CAT insertions within a CAG repeat sequence appears to greatly stabilize the trinucleotide repeat against expansions into the pathological range during replication (Klement et al., 1998; Bauer et al., 2005). In addition, however, the insertions appear to play a role at the protein level to reduce disease risk. Indirect evidence consistent with this is the observation that all expanded CAG repeats above the pathological range in patients presenting with SCA1 are devoid of CAT insertions (Zoghbi and Orr, 1995; Matsuyama et al., 1999). More direct evidence is that some individuals who are disease-free in spite of possessing a SCA1 allele that contains an expansion (44 codons) above the pathological threshold (approximately 40) also have four CAT codons within the CAG tract (Quan et al., 1995).
Disease tissues from expanded CAG repeat diseases exhibit the presence of polyQ-containing inclusions in neurons (Bates and Benn, 2002), and expansion of polyQ sequences has been demonstrated in vitro (Scherzinger et al., 1997; Chen et al., 2001; Klein et al., 2007) and in vivo (Krobitsch and Lindquist, 2000; Morley et al., 2002; Apostol et al., 2003) to greatly enhance the tendency of these sequences to aggregate. While there is much to be done, in each of these diseases, to characterize the nature of the aggregated species and their place in the disease mechanism, the general association of pathogenicity with aggregation propensity is well established. An attractive hypothesis for SCA1 is therefore that the effect of CAT codon interruption on expanded CAG pathology at the protein level may be owing to some kind of suppressing effect of inserted His residues on polyQ aggregation. Supporting this hypothesis, histidine residues in some other amyloid-forming proteins are thought to play important roles in controlling aggregation (Fraser et al., 1994; Abedini and Raleigh, 2005). In a preliminary SCA1-related study, Sharma et al. found that the solubility of a model Q22 peptide was improved when interrupted with His residues (Sharma et al., 1999). This group went on to show that four His residues added within the polyQ tract, in an identical pattern to that found to block disease in a SCA1 allele (Frontali et al., 1999), do not alter the random coil circular dichroism spectra of monomeric polyQ in native, aqueous buffer, but do qualitatively retard the aggregation kinetics of the peptides into β-sheet-rich aggregates (Sen et al., 2003). A structural model was proposed in which the His residues are placed in reverse turns in the β-sheet network of the resulting aggregates (Sen et al., 2003). However, no data supporting this model were provided.
Previously, our group characterized in detail the aggregation of the simple polyGln sequences that are the only obvious ubiquitous feature of the nine expanded CAG repeat disease proteins (Chen et al., 2001; Chen et al., 2002a, b; Bhattacharyya et al., 2005; Slepko et al., 2006). We found that the aggregation rate increases as repeat length increases, consistent with a role for polyQ aggregation in controlling disease risk and severity (Chen et al., 2001). We found that the spontaneous aggregation of simple polyQ sequences occurs via a nucleated growth polymerization mechanism with a critical nucleus of one (Chen et al., 2002b). We also developed a method to determine the second-order elongation rate constant for aggregation and thereby extract the nucleation equilibrium constant, which proves—as expected—to be a very low number (Bhattacharyya et al., 2005) confirming the expected thermodynamic instability of the nucleus (Ferrone, 1999). More recently we found that sequences flanking the polyQ sequence in the disease protein can have profound effects on the rate of aggregation, the stability/solubility of the aggregates formed, and even the aggregation mechanism (Bhattacharyya et al., 2006; Thakur et al., 2009).
In this paper we apply these methods to a study of the effects of inserted His residues within a polyQ sequence on various aspects of aggregation. We find that His insertion generally diminishes aggregation kinetics by different mechanisms depending on pH. These effects appear to be realized primarily through modulation of the thermodynamics of nucleus formation, and hence have implications for the nature of the critical nucleus. We also find that His insertion decreases the stability of the final aggregates. We also provide data on aggregate structure, some of which supports a previous structural model placing the His residues in solvent-exposed reverse turns within the aggregate. These results, along with previous studies, suggest constraints on the molecular mechanism for how insertions of two to four CAT codons in expanded CAG repeat sequences might temper toxicity operating at the protein level.
Materials and methods
Materials
PolyQ peptides such as K2Q30K2, K2Q15HQHQ15K2 and PEG-Biotin-labeled K2Q30K2 (Osmand et al., 2006) were synthesized at the Keck Biotechnology Center, Yale University. Peptides were designed to contain flanking pairs of Lys residues to improve handling ability. PolyQ peptides were purified and subjected to a disaggregating protocol (O'Nuallain et al., 2006).
Aggregation kinetics
For the spontaneous aggregation kinetics of polyQ, reactions were initiated using disaggregated peptides. All aggregation reactions were carried out at various pH values of phosphate-buffered saline containing 0.05% sodium azide (‘PBSA’). The aggregation reaction was monitored through determining the concentration of monomeric polyQ remaining in solution at each time point using reverse-phase high-performance liquid chromatography (HPLC) on centrifugation supernatants, and subjecting reaction mixture aliquots to Thioflavin T fluorescence measurements (O'Nuallain et al., 2006). For seeded aggregation reactions, 5% of preformed aggregates (w/w) were used. Biotin-labeled K2Q30K2 was incubated with aliquots of the aggregate stock suspensions used for seeding kinetics to determine the number of growing ends. Nucleation kinetics analysis was carried out to determine the critical nucleus (n*), the nucleation equilibrium constant (Kn*), and the second-order elongation rate constant (k+) using the equation Δ = ½Kn* k+2Cn*+2t2 as described previously (Bhattacharyya et al., 2005; O'Nuallain et al., 2006). Briefly, a log–log plot of the initial aggregation kinetics versus concentration yields a slope from which can be calculated the critical nucleus (n* = slope − 2) and a complex parameter composed of both Kn* and k+. Independent determination of the pseudo-first-order elongation rate constant k*, and the molar concentration of viable growth points in the seed fibrils, allows calculation of the second-order elongation rate constant, k+. This value can then be used to calculate Kn*.
Covalent modification of histidine imide group at the fibrillar level
Monomeric peptide and aggregates were treated with the alkylating agent iodoacetate to modify the side chain imide group of histidine. Samples with a peptide concentration of 1 mg/ml were incubated over a period of time in argon-purged 10 mM PBSA buffer at 37°C with 10 mM iodoacetate for 3 days at pH 6.0 (Crestfield et al., 1963; Shivaprasad and Wetzel, 2006). Aggregate reaction mixtures were centrifuged and the pellets dissolved in 20% formic acid, and immediately analyzed by HPLC-mass spectrometry. Monomer reaction time points were directly dissolved in 20% formic acid and similarly analyzed. Using Agilent software, the mass distribution data at various HPLC elution times were deconvoluted to allow extraction of relative abundances of unmodified, singly-modified, and doubly-modified peptides. These relative abundances were used to calculate percent modification.
FTIR spectroscopy
An MB series spectrophotometer with PROTA software (ABB Bomem) was used for Fourier transform infrared spectrum of polyQ fibrils at room temperature. Samples were prepared by centrifuging at 14 000 rpm and re-suspending in PBSA. Spectra of 400 scans were recorded at 4 cm−1 resolution. The fibril spectra were corrected for buffer and the area of the amide I region (1600–1700 cm−1) was normalized to one for the primary spectra by using PROTA software. The second derivative spectra for the amide I region were calculated from the primary spectra using the PROTA software. FTIR secondary structure assignments are from reference (Jackson and Mantsch, 1995).
Electron microscopy
A 3 µl suspension of polyQ fibrils were placed on a freshly glow-charged carbon-coated 400 mesh size copper grid and allowed to adsorb for 2 min, then washed with deionized water. The grids were then negatively stained with 2 µl of 1% uranyl acetate visualized under Tecnai T12 microscope with a magnification of ×30 000. The TEM images were captured with an UltraScan 1000 CCD camera (Gatan, Pleasanton, CA, USA) with post-column magnification of ×1.4.
Data analysis
Aggregation kinetics data were fitted in Sigma plot (version 10) to either three- or four-parameter equations (exponential decay, exponential rise to maximum or sigmoidal) or linear. Reported R2 values and standard deviation are from the Sigma plot-fits.
Results
The SCA1 gene encodes polyQ repeats typically interrupted by one to two His residues (Chung et al., 1993; Orr et al., 1993). We therefore investigated the effect of His-interruption on polyQ aggregation, using a model peptide sequence K2Q15HQHQ15K2, with a K2Q30K2 peptide used as control. As in all of our studies of simple polyQ aggregation, the polyQ sequence is flanked by pairs of lysine residues in order to confer kinetic solubility onto the peptides, so that their aggregation kinetics can be studied from a soluble monomeric state (Chen et al., 2001; Chen et al., 2002a, b; O'Nuallain et al., 2006; Wetzel, 2009).
Basic trends in aggregation kinetics
We assessed the aggregation of both peptides in PBSA solutions at different pH values at 37°C by monitoring the time-dependence of both the loss of monomer from solution (Fig. 1A) and the development of a ThT signal (Fig. 1B) (O'Nuallain et al., 2006). Both measures show that K2Q15HQHQ15K2 aggregates more slowly than K2Q30K2 at all pH values investigated. Thus, at pH 7.5, a 37 µM solution of K2Q30K2 (open triangles) is about 75% aggregated after 4 days, while an identical concentration of K2Q15HQHQ15K2 (filled triangles) aggregates only about 10% to completion over the same period (Fig. 1A and B). For both K2Q30K2 (open symbols) and K2Q15HQHQ15K2 (closed symbols), aggregation rate systematically drops as pH decreases from 8.5 (open circles, closed circles) to 6.0 (open squares, filled squares). Neither peptide aggregates appreciably at pH 5.5 (data not shown).
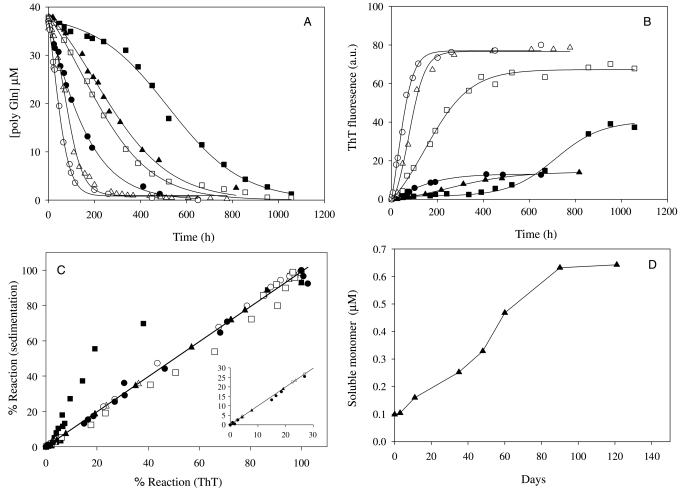
Fig. 1.
Aggregation kinetics of polyQ peptides. Spontaneous aggregation of polyQ peptides incubated at approximately 37 µM in PBSA at 37°C. (A) Monitored by the decrease in the concentration of soluble peptide of K2Q30K2 at pH 6.0 (open squares, R2 = 0.9975, SD = ±0.80), pH 7.5 (open triangles, R2 = 0.9968, SD = ±0.82) and pH 8.5 (open circles, R2 = 0.9984, SD = ±0.66), and of K2Q15HQHQ15K2 at pH 6.0 (filled squares, R2 = 0.9960, SD = ±0.90), pH 7.5 (filled triangles, R2 = 0.9949, SD = ±0.86) and pH 8.5 (filled circles, R2 = 0.9984, SD = ±0.57). (B) Monitored by increase in ThT signal for K2Q30K2 at pH 6.0 (open squares, R2 = 0.9898, SD = ±3.03), pH 7.5 (open triangles, R2 = 0.9936, SD = ±3.04) and pH 8.5 (open circles, R2 = 0.9965, SD = ±2.20), and for K2Q15HQHQ15K2 at pH 6.0 (filled squares, R2 = 0.9930, SD = ±1.23), pH 7.5 (filled triangles, R2 = 0.9980, SD = ±0.28) and pH 8.5 (filled circles, R2 = 0.9961, SD = ±0.33). (C) Plot of extent of reaction monitored by sedimentation assay (A) versus extent of reaction as monitored by ThT (B) for the six combinations of peptide and pH shown in parts (A) and (B). (D) Dissociation kinetics of the pH 7.5 K2Q15HQHQ15K2 aggregate incubated in PBSA at 37°C, as monitored by the high-performance liquid chromatography sedimentation assay. At the start of incubation, the aggregate suspension contained an equivalent of about 1.5 µM K2Q15HQHQ15K2. The plateau of monomer concentration (Cr) is 0.63 ± 0.07 µM.
Importantly, the rates of progression of the aggregation reactions at pH 7.5 and 8.5 are remarkably similar, whether monitored by ThT or by the sedimentation assay. Thus, a plot of reaction progress as measured by ThT versus reaction progress measured by sedimentation, for reactions for both peptides at pH 7.5 and 8.5, yields a straight line with no data points significantly off the line (Fig. 1C). This parallel development of the two signals, even at early time points (Fig. 1C inset), is an indication of no significant non-fibrillar intermediates in the nucleated growth pathway, a feature noted previously for the spontaneous nucleation of various polyQ peptides at pH 7.5 (Chen et al., 2002a, b). In contrast, there is a dramatic non-linearity in Fig. 1C for K2Q15HQHQ15K2 at pH 6 (filled squares). In this reaction, development of the ThT signal significantly lags behind that of the sedimentation assay, suggesting the presence of an intermediate with a relatively poor response to ThT. We reported a similar result recently for polyQ sequences that include the 17 amino acid huntingtin N-terminus (Thakur et al., 2009). Consistent with a less ThT-sensitive intermediate, EM grids from early time points of the pH 6.0 reaction of the His peptide exhibit a mixture of fibrillar and spherical oligomeric structures (not shown). The Q30 peptide also exhibits some irregularity in its aggregation at pH 6.0, with reaction extent measured by ThT in this case developing somewhat more rapidly than measured by sedimentation (Fig. 1C; □). The significance and basis of this relatively small effect, only observed at pH 6.0, is not clear.
The ThT data (Fig. 1B) also suggest some structural differences among these aggregates. Since all six of these reactions were initiated with essentially the same concentration of peptide, and since they all proceeded essentially to completion, we can be sure that the ThT signal measured at the end of the reactions reflect essentially the same mass of aggregates. It is clear, therefore, that while the aggregates formed by Q30 at pH 7.5 and 8.5 are identical, with respect to ThT fluorescence yield, the pH 6.0 Q30 aggregate may have an altered structure with a somewhat lower ThT response. More dramatically, while K2Q15HQHQ15K2 gives apparently identical aggregates grown at pH 7.5 and 8.5, the aggregates grown at pH 6.0 are substantially different, yielding a much higher ThT response. In general, the Q30 aggregates give much higher ThT responses than K2Q15HQHQ15K2 aggregates, but whether this is because of different aggregate folded structures, or the presence of surface His residues that might influence ThT binding, cannot be determined. Point mutations in amyloidogenic peptides can have dramatic differences on the ThT fluorescence yields of their aggregates (Shivaprasad and Wetzel, 2006). In addition, however, polymorphic forms of the same peptide sequence can also exhibit strong ThT differences (R. Kodali and R. Wetzel, manuscript submitted).
Critical concentration for fibril assembly
K2Q15HQHQ15K2 incubated at pH 7.5 at 37°C exhibits an easily measured amount of monomer in solution (approximately 2.5 µM) even after 800 h reaction time (Fig. 1A). The K2Q30K2 peptide, in contrast, has about 0.2 µM of monomer remaining. To probe the explanation of the 2.5 µM monomer levels, we diluted a sample of this reaction mixture to a total peptide concentration (i.e. monomer plus aggregates) in PBSA of 1–2 µM and incubated at 37°C. We observed a very slow off-rate of K2Q15HQHQ15K2 monomer over a 100-day period to a plateau of 0.63 µM (Fig. 1D). This value for Cr, the monomer concentration in equilibrium with aggregates, is the reciprocal of the equilibrium constant for fibril elongation (O'Nuallain et al., 2005) which, by calculation, is thus 1.59 × 106 M. Because of its low value, we were not able to accurately determine a Cr for K2Q30K2 under these conditions, but clearly it can be no greater than 0.2 µM. Fibrils of K2Q15HQHQ15K2 are thus at least three to four times less stable against dissociation than K2Q30K2 fibrils.
Nucleation kinetics analysis
To probe the basis of the different aggregation kinetics seen at pH 7.5 and 8.5 in Fig. 1, we studied the nucleation kinetics for aggregation of these two peptides. Although spontaneous amyloid growth reactions are often said to exhibit a lag phase, true extended baseline lag phases are normally only observed when nucleation is exaggerated by secondary nucleation effects (Ferrone, 1999). While such secondary nucleation events generate dramatic and clear lag phases, they can also complicate kinetic analysis of the nucleation mechanism (Collins et al., 2004) by obscuring the primary nucleation event. For reasons that are structurally unclear, polyQ fibrils are consistently resistant to such secondary nucleation phenomena. This fortuitous property allows analysis of the early aggregation kinetics to probe the basis of the more obscure primary nucleation event (O'Nuallain et al., 2006).
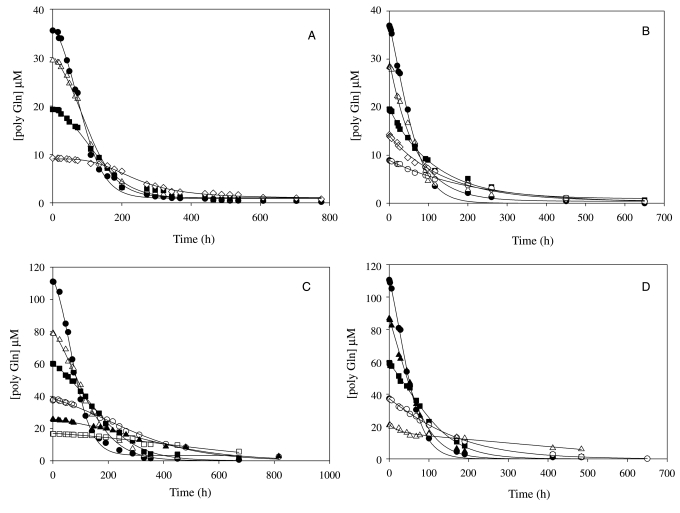
Using the sedimentation assay, we determined the concentration dependence of aggregation at pH 7.5 and 8.5 for rigorously disaggregated samples of Q30 peptides with and without the inserted His-Gln-His (Fig. 2). The initial time points were analyzed by a plot of peptide concentration versus time2 (Supplementary data are available at PEDS online, Figure S1). The rates were plotted versus starting concentration on a log–log plot (Fig. 3A). These data gave very good linear fits with slopes in the range of 2.5–3, as found for the analysis of other polyQ peptides (Chen et al., 2002b; Thakur and Wetzel, 2002; Bhattacharyya et al., 2005; Bhattacharyya et al., 2006; Slepko et al., 2006; Wetzel, 2009). These slopes correspond to a critical nucleus of approximately 1 for each peptide and pH analyzed. Pseudo-first-order elongation rate constants (k*) were determined for the self-seeded elongation of both peptides at both pH 7.5 and 8.5 (Fig. 3B), the molar concentrations of the growing ends of the seeds employed were titrated using biotinyl-Q30 (Bhattacharyya et al., 2005) (Fig. 3C), and the values combined to calculate the k+ values (Table I). These values were used to determine Kn* (Table I). As suggested for the visually parallel lines in Fig. 3A and confirmed by the values in Table I, there appear to be no fundamental differences in mechanism for peptides with or without a His insertion in the pH 7.5–8.5 range.
Fig. 2.
Concentration-dependent aggregation in PBSA at 37°C of K2Q30K2 and K2Q15HQHQ15K2 at pH 7.5 and 8.5 for nucleation kinetics analysis. (A) K2Q30K2, pH 7.5 (filled circles, 35.58 µM, R2 = 0.9965, SD = ±0.86; open triangles, 29.42 µM, R2 = 0.9930, SD = ±0.95; filled squares, 19.29 µM, R2 = 0.9956, SD = ±0.53; open diamonds, 9.23 µM, R2 = 0.9964, SD = ±0.21); (B) K2Q30K2, pH 8.5 (filled circles, 36.96 µM, R2 = 0.9977, SD = ±0.76; open triangles, 28.44 µM, R2 = 0.9930, SD = ±1.01; filled squares, 19.57 µM, R2 = 0.9964, SD = ±0.44; open diamonds, 14.26 µM, R2 = 0.9964, SD = ±0.30; open circles, 8.94 µM, R2 = 0.9965, SD = ±0.18); (C) K2Q15HQHQ15K2, pH 7.5 (filled circles, 111.02 µM, R2 = 0.9945, SD = ±2.73; open triangles, 79.25 µM, R2 = 0.9760, SD = ±1.96; filled squares 60.22 µM, R2 = 0.9974, SD = ±1.15; open circles, 37.45 µM, R2 = 0.9949, SD = ±0.86; filled triangles, 25.9 µM, R2 = 0.9898, SD = ±0.74; open squares, 16.67 µM, R2 = 0.9543, SD = ±0.72); (D) K2Q15HQHQ15K2, pH 8.5 (filled circles, 110.57 µM, R2 = 0.9986, SD = ±1.76; filled triangles, 86.84 µM, R2 = 0.9964, SD = ±2.16; filled squares, 59.45 µM, R2 = 0.9961, SD = ±1.44; open circles, 37.2 µM, R2 = 0.9984, SD = ±0.57; open triangles, 20.83 µM, R2 = 0.975, SD = ±0.83). (For time2 plots of the initial time points, see Supplementary data are available at PEDS online.)
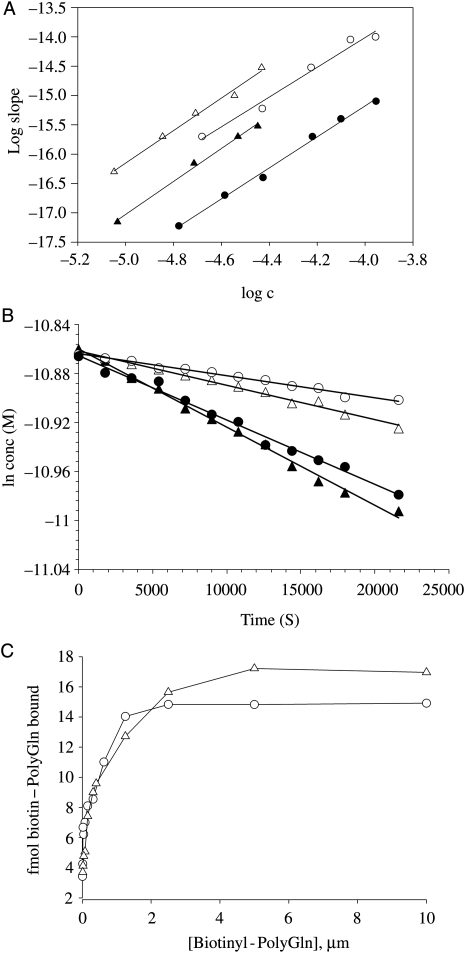
Fig. 3.
Nucleation kinetics analysis for peptides at pH 7.5 and 8.5. (A) Log–log plots of initial reaction rates (from time2 plots; see Supplementary data are available at PEDS online) versus initial concentration for K2Q15HQHQ15K2 at pH 7.5 (filled circles, slope = 2.64, R2 = 0.9950, SD = ±0.06) and pH 8.5 (open circles, slope = 2.54, R2 = 0.9799, SD = ±0.12), and for K2Q30K2 aggregation at pH 7.5 (filled triangles, slope = 2.81, R2 = 0.9951, SD = ±0.06), and pH 8.5 (open triangles, slope = 2.77, R2 = 0.9907, SD = ±0.07). (B) Determination of pseudo-first-order elongation rate constants (k*) from elongation kinetics of 20 µM monomers seeded with 5% (w/w) aggregates. K2Q30K2, pH 7.5 (open triangles), R2 = 0.9885, k* = 3 × 10−6 s−1; K2Q15HQHQ15K2, pH 7.5 (open circles), R2 = 0.9867, k* = 2 × 10−6 s−1; K2Q30K2, pH 8.5 (filled triangles), R2 = 0.9953, k* = 6 × 10−6 s−1; K2Q15HQHQ15K2), pH 8.5 (filled circles), R2 = 0.9895, k* = 5 × 10−6 s−1. (C) Titration of growth site concentrations in seed fibril suspensions used in part (B), using biotinylated polyQ as described (Bhattacharyya et al., 2005). Different concentrations of biotinyl-polyQ were incubated in 200 ng/ml aggregate at 25°C for 30 min and the aggregates washed by centrifugation then exposed to europium–streptavidin, washed and the europium fluorescence determined (Bhattacharyya et al., 2005; O'Nuallain et al., 2006). Growing end concentrations for these 200 ng/ml suspensions were determined to be 3.72 × 10−10 M for K2Q30K2 aggregates (open triangles) and 3.6 × 10−10 M for K2Q15HQHQ15K2 aggregates (open circles). Values were used to calculate second-order elongation rate constants from the above k* values (Table I).
Table I.
Thermodynamic and kinetic parameters of polyQ aggregation
| Peptide | fmol biotin-Q30 | fmol/µg aggregate | [Growing end], M | k+ (M−1, s−1) | Log–log Y-axis intercept | Kn* | ΔGn* (kcal/mol) | n* |
|---|---|---|---|---|---|---|---|---|
| K2Q47K2 (pH 7.5)a | 21.1 | 221 | 4.2 × 10−10 | 1.14 × 104 | −0.76 | 2.6 × 10−9 | 12.2 | 0.87 |
| K2Q30K2 (pH 7.5) | 17 | 85 | 3.72 × 10−10 | 0.81 × 104 | −2.985 | 3.18 × 10−11 | 14.31 | 0.81 |
| K2Q15HQHQ15K2 (pH 7.5) | 15 | 75 | 3.6 × 10−10 | 0.56 × 104 | −4.869 | 8.61 × 10−13 | 16.45 | 0.57 |
| K2Q30K2 (pH 8.5) | 17 | 85 | 3.72 × 10−10 | 1.61 × 104 | −2.3047 | 3.82 × 10−11 | 14.20 | 0.77 |
| K2Q15HQHQ15K2 (pH 8.5) | 15 | 75 | 3.6 × 10−10 | 1.39 × 104 | −3.93 | 1.20 × 10−12 | 16.25 | 0.56 |
aFrom Bhattacharyya et al. (2005). See this reference as well for detailed explanation of the parameters listed in this table.
Using the above approach, we previously reported a Kn* value of 2.6 × 10−9 for K2Q47K2, corresponding to a free energy of 12.2 kcal/mol (using the expression of ΔG = −RT ln Keq) for the pre-equilibrium between the ground state monomer and the monomeric nucleus. As expected (Chen et al., 2002b), the values for Kn* determined here for K2Q30K2 are lower than for K2Q47K2, in the range of 10−11 at both pH 7.5 and 8.5. In contrast, the k+ values for K2Q30K2 at the two pH values are in the same range (approximately 104 M−1s−1) as that for K2Q47K2 (Table I). Thus, although in general the efficiency of nucleation depends on k+ as well as Kn* (Slepko et al., 2006), the slower spontaneous aggregation for K2Q30K2, compared with K2Q47K2, is due mostly to the more efficient Kn*.
Similarly, the values in Table I reveal details of the underlying parameters on how pH changes and His insertions influence aggregation. For example, the effect on the Kn* value appears to determine the reduced spontaneous aggregation rate in polyQ-containing His residues. Thus, the effect of His insertion on second-order elongation rate constant is modest, with ratios of the k+ values for K2Q30K2 to K2Q15HQHQ15K2 of 1.4 at pH 7.5 and 1.2 at pH 8.5. In contrast, the Kn* value for K2Q30K2 is approximately 21-fold higher than that for K2Q15HQHQ15K2 at pH 7.5, and approximately 28-fold higher at pH 8.5. The effect of these apparently conservative mutations on the ability of a polyQ peptide to form a nucleus may hold important clues into the nature of nucleus formation and structure (see Discussion).
How the nucleation kinetics parameters underlie changes in the rates of spontaneous aggregation for each peptide when comparing pH 7.5–8.5 are illustrative of the subtle interplay between Kn* and k+ in determining overall nucleation efficiency. Elongation rate constant plays an important role in nucleation efficiency, since it helps determine the percentage of nuclei that go forward to develop into a growing fibril (Slepko et al., 2006). An increase in elongation rate, due either to an increase in the rate constant, or an increase in the concentration of total polyQ, can improve nucleation efficiency without changing Kn* (Slepko et al., 2006). Both K2Q30K2 and K2Q15HQHQ15K2 aggregate more rapidly at pH 8.5 than at 7.5. The K2Q30K2 peptide achieves this through more favorable values for both Kn* and k+ (Table I). In contrast, K2Q15HQHQ15K2 achieves this purely by an increase in k+, since the Kn* value for pH 8.5 is actually slightly less favorable than the pH 7.5 value (Table I).
Aggregate structures and properties
The reduced aggregation kinetics observed for the K2Q15HQHQ15K2 may be responsible for the apparently decreased toxicity of His-interrupted, expanded polyQ sequences (Quan et al., 1995). However, there may also be a role for aggregate structure. We compared the structures and properties of various K2Q30K2 and K2Q15HQHQ15K2 aggregates by a number of methods.
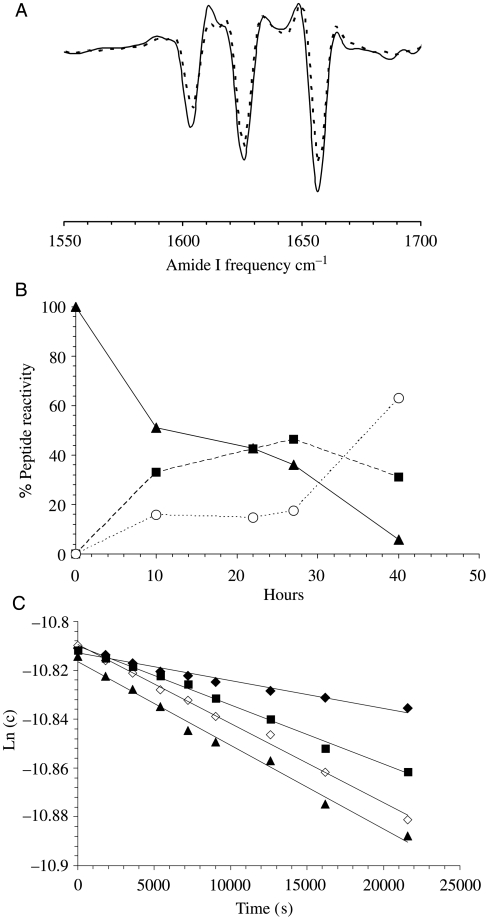
The secondary structures of these aggregates were analyzed by FTIR spectroscopy. We found that aggregates of K2Q30K2 and K2Q15HQHQ15K2 exhibited very similar, quite simple FTIR spectra (Fig. 4A), giving major bands at wavelengths 1606.5 cm−1 (Gln side chain NH2 deformations), 1625.8 cm−1 (β-sheet) and 1656.7 cm−1 (Gln side chain C = O stretch).
Fig. 4.
Structures and properties of aggregates. (A) Second derivative FTIR spectra of fibrils, K2Q15HQHQ15K2 (continuous line) and K2Q30K2 (broken line). (B) Reaction of His residues in K2Q15HQHQ15K2 fibrils with iodoacetate, determined by quantifying peptides by LC-MS from dissolved fibrils at various time points: unmodified peptide (filled triangles); singly-modified peptide (filled squares); doubly-modified peptide (open circles). (C) Cross-seeding efficiencies between polyQ peptides in PBSA at 37°C. Aggregate suspensions described in Fig. 3C were used to K2Q30K2 and K2Q15HQHQ15K2 monomers at 5% (w/w). Elongation of monomeric K2Q30K2 seeded with K2Q30K2 aggregates (filled triangles, R2 = 0.991) or K2Q15HQHQ15K2 aggregates (open diamonds, R2 = 0.9949). Elongation of K2Q15HQHQ15K2 monomers seeded with K2Q15HQHQ15K2 aggregates (filled squares, R2 = 0.9934) or K2Q30K2 aggregates (filled diamonds, R2 = 0.9681).
It is particularly important to understand how the polyQ aggregate structure accommodates His side chains. Since His has only a few more atoms than Gln, is equally capable of H-bonding, and at neutral pH, is—like Gln—uncharged, it is possible that His might be readily incorporated into the polyQ β-sheet core. To test this, we addressed the accessibility of the His side chains to alkylation by iodoacetate. Previously we used alkylation of Cys residues to determine solvent accessibility of different residue positions in Aβ amyloid fibrils (Shivaprasad and Wetzel, 2006). Here, we isolated K2Q15HQHQ15K2 aggregates, resuspended them in buffer, and challenged them with 10 mM iodoacetate. Aggregates were periodically isolated and analyzed by LC-MS. The results (Fig. 4B) show that both His residues are solvent-accessible, but are modified at different rates. One His residue per peptide is modified within 10 h of incubation, with the remaining His residue requiring an additional 30 h of incubation to be modified. (Under similar conditions, complete modification of the His residues in the monomer occurs after about 1 h; data not shown.) It is not clear whether this kinetic discrimination between the two His residues is because they are intrinsically in different environments in the aggregate, or modification of one His alters the reactivity of the other His (Crestfield et al., 1963). In any case, it seems very likely that both are located in solvent-exposed structure within the fibril. While, in principle, it is possible that some alkylation within the aggregate might occur by monomer dissociation, modification and reassociation, we think this can account, at best, for very little incorporation of carboxymethyl groups, because it would be limited by the very slow monomer dissociation rate of these aggregates (Fig. 1D).
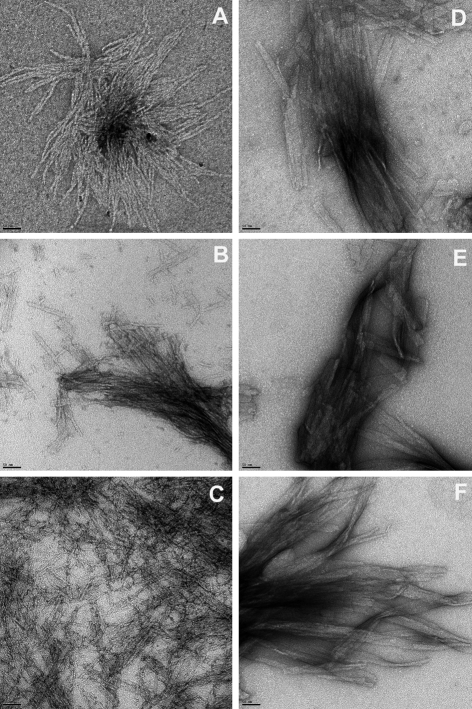
We also obtained negative stain electron micrographs for the final fibrils of each of the six aggregation reactions described here. We found that K2Q30K2 aggregates produced in PBSA at pH 6–8.5 at 37°C (Fig. 5D–F) are ribbon/plate-like structures very similar to those we described previously for K2Q20K2 and K2Q37K2 aggregates grown in pH 7.5 PBSA (Chen et al., 2002a). Although all aggregates of K2Q15HQHQ15K2 are also basically filamentous, when formed at different pH values they each have their own individual attributes. Aggregates of K2Q15HQHQ15K2 grown at pH 7.5 exhibit two forms, one consisting of small plates, and the other, large bundles of long, single filaments (Fig. 5B). Both are morphologies previously seen for other polyQ aggregates (Chen et al., 2002a; Thakur and Wetzel, 2002). Aggregates of K2Q15HQHQ15K2 grown at pH 8.5 are exclusively short rods and small, fragmented plates (Fig. 5C). Aggregates of K2Q15HQHQ15K2 grown at pH 6.0 exhibit a very strong and different fibrillar morphology. These straight, rather thick fibrils are not normally seen in aggregates of simple polyQ peptides, but we recently reported very similar morphologies for aggregates of huntingtin (htt) exon1 peptides composed of polyQ flanked by a 17 amino acid N-terminal sequence (that contains no histidine residues) (Thakur et al., 2009). While the basis of this morphological similarity is not clear, it is interesting that both htt exon1 peptides aggregated at pH 7.5, and K2Q15HQHQ15K2 aggregated at pH 6.0, are unusual in exhibiting a low ThT-binding intermediate prior to fibril nucleation.
Fig. 5.
Electron micrographs of fibrils harvested at the times indicated from reactions of polyQ peptides K2Q15HQHQ15K2 [pH 6.0, 648 h (A); pH 7.5, 412 h (B); pH 8.5, 364 h (C)] and K2Q30K2 [pH 6.0, 512 h (D); pH 7.5, 240 h (E); pH 8.5, 162 h (F)]. Images were collected on a Technai 12 electron microscope on samples adsorbed onto freshly glow-charged carbon grids and stained with 1% uranyl acetate solution. Scale bar=50 nm.
The above evidence for some effects of His insertions on aggregation structure is consistent with the possibility that reduced toxicity may derive, not from reduced aggregation rates, but because of some critical change in aggregate properties. Although much remains to be learnt about how various aggregated states disrupt cellular function, one attractive mechanism for polyQ toxicity is the recruitment–sequestration mechanism (Huang et al., 1998; Perez et al., 1998; Preisinger et al., 1999; McCampbell et al., 2000; Chen et al., 2001; Nucifora et al., 2001) according to which recruitment by polyQ aggregates reduces the levels of important polyQ-containing cellular factors, leading to toxic consequences. We therefore investigated the efficiency of cross-seeding, by which aggregates of one polyQ sequence can act as seeds for elongation of the other sequence. We found that aggregates of both K2Q30K2 (filled triangles) and K2Q15HQHQ15K2 (filled squares) are very good seeds for elongation of their self-peptides (Fig. 4C). We also found that K2Q15HQHQ15K2 (open diamonds) is essentially identical in its seeding ability for monomeric K2Q30K2 as are K2Q30K2 fibrils (filled triangles). Since the seed populations used for these experiments were matched in their concentrations of growing ends (Table I), these results can be taken directly as an indication of essentially equivalent molar seeding efficiencies of these two aggregates. This suggests that the ability of His insertion to suppress expanded polyQ toxicity does not lie in a reduced recruitment activity of the resulting amyloid aggregates.
Discussion
Different expanded CAG repeat diseases exhibit varying repeat length disease thresholds (Bates and Benn, 2002). Our group is exploring one possible explanation for these differences—that unique sequence features local to the polyQ in the disease proteins might modulate the aggressiveness of aggregation. For example, we found that the Pro-rich sequence on the C-terminal side of the polyQ segment in htt exon1 slows down aggregation and reduces aggregate stability, while not altering the fundamental aggregation mechanism (Bhattacharyya et al., 2006). In contrast, the N-terminal 17 amino acid long peptide of htt exon1 tremendously increases aggregation rate, due primarily to a dramatic change in aggregation mechanism (Thakur et al., 2009). Other polyQ disease proteins differ in their sizes and predicted structures, their intracellular locations and concentrations, the locations within the protein of their polyQ sequences, and their polyQ flanking sequences. Unique to these disease proteins is the sequence of AT-1, in which His residues are often found embedded within the polyQ sequences of benign proteins, but are absent from the expanded polyQ sequences found in disease-associated AT-1 proteins (Zoghbi and Orr, 1995). In one family, however, unaffected individuals contain His residues in polyQ stretches well above the pathological cutoff (Quan et al., 1995). We and others (Sen et al., 2003) are interested in the mechanism implied by this finding, in which His residues suppress polyQ toxicity.
Histidine and its imidazole side chain have a nitrogen atom with a pKa of around 6.4 in a small molecule (Matthew, 1985) and a range of 5.5–8.1, with an average of 6.58, in two folded proteins (Matthew, 1985). There are several reports of aggregation kinetics being affected by histidine. Mulkerrin and Wetzel reported that in human IFN-γ, the side chains of non-protonated His residues can stimulate aggregation of the unfolded protein, and this can be suppressed by protonation, chemical modification or replacement of His residues (Mulkerrin and Wetzel, 1989). Similarly, Abedini and Raleigh showed that an islet amyloid polypeptide molecule, in which the only titratable group is a His residue, aggregates sluggishly at low pH and rapidly at high pH (Abedini and Raleigh, 2005). One mechanism by which unprotonated His residues are capable of mediating aggregation is by their ability to coordinate with bivalent metals like Zn+2, as shown for fragments of the Alzheimer's plaque peptide Aβ (Morgan et al., 2002) and the human prion protein (Jobling et al., 2001). Owing to its aromatic ring structure, non-protonated histidine might contribute not only to generalized hydrophobic interactions, but also to more specific aromatic interactions, such as π–electron cloud effects (Burley and Petsko, 1986).
Consistent with the preliminary kinetic studies of Sen et al. (Sen et al., 2003), we show here that a His-Gln-His segment inserted into the middle of a polyQ tract significantly reduces—but does not eliminate—aggregation rates (Fig. 1). Interestingly, the degree by which K2Q15HQHQ15K2 aggregation is suppressed when compared with K2Q30K2 is approximately the same at all pH values tested: 6.0, 7.5 and 8.5. Although the pKa values of the His residues in monomeric K2Q15HQHQ15K2 are not known, it is almost certain that they are both essentially fully deprotonated at pH 8.5, where this peptide continues to aggregate more slowly than K2Q30K2. It was not possible to meaningfully carry the pH series any lower than 6.0, because we found that aggregation of even the K2Q30K2 peptide is nil at pH 5.5 (data not shown).
Detailed analysis of nucleation kinetics reveals that the major impact of inserted His residues in the pH 7.5–8.5 range is on the nucleation equilibrium constant, Kn*. This value at pH 7.5 is diminished by 20.6-fold (Table I), corresponding to a ΔΔGn* of 1.86 kcal/mol. In contrast, the second-order elongation rate constant k+, which also significantly contributes to overall nucleation efficiency (Slepko et al., 2006), is reduced at pH 7.5 only by 30% with the insertion of the His-Gln-His sequence into K2Q30K2 (Table I). The modest effect on the elongation rate constant suggests that the presence of the His residues does not greatly impair the K2Q15HQHQ15K2 peptide's ability to undergo the ‘dock and lock’ (Esler et al., 2000; Nguyen et al., 2007) elongation cycle compared with unbroken polyQ.
His protonation is very likely to play a role in the aggregation of K2Q15HQHQ15K2 as the pH is shifted from 7.5 to 6.0, where there appears to be a fundamental change in the mechanism of aggregation. At pH 6.0, compared with pH 7.5, there is a dramatic discontinuity between the ThT determination of reaction progress and the sedimentation assay assessment (Fig. 1C). This is qualitatively similar to the aggregation of htt exon1 peptides, in which formation of a ThT-negative, spherical oligomeric intermediate by downhill aggregation precedes the nucleation of an amyloid center (Thakur et al., 2009). In fact, the EM morphology of the pH 6.0 aggregates of K2Q15HQHQ15K2 (Fig. 5A) is remarkably similar to that for htt exon1 aggregates grown at pH 7.5 (Thakur et al., 2009). The ThT response per mass of aggregate is also much higher for pH 6.0 K2Q15HQHQ15K2 aggregates than for pH 7.5 aggregates (Fig. 1B), likewise consistent with a different morphology. The main difference between the pH 6.0 versus pH 7.5 aggregation of K2Q15HQHQ15K2 and the effect of htt exon1 sequence context on simple polyQ aggregation (Thakur et al., 2009) is that, in the latter, the new mechanism occurs at a dramatically accelerated rate, while, in the former, the new mechanism occurs at a much slower rate. This suggests that, whatever the details of the pH 6.0 mechanism for K2Q15HQHQ15K2 aggregation, one contributing factor must be the ability of His protonation to suppress the normal, neutral pH mechanism.
Can these kinetic effects of histidine insertion tell us anything about the nature of the critical nucleus? All we know about the nucleus is that it is a rare monomeric state. The ground state for polyQ monomers in water is a collapsed coil (Crick et al., 2006) that is not, however, as compact as a folded, globular protein (Thakur et al., 2009). There are two possible kinds of states one can imagine that might be (a) of higher energy than a collapsed coil and (b) empowered to initiate amyloid formation. In Model A, elements of the polyQ chain might transiently snake out of the collapsed coil to achieve a more extended, solvated chain that would be available for forming intermolecular H-bonds with other molecules. In Model B, the collapsed polyQ could transiently organize internally, through formation of intramolecular H-bonds, into more defined structures with β-sheet content capable of undergoing an initial elongation step that would be very similar to the fibril elongation steps later.
As described above, at pH values where His in the monomer is unlikely to be protonated, His insertion significantly destabilizes the nucleus. His and Gln have roughly the same side chain volumes and H-bonding capabilities, and non-protonated His is also similar to Gln in free energies of transfer from organic solvents to water (Radzicka et al., 1988) and in virtual free energies in distribution from the interior to the exterior of folded proteins (Chothia, 1976). According to this analysis, the hypothetical “collapsed to extended” transition in Model A for the nucleus would be expected to be relatively unaffected by His insertion into polyQ. It is difficult to predict the effect of His insertion on the stability of a folded nucleus, as in Model B, without knowing the details of this organized structure.
As described above, at pH 6.0 where His in the monomer is likely to be at least partially protonated, the aggregation reaction (a) slows down and (b) occurs by a different mechanism. This suggests that either the nucleation equilibrium or the elongation rate constant for the normal, neutral pH mechanism must be significantly diminished by His protonation. If the nucleus state is as described in Model A, one would expect protonation to actually favor nucleation, rather than suppress it; we have no data to address the possible effect of pH 6.0 on K2Q15HQHQ15K2 aggregate elongation. It is again difficult to predict the effect of protonation on formation of the Model B nucleus, although intuitively this might be expected to be minor if the folded nucleus resembles the folded fibril with its solvent-exposed His residues.
While no firm conclusions can be drawn from our data about the nature of the kinetic nucleus for polyQ aggregation, we believe that the above analysis suggests the feasibility of using classical protein structure–function approaches, coupled with nucleation kinetics analysis, to address this question.
It is also possible to estimate the thermodynamic impact of the His-Gln-His insertion on the equilibrium position of the aggregation reaction, a parameter that, like Kn*, is a measure of the ΔG between the monomer ensemble and another state. The Cr for K2Q15HQHQ15K2 aggregation at pH 7.5 is 0.63 ± 0.07 µM (Fig. 1C), corresponding to a ΔG for the aggregation reaction of −8.79 kcal/mol. In contrast, the Cr for K2Q30K2 aggregation at pH 7.5, estimated from the last time points of the forward aggregation reaction (Fig. 1A), is 0.21 µM. This corresponds to a ΔΔGagg between the K2Q30K2 and K2Q15HQHQ15K2 aggregation reactions of −0.71 kcal/mol. (It is possible, even likely, that the Cr for K2Q30K2 is lower than 0.2 µM.) Thus, the impact of the His-Gln-His insertion into polyQ is somewhat greater on the nucleation equilibrium at pH 7.5 (approximately 21-fold) than on the final aggregate assembly equilibrium (approximately 3-fold). While nucleation thermodynamics is determined by the folding of the solvated monomer into a solvated nucleus, elongation thermodynamics is determined by the folding of the monomer onto a fibril template, where some stabilization energy is expected to be gained from intermolecular contacts that are not available in nucleus formation. It seems reasonable, then, that the impact of a mutation might be greater at the monomeric nucleation stage, even if the folding of the peptide is similar in the nucleus and as a subunit of a fibril. Of course, the structure of polyQ in either state is not known in any detail, although the rough organization within the fibril is well-documented (Sharma et al., 2005).
Here, we show that the His residues are surface-exposed and accessible to alkylation (Fig. 4B) in the final aggregate structure. This is entirely consistent with the hypothesis that the His residues are placed in reversed turns and excluded from the packed β-sheets in the assembled amyloid fibril (Sen et al., 2003); however, the possibility that His residues are in β-sheet, but with side chains projecting into solvent, cannot be formally excluded by our data. While much remains to be learned about the role of aggregation in expanded polyQ repeat diseases, it is interesting that the results presented here are yet another example of a subtle change in a polyQ protein or its environment that affects aspects of both aggregation and toxicity. Besides the exhaustively reported data on the toxic and aggregation effects of repeat length increases themselves, other examples include the ability of the Pro-rich domain of htt exon1 to suppress both intracellular aggregate formation and toxicity in a yeast model (Duennwald et al., 2006), and the ability of co-expressed short polyQ peptides to enhance both aggregation and toxicity in a Drosophila model (Slepko et al., 2006).
Individuals with expanded polyQ sequences in AT-1 that are, however, interrupted with His residues do not develop SCA1 (Quan et al., 1995). This suggests that the His interruptions somehow compromise the normal toxicity associated with expanded polyQ. It is possible that this effect has something to do with protein aggregation. We describe here the results of a number of comparisons of unbroken polyQ with polyQ containing a central HQH motif. At neutral pH, inserted His residues suppress aggregation by decreasing the thermodynamic stability of the kinetic nucleus. Inserted His residues also somewhat decrease the stability of aggregates in the elongation equilibrium. There does not appear to be a huge effect on structure, and the ability of aggregates of His-containing polyQ to seed elongation by unbroken polyQ, as required for a recruitment mechanism of toxicity, is not significantly altered. At pH 6.0, not only does His insertion reduce aggregation rate, it also appears to change the aggregation mechanism and alter the final aggregate structure. This mechanism and rate change by a modest pH shift might play a role in vivo, if AT-1 in its cellular life were to experience a pH 6.0 environment. Taken together, the ability of inserted His residues to reduce aggregation kinetics at pH values relevant to cell biology is consistent with SCA1 genetics which suggest that His insertions in expanded polyQ sequences in the AT-1 protein block toxicity.
Funding
This work was supported by the National Institutes of Health (R01 AG019322 to R.W.) and by a research contract from the Hereditary Disease Foundation (to R.W.).
Supplementary Material
Acknowledgements
We thank Ashwani Thakur and Anusri Bhattacharyya for help with the nucleation kinetics analysis.
Footnotes
Edited by Steve Bottomley
References
- Abedini A., Raleigh D.P. Biochemistry. 2005;44:16284–16291. doi: 10.1021/bi051432v. [DOI] [PubMed] [Google Scholar]
- Apostol B.L., et al. Proc. Natl Acad. Sci. USA. 2003;100:5950–5955. doi: 10.1073/pnas.2628045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates G.P., Benn C. In: Huntington's Disease. Bates G.P., Harper P.S., Jones L., editors. Oxford, UK: Oxford University Press; 2002. pp. 429–472. [Google Scholar]
- Bauer P.O., Matoska V., Zumrova A., Boday A., Doi H., Marikova T., Goetz P. J. Appl. Genet. 2005;46:325–328. [PubMed] [Google Scholar]
- Bhattacharyya A.M., Thakur A.K., Wetzel R. Proc. Natl Acad. Sci. USA. 2005;102:15400–15405. doi: 10.1073/pnas.0501651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Thakur A.K., Chellgren V.M., Thiagarajan G., Williams A.D., Chellgren B.W., Creamer T.P., Wetzel R. J. Mol. Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- Burley S.K., Petsko G.A. FEBS Lett. 1986;203:139–143. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- Chen S., Berthelier V., Yang W., Wetzel R. J. Mol. Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- Chen S., Berthelier V., Hamilton J.B., O'Nuallain B., Wetzel R. Biochemistry. 2002;a 41:7391–7399. doi: 10.1021/bi011772q. [DOI] [PubMed] [Google Scholar]
- Chen S., Ferrone F., Wetzel R. Proc. Natl Acad. Sci. USA. 2002;b 99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. J. Mol. Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- Chung M.Y., Ranum L.P., Duvick L.A., Servadio A., Zoghbi H.Y., Orr H.T. Nat. Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- Collins S.R., Douglass A., Vale R.D., Weissman J.S. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crestfield A.M., Stein W.H., Moore S. J. Biol. Chem. 1963;238:2421–2428. [PubMed] [Google Scholar]
- Crick S.L., Jayaraman M., Frieden C., Wetzel R., Pappu R.V. Proc. Natl Acad. Sci. USA. 2006;103:16764–16769. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald M.L., Jagadish S., Muchowski P.J., Lindquist S. Proc. Natl Acad. Sci. USA. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esler W.P., Stimson E.R., Jennings J.M., Vinters H.V., Ghilardi J.R., Lee J.P., Mantyh P.W., Maggio J.E. Biochemistry. 2000;39:6288–6295. doi: 10.1021/bi992933h. [DOI] [PubMed] [Google Scholar]
- Ferrone F. Methods Enzymol. 1999;309:256–274. doi: 10.1016/s0076-6879(99)09019-9. [DOI] [PubMed] [Google Scholar]
- Fraser P.E., McLachlan D.R., Surewicz W.K., Mizzen C.A., Snow A.D., Nguyen J.T., Kirschner D.A. J. Mol. Biol. 1994;244:64–73. doi: 10.1006/jmbi.1994.1704. [DOI] [PubMed] [Google Scholar]
- Frontali M., Novelletto A., Annesi G., Jodice C. Phil. Trans. R. Soc. Lond. B Biol. Sci. 1999;354:1089–1094. doi: 10.1098/rstb.1999.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.C., Faber P.W., Persichetti F., Mittal V., Vonsattel J.P., MacDonald M.E., Gusella J.F. Somat. Cell Mol. Genet. 1998;24:217–233. doi: 10.1023/b:scam.0000007124.19463.e5. [DOI] [PubMed] [Google Scholar]
- Jackson M., Mantsch H.H. Crit. Rev. Biochem. Mol. Biol. 1995;30:95–120. doi: 10.3109/10409239509085140. [DOI] [PubMed] [Google Scholar]
- Jobling M.F., et al. Biochemistry. 2001;40:8073–8084. doi: 10.1021/bi0029088. [DOI] [PubMed] [Google Scholar]
- Klein F.A., Pastore A., Masino L., Zeder-Lutz G., Nierengarten H., Oulad-Abdelghani M., Altschuh D., Mandel J.L., Trottier Y. J. Mol. Biol. 2007;371:235–244. doi: 10.1016/j.jmb.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Klement I.A., Skinner P.J., Kaytor M.D., Yi H., Hersch S.M., Clark H.B., Zoghbi H.Y., Orr H.T. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- Krobitsch S., Lindquist S. Proc. Natl Acad. Sci. USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama Z., Izumi Y., Kameyama M., Kawakami H., Nakamura S. J. Med. Genet. 1999;36:546–548. [PMC free article] [PubMed] [Google Scholar]
- Matthew J.B. Annu. Rev. Biophys. Biophys. Chem. 1985;14:387–417. doi: 10.1146/annurev.bb.14.060185.002131. [DOI] [PubMed] [Google Scholar]
- McCampbell A., et al. Hum. Mol. Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- Morgan D.M., Dong J., Jacob J., Lu K., Apkarian R.P., Thiyagarajan P., Lynn D.G. J. Am. Chem. Soc. 2002;124:12644–12645. doi: 10.1021/ja0273086. [DOI] [PubMed] [Google Scholar]
- Morley J.F., Brignull H.R., Weyers J.J., Morimoto R.I. Proc. Natl Acad. Sci. USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkerrin M.G., Wetzel R. Biochemistry. 1989;28:6556–6561. doi: 10.1021/bi00442a005. [DOI] [PubMed] [Google Scholar]
- Nguyen P.H., Li M.S., Stock G., Straub J.E., Thirumalai D. Proc. Natl Acad. Sci. USA. 2007;104:111–116. doi: 10.1073/pnas.0607440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora F.C., Jr, et al. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- O'Nuallain B., Shivaprasad S., Kheterpal I., Wetzel R. Biochemistry. 2005;44:12709–12718. doi: 10.1021/bi050927h. [DOI] [PubMed] [Google Scholar]
- O'Nuallain B., Thakur A.K., Williams A.D., Bhattacharyya A.M., Chen S., Thiagarajan G., Wetzel R. Methods Enzymol. 2006;413:34–74. doi: 10.1016/S0076-6879(06)13003-7. [DOI] [PubMed] [Google Scholar]
- Orr H.T., Chung M.Y., Banfi S., Kwiatkowski T.J., Jr, Servadio A., Beaudet A.L., McCall A.E., Duvick L.A., Ranum L.P., Zoghbi H.Y. Nat. Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- Osmand A.P., Berthelier V., Wetzel R. Methods Enzymol. 2006;412:106–122. doi: 10.1016/S0076-6879(06)12008-X. [DOI] [PubMed] [Google Scholar]
- Perez M.K., Paulson H.L., Pendse S.J., Saionz S.J., Bonini N.M., Pittman R.N. J. Cell. Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger E., Jordan B.M., Kazantsev A., Housman D. Phil. Trans. R. Soc. Lond. B Biol. Sci. 1999;354:1029–1034. doi: 10.1098/rstb.1999.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan F., Janas J., Popovich B.W. Hum. Mol. Genet. 1995;4:2411–2413. doi: 10.1093/hmg/4.12.2411. [DOI] [PubMed] [Google Scholar]
- Radzicka A., Pedersen L., Wolfenden R. Biochemistry. 1988;27:4538–4541. doi: 10.1021/bi00412a047. [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G.P., Davies S.W., Lehrach H., Wanker E.E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Sen S., Dash D., Pasha S., Brahmachari S.K. Protein Sci. 2003;12:953–962. doi: 10.1110/ps.0224403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Sharma S., Pasha S., Brahmachari S.K. FEBS Lett. 1999;456:181–185. doi: 10.1016/s0014-5793(99)00933-3. [DOI] [PubMed] [Google Scholar]
- Sharma D., Shinchuk L.M., Inouye H., Wetzel R., Kirschner D.A. Proteins. 2005;61:398–411. doi: 10.1002/prot.20602. [DOI] [PubMed] [Google Scholar]
- Shivaprasad S., Wetzel R. J. Biol. Chem. 2006;281:993–1000. doi: 10.1074/jbc.M505091200. [DOI] [PubMed] [Google Scholar]
- Slepko N., Bhattacharyya A.M., Jackson G.R., Steffan J.S., Marsh J.L., Thompson L.M., Wetzel R. Proc. Natl Acad. Sci. USA. 2006;103:14367–14372. doi: 10.1073/pnas.0602348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A., Wetzel R. Proc. Natl Acad. Sci. USA. 2002;99:17014–17019. doi: 10.1073/pnas.252523899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A.K., et al. Nat. Struct. Mol. Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel R. In: Protein Misfolding Diseases: Current and Emerging Principles and Therapies. Dobson C.M., Kelly J.W., Ramirez-Alvarado M., editors. New York: Wiley; 2009. in press. [Google Scholar]
- Zoghbi H.Y., Orr H.T. Semin. Cell. Biol. 1995;6:29–35. doi: 10.1016/1043-4682(95)90012-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.