Abstract
Amyloid fibrils are proteinaceous nano-scale linear aggregates. They are of key interest not only because of their association with numerous disorders, such as type II diabetes mellitus, Alzheimer's and Parkinson's diseases, but also because of their potential to become engineered high-performance nano-materials. Methods to characterise the length distribution of nano-scale linear aggregates such as amyloid fibrils are of paramount importance both in understanding the biological impact of these aggregates and in controlling their mechanical properties as potential nano-materials. Here, we present a new quantitative approach to the determination of the length distribution of amyloid fibrils using tapping-mode atomic force microscopy. The method described employs single-particle image analysis corrected for the length-dependent bias that is a common problem associated with surface-based imaging techniques. Applying this method, we provide a detailed characterisation of the length distribution of samples containing long-straight fibrils formed in vitro from β2-microglobulin. The results suggest that the Weibull distribution is a suitable model in describing fibril length distributions, and reveal that fibril fragmentation is an important process even under unagitated conditions. These results demonstrate the significance of quantitative length distribution measurements in providing important new information regarding amyloid assembly.
Keywords: bias correction, brittleness, fibril fragmentation, single-molecule method, size distribution
Introduction
Amyloid fibrils are highly ordered proteinaceous assemblies commonly regarded as the end products of nucleated polymerisation (Ferrone, 1999). These self-assembled aggregates are of key biological interest because of their association with numerous disorders, such as type II diabetes mellitus, Alzheimer's and Parkinson's diseases (Chiti and Dobson, 2006). Amyloid fibrils share a common core cross-beta molecular architecture (Sunde et al., 1997), and appear usually as unbranched filaments up to several micrometres in length, despite having a width in the order of only ∼10 nm (Knowles et al., 2007; White et al., 2009). The strong mechanical properties of these stable assemblies suggest that they are potential candidates to become engineered high-performance nano-materials (Smith et al., 2006b; Knowles et al., 2007).
To further our understanding of the complex mechanisms involved in the formation of amyloid fibrils, as well as the biological impact of amyloid assembly in disease, it is important to determine the precise length distribution of fibril samples. For example, the ability for a fibril sample to seed the growth of new fibrils is strongly dependent on the extent to which fibrils have been fragmented (Collins et al., 2004; Xue et al., 2008), with samples containing shorter fibrils being more effective in seeding due to the increased number of extension sites per weight of fibril material, compared with long fibrils. Similar to the ability to seed, the dynamic equilibrium between fibrils and soluble, potentially cytotoxic, species is also dependent on the length distribution of the fibrils (Carulla et al., 2005). Fibril fragmentation, a mechanism that significantly reduces fibril length depending on the length distribution of the fibrils being fragmented (Hill, 1983), has been show to be an essential factor in the replication and the phenotype strength of prions (Tanaka et al., 2006). Akin to other nano-scale materials (Colvin, 2003; Lynch et al., 2006), amyloid fibrils may also assert other distinct biological properties depending on their dimensions. Thus, methods to characterise fibril length distributions could have important implications in the development of therapies against amyloid disease by providing information about the mechanism of fibril formation (Sun et al., 2008; van Raaij et al., 2008; Bernacki and Murphy, 2009), as well as fibril length-dependent factors in amyloid disease. Methods to characterise fibril length distributions are also important in characterising the mechanical properties and the biological impact of artificial amyloid-like fibrils designed as potential nano-materials.
Tapping-mode atomic force microscopy (TM-AFM) is a direct and model-free method for characterising particle size distributions. This imaging method does not suffer from the need to separate the size distribution information from other complicating factors, such as solution viscosity, particle density and particle shape, as in the case of sedimentation or light scattering measurements. Being inherently a single-particle method, TM-AFM image analysis also provides detailed distribution information that cannot easily be obtained from ensemble methods. However, because TM-AFM is a surface-based technique, the observed size distribution of particles may not reflect the bulk sample size distribution, because of unequal probability of detecting different species. Here, exemplified by the detailed characterisation of the length distribution of samples containing long-straight fibrils formed in vitro from human β2-microglobulin (β2m) (Gosal et al., 2005; White et al., 2009), we present a quantitative TM-AFM and single-particle image analysis method for characterising the length distribution of amyloid fibrils. The method described includes a novel approach to detect and correct for length-dependent bias, which is a common problem associated with the surface-based nature of AFM imaging techniques. Using this method, we show that the length distribution of long-straight β2m fibrils (Gosal et al., 2005) cannot be adequately described by the normal distribution, whereas the Weibull distribution (Weibull, 1951) could be used instead as a suitable distribution model for fibril length. Our analysis of β2m fibrils also suggests that fragmentation is an important process even under unagitated conditions, highlighting the significance of fragmentation in determining the rate of fibril propagation and the properties of fibrils formed under ambient conditions.
Results
Extracting observed fibril length probability distributions from TM-AFM images
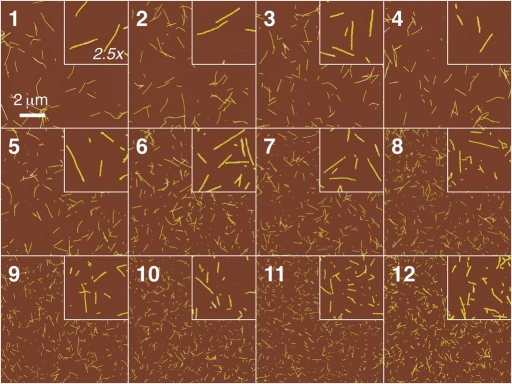
TM-AFM images of long-straight β2m fibrils deposited on freshly cleaved mica surfaces were collected at a resolution of 1024 × 1024 pixels over 10 × 10 µm areas as described in the Materials and methods section. A total of 76 height images were collected from 12 different samples containing fibrils of identical morphology but of varying length. To ensure that the same amount of fibrils in terms of initial monomer concentration or weight is present in the samples, each sample was carefully prepared by applying agitation subsequent to seeded fibril growth (described in the Materials and methods section). Under the solution conditions employed, small soluble oligomers or large non-fibrillar aggregates are not observed in the fibril samples (Smith et al., 2006a; Xue et al., 2008), and virtually all (>95%) of the initial monomers are incorporated into fibrils (Smith et al., 2006a), further ensuring equal mass concentration of fibrils present in every sample. Figure 1 shows typical height image of each of the 12 samples. From these height images, the length and the height along the highest ridge of individual fibrils, unambiguously traced according to criteria described in the Materials and methods section, were measured. Depending on the length of the fibrils in each sample, 4–16 images were collected and 374–1298 fibrils were successfully traced for each sample, with 20–340 fibrils successfully traced on each image (again depending on fibril length). A total of 9298 fibrils were traced and analysed. In Fig. 2A, the measured length L of traced fibrils for samples 1, 2, 6 and 12, as examples, is plotted in unbinned frequency histograms to illustrate the connection between the raw fibril length data and the probability density of the observed length probability distribution in each case. For each sample, the measured length of fibrils, L, can be regarded as a continuous random variable independently drawn from an underlying probability distribution characterised by its cumulative distribution function, FL(l) = P(L≤l), or its probability density function, fL(l) = dFL(l)/dl, where l represents the length and P the probability. The goal of the length distribution analysis described herein is thus to find FL(l) and fL(l) of a probability distribution model that can empirically describe the probability of finding fibrils with length L in each sample analysed.
Fig. 1.
TM-AFM height images of samples with long-straight fibrils formed from β2m in vitro at pH 2.0. Images of 1024 × 1024, 10 × 10 µm size, are shown together with zoomed in 2 × 2 µm sections. Samples are ordered and numbered (used throughout the text) approximately according to their fibril length.
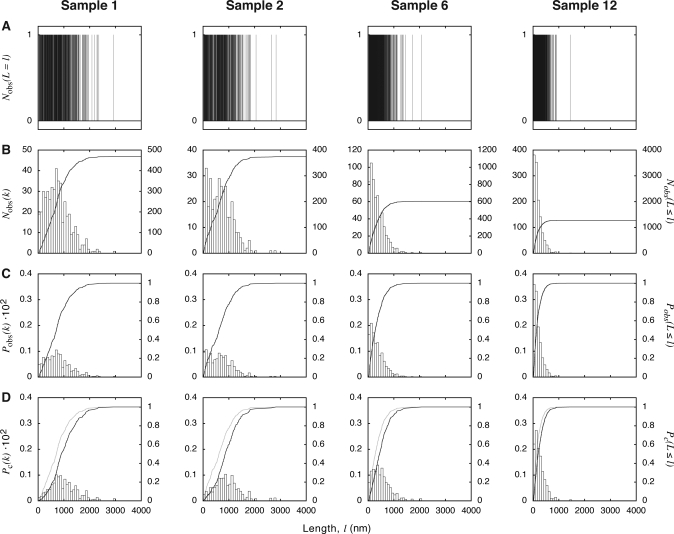
Fig. 2.
Processing of the fibril length data obtained from height images exemplified by samples 1, 2, 6 and 12. (A) Frequency histograms of observed, unbinned fibril length data, illustrating the probability density of the observed length distributions. (B) Frequency histograms shown together with the cumulative frequency plots of the observed fibril lengths. (C) Unit area histograms of the observed fibril lengths, obtained by normalising the frequency histograms in B by sample size and bin size, shown together with the cumulative probability plot of the observed fibril length, obtained by normalising the cumulative frequency plots in B with sample size. (D) Unit area histograms and cumulative probability plot of fibril length after bias correction for detection of fibrils of different length using the method outlined in the text. The cumulative probability of the observed lengths (the same as in C) is also shown as grey lines for comparison.
Figure 2B shows binned frequency histograms for the same examples as in Fig. 2A, with each bar of bin k having a value corresponding to the number of observed fibrils N(k) with fibril length L that satisfies l(k)≤L<l(k)+Δlk, where l(k) is the lower boundary length value of bin k and Δlk the bin size (Δlk = 83.3 nm in Fig. 2). The cumulative frequency plots of the number of fibrils with fibril length Lequal or shorter than l, N(L≤l), are also shown for comparison in the same graphs. These cumulative functions are bin size independent and the value at l equal to or larger than the longest fibril observed indicates the total number of fibrils measured for each sample. To facilitate direct comparison between the length distributions of different samples, the probability density, and the cumulative probability of the observed length probability distributions, was evaluated. Figure 2C, for samples 1, 2, 6 and 12, shows unit area histograms that represent estimation of the observed length probability density functions. The probability density of each bin, Pobs(k), on these unit area histograms was obtained by normalising N(k) with the number of observations made (in this case, the number of fibrils measured for each sample), ∑Nobs, and the bin size, Δlk, so that the total area of all bars is equal to 1:
| (1) |
On the same plots, the observed cumulative probability, Pobs(L≤l), is also plotted for each sample:
| (2) |
As shown in Eq. (2), the observed cumulative probability is obtained by normalising Nobs(L≤l) with the total number of observations made.
Detection of length-dependent bias
Fibrils of different length may not be detected by TM-AFM imaging with identical efficiency due to length-dependent differences in their surface deposition efficiency. Length measurements of fibrils may be further biased due to length dependence in the frequency at which a fibril can be unambiguously traced in image analysis. To detect whether the observed length probability distributions are length biased, the relationship between the observed weight average length and the observed total length of fibrils successfully traced on each image was analysed.
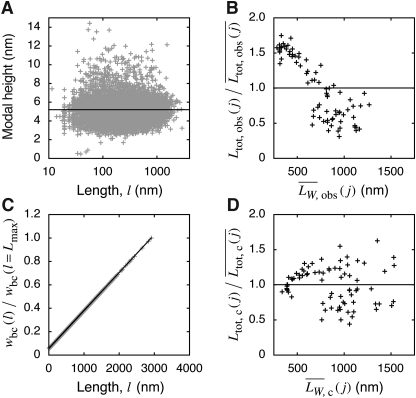
The modal height of all analysed fibrils plotted against their length is shown in Fig. 3A, with the horizontal line indicating the average modal height of 5.2 nm [consistent with previous TM-AFM measurements (Gosal et al., 2005)]. As shown in Fig. 3A, there is a considerable variation in the observed height of fibrils, which is to be expected due to variations in the twist of the fibrils and the orientation of deposition on the surface. Importantly, however, there is no significant length dependence of fibril height, consistent with the fact that fibrils of the same morphology were analysed. This confirms that the average width of these fibrils is not related to their length in the samples analysed. Given the same fibril width, the observed weight average length, 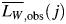 , of analysed fibrils on each image j can then be calculated because the mass, mi,j, of a fibril i is proportional to its length Li,j:
, of analysed fibrils on each image j can then be calculated because the mass, mi,j, of a fibril i is proportional to its length Li,j:
Fig. 3.
Bias correction of the observed fibril length data. (A) Modal height of fibrils plotted against their length, illustrating that the width of the fibrils in analysed samples is not length-dependent. The black line denotes the average modal height of 5.2 nm. (B) The observed total length of traced fibrils on each image normalised by its average value over all images analysed plotted against the observed weight average length of traced fibrils on each image. The relationship plotted demonstrates significant length-dependent bias in the detection efficiency for fibril length measurements. (C) The experimentally determined bias correction weighting function wbc(l) obtained for the observed data shown in A and B using the method outlined in the text and Eqs. (4–6). The y-axis is normalised with the value of obtained wbc(l) for the longest fibril measured in the entire data set (l = Lmax). (D) The same plot as in B but with the data bias corrected using the wbc(l) function shown in C.
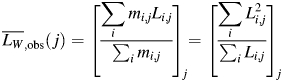 |
(3) |
Because all samples imaged contain identical weight or monomer equivalent concentration of fibrils, all fibrils analysed have the same morphology and width, and an identical protocol to deposit fibrils onto mica surfaces was employed for all samples, the total length of fibrils successfully traced on each image should (on average) be independent of the length of fibrils imaged, if there is no length-dependent bias. However, the total length of fibrils successfully traced on each image relative to the average total length over all images, 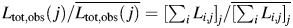 , plotted against the weight average length of fibrils successfully traced on each of the 76 images analysed,
, plotted against the weight average length of fibrils successfully traced on each of the 76 images analysed, 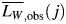 (Fig. 3B) shows the contrary for imaged long-straight β2m fibrils. The total length of traced fibrils on images with shorter fibrils is greater than traced fibrils on images with longer fibrils (Fig. 3B). This behaviour indicates that a greater mass of fibrils are effectively detected for the purpose of length measurements in samples containing short fibrils compared with their longer counterparts, despite all samples containing identical bulk mass concentration of fibrils. The observed length probability distribution is therefore biased towards short fibrils. This bias reflects the more favourable net effect of mass transport (more favourable diffusion, more favourable number concentration gradient and less favourable sedimentation) and binding (likely to be less favourable) to the mica surface during surface preparation for shorter β2m fibrils under the conditions employed compared with their longer counterparts. The difference in the deposition efficiency is evident from the fact that the total mass of fibrils deposited on the mica surfaces, estimated by the total number of pixels per image with height >2 nm, is two to three times greater for sample 12 than for sample 1 or 2 (Fig. 1), despite identical image size. The bias towards short fibrils may also reflect the fact that the frequency of tracing long fibrils successfully is lower than for short fibrils due to more frequent cases of cut off by image boundaries and/or fibril overlap.
(Fig. 3B) shows the contrary for imaged long-straight β2m fibrils. The total length of traced fibrils on images with shorter fibrils is greater than traced fibrils on images with longer fibrils (Fig. 3B). This behaviour indicates that a greater mass of fibrils are effectively detected for the purpose of length measurements in samples containing short fibrils compared with their longer counterparts, despite all samples containing identical bulk mass concentration of fibrils. The observed length probability distribution is therefore biased towards short fibrils. This bias reflects the more favourable net effect of mass transport (more favourable diffusion, more favourable number concentration gradient and less favourable sedimentation) and binding (likely to be less favourable) to the mica surface during surface preparation for shorter β2m fibrils under the conditions employed compared with their longer counterparts. The difference in the deposition efficiency is evident from the fact that the total mass of fibrils deposited on the mica surfaces, estimated by the total number of pixels per image with height >2 nm, is two to three times greater for sample 12 than for sample 1 or 2 (Fig. 1), despite identical image size. The bias towards short fibrils may also reflect the fact that the frequency of tracing long fibrils successfully is lower than for short fibrils due to more frequent cases of cut off by image boundaries and/or fibril overlap.
Correction of length-dependent bias
To correct for the length-dependent bias in the observed length probability distributions, a weighting function, wbc(l), was proposed, which allows the probability of finding a fibril with length l = L in the bulk samples, Pc(l), to be obtained from the probability of successfully tracing a fibril of the same length on TM-AFM images of the sample, Pobs(l), such that:
| (4) |
To find a suitable function wbc(l) that could satisfy Eq. (4), the criterion that the total length of traced fibrils on each image after bias correction should be the same, on average, was used. This criterion reflects the reasonable assumption that if there is no length-dependent bias, the same total mass of fibrils should be traceable for the purpose of length measurements, given that the same mass concentration of fibrils is present in each sample. For the purpose of bias correction in accordance with Eq. (4), wbc(l) could be an empirical function of any suitable functional form. Different functional forms were therefore tested to find a weighting function that is low in complexity, but effective in correcting the observed bias. A power function in this case is a good starting point as the relative fibril detection efficiency for length measurements may be governed by an underlying power law:
| (5) |
Parameters a of possible wbc(l) function(s) that satisfy the bias correction criterion, such as Eq. (5), could be found using a least-squares minimisation approach. In this case, the residual sum of squares to be minimised, RSS, corresponds to the sum of squares of the relative difference between the total length of fibrils i traced on each image j and the average of the total length of fibrils traced on each image over all images j of the data set:
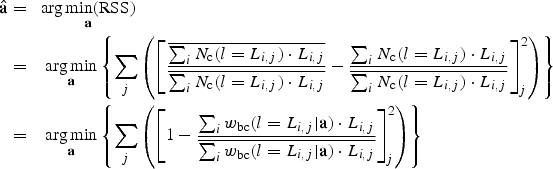 |
(6) |
Estimates 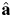 of adjustable parameters a are then found when the RSS function is at its minimum. In Eq. (6), Nc(l=Li,j) represents the corrected detection frequency of a fibril with length Li,j, obtained by correcting the observed frequency [in this case, Nobs(l=Li,j)=1, Fig. 2A], according to Eq. (4):
of adjustable parameters a are then found when the RSS function is at its minimum. In Eq. (6), Nc(l=Li,j) represents the corrected detection frequency of a fibril with length Li,j, obtained by correcting the observed frequency [in this case, Nobs(l=Li,j)=1, Fig. 2A], according to Eq. (4):
| (7) |
Least-squares fitting using Eqs. (5) and (6) to the β2m fibril length data set (Fig. 3B) yielded an exponent parameter a2 = 0.99, indicating that a linear function may be appropriate to use as wbc in this case. Fitting with a2 fixed at 1 (effectively fitting a linear function) resulted in largely unchanged results as expected, suggesting that the relative efficiency of tracing fibrils depends linearly on their length in this case. Figure 3C shows the resulting linear wbc(l) function versus fibril length. Figure 3D shows the corrected total length of traced fibrils relative to the average corrected total length over all 76 images, 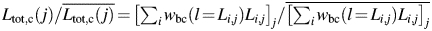 , plotted against the corrected weight average length of fibrils successfully traced on each image,
, plotted against the corrected weight average length of fibrils successfully traced on each image, 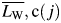 , where:
, where:
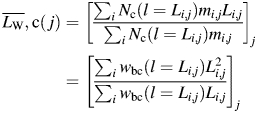 |
(8) |
As shown in Fig. 3D, the total length of fibrils successfully analysed on each image, corrected by wbc(l), on average, is no longer dependent on the length of fibrils analysed, as specified by the criterion of bias correction. The resulting function wbc(l) in this case, as seen in Fig. 3C, suggests for the β2m fibrils analysed and under the conditions employed that the ratio between the bulk number concentration of fibrils of 3 µm in length and the frequency of which these fibrils were unambiguously traced on images is ∼14 times lower than fibrils of 50 nm. This information could then be used to quantitatively correct the length distribution of fibrils observed experimentally by TM-AFM for the length-dependent bias using Eqs. (4) and (7). Figure 2D shows examples of unit area histograms Pc(k) and cumulative probabilities Pc(L≤l) of corrected fibril length probability distribution of samples 1, 2, 6 and 12. Compared with the observed length probability distributions (Fig. 2C and grey lines in Fig. 2D), the bias-corrected distributions are shifted towards longer length. The difference between the observed and the corrected length distributions is, however, sample-dependent, with bias having the most pronounced impact on samples with broad length distributions and longer fibril length, such as in the case of samples 1 and 2 where the median lengths are shifted by as much as over 200 nm. Thus, the length-dependent bias may have a significant impact on the quantitative measurements of amyloid fibril length distributions, and should be taken into account for any analysis of fibril length or length dependence in biological or physical properties of the amyloid fibrils.
Fitting probability distribution models to the length distribution data
To examine whether the bias-corrected fibril length probability distributions can be described by a probability distribution model, a set of five continuous probability distributions that are defined between 0 and +∞ were tested against the bias corrected experimental data shown in Fig. 2D, including log-normal, exponential, gamma, Rayleigh and Weibull distributions. Although not appropriate from a physical point of view since it allows probability density at negative length values, the normal distribution was also fitted to the data for comparison. To ensure that the probability distributions tested were fitted to the data in a manner independent of bin size that appropriately reflect the probability distribution of each tested model, parameter estimations 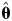 of each tested distribution model were obtained by using the maximum likelihood estimation (MLE) (Press, 2002) method:
of each tested distribution model were obtained by using the maximum likelihood estimation (MLE) (Press, 2002) method:
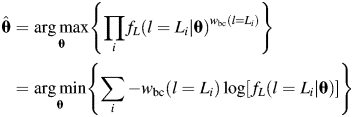 |
(9) |
In Eq. (9), each fibril length measurement in a sample is assumed to be an independent observation from an identical length probability distribution. Best-fit parameters 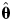 of the tested distribution can then be found when the likelihood of observing the measured lengths, i.e. the product of the probability densities fL(l) of each observation of the sample, is maximised. Since length observations in this case are biased towards short fibrils as shown in Fig. 3B, the probability density of each observation is corrected by modifying the frequency of measuring a fibril of length Li with the experimentally determined bias correcting function [wbc(l=Li) in Eq. (9) using Eq. (7)] in order to effectively fit distribution models to the bias-corrected data.
of the tested distribution can then be found when the likelihood of observing the measured lengths, i.e. the product of the probability densities fL(l) of each observation of the sample, is maximised. Since length observations in this case are biased towards short fibrils as shown in Fig. 3B, the probability density of each observation is corrected by modifying the frequency of measuring a fibril of length Li with the experimentally determined bias correcting function [wbc(l=Li) in Eq. (9) using Eq. (7)] in order to effectively fit distribution models to the bias-corrected data.
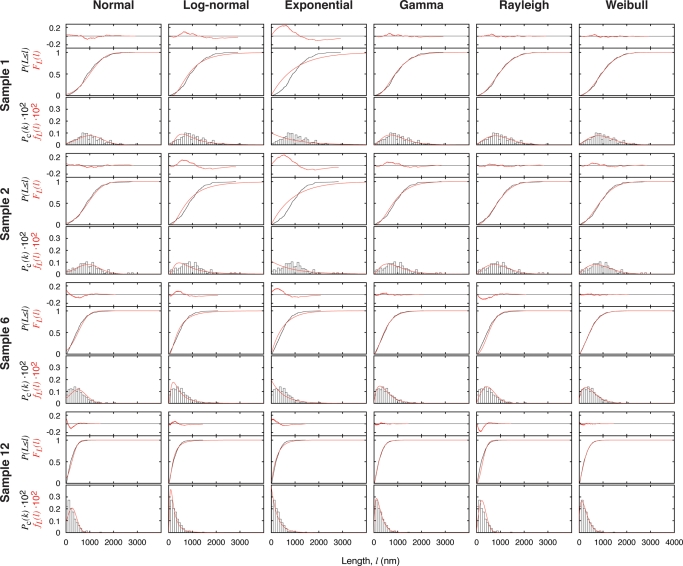
Typical results of the MLE fitting of the tested distribution models to the length probability distribution data are shown in Fig. 4, exemplified again using samples 1, 2, 6 and 12. For each model and each sample in Fig. 4, the cumulative distribution function of the fitted model, FL(l) (red line, middle graph), and the data, Pc(L≤l) (black line, middle graph), as well as the probability density function of the fitted model, fL(l) (red line, bottom graph), and the unit area histogram, Pc(k) (bars, bottom graph), are shown for comparison. The difference between the cumulative distribution function of the fitted model and the data, FL(l)− Pc(L≤l) (red line, top residual plot), indicating the quality of the fit, is also shown in Fig. 4 for each model and each sample. In the case of probability distribution fitting, the maximum difference between the cumulative distribution function of the fitted model and the data, i.e. max(|FL(l)− Pc(L≤l)|) or the maximum absolute value of the red line in the residual plots in Fig. 4, quantitatively describes the goodness of the fits (Table I). This quantity, given sufficient sample size as the case presented here (for every sample analysed, Nobs ≫ the number of fitted parameters, i.e. two parameters for the normal, log-normal, gamma and Weibull distributions and one for the exponential and Reyleigh distributions), can be used in Kolmogorov–Smirnov test (Press, 2002), which determines whether it is likely that the measured lengths with a given sample size are consistent with being drawn from a probability distribution equal to the fitted distribution. Table I summarises the quality of the fits of each of the six tested models to the data of each of the 12 samples, with italicised entries corresponding to the cases where the Kolmogorov–Smirnov test rejected the null hypothesis in favour of the alternative hypothesis that the data, with 95% significance, are not consistent with the fitted distribution model. As shown in Table I, all tested models except Weibull and gamma distributions fitted significantly poorly to the majority of the measured sample distributions as judged by the Kolmogorov–Smirnov test. Visual comparison of the fits and the residual plots in Fig. 4 also showed that both the Weibull and the gamma distributions consistently yielded fitted distributions that have the smallest differences to the data across different samples, corroborating with the Kolmogorov–Smirnov test results in Table I. Out of all six tested models, the Weibull distribution is least inconsistent with the data (not significantly inconsistent with 11 out of 12 samples, Table I), suggesting that the Weibull probability distribution could be a suitable model describing the measured fibril length probability distributions. More importantly, results shown in both Table I and Fig. 4 indicate that commonly used distribution models such as normal and exponential distributions do not represent satisfactory models in the case of fibril length distribution.
Fig. 4.
Fitting of a set of probability distribution models to the bias-corrected data using the MLE method. A set of six probability distribution models were tested and the results of the analysis on samples 1, 2, 6 and 12 are shown as examples. For each model and each sample, the result of the MLE is visualised by the cumulative distribution function of the fitted model, FL(l) (red line, middle graph), and the data, Pc(L ≤ l) (black line, middle graph), and the probability density function of the fitted model, fL(l) (red line, bottom graph), and the unit area histogram of the data, Pc(k) (bars, bottom graph). The difference between the fitted cumulative distribution function and the data, FL(l) − Pc(L ≤ l), is shown above the cumulative plots in each case (red line, top graph).
Table I.
Goodness of fit testing of the MLE results by the Kolmogorov–Smirnov test
| Sample | Tested probability distributions |
|||||
|---|---|---|---|---|---|---|
| Normal | Log-normal | Exponential | Gamma | Rayleigh | Weibull | |
| 1 | 0.074 | 0.102 | 0.248 | 0.064 | 0.051 | 0.046 |
| 2 | 0.055 | 0.126 | 0.244 | 0.079 | 0.036 | 0.056 |
| 3 | 0.085 | 0.090 | 0.217 | 0.047 | 0.052 | 0.032 |
| 4 | 0.040 | 0.141 | 0.207 | 0.094 | 0.053 | 0.053 |
| 5 | 0.073 | 0.124 | 0.169 | 0.075 | 0.088 | 0.053 |
| 6 | 0.088 | 0.084 | 0.141 | 0.039 | 0.111 | 0.026 |
| 7 | 0.076 | 0.097 | 0.140 | 0.060 | 0.117 | 0.044 |
| 8 | 0.097 | 0.066 | 0.139 | 0.039 | 0.142 | 0.041 |
| 9 | 0.096 | 0.080 | 0.110 | 0.039 | 0.147 | 0.036 |
| 10 | 0.102 | 0.042 | 0.161 | 0.038 | 0.120 | 0.040 |
| 11 | 0.107 | 0.049 | 0.146 | 0.036 | 0.156 | 0.043 |
| 12 | 0.113 | 0.060 | 0.110 | 0.031 | 0.179 | 0.033 |
The number listed for each sample and each model indicates the test quantity, which is the maximum difference between the cumulative distribution function of the fitted model and the data, max(|FL(l) − Pc(L ≤ l)|), with a lower value suggesting a better fit given the same sample. Italicised entries indicate that the quality of the fit is sufficiently low to favour the alternative hypothesis in the Kolmogorov–Smirnov test with 95% significance, indicating that the tested distribution model is significantly inconsistent with the data.
Discussion
TM-AFM represents a powerful technique to visualise nano-scale linear assemblies such as amyloid fibrils. Here, a method to quantitatively extract the length distribution of amyloid fibrils from TM-AFM images is presented. This method is composed of three key components: (i) conversion of the raw length–frequency distribution data to length probability distributions, represented by both their cumulative probability and probability densities. This facilitates sample comparison in a sample size and bin size-independent manner. Here, the probability density is estimated by unit area histograms obtained by normalising frequency histograms with sample size and bin size. Histograms estimate probability density in a highly bin size-dependent manner, in that the apparent ‘noise’ seen in the histograms varies with changes in the bin size. Therefore, they do not readily reflect the quality of the data. It is therefore important that the data are also represented by their cumulative probability plot where each individual observation is visualised more effectively. (ii) Given that a length-dependent bias is found in the observed length probability distributions, this is corrected by an experimentally determined weighting function in order to estimate the true bulk sample length probability distributions from the observed data. This method used to determine the bias correction weighting function relies on the assumption that the mass of effectively traced fibrils per unit image area, independent of the sample length distribution, should be the same after bias correction for samples containing equal mass concentration of fibrils. As TM-AFM imaging is a surface-based method, the effective detection of fibrils for the purpose of length measurements is dependent on the surface deposition efficiency and the image analysis efficiency, both of which are likely to be length dependent, as demonstrated here for long-straight β2m fibrils. It is therefore crucial that the observed length distribution is not taken directly as the bulk length distribution without testing and correcting for length-dependent bias. The bias correction weighting function could be readily determined for other types of fibrils, correcting for bias originating from similar sources but with the extent of the bias likely to be dependent on the type of sample material, the type of surface and the protocol of deposition. Such an approach is therefore generically applicable and could be used for fibrils formed from different peptide or protein precursors, deposited onto different surfaces under different conditions, and imaged using entirely different techniques such as electron microscopy. (iii) Probability distribution models are fitted to the bias-corrected data using the MLE method to find distribution model(s) that can describe the data in a bin size-independent manner appropriate and consistent with the fitted model. Because any form of model fitting should be performed against the raw data if possible, it is not ideal to fit distribution models by least-squares fitting probability density functions to binned data in the form of histograms. Here, by using the MLE (Press, 2002) method, distribution fitting is performed directly on the raw observations, based on the probabilities calculated from the fitted distribution model. The goodness of fit can then be quantitatively assessed by performing the Kolmogorov–Smirnov test (Press, 2002) to check if the fitted distribution model is reasonable. The outlined fitting procedure is general, and could readily be applied to the analysis of other fibrillar samples, including amyloid fibrils as well as other linear polymers.
The method we have outlined here was exemplified by the analysis of 12 samples containing long-straight fibrils of human β2m with a spectrum of lengths formed in vitro by controlled mechanical agitation. Results of this analysis suggest that the normal distribution does not provide good description of fibril length distribution data. Instead, the Weibull distribution (Weibull, 1951) provides a satisfactory distribution model in describing fibril length distributions, potentially providing critical constraint for future mechanistic studies of fibril formation. More importantly, samples 1 and 2 (Figs 2 and 4) show similar length distributions, despite the fact that these samples are formed under quiescent condition by seeding a monomer solution with 0.1% (w/w) or 10% (w/w) fibrillar seeds taken from an identical solution of preformed fibrils, respectively. Since long-straight β2m fibril growth from preformed extension sites under the conditions employed proceeds orders of magnitude more rapidly than the creation of new extension sites by nucleation (Xue et al., 2008), the length of fibrils extended from 0.1% (w/w) fibril seeds is expected to be up to two orders of magnitude longer on average compared with growth from 10% (w/w) seeds. The observed similarity in the length distribution of samples 1 and 2 therefore suggests that fibril fragmentation (Collins et al., 2004; Smith et al., 2006b; Xue et al., 2008) must be a significant process even when fibril samples are not agitated, such that the resulting fibrils do not extend beyond a few micrometres in length, independent of the amount of seeds added. These conclusions highlight the important information contained within fibril length distribution data and show how critical insights can be derived about the properties of fibril formation mechanisms from these data. Thus, analysis of the mechanism of amyloid assembly and the biological impact of amyloid in disease could benefit significantly from data obtained through quantitative measurements of fibril length distributions. As a whole, the method presented herein offers a quantitative approach to the experimental determination of the length distribution using TM-AFM that could be applied not only to amyloid fibrils but also to other linear polymers. The distribution analysis approach outlined here should also be useful for other types of single-particle or single-molecule techniques that probe the underlying distributions of measured properties.
Materials and methods
Fibril sample preparations
Wild-type β2m was expressed and purified as previously described (Kad et al., 2001; Gosal et al., 2005). Long, straight fibrils of β2m were formed at 120 µM initial monomer concentration in a reaction buffer containing 10 mM sodium di-hydrogen phosphate and 50 mM NaCl, adjusted to pH 2.0 using HCl. The fibril growth reaction was initiated by dissolving lyophilised protein into the reaction buffer using the method previously described (Xue et al., 2008), and was performed by stirring a 500 µl fibril sample in a 1.5 ml glass vial containing a 3 × 8 mm PTFE-coated magnetic stirring bar. Stirring was carried out using a custom-made precision stirrer with accurate rpm readout provided by a rev-counter on the rotor axis (custom built by the workshop of the School of Physics and Astronomy, University of Leeds) at 1000 rpm, 25°C. The above sample was subsequently used as seed to generate all fibril samples imaged in this study to ensure an identical morphology of the fibrils produced. The identities of the samples imaged are the following: (i) fibril solution of 120 µM initial monomer concentration containing 0.1% (w/w) seed grown under quiescent growth condition at 25°C for 2 days; (ii) same as sample 1 but with 10% (w/w) seed; (iii) sample 1 subsequently stirred for 0.77 h; (iv) sample 2 subsequently stirred for 1.1 h; (v) sample 1 subsequently stirred for 2.2 h; (vi) sample 1 subsequently stirred for 4.6 h; (vii) sample 2 subsequently stirred for 4.2 h; (viii) sample 1 subsequently stirred for 10.8 h; (ix) sample 2 subsequently stirred for 9.8 h; (x) sample 1 subsequently stirred for 23 h; (xi) sample 1 subsequently stirred for 30 h; (xii) sample 2 subsequently stirred for 25 h.
Tapping-mode atomic force microscopy image acquisition
The long-straight β2m fibrils were deposited onto freshly cleaved mica surfaces. To ensure optimum surface coverage and dispersion of fibrils, each sample was diluted to 0.4 µM (monomer equivalent concentration) with sterile-filtered deionised water immediately before a 20 µl drop was deposited onto the mica surface. For each sample, the surface was incubated with the sample for 5 min before washing with 1 ml sterile-filtered deionised water and drying under a gentle stream of N2 gas. The samples were imaged using a Dimension 3100 Scanning Probe Microscope (Veeco Instruments) and PPP-NCLR silicon cantilever probes (Nanosensors, Neuchatel, Switzerland) with nominal force constant of 48 N/m. Height images of 1024 × 1024 pixels in size, covering surface areas of 10 × 10 µm, were acquired under ambient environmental conditions. The acquisition of images with large size is aimed to reduce the relative number of fibrils that are cut off by image boundaries and increase the number of fibrils that can be traced on each image. Images were processed using supplied software NanoScope 6.13r1 to remove sample tilt and scanner bow before analysis.
Analysis of fibril length and height
The AFM images were analysed using automated scripts written in Matlab (Mathworks). To ensure objective analysis, fibril particles suitable for analysis were traced automatically from each image using the following criteria: (i) fibrils must lie entirely within the boundaries of the image; (ii) fibrils must be unambiguously traceable by the scripts in a way that is unaffected by any overlapping fibrils; and (iii) fibrils must be 2 pixels or more in length (after the apparent width of fibrils on images caused by cantilever tip convolution has been taken into account). The length and the height along the highest ridge of each fibril were subsequently extracted for fibrils that were unambiguously traced by the scripts.
Funding
This study was funded by the Wellcome Trust grant no. 075675. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Acknowledgements
We thank the members of the Radford and the Homans groups for helpful comments. We would also like to thank Neil Thomson for critical reading of the manuscript and Simon Connell for technical help with AFM.
Footnotes
Edited by Regina Murphy
References
- Bernacki J.P., Murphy R.M. Biophys. J. 2009;96:2871–2887. doi: 10.1016/j.bpj.2008.12.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulla N., Caddy G.L., Hall D.R., Zurdo J., Gairi M., Feliz M., Giralt E., Robinson C.V., Dobson C.M. Nature. 2005;436:554–558. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Collins S.R., Douglass A., Vale R.D., Weissman J.S. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin V.L. Nat. Biotechnol. 2003;21:1166–1170. doi: 10.1038/nbt875. [DOI] [PubMed] [Google Scholar]
- Ferrone F.A. Methods Enzymol. 1999;309:256–274. doi: 10.1016/s0076-6879(99)09019-9. [DOI] [PubMed] [Google Scholar]
- Gosal W.S., Morten I.J., Hewitt E.W., Smith D.A., Thomson N.H., Radford S.E. J. Mol. Biol. 2005;351:850–864. doi: 10.1016/j.jmb.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Hill T.L. Biophys. J. 1983;44:285–288. doi: 10.1016/S0006-3495(83)84301-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kad N.M., Thomson N.H., Smith D.P., Smith D.A., Radford S.E. J. Mol. Biol. 2001;313:559–571. doi: 10.1006/jmbi.2001.5071. [DOI] [PubMed] [Google Scholar]
- Knowles T.P., Fitzpatrick A.W., Meehan S., Mott H.R., Vendruscolo M., Dobson C.M., Welland M.E. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- Lynch I., Dawson K.A., Linse S. Sci. STKE. 2006;2006:e14. doi: 10.1126/stke.3272006pe14. [DOI] [PubMed] [Google Scholar]
- Press W.H. Numerical Recipes in C++: The Art of Scientific Computing. 2nd edn. Cambridge, New York, UK: Cambridge University Press; 2002. [Google Scholar]
- Smith A.M., Jahn T.R., Ashcroft A.E., Radford S.E. J. Mol. Biol. 2006;a 364:9–19. doi: 10.1016/j.jmb.2006.08.081. [DOI] [PubMed] [Google Scholar]
- Smith J.F., Knowles T.P., Dobson C.M., Macphee C.E., Welland M.E. Proc. Natl Acad. Sci. USA. 2006;b 103:15806–15811. doi: 10.1073/pnas.0604035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Makarava N., Lee C.I., Laksanalamai P., Robb F.T., Baskakov I.V. J. Mol. Biol. 2008;376:1155–1167. doi: 10.1016/j.jmb.2007.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde M., Serpell L.C., Bartlam M., Fraser P.E., Pepys M.B., Blake C.C. J. Mol. Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Collins S.R., Toyama B.H., Weissman J.S. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- van Raaij M.E., van Gestel J., Segers-Nolten I.M., de Leeuw S.W., Subramaniam V. Biophys. J. 2008;95:4871–4878. doi: 10.1529/biophysj.107.127464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibull W. J. Appl. Mech. Trans. ASME. 1951;18:293–297. [Google Scholar]
- White H.E., Hodgkinson J.L., Jahn T.R., Cohen-Krausz S., Gosal W.S., Muller S., Orlova E.V., Radford S.E., Saibil H.R. J. Mol. Biol. 2009 doi: 10.1016/j.jmb.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W.-F., Homans S.W., Radford S.E. Proc. Natl Acad. Sci. USA. 2008;105:8926–8931. doi: 10.1073/pnas.0711664105. [DOI] [PMC free article] [PubMed] [Google Scholar]