Abstract
We have employed pramlintide (prAM) as a surrogate for hAM in CD and NMR studies of the conformational preferences of the N-terminal portion of the structure in media which do not provide long-lived monomeric solutions of hAM due to its rapid conversion to preamyloid β aggregate states. Direct comparison of hAM and prAM could be made under helix-formation-favoring conditions. On the basis of CD and NMR studies: (i) the Cys2–Cys7 loop conformation has a short-span of helix (Ala5–Cys7); (ii) the extent to which this helix propagates further into the sequence is medium-dependent; a helix from Ala5 through Ser20 (with end fraying from His18 onward) is observed in aqueous fluoroalcohol media; (iii) in 12+ vol.% HFIP, the amyloidogenic region of hAM forms a second helical domain (Phe23–Ser29); (iv) the two helical regions of hAM do not have any specific geometric relationship as they are connected by a flexible loop that takes different conformations and (v) although the extreme C-terminus is essential for bioactivity, it is found to be extensively randomized with conformer interconversions occurring at a much faster rate than that is observed in the remainder of the peptide sequence. Two NMR-derived structures of the 1–22 sequence fragment of hAM have been derived. The work also serves to illustrate improved methods for the NMR characterization of helices. A detailed quantitative analysis of the NOE intensities observed in aqueous HFIP revealed alternative conformations in the C-terminal portion of the common amylin helix, a region that is known to be involved in the biorecognition phenomena leading to amyloidogenesis. Even though the SNN sequence appears to be a flexible loop, the chemical shifts (and changes induced upon helix structuring) suggest some interactions between the loop and the amyloidogenic segment of hAM that occur on partial helix formation.
Keywords: amyloidogenesis, CD spectroscopy, chemical shift deviations, helix formation, NMR
Introduction
No solution-state 3D structure of hAM (aka IAPP, islet amyloid polypeptide) that produces the islet amyloid plaques associated with adult onset (Type II) diabetes (Westermark and Wilander, 1978; Cooper et al., 1987; Westermark et al., 1987a) has been reported to date. Early CD studies suggested a partially helical structure in SDS micelles and a β-sheet structure with dimyristoyl PC vesicles and in trifluoroethanol (McLean and Balasubramaniam, 1992; Matsuura and Manning, 1993). Structure predictions based on model building and homology (Saldanha and Mahadevan, 1991) as well as similarities (Hubbard et al., 1991) to other bioactive peptides (CGRPs) (Westermark et al., 1986), with an amphipathic helix attached to a Cys–(Xaa)n–Cys loop (n = 4 or 5) (Breeze et al., 1991; Meadows et al., 1991; Meyers et al., 1991), appeared nearly two decades ago. In 1994, we reported (Cort et al., 1994), in conjunction with the CD studies of preamyloid states of hAM produced upon the addition of HFIP to 6–9 vol.% in aqueous buffer, the structuring shifts (as CSDs) that result for hAM and the non-amyloidogenic analog found in the rat (rAM). The CSDs reported at that time have been incorporated in Fig. 1 for comparison with similar reports for hAM (Yonemoto et al., 2008) and rAM (Williamson and Miranker, 2007) that appeared recently. With the exception of a few hAM fragments (e.g. Andersen et al., 1996b; Mascioni et al., 2003), to our knowledge, no definitive NMR studies of monomeric amylins appeared in the 1995–2006 period.
Fig. 1.
Amylin Hα-CSD plots from the literature, in the following order at each residue: rAM and hAM in 25% aqueous HFIP as reported in Cort et al. (1994), rAM in water (Williamson and Miranker, 2007) and hAM in water (Yonemoto et al., 2008).
In the interim, biophysical studies of hAM and its amyloidogenic fragments, e.g. hAM(20–29) (Ashburn and Lansbury, 1993) and hAM(8–37) (Goldsbury et al., 2000; Koo and Miranker, 2005; Abedini and Raleigh, 2006), the latter viewed as a fully competent fibril forming species that can be used as a surrogate for WT hAM, focused on the discovery of amyloidogenesis inhibitors (Kapurniotu et al., 2002; Porat et al., 2004; Yan et al., 2006; Abedini et al., 2007), determining the mechanistic details of the stages of amyloidogenesis (Padrick and Miranker, 2002; Ruschak and Mirander, 2007) and establishing models of peptide geometry (Padrick and Miranker, 2001; Kajava et al., 2005; Luca et al., 2007; Madine et al., 2008) in the fibril state and its precursors. Early mutational studies and species comparisons (Glenner et al., 1988; Asai et al., 1989; Westermark et al., 1990), as well our report in 1994 (Cort et al., 1994), focused attention on the FGAILS sequence as the most amyloidogenic patch in the hAM structure; this region remains unstructured in rAM but becomes a partially formed helix in hAM (Fig. 1). Our further studies focus on the Pro25,28,29 mutant of hAM, pramlintide. Pramlintide (prAM) is now a widely prescribed amylin replacement therapy in the U.S. for patients taking insulin for the treatment of both type I and type II diabetes.

The helicity of amylins and structural features outside of the hAM(20–29) sequence span have been under intense investigation recently. Miranker and coworkers (Knight et al., 2006) have suggested that partial helix formation is an important feature in both amyloid formation and toxicity, not only for hAM but that this might be a general feature for amyloidogenic peptides. CD and NMR CSDs continue as the primary measures of amylin peptide helicities. Representative CSD plots along the sequences of rAM and hAM under aqueous conditions and helix-favoring media are shown in Fig. 1.
The Hα-CSD data for aqueous 200 µM hAM-free acid reported by Yonemoto et al. (2008) in 100 mM aqueous NaCl (pH 6) stand out as being particularly helical, with many upfield shifts that exceed those we reported for hAM in 25% HFIP (Cort et al., 1994), a particularly good helix-inducing solvent. Increased upfield Hα shifts were particularly evident in the 20–25 residue span and from Thr30 to the C-terminus.
Mutations outside of the 20–29 residue span are of increasing recent interest. Abedini and Raleigh (2006) established that proline substitutions at residues 17, 19 and 30 greatly reduce fibril formation and that the resulting fibrils dissociate on dilution. Koo et al. (2008) recently reported a survey of the effects of mutations of Asn sites in hAM(8–37) upon amyloid formation kinetics: the N14L and N21L mutants were notably less amyloidogenic and fibril formation could not be seeded with WT hAM(8–37). The same report also notes that the A25P, S28P and S29P mutants of hAM(8–37), as reported by Green et al. (2003), is still amyloidogenic. These results were viewed as further evidence for the importance of features in the helical region of amylins (particularly N14) to amyloid assembly. The importance of the portion of the amylin sequence (5–20) with a helical propensity to the biorecognition phenomena in amyloidogenesis is also supported by the observation that the most potent inhibitors of WT hAM aggregation, although bearing β-structure breaking mutations in the amyloidogenic patch, I26P (Abedini et al., 2007) or N-methylation at G24 and I26 (Yan et al., 2006), retain the entire N-terminal portion of the hAM sequence. We found (data of K.N.L.H., reported at the 30th EPS) that a shorter sequence fragment (V32Y)-hAM(17–33), which mimics WT hAM in it amyloidogenesis assays, is not converted into an inhibitor by either an I26P or F23P mutation. Studies indicating that sites in the 11–20 residue span are key to molecular recognition and self-assembly have appeared: e.g. LANFLV is able to accelerate amylin self-assembly and hAM-induced cytotoxicity (Mazor et al., 2002; Scrocchi et al., 2003). The present study provides structural data for this span.
NMR structures of hAM(1–19) and rAM(1–19) in DPC micelles were reported recently (Nanga et al., 2008); these were prompted by the observation (Brender et al., 2008b) of membrane disruption by hAM(1–19) but not rAM(1–19). Both amylin fragments were predominantly helical in the micelle-bound state but there were differences in both the C-terminal turn and the disulfide loop and in the manner of insertion in the micelle. Very recently, NMR structures were reported for full-length rAM and its complex with insulin; residual helical structure was observed in the 11–18 residue span with features in this span also placed at the amylin/insulin interface (Wei et al., 2009). Wei et al. also reported that hAM has a lower affinity for insulin. The importance of the H18R mutation is also clear from the observation that (H18R)-rAM, unlike rAM, forms amyloid fibrils (Green et al., 2003). The results outlined here, and in the two prior paragraphs, prompt us to report our NMR studies of hAM, (A25P)-hAM and prAM. Our choice of prAM as the primary less-amyloidogenic amylin for these studies allows us to examine a system with the hAM sequence from residues 1–24. The improved solubility characteristics of prAM facilitated NMR-structural studies in a wide variety of media. These studies have provided the secondary structural preferences of the amylins in different media, revealed a slow conformational interconversion in the disulfide loop and yielded NMR structure ensembles for hAM and prAM in a medium that stabilizes the helical domain(s) of the monomeric species. These structures provide additional insights into the structural features that may be involved in the earliest stages of amyloidogenesis as well as models of a hAM structure that should contain the pharmacophore features required for its essential biological activities.
Methods and materials
Peptide samples
With the exception of 15N-prAM-free acid, all peptide samples were synthesized by automated solid phase synthesis using standard FMOC protocols and purified by HPLC (acetonitrile/0.1% aq. TFA gradient). The resulting lyophilizates contained the expected equivalents of TFA and ca. 10% (by weight) water. Several samples of prAM were further purified (98+% by HPLC) and converted to the acetate salt. The sample (Amylin lot # MSAAC137) employed for the definitive NMR studies was 85.5% peptide (by amino acid analysis), 8% water by weight and contained 4.8 equivalents of acetate. A sample of prAM trifluoroacetate was prepared with the three leucine universally 13C labeled by repeating the standard synthesis employing U-13C-Leu-FMOC reagent. For all species, amino acid content and sequence were confirmed by mass spectrometry and by the observation of the expected inter-residue NOE connectivities in 2D NMR studies reported herein.
Universally 15N-labeled pram-free acid
The coding region for prAM sequence was inserted into a vector for intracellular expression under the control of the T7 promoter in E. coli strain BL21. The cells were cultured in a minimal medium containing 1.5 g/l 96–98% U-15N algal whole hydrolyzate and 2.5 g/l 99% U-15N ammonium sulfate (Cambridge Isotopes Lab). Expressed prAM was obtained from inclusion bodies by cell lysis, solubilization and removal of the fusion leader using a cyanogen bromide cleavage step. Positive electrospray ionization MS (VG Trio 2000 Spectrometer) indicated 93% 15N incorporation based on relative intensities of the (M + 4H)+4 molecular ion peak (m/z = 1000.0, z = 4) of the product and co-injected unlabeled synthetic standard. A Waters 625 HPLC System with a Vydac 214TP510 (C4, 1.0 × 25 cm) column was used for purification with a gradient from 15% to 50% acetonitrile (0.1% TFA) over 45 min at a flow rate of 5 ml/min. Analytical HPLC (detection by UV at 214 nm) using a Vydac 208HS3405 column with a 1.5 ml/min flow rate and a 10 min linear gradient from 2% to 35% acetonitrile (0.1% TFA) revealed that the labeled peptide was 98% pure (tR = 10.71 min).
UV and CD spectroscopy
All ε-values are given in the units of cm2/mmol. Two lots of hAM were employed for the determination of the extinction coefficient—ε274 − ε310 = 1533 (±86) cm2/mmol (n = 9) in 25 vol.% HFIP at 200–600 µM peptide (Cort et al., 1994). The reference UV spectrum of prAM was obtained using the high-purity acetate salt samples (n = 6): ε274 = 1554 ± 18 (a maximum) and ε250 = 904 ± 24 (a minimum), at 250 µM peptide in pH 4, 20 mM aqueous acetate buffer. These values are in full accord with the values predicted (1547 at 274 nm, 892 at 250 nm) using the expected contributions (taken from Mihalyi, 1969) for the chromophores present (one disulfide, two Phe residues and one Tyr residue). A series of 15-fold dilution experiments based on a 600 µM solution of prAM TFA salt indicated that ε274 is concentration-independent and varies by <3% over the pH range of 2–6 in 0–35 vol.% HFIP.
CD spectra were recorded as described previously (Andersen and Palmer, 1994; Andersen et al., 1996a, 1996b, 2002) on a JASCO J-720 spectropolarimeter which had been calibrated with NH4+ (+)-10-camphorsulfonate. Spectra were terminated in the far UV at a point where the dynode voltage of the photomultiplier tube was still low enough to ensure reliable ellipticity measurements. Typical spectral accumulation parameters were: time constant, 0.25 s; scan rate, 100 nm/min with a 0.2 nm step resolution over the range 178–270 nm; with 12–16 scans averaged for each spectrum. All CD spectral values for peptides are expressed in units of residue molar ellipticity (deg cm2/residue-dmol) based on a 37-residue count. For hAM (and its Pro25 analog), concentrations were determined by the absorbance of a ca. 500 µM stock solution in 25% aq. HFIP assuming that the extinction coefficient determined for hAM, ε274 − ε310 = 1533 cm2/mmol applies for both species. For prAM, a pH 4 aqueous medium stock solution was employed—ca. 250 µM, with the exact concentration based on ε274 − ε310 = 1554. Cosolvent titrations were accomplished by making appropriate dilutions of aliquots from these stock solutions: solvent compositions are reported as final volume-% of the cosolvent, the aqueous component was at pH = 4 (20 mM acetate) unless otherwise specified. For prAM, the thermal dependence of the CD in this buffer could be obtained: [θ]201 = −17 150 + 38 T and [θ]221 = −3800 − 8 T, the latter over the 0–25°C range. For prAM, the thermal gradient (Δ[θ]221/ΔT) increases at higher temperatures. Upon addition of GdmCl, [θ]221 at the low temperature limit becomes less negative and the gradient becomes more negative: −2050 − 33 T (5 M), −1065 − 40 T (7.5 M GdmCl). However, even more positive values and more negative gradients are expected for a fully disordered version of this sequence—[θ]221 = 1830 − T (56 ± 10), based on previously published prediction algorithms (Andersen and Tong, 1997). This suggests that prAM (and presumably other hAM analogs) retains some helicity even under highly denaturing conditions. For hAM, only limited data could be obtained in a strictly aqueous medium due to rapid aggregation; a shoulder at 222 nm ([θ] = −3900) in the initial spectra does suggest a slightly larger extent of helicity for its monomeric state; particularly by the distinctness of the shoulder rather than the observed magnitude, which could be diminished by partial aggregation.
NMR spectroscopy and analysis
NMR spectra obtained at 500 MHz (Bruker AM-500) using methods previously described in full (Andersen et al., 1992, 1993), the specific experiments performed at 750 MHz are described or referenced. Deuterated DMSO (containing 0.03% TMS), HFIP and 99.96+% D2O were purchased from Cambridge Isotope Laboratories. Sodium 3-trimethylsilyl-propionate was used as an internal reference in all aqueous samples; these were examined at an exchangeable D/H ratio of 0.1–0.15 or in 99.9+% D2O with prior exchange of the peptide NH/OH signals. The previously reported spectra of hAM and rAM TFA salt (Cort et al., 1994) were employed with additional 500 MHz spectra at 35% HFIP (1.2 mM peptide, 65 mM DCO2D) and 25% HFIP (1.0 mM peptide, 4 mM DCO2D). Spectra of [Pro25]-hAM (1.2 mM) were recorded at 25% HFIP with ca. 100 mM added CD3CO2D. For prAM acetate, a ROESY spectrum (2.3 mM peptide) in aqueous buffer (final concentrations: 11 mM acetate, 9 mM d-formate, 6 mM TFA, pH = 4.0) at 285 K employing a train of small flip angle pulses followed by a 180° pulse for a mixing time of 250 ms at a spin lock field of 12.5 kHz (Zagorski, 1990) was used for the sequential assignment, with additional data (TOCSY, COSY) recorded at 275–305 K (20 mM d3-acetate, 20 mM d-formate, 11 mM TFA, pH = 3.2) used to resolve overlap ambiguities. At 25% and 35% HFIP, the prAM concentration was 3.6 mM and the final buffer concentrations depended on whether the acetate or TFA salt was used to prepare the solutions (17 mM acetate, 10 mM d-formate, 6.7 mM TFA or 15 mM d3-acetate, 15 mM d-formate, 14 mM TFA). The time course of amide NH exchange for hAM in 25% HFIP at 302 K (pH* = 3) was determined by recording 1D spectra repeatedly over a 20 h period when the protic TFA salt is dissolved in 99.9+% D2O medium. Protection factors were calculated assuming that the exchange half-life displayed by Val32 (ca. 7 min at pH* = 3, 29°C) corresponds to an unprotected NH and that the Molday factors reported for water (Bai et al., 1993) apply to aqueous HFIP, the protection factors thus derived are: Gln10 (>8, ∼24), Arg11 (>10, ∼22), Leu12 (≥26), Asn14 (≥30, ∼70), Ala13–Phe15–Leu16 (>90), Val17 (>30), His18 (>4, ∼15), Ala25 (>13), Ile26 (≥5), Leu27 (∼10) and Ser28 (∼15-fold protection)—in cases where assuming specific acid versus base catalyzed exchange yield different estimates, both are included.
NMR data recorded at 750 MHz were used to confirm the 35% HFIP assignments and to obtain NOE constraints at shorter mixing times (τm = 90 ms at 275 K for 3.6 mM prAM acetate) or under non-aggregating conditions (τm = 110 ms at 295 K for 0.7 mM hAM). Both of the NOESY spectra employed the WATERGATE sequence (Piotto et al., 1992). In the case of prAM, the universally 15N-labeled form of the C-terminal free acid (which displays essentially identical chemical shifts) was employed for several gradient-tailored 3D 15N NOESY–HMQC spectra (the pulse sequence was based on the corresponding HSQC experiment reported by Sklenar et al., 1993). The temperature dependence of the 15N1H resonances of prAM-free acid (2.1 mM) in 35% HFIP (pH* 4) was obtained from a set (T = 268, 275, 285, 300 and 315 K) of 256 by 4 K complex points 15N-HMQC spectra recorded serially.
Complete assignments were also obtained for the hAM and rAM TFA salts (4 mM) in a 88:11:1 volume mixture of d6-DMSO/CD3CO2D/H2O from COSY, TOCSY and NOESY spectra recorded at 310 K. Spectra of prAM acetate (3.1 mM) were recorded in d6-DMSO initially containing 1% H2O and 2 mM CD3CO2D. The changes in resonance shifts were recorded as the solution was acidified, first with additional CD3CO2D, then with DCO2D and finally by the addition of TFA.
The 13C shift assignments for [U-13C-Leu12,16,27]-prAM were obtained in the 35% HFIP medium using 13C-HMQC spectra with and without an added RELAY sequence based on an inverse-detected 13C-INADEQUATE experiment (Pratum and Moore, 1993). These spectra provided, in addition to the C′, Cα, Cβ and Cγ shifts, differentiation of the β, β′ and γ proton resonances for the three leucines.
Sequence histograms of inter-/intra-residue Hα–HN NOE ratios are calculated and displayed as described by Lee et al. (1994). Structuring shift comparisons, as sequence plots of CSDs, CSD = δobs − δref, with the then current ‘coil reference values’ (Andersen et al., 1997) and nearest neighbor correction only for proline. With very few exceptions, these coil values are within 0.07 ppm of more recently refined values (Fesinmeyer et al., 2004, 2005; Andersen et al., 2006; Eidenschink et al., 2009) and those used by others reporting CSDs for amylin peptides. For data collected in the DMSO medium, we employed the Hα reference values of Bundi et al. (1975) with the exception of Pro (4.36, no previous value), Val (4.19 versus 4.26) and Ile (4.22 versus 4.25) and include the same correction for the random coil reference values of Hα of Xaa in Xaa-Pro (+0.29, but +0.17 for Gly–Pro) which were used for the aqueous medium. For a +H3N-terminated peptide in DMSO, the correction for the Hα of the first residue is −0.40, and the ‘2’-effect (+0.54) and ‘3’-effect (+0.26) are assumed for NH reference values (Andersen et al., 1995). Our 1997 publication also introduced a +0.075 ppm correction to Hα coil shifts in 25–35 vol.% HFIP which was employed in the present study, but not in the preliminary report of Cort et al. (1994).
NMR structure ensemble calculations
NOESY data (as peak intensities) were converted to DCs of the form, rij = (dij − d−) − (dij + d+), essentially as described previously (Andersen et al., 1992, 1993; Neidigh et al., 2001) with a three-stage XPLOR (Brünger, 1992) SA protocol utilized to obtain an NMR ensemble. The observed NOE intensities were corrected for relative peak width in the directly detected dimension and for the H/D ratio for peaks involving NHs. Four features deserve note: (i) the intensity scale was calibrated using the following reference distances: smallest αiNi+1 NOE = 3.3–3.8, largest αiNi+1 NOE = 2.0–2.4, largest NiNi+1 NOE in the helical span = 2.45–2.9, and <αiNi> = 2.8 Å with the high- and low-distance bounds (d− and d+) derived from a range of NOE intensities in each class; (ii) the constraints involving rotation symmetry equivalent aromatic hydrogens were r−6 averaged; (iii) a limited number of backbone dihedral constraints (derived from chemical-shift indices and local NOE ratios) were included in the first high-temperature dynamics of each stage—these were given an ineffectively small weighting in the subsequent dynamics and during minimizations; and (iv) during the cool-down dynamics and Powell minimization of the last stage, the Erepel function was replaced by a standard Lennard–Jones van der Waals function.
NOE DCs were placed in three categories which were accorded different weighting during the protocol. In the terminal portion of the cool-down and subsequent minimization of the final stage, the NOE force constants were 32, 14 and 7 kcal/Å2/mol, respectively, for the three categories. A fourth ‘trial’ category with even lower weighting which employed soft-square, rather than a biexponential energy function, was used for distances derived from NOEs that could be predominantly secondary and for alternate attributions of NOE peaks that had two or more potential assignments. During the course of the refinement, those ‘trial’ distances that could be satisfied without increasing the ENOE term due to the unambiguously determined DCs were incorporated into the other categories. This procedure affords relatively tight DCs; for example, the average range (d−+d+) for a constraints with an upper bound of 3.4–3.9 Å was 0.76 ± 0.21 (n = 57, for the hAM constraint file) and 0.80 ± 0.18 Å (n = 80, for the prAM constraint file), corresponding to a DC of 2.9–3.7, not 2.0–3.5 Å, as has often been used for a typical medium-strength NOE. When inter-residue NOEs that would be diagnostic for a particular secondary structure were completely absent, LBO constraints, also known as ADCs ‘ADCs’ (Brushweiler et al., 1991), were employed. An alternative ‘loose bounds set’ was created by increasing each upper bound by 4% and setting the lower bound to the larger of 0.8 (dij−d−), (dij−1 Å) or 1.9 Å; these changes completely eliminate the influence of the LBO constraints. Structure ensembles generated using the ‘loose bounds’ are designated as ‘conservative’; those generated using the tight constraints are designated as ‘aggressive’ ensembles. Hydrogen-bonding constraints for an NiHi → O=Cj interaction supported by NH exchange protection and spatial proximity in 80% of the structures in an ensemble calculated without H-bond constraints took the form: r(HiOj) = 1.71–2.01 and r(NiOj) = 2.78–3.03 Å or r(HiOj) = 1.71–2.21 and r(NiOj) = 2.65–3.25 Å, depending on whether a large or modest the NH exchange protection factor was observed. For stereospecific assignments of Cβ methylenes, anti/gauche assignments were made from the TOCSY spectra (Driscoll et al., 1989) recorded in d2-HFIP/D2O and confirmed by the αβ/αβ′ NOE ratios observed in the short mixing time NOESY. The α/ββ′ anti/gauche relationships were expressed as DCs (2.8–3.15 and 2.2–2.7 Å, respectively) and both possible assignments were examined for these and all βiNi+1/βiNi/βiαi−3 constraints. If one assignment produced lower ENOE values for a large majority of the structures in the ensemble, the stereospecific version of the constraints were used in the final stage of the refinement.
In the case of the hAM refinements, both ensembles had acceptable values of Eimpr (1.0 kcal/residue) and EvdW (−4.01 kcal/residue for the ‘aggressive’ versus −4.14 kcal/res for the ‘conservative’ ensemble). Even in the aggressive application of the constraints, the LBO constraints accounted for only 1.7% of the NOE penalty. The ENOE term for the aggressive ensemble was 51.1 ± 6.4 kcal/mol which was reduced to 9.1 ± 2.1 kcal/mol when the ‘aggressive’ ensemble was evaluated against the ‘loose’ constraint set. The latter was significantly lower than the ENOE for the ‘conservative’ ensemble evaluated similarly (16.1 ± 3.7 kcal/mol). The structural conclusions have been confirmed by using the same constraints in our current CNS-based structure refinement protocol, but we are presenting the prior XPLOR ensembles since they are supported by statistics of a larger number of runs.
Evaluation of convergence and structural deviations in NMR ensembles
The standard output from XPLOR was used to select the best-fitting low-energy structures from the complete ensembles. Eimpr and EvdW provide the best measures of strain and conflict between NOEs and low-energy conformations, and ENOE provided the initial measure of fit to the DCs. Typically, 60–80% of the structures had values of all three energy terms that were within 15 kcal/mol of the lowest values observed in that ensemble for each energy term; these constitute the ‘accepted structures’. Backbone RMSD measures of convergence are pairwise over the consensus conformation or ensemble; the carbonyl oxygen is included in the definition of backbone for RMSD measures.
Results
Circular dichroism
CD provides a well-proven method for determining percent helicity and has been used to estimate the ‘α/β/coil’ composition of proteins (Yang et al., 1986; Johnson, 1991) for many years. In a preliminary account of a portion of the present study, we reported that the aggregation/gelling process of hAM was more readily, and reproducibly, examined in aqueous medium containing 6–9 vol.% HFIP (Cort et al., 1994). Two distinct ‘β-sheet’ CD spectral signals were routinely observed during these experiments; neither was observed with prAM; rather, prAM displays a partially formed helix with maximal helicity at 25–35°C which unfold upon both warming and cooling. This was the discovery system for the cold-denaturation of peptide structures in HFIP/H2O media (Andersen et al., 1996a; Andersen et al., 1999); both the thermal melting and cold-induced unfolding transitions of rAM and prAM are concentration-independent indicating that the helical state involved is monomeric. CD spectral comparisons for hAM and prAM shown in Fig. S1 (Supplementary data are available at PEDS online). The β-structuring distinction between hAM and prAM was also observed in octyl-glucoside micelles; hAM yields a classic β CD signature, whereas the CD of prAM is random coil with a shoulder indicative of a minor α-helical contribution. In the absence of micelles and fluoroalcohol, hAM displayed a spectrum very similar to that of prAM in octyl-glucoside micelles.
Cosolvent-induced helix formation
CD measures of helicity, [θ]221 and the two diagnostic extrema ratios increase on HFIP addition leveling off at ca. 16% HFIP at ≤100 µM peptide concentrations for all of the amylin examined. The effects of other cosolvents shown in Fig. S4 (Supplementary data are available at PEDS online); HFIP was the most effective helix-inducing cosolvent. However, we found that hAM (and [Pro25]-hAM) displayed better solubility in 25–35% HFIP. Since we planned to use NMR to locate the residues that are responsible for the differences in net helicity, detailed CD comparisons were made in the 25% HFIP medium. The spectral parameters observed are given in Table I. PrAM-free acid was included in the survey, C-terminal amidation has no effect on the helicity of amylins. The CD spectrum observed for hAM in 25% HFIP is clearly that of a substantially helical peptide. By all of the usual criteria, both hAM and [Pro25]-hAM are more helical than rAM and prAM.
Table I.
CD comparison of amylin peptides in 25 vol.% aqueous HFIP
| Peptide | Residue molar ellipticities (and their temperature gradients) |
|||
|---|---|---|---|---|
| n→π* (220 nm)a | π→πx*b | π→πy* (191 nm) | R1/R2c | |
| hAM | −23 150 (+142) | −24 720 (+111) | +48 300 (−276) | −1.883/+0.893 |
| −1.95/+0.94 (0°C) | ||||
| [Pro25]-prAM | −21 900 (+130) | −21 150 (+85) | +40 800 (−268) | −1.75/+0.96 |
| −18 200 (+97) | −21 600 (+69) | +33 500 (−190) | −1.44/+0.795 | |
| −1.55/+0.844 (0°C) | ||||
| free-acid rAM | −17 000 (+86) | −20 300 (+67) | +31 400 (−196) | −1.42/+0.795 |
| −18 800 (+115) | −23 200 (+100) | +33 100 (−225) | −1.33/+0.760 | |
The residue ellipticity at each extreme is expressed as the best fit (over T = 0–35°C) to the following linear relationship: [θ]λ(T°C) = [θ]λ(0°C) + Δ[θ]λ(T°C).
aThis band is shifted to 221 nm for hAM and [Pro25]-hAM, an additional indication of increased helicity.
bThe location of the minimum (207 nm) indicates the reduced helicity of the analogs with [Pro25,28,29]-substitution. For hAM and [Pro25]-hAM, it appears at 208 nm.
cThe ellipticity ratios, R1 = [θ]max or 191/[θ]min and R2 = [θ]221/[θ]min) (Bruch et al., 1991), reported are the averages of triplicate (or greater) determinations at 25°C unless otherwise indicated. In aqueous HFIP (and 40+% TFE), the n→π* band appears closer to 220 nm (rather than 222 nm, the wavelength at which helicity is usually quantitated), this reflects a blue shift associated with solvent polarity which is more pronounced for the n→π* transition than for the π→π* couplet (Andersen and Palmer, 1994; Andersen et al., 1996a,b). These solvent-induced shifts of the helix CD signature are also observed for helical proteins, e.g. the extrema for myoglobin (222.0, 209.2 and 192.8 nm in aqueous buffer) shift to 220.4, 207.8 and 191.8 nm in 25% HFIP and 40% TFE (Cort and Andersen, 1997). For the SDS-micelle-associated state of hAM, which displays band-intensity ratios that indicate a somewhat lower fractional helicity (R1 = −1.63 and R2 = 0.84 at 25°C), these bands appear at the normal wavelengths (data not shown). The solvent-induced shifts are taken into account in selecting [θ]λ values for the calculation of R1 and R2.
Preliminary NMR spectroscopic comparisons
Owing to the poor aqueous solubility of hAM, our initial comparisons with a [Pro25,28,29] analog (rAM) were carried out using an 88/11/1 vol/vol mixture of d6-DMSO/CD3CO2D/H2O, a presumably denaturing medium. We were able to derive complete assignments; the chemical shifts will be provided upon request; the Hα and HN-CSD histograms are shown in Fig. 2.
Fig. 2.
CSD comparisons for fully protonated hAM and rAM in an 88/11/1 volume ratio of d6-DMSO/CD3CO2D /H2O, large structuring shifts are only observed for the NHs in the disulfide loop (lower panel). Human versus rAM CSD differences >0.05 ppm are observed only at the sites of residue substitution and a few other sites within 20–30 residue span.
The high degree of similarity between rAM and hAM over all regions except the immediate vicinity of the Pro substitutions can be seen in this comparison of the backbone hydrogen chemical shifts. On the basis of Fig. 2, the only substantially structured region is the disulfide loop that displays shift deviations from coil values as large as 0.6 ppm. The amyloidogenic species, hAM, displays only a few weakly positive Hα shift deviations in the amyloidogenic span. A string of negative Hα values is observed from Thr9 to Val17; assuming that the secondary structure shift trends observed in water (Wishart et al., 1992) apply in DMSO, this is evidence for a partially helical region.
In a similar study of prAM acetate, the initial solution included only stoichiometric quantities of CH3CO2H, insufficient to protonate the N-terminus, and changes in the spectrum were monitored as the DMSO/H2O solution was progressively acidified—first with CH3CO2H (pKa = 12.3 in DMSO), then DCO2D and finally TFA (pKa = 3.5). Not until acids more acidic than acetic were added, did the NHs for the loop region (particularly those of Thr4,6) sharpen and shift toward the characteristic δ values observed in previous samples. The shifts were fully reproduced when TFA was added. Protonation of the N-terminus (estimated pKa ≈ 8.5 in DMSO) apparently leads to either a significant conformational change or has large Coulombic contributions to the chemical shifts of sequence remote NH signals. Although further studies are required to define the changes occurring, our analysis suggests that the two-protonation states are not in rapid equilibrium. In retrospect, this was the first evidence suggesting the existence of multiple conformations of the loop which are in slow exchange.
NMR parameter comparisons in aqueous HFIP
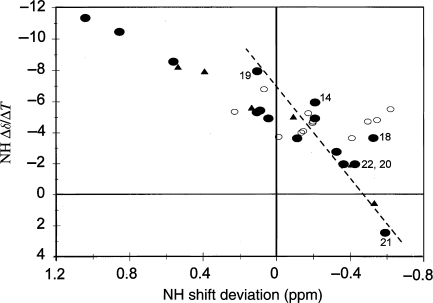
As indicated previously, solubility considerations led us to select 25–35 vol.% HFIP as the medium for a detailed NMR comparison of the amylins. The essentially complete set of αiNi+3 and αiβi+3 connectivities for i = 5→18, which appeared for all four amylins, greatly facilitated the assignment process and served to indicate a common helical segment. Several annotated spectra of prAM in the HFIP-containing medium, together with the complete assignment for prAM-terminal amide and prAM-free acid (Table SI, Supplementary data are available at PEDS online), and the NH shift temperature gradients (ΔδNH/ΔT, derived from a series of 15N-HMQC spectra spanning the temperature range 268–315 K) appear in the Supplementary data available at PEDS online. Figure 3 shows the correlation of ΔδNH/ΔT values with NH-CSDs observed for prAM. The excellent correlation coefficient suggests that the helical structure melts with a significant degree of cooperativity (Andersen et al., 1997).
Fig. 3.
Correlation of NH shift temperature gradients (Δδ/ΔT, in ppb/°C) and the CSDs observed at 285 K for prAM in 35% HFIP. Different symbols are used for the discreet sections of the structure: the helix (filled circles, for residues 8–22), the disulfide loop (filled triangles, residues 2–7) and the remaining residues (open circles, which do not display a correlation). The two most highly exchange protected NHs are the points with the largest negative Δδ/ΔT values. The steeper slope for the S19–N22 points (dashed line) indicates enhanced thermal fraying.
The series of 15N-HMQC spectra at 268–315 K also provided evidence of different time scales of conformational averaging in different portions of the sequence (Fig. 4). The 15N-1H correlation peaks for residues 3–5 broaden significantly on cooling and disappear from the HMQC spectra recorded by 275 K. The residues 6–9 peaks disappear into the noise at 268 K. In contrast, the correlations for residues 30–37 are relatively more intense at the lower temperatures. Segmental motion more rapid than molecular tumbling is suggested for the C-terminal segment. The broadening of the peaks for the loop region can be attributed to the interconversion of loop conformers with >50 µs lifetimes at the low temperature limit and divergent chemical shifts (Olsen et al., 2005).
Fig. 4.
Gradient enhanced 15N-HMQC spectra recorded for U-15N-prAM-free acid in 35% HFIP at 275 (left panel) and 300 K (right panel) appear with the correlations labeled by residue number. The extrapolated locations of the correlations for residues 3–5 and 7, all of which have broadened into the base plane at the colder temperature, are shown as parenthetic residue numbers (#); also, the peaks for residues 6, 8 and 9 are barely visible at the lower temperature.
The Hα-CSD histograms for the four amylins shown in Fig. 5. The histograms are nearly identical from the N-terminus to Ser20 and from Thr30 to the C-terminus. The one exception to this is rAM, which has an H18R mutation. The sequence histogram for rAM differs from the others at Asn14, Phe15 and Asn21 reflecting the expected periodicity of an α helix about the site of the mutation. With the exception of Leu12, there is a continuous string of upfield shifted Hα resonances from Ala5 to Ser20 for all four amylins, indicating a common helical span. A second helix (Phe23 to Ser29) is suggested for hAM and [Pro25]-hAM. The latter feature provides a rationale for both the greater net helicity of these species and for the facile melting of this ‘additional’ helicity: the shorter helix would be expected to end-fray and melt more readily. The pattern of small alternating upfield and downfield shifts in the C-terminal segment does not correspond to any known regular secondary structural feature, but could be rationalized by a set of conformers with β-turns at different loci in this span.
Fig. 5.
The CSDs of hAM, [Pro25]-hAM, prAM and rAM in the 25% HFIP medium (pH 3.7 ± 0.3, T = 25 ± 3°C) are displayed in that order at each position along the sequence. Upfield shifts appear as negative values; a Hα-CSD value of −0.40 is generally equated with a fully populated helical conformation (Wishart and Sykes, 1994). The random coil values and Xaa-Pro correction employed for these correlations were published in 1997 (Andersen et al., 1997).
The lack of an upfield Hα shift at Leu12 is attributed to a ring current effect due to the Phe15 sidechain (vide infra). Plots of inter/intra-Hα–HN NOE ratios (αiNi+1/αiNi, Lee et al., 1994) for hAM (Fig. S6, Supplementary data are available at PEDS online), and prAM do not show any interruption in the helical ψ values through the 6–20 residue span. As a final proof of this point, the 13C shifts for the three leucines were obtained for a sample of prAM prepared using U-13C-Leu. Leu12,16 were clearly helical, δ′C/δCα = 178.4/57.6 and 180.7/58.1 ppm, respectively. The corresponding shifts, 173.8 and 51.7 ppm, for Leu27 confirmed an extended, more β-like conformation in this residue span.
An H/D exchange experiment (Fig. S7, Supplementary data are available at PEDS online) performed on hAM serves to confirm both of the helical segments suggested by the CSD histograms. The NHs of residues 16, 17 and 13 were the slowest exchanging (and also are shifted far downfield) indicating particularly strong H-bonds—see the HN-CSD histograms for hAM and prAM, Fig. S5 (Supplementary data are available at PEDS online). Exchange at the NHs of residues 12, 14, 15, 26 and 27 was also significantly retarded. Protection factors greater than 50 were observed for the Ala13–Leu16 span. Decreased protection was observed at His18 and in the AILS span. However, both the NH exchange protection and CSD data imply that AILS sequence is in an α helix rather than in some nascent helical state (Dyson and Wright, 1991).
The assignment of prAM in aqueous buffer
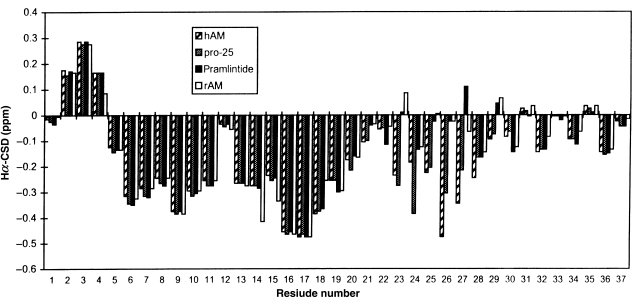
We selected prAM for NMR studies in aqueous buffer (pH 4). The ROESY spectrum (Fig. S3, Supplementary data are available at PEDS online) was the key to the assignment. The rich web of i/i + 3 connectivities for i = 8–18 observed in aqueous HFIP are either absent or present at barely detectable levels in the ROESY spectrum recorded in the absence of added HFIP. Another significant difference is that the correlation of ΔδNH/ΔT values with NH-CSDs observed in aqueous HFIP (Fig. 3) is entirely absent in aqueous buffer alone (Fig. S8, Supplementary data are available at PEDS online). The lack of correlation implies the absence of any cooperatively melting secondary structure feature in the absence of the fluoroalcohol cosolvent (Andersen et al., 1997). A comparison of the Hα shift deviation histograms of amylin in the DMSO, aqueous buffer and aqueous HFIP media shown in Fig. 6. The complete assignment of prAM in aqueous buffer also given in Table SII (Supplementary data are available at PEDS online). The Hα-CSD histogram for aqueous buffer (and in the DMSO medium) provides a rationale for the residual helicity which was observed by CD even in the presence of high concentrations of denaturant. The disulfide closed loop retains a very similar conformational preference throughout. This is even more apparent in the HN-CSD histograms (see Fig. 2B, for the DMSO data). Parallel studies of salmon calcitonin and CGRP (data not shown) revealed, as comparable HN-CSDs in the disulfide loop, a similar N-terminal nucleation of a helix.
Fig. 6.
A comparison of the alpha methine CSDs of amylins in three media. At each residue, the CSD values are shown in the order (left to right): aqueous buffer, DMSO and 35% HFIP. The data for the aqueous buffer and 35% HFIP states are that derived from the studies of prAM. The data for DMSO are a composite of hAM (residues 1–20 and 31–37) and rAM (residues 21–30).
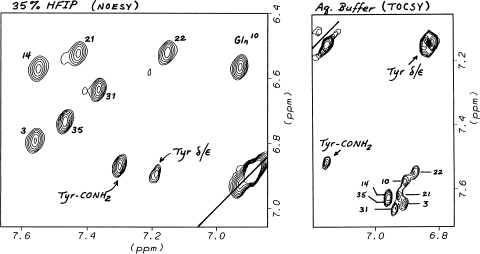
The comparison of prAM chemical shifts in water, with and with the addition of HFIP, revealed sidechain shift changes that that could not be rationalized as the expected result of increased helicity. Of these, the increased shift dispersion of –CONH2 resonances (Fig. 7) was notable; in aqueous buffer, both the E and Z-NHs of the seven carboxamide units are all within 0.06 ppm of their coil reference values; in the 25+% HFIP, the signals due to the E-NHs (δ2 & ε2) are spread over a 0.64 ppm range, with the Q10ε2, N14δ2, N21δ2 and N22δ2 moving upfield by 0.63, 0.06, 0.18 and 0.39 ppm, respectively. hAM displays similar CH2CONH2 shifts, but N22δ2 appears 0.47 ppm upfield with the β-CH2 shifted downfield 0.18 ppm versus prAM. Two truncated hAM analogs, hAM(1–22)G-NH2 (cyclo- KCNTATCATQRLANFLVHSSNNG-NH2) and a hAM-loop model (hAM-LM, GR-Aib-ANFLVHSSNNFGAK), were examined to determine which neighboring sequence features are the requisites for these shifts. The helix-forming hAM(1–22)GY-NH2 analog nearly reproduced, upon HFIP addition, the Q10ε2 shift, but N14δ2 and N21δ2 were still at the shift observed in the absence of HFIP. The hAM-LM construct appears to be a better structural mimic; the inclusion of an Aib residue results in measurable helicity (including H-bonding by the Leu and Val which results in deshielding) upon HFIP addition, and upfield shifts are observed for the asparagine Hδ2 sites upon HFIP addition. These shifts (0.27, 0.11 and 0.33 ppm, respectively, for N14, 21 and 22) reproduced the relative shifts for N21 and N22 in prAM, but N14 was further upfield in the loop model.
Fig. 7.
A comparison of the CONH2 shifts of prAM with and without HFIP addition. Opposite sides of the diagonal are shown in the two segments. The Asn/Gln side-chain sites are labeled by residue number. The terminal amide unit displays upfield shifts 0.24 ± 0.03 ppm for both NHs upon HFIP addition.
Structural details of the helical state in 25–35% HFIP, NOE-based refinement
Pramlintide
The greater solubility of prAM and the availability of 15N NOESY–HMQC data resulted in a rich web of NOE distances. In excess of 450 distances were derived, largely from the 2D (τm = 90 ms, T = 275 K) and 3D NOESY (τm = 110 ms, T = 295 K) spectra recorded at 750 MHz. The final refinement protocol employed 404 NOE distances (of which 75 were LBO constraints) and helical (to the i − 4 C=O) H-bond constraints for HN of residues 12–17 (which were demonstrably exchange protected in hAM). Significant constraint violations were found only in the disulfide loop and at the C-terminus of the residue 5–19 (22) helix. The decreasing intensity of the αiβi+3 peaks at the C-terminus of the helix is shown in Fig. 8.
Fig. 8.
An annotated segment of the NOESY spectrum (at 17.6 T, τm = 90 ms) of prAM in 35% HFIP showing the intense αiβi+3 NOEs in the central portion of the helix (filled squares), open triangle is the 17β/14α peak partially obscured by the acetate methyl streak, and the diminished intensity αiβi+3 peaks at the frayed C-terminus (asterisks). The weak 32α/35β peak is also labeled. The Hα lines are labeled by residue number.
We attribute this to helix fraying and to the contributions of alternate C-capping conformations. A quantitative analysis, using the intensities of the intra-residue αiNi NOEs as a calibration confirmed this: the sequence dependence of the ratios of diagnostic NOEs appear below. The values expected for the well-converged 7–14 residue span (ϕ = −60 ± 8°, ψ = −43 ± 11° over residues 7–17) were also calculated based on the distances in the NMR structure ensemble employing default order parameters with a 3.2 ns correlation time.(Andersen et al., 1990; Lai et al., 1993).
| Relative NOE intensities | αiNi | αiNi+1 | αiNi+3 | αiβi+3 |
|---|---|---|---|---|
| For i = 7–14 | 1.00 | 0.35 ± 0.11 | 0.42 ± 0.12 | 0.83 ± 0.26 |
| Calculated | 1.00 | 0.32 (±0.04) | 0.41 (±0.12) | 1.03 (±0.44) |
| For i = 18 | 1.00 | 0.69 | 0.08 | 0.24 |
| For i = 19 | 1.00 | 0.80 | 0.04 | 0.13 |
| For i = 20 | 1.00 | 1.08 | <0.04 | <0.10 |
Over the well-converged portion of the helix, the experimental NOE ratios are in excellent quantitative agreement with those calculated from the NMR structure ensemble. The only disagreement is a somewhat diminished experimental αiβi+3 intensity, presumably reflecting sidechain fluxionality that is greater than that of the backbone. The loss of i/i + 3 NOE intensities at the C-terminus is coupled with an increase in the αiNi+1 NOE intensities. For a statistical coil state, the αiNi+1/αiNi ratio would be 1.7, whereas this value is never reached, the increase observed does imply significant populations of more extended structures in this span.
A plot of ϕ/ψ values along the sequence revealed two families of structures—one in which the helix ends abruptly with Ser20 taking on backbone torsion values of ϕ20 = −112 ± 25 and ψ20 = +179 ± 15° and in the other, the ϕ/ψ values from Ser19 to Asn22 deviate more gradually from the helical norms. In order to obtain unstrained structures of both conformational families, the final stage of constrained-MD refinement was repeated with the following modification: for the long-helix family, the αiNi+1 distances were accorded lower weight, and the low bounds for the i/i + 3 constraints for i = 17–20 were relaxed; for the ‘C-capped helix’ refinement, the vicinal constraints retained high weighting and the original i/i + 3 constraints for i = 17–20 were moved to the lowest weight category. The ϕ/ψ plots for the two consensus ensembles that resulted are shown in Fig. 9, the violation and structure statistics for the complete ensembles appear in the Supplementary data available at PEDS online. Both ensembles displayed convergence of the residue 5–18 domain: the backbone RMSDs were, for the long frayed helix, 0.84 ± 0.28 Å over residues 5–21; for the C-capped conformation, 0.63 ± 0.22 Å over residues 5–18. The C-capped helix ensemble displays marginally better fits to the complete set of NOE intensities; the difference, however, is not statistically significant.
Fig. 9.
The ϕ (upper panel) and ψ (lower panel) versus residue number plots for two NMR structure ensembles derived for prAM in 35% HFIP. The blue traces correspond to 13 representative structures from the ‘C-capped helix’ ensemble. The red traces are 14 representative structures from the ‘long helix’ ensemble.
Human amylin
The shift assignments for hAM in 35% HFIP given in Table SIII (Supplementary data are available at PEDS online). The majority of the NOE DCs employed for this structure elucidation were derived from a short mixing time (110 ms) NOESY experiment at 750 MHz performed under non-aggregating conditions (0.7 mM peptide). A few longer distances were derived from NOEs that were only evident in 250 ms mixing time NOESYs recorded at 500 Mz. Only 2D data were available for hAM; as a result, a number of observed NOESY peaks corresponded to the sum of two or more interproton cross-relaxation processes. Since all of the backbone chemical shifts and NOE ratios are essentially identical for hAM, prAM and a variety of abbreviated analogs (data not shown) over the common residues 2–19, we assume that clarifications of the partitioning of these overlapped NOEs in the 15N-dispersed NOESY spectra of prAM and in the analogs apply to hAM as well. One example of the use of an analog in this process is given here. In the hAM(1–20)GYNH2 analog, with residues 21–36 replaced by a single glycine, Hα and Hβ of Thr4 are also shift coincident in both aqueous buffer and in 20+% aqueous HFIP. This is an unfortunate overlap since Thr4-Hα/Hβ displays significant NOEs to a number of sites including the NHs of Ala5 and Thr6. This shift coincidence is broken in 15% aqueous TFE and also in analogs with an A8V mutation. Studies of these systems revealed a constant ratio of NOEs with, for example, a very dominant Hβ4/HN6 and a major Hα4/HN5 contribution. The NOE ratios observed in the analogs were assumed to apply to both prAM and hAM.
The NMR structure ensemble for hAM in 35% HFIP is based on 271 DCs (of which 24 were LBO constraints) and five H-bond constraints (from HN of residues 12–17). Throughout the serial refinement, we observed two conformers of the disulfide-linked loop—the ϕ values at the Thr4, falling into two tight clusters. Neither loop conformer provided a satisfactory fit to all of the NOE constraints; however, assuming a mixture of the two conformers improved the fit. We conclude that the observed NOEs represented short distances in two distinct conformers. The chemical shifts of the NHs of residues 3–5 would be very different in the two loop conformers (Fig. S9, Supplementary data are available at PEDS online) that arose from the refinement. Thus, these two conformers would, if they are relatively long-lived at the lower temperatures, explain the disappearance of these peaks in the low-temperature panel in Fig. 4.
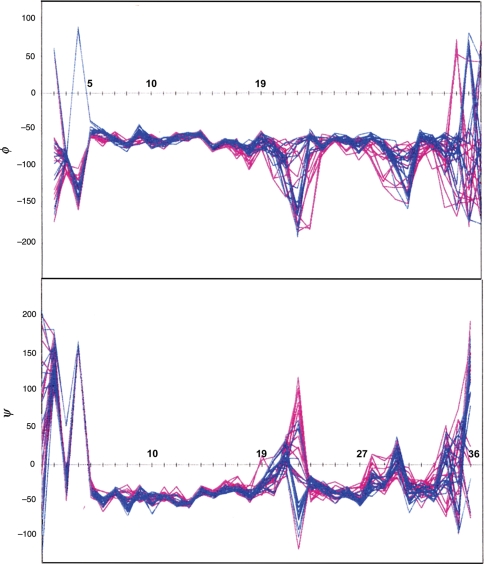
In order to assess the potential effects of DC precision, the three-stage refinement was repeated twice with the final constraints on a fresh set of random starting structures—in the one case retaining the original constraint precision, in the other converting to ‘looser’ constraints (see Methods and Materials). A sequence plot of the ϕ/ψ values of the 15 structures of the ‘aggressive ensemble’ which display the lowest NOE penalty appear as blue lines in Fig. 10; the two minor loop conformers met the ENOE criterion. The corresponding plots for the ‘conservative ensemble’ obtained using ‘loose’ constraints appear as red lines. The ϕ/ψ values for the complete ‘aggressive ensemble’ are tightly defined from residue 5–18 (pairwise backbone RMSD = 0.57 ± 0.22 Å) and 23–29 (0.38 ± 0.15 Å), the two helical domains previously revealed by the sequence CSD histograms. Considerable variation is seen at residues 20–22, suggesting a flexible hinge region, and the extreme C-terminus is either under-determined or conformationally averaged. The XPLOR statistics (average violations, ENOE, EvdW etc.) for both ensembles, and an evaluation of the ‘aggressive ensemble’ against the loose constraints, given in Table SIII (Supplementary data available at PEDS online). The observation that the ‘aggressive ensemble’ displayed a better fit to the loose constraints than the ensemble that was generated with those constraints (without a significant increase in the Eimpr term) indicates that the tighter constraints did not generate any unusual features as a result of assigning an inappropriately high prevision to NOE distances; rather, the slightly larger ENOE terms just served to define the structure more accurately.
Fig. 10.
The ϕ (upper panel) and ψ (lower panel) versus residue number plots for two NMR structure ensembles derived for hAM in 35% HFIP. The blue traces correspond to 15 structures from the ‘aggressive’ ensemble with the lowest ENOE values. The red traces are 15 structures similarly selected from the ‘conservative’ ensemble.
Discussion
All four of the amylins examined display some local helicity in aqueous buffer and in highly denaturing media. NMR studies in DMSO and aqueous buffer indicate that the conformational preference of the disulfide loop locks in a single turn of helix, Ala5–Cys7, and that this preference propagates, in part, into the Ala8–Val17 segment in these media. Partial retention of helicity in DMSO has also been noted for another peptide hormone (Hudson and Andersen, 2004). The absence of measurable i/i + 3 NOEs in this span suggests that this represents a nascent helix (Dyson and Wright, 1991), rather than small populations of a cooperatively formed helices spanning a significant segment of the Ala8–Val17 span. This conclusion also applies to the non-aggregatory monomeric state of prAM and rAM in aqueous media and presumably to aqueous hAM prior to the onset of the amyloidogenesis process. Upon addition of HFIP, CSDs, inter/intra-residue αN NOE ratios and the intensity of the i/i + 3 NOEs all indicate a fully formed helix that spans from Ala5 to Arg/His18 and extends in a frayed form to Ser20 or slightly further. In hAM and [Pro25]-hAM, a second helix is found over the Phe23–Ser29 span.
The extent of helical structuring for hAM and prAM found by NMR are fully consistent with the CD data at 3–600 µM peptide concentrations in 25–35 vol.% HFIP reported herein, −[θ]220: 23 150° (hAM) and 18 500° (rAM and prAM). But there are some disagreements with literature reports for other helical states. A recent report for hAM in SDS micelles (Patil et al., 2009) illustrates a CD with a −[θ]220 value in excess of 32 000°, even though the NMR structure reported for this medium is very similar (vide infra) to the one derived herein. (Data recorded in this lab suggest that this may be a calibration or concentration error; we observed that CD helicity measures are slightly smaller for the SDS micelle state than those reported for hAM in Table I.) Knight et al. (2006) reported fully bound −[θ]220 values hAM (17 000°) and rAM (13 000°) associated with 1,2-dioleoyl-sn-glycero-3-phosphocholine liposomes that are not as helical as the data reported herein, but still reflect additional helicity for hAM. In the absence of lipids or fluoroalcohols, −[θ]220 values on the order of 4000° (as observed in the present study), suggesting percent helicities <15%, have been reported for both hAM and rAM by numerous researchers (Goldsbury et al., 2000; Knight et al., 2006; Williamson and Miranker, 2007; Patil et al., 2009). Yonemoto et al. (2008) also report [θ]220 = −4000° for monomeric 50 µM hAM-free acid in 100 mM aqueous NaCl (pH 6), even though the Hα-CSDs reported for a 200 µM hAM concentration in the same medium (Fig. 1) were the most helical reported to date. The authors did note that although they observed (with the exception of T4/A5 connectivity, also absent in our data) NiNi±1 NOEs throughout the sequence, no i to i + 3 NOEs were observed. This prompted us to examine the NMR data of Yonemoto et al., as they appear in the supporting materials. The 2D spectra did not display the shift dispersion we observed for the solution-state helix, notably the HN shifts over the 5–22 residue span resembled prAM in water rather than 35% HFIP. Figure 11 shows a comparison of the CSDs based on the data of Yonemoto et al. and our data for 35% HFIP using the same coil values and sequence corrections.
Fig. 11.
A Hα-CSD comparison, hAM in water with and without added HFIP, using the same coil values and sequence corrections. The data for aqueous hAM are taken from the supporting material from Yonemoto et al. (2008).
In this direct comparison, hAM in water is seen to be significantly less helical than the monomeric helical state that forms upon HFIP addition. It appears that the coil values and sequence corrections, based on shifts observed in a (GGXGG)-context (Schwarzinger et al., 2000, 2001), tend to produce more negative CSDs than our method based on KAXAA and GAXAA sequence contexts. We have previously noted (Eidenschink et al., 2009) problems defining secondary structural features using CSD determinations based on the data of Schwarzinger et al.
The NOE-derived structure ensemble (Fig. 10) generated for hAM confirmed both helices but also produced structures (Fig. S10, Supplementary data are available at PEDS online) that are inconsistent with the structural conclusions derived from the Hα-CSD and NOE ratio sequence histograms. The NOE ensemble contains structures with more extensive helical domains, including helices that run through the Ser19 to Asn22 patch and others that extend the second helix nearly to the C-terminus; we view these as artifacts of the NOE distance-based structure refinement. Throughout the Thr30–Thr36 span, the NiNi+1 NOEs are much larger than that would be expected for a statistical coil peptide state. There is more than a scattering of αiNi+2 (i = 29, 30, 33, 34), αiNi+3 (i = 29, 31) and even some very small αiβi+3 NOEs (i = 29, 31, 32) in this span. The CSD sequence histogram over this region shows little evidence of helicity, not even a string of small negative deviations, as was observed for the nascent helix present over the Ala8–Val17 span in water and the DMSO media. The NOE-ratio plot (Fig. S6, Supplementary data are available at PEDS online) suggests that helicity is completely interrupted at residues 21 and 22, and stops again at residue 31. In the case of prAM-free acid, which shows a similar CSD histogram in these spans, the temperature dependence of the 15N-HMQC intensities (Fig. 4) confirms that most of the C-terminal segment is undergoing rapid segmental motion. The ϕ/ψ plots (Figs 9 and 10) suggest, as the only possible structural feature in this span, a type III turn locus at 32/33. Conformer interconversion at this site could rationalize the disappearance of the Val32 NH in the low-temperature panel of Fig. 4. As a result, we view hAM as a peptide that lacks tertiary structure even under the most structure-favoring conditions. A selection of structures from the aggressive hAM ensemble that illustrate alternative bent conformations appears in Fig. 12.
Fig. 12.
Alternative bent conformations of hAM.
A quite similar NMR structure for hAM has just appeared for the SDS micelle-associated state (Patil et al., 2009). The authors present the structure as two helices, residues 6–17 and 18–27 with an inter-helical angle of 30 ± 18°; although this differs somewhat from our model (Fig. 12) for the solution state in 35% HFIP, we and Patil et al. both indicate that the flexibility and helix break are located in the 18–22 residue span. On the basis of relaxation studies, the 5–17 residue segment is imbedded in the SDS micelle, and the authors suggest that the interfacial location of the 20–29 residue segment may mediate the transition to a toxic structure.
The results presented herein should serve as yet another caution against using NOE distances for structure elucidation of peptides that display conformational averaging. In our view, the CSD histograms and a careful examination of backbone NOE ratios and NH protection factors provides a more dependable diagnosis of the extent of secondary structure formation in such peptides. In the case of [Pro25]-hAM, the CSD histogram indicates that proline can be incorporated at position N3 of a 6 residue helix in aqueous fluoroalcohol media. However, the FGAILS (or FGPILS) helix is, even under these conditions (25–35 vol.% HFIP), incompletely populated. Turning to the longer α-helical domain (Ala5–His18) and its spatial relationship to the C-terminal part of the structure and the amyloidogenic patch, CSD histograms also appear to provide insights into which backbone NHs have the strongest H-bonding and loci of unfolding. The NHs of Ala13, Leu16 and Val17, which display the largest protection factors, are far downfield. In the absence of added fluoroalcohol, Ala8 is a locus of disorder. The NOE-derived ensemble for the state present in 25–35% HFIP fails to display disorder at this point in the sequence. However, the absence of NH exchange protection at Thr9 of hAM suggests that this conclusion applies to the more helical state as well. The Hα-CSD histogram for the monomeric state in aqueous HFIP provides a clearer picture of the increasing extent of end-fraying that occurs from His18 onward. The implied residue-specific fractional helicities predict the relative intensities of the αiβi+3 NOEs at i ≥ 17 to within experimental error.
The NOE-derived structure ensembles for the ‘state’ favored in aqueous HFIP do, however, provide additional insights for the loop and first helical domain. These will be illustrated with prAM, the system which had the denser web of NOEs. With regard to the 1–22 residue span, the conclusions should also apply to hAM. The decreasing intensity of the αiβi+3 NOEs at i ≥ 17 (Fig. 8) and the increasing intensities of the αiNi+1 NOEs through this span indicate that the helix ends abruptly at Ser20 in the dominant conformer, but extends further (to as far as Asn22) in minor conformers. Figure 13 shows the polar and hydrophobic faces of a representative of both the C-capped (left-hand views) and extended helix (right-hand views) ensembles.
Fig. 13.
The hydrophobic (upper view) and polar (lower view) faces of a representative of the ‘C-capped helix’ ensemble (left panel) and the ‘extended helix’ ensemble (right panel). In each case, all atoms of residues 8–22 are displayed with the C-terminus of the helix shown to the right. The Leu12 α-methine is shown as dark lavender CPK surface that can be located along the long axis of the phenyl ring (Phe15) is in a ring current deshielded location in >90% of the structures in each ensemble.
The polar face of the helix displays interactions between the Asn14–His18–Asn21 sidechains. The chemical shifts of the CONH2 units of hAM are highly state-dependent, in SDS micelles (Patil et al., 2009), the Q10/N14/N21/N22 E-NHs all appear at 7.40 ± 0.04 ppm rather than being spread over a 0.64 ppm range as they are for prAM in 35% HFIP. In both forms, the helix is amphipathic with a well-packed continuous patch of fatty sidechains including those of Leu12, Phe15, Leu16, Val17 and extending to include the methyl of Thr9 in many of the structures in the ensembles. The amphipathic character of the helix rationalizes the downfield shifts and greater protection at HN of Ala13, Leu16 and Val17. These downfield shifts are not observed for hAM in the SDS micelle-associated state, consistent with the lipid burial of this portion of the helix as reported by Patil et al.
Do these structural conclusions have biological relevance? Here, there are a number of considerations: the hormone-like activities of hAM, the direct interaction of hAM and insulin (Wei et al., 2009), hAM amyloidogenicity [leading to toxicity presumably related to an intermediate (Butler, 2004; Haataja et al., 2008) along the amyloid fibril formation pathway] and the cytotoxicity also observed with amylin fragments (Brender et al., 2008a, 2008b) that appears to be related to membrane disruption. It has been suggested that the amphipathic helices of amylins, calcitonins and CGRPs are an important recognition element for these signaling peptides (Boulanger et al., 1996; Deftos, 1997; Carpenter et al., 2001; Howitt et al., 2003). In the case of hAM, the present study reveals a helical preference over the Ala5–Val17 span that is present to at least some extent, in all media examined. It would appear prudent to include this element in the pharmacophore model. Such a pharmacophore model has provided a basis for rationalizing the bioactivity changes associated with residue mutations (unpublished data), and alternative helix nucleation and stabilization strategies can restore activity to mutants showing diminished helicity. The amphipathic helix is also the feature that is associated with membrane interactions. The second helix observed for hAM (Phe23–Ser29), which is observed in aqueous HFIP and in the SDS micelle-associated state (Patil et al., 2009), is unlikely to figure in the bioactive state, but local conformation changes in this span could be a significant determinant of amyloidogenicity.
Biological roles for the flexible patch observed from Ser19 to Asn22 in the amylins can also be proposed. Modifications at both extreme termini of hAM, including deamidation at Tyr37, abolish activity. Any pharmacophore model which places the termini in proximity requires a unit that allows chain reversal. Calcitonin/amylin hybrids, which have a six-residue deletion in this span, retain significant cross-reactivity at amylin receptors (Gebre-Medhin et al., 2000; Poyner et al., 2002). In this regard, it should be noted that an NMR study of hAM(20–29) in SDS micelles suggested a type I β turn at the F23–G24 locus (Mascioni et al., 2003) and that through-space interactions between N21 and S28 have been identified in fibrils incorporating 13C-labeled hAM(20–29) fragments (Madine et al., 2008). The S19SNN22 unit is the most conserved sequence segment in mammalian amylin structures (Knight et al., 2006). The turn-forming propensity of this unit may be the essential element that allows the formation of the tertiary structure of the bioactive state. It has also been established that the N21 (as well as N14) sidechains present ‘specific interactions that are central to the ordered fibrous states of IAPP’ (Koo et al., 2008). Fluoroalcohol- and helix-formation-induced conformational changes produce notable CSDs for the Asn residues throughout the sequence (vide supra). These shifts are different for hAM and rAM and also different for sequence fragments bearing the carboxamide functions singly or in pairs. Our loop model, hAM-LM, is to our knowledge the best mimic of these features and may prove to be a useful model system and basis for amyloidogenesis inhibitor design.
Turning to amyloid fibril formation by hAM in its biological milieu, insulin (with which it is co-secreted in pancreatic β cell granules) has been established to have an inhibitory effect on amyloid fibril formation in vitro (Westermark, 1996; Kudva et al., 1998) and in the membrane-associated state (Knight, 2008). The interaction of amylins with insulin is now known (Wei, 2009) to involve portions of the common helical segment defined in the present study. The concentration of hAM at its site of synthesis is in excess of 800 µM (Nishi, 1990; Wei, 2009); conditions under which helix formation and association could be expected. Helix-bundle formation could serve as a protective sequestration of key amyloidogenic features, but it has also been suggested (Knight et al., 2006) that helix association at membrane interfaces (and other peptide binding sites) is an essential feature for the accelerated production of amyloid fibril intermediates. The stereochemical features of the monomeric helical states of hAM and prAM should provide insights into all of these processes; the structures will be made available on the Andersen laboratory website (http://andersenlab.chem.washington.edu/structures) upon publication of this account.
Funding
The US National Science Foundation and National Institutes of Health provided funds for the upgrades of NMR instrumentation in the UW Chemistry Department facility.
Supplementary Material
Acknowledgements
Researchers at U.W. thank Amylin Pharmaceuticals for providing materials and funding essential to the studies described herein.
Footnotes
Edited by Mikael Akke
References
- Abedini A., Raleigh D.P. J. Mol. Biol. 2006;355:274–281. doi: 10.1016/j.jmb.2005.10.052. [DOI] [PubMed] [Google Scholar]
- Abedini A., Meng F., Raleigh D.P. J. Am. Chem. Soc. 2007;129:11300–11301. doi: 10.1021/ja072157y. [DOI] [PubMed] [Google Scholar]
- Andersen N.H., Palmer R.B. Bioorg. Med. Chem. Lett. 1994;4:817–822. [Google Scholar]
- Andersen N.H., Lai X., Hammen P.K., Marschner T.M. In: NMR Applications in Biopolymers. Finley J.W., et al., editors. New York: Plenum; 1990. pp. 95–134. [Google Scholar]
- Andersen N.H., Chen C.P., Marschner T.M., Krystek S.R., Jr, Bassolino D.A. Biochemistry. 1992;31:1280–1295. doi: 10.1021/bi00120a003. [DOI] [PubMed] [Google Scholar]
- Andersen N.H., Cao B., Rodríguez-Romero A., Arreguín B. Biochemistry. 1993;32:1407–1422. doi: 10.1021/bi00057a004. [DOI] [PubMed] [Google Scholar]
- Andersen N.H., Chen C., Lee G.M., Liu Z., Tong H. Protein Pept. Lett. 1995;4:215–222. [Google Scholar]
- Andersen N.H., Cort J.R., Liu Z., Sjoberg S.J., Tong H. J. Am. Chem. Soc. 1996;a 118:10309–10310. [Google Scholar]
- Andersen N.H., Liu Z., Prickett K.S. FEBS Lett. 1996;b 399:47–52. doi: 10.1016/s0014-5793(96)01279-3. [DOI] [PubMed] [Google Scholar]
- Andersen N.H., Neidigh J., Harris S.M., Lee G.M., Liu Z., Tong H. J. Am. Chem. Soc. 1997;119:8547–8561. [Google Scholar]
- Andersen N.H., Tong H. Protein Sci. 1997;6:1920–1936. doi: 10.1002/pro.5560060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen N.H., Dyer R.B., Fesinmeyer R.M., Gai F., Liu Z., Neidigh J.W., Tong H. J. Am. Chem. Soc. 1999;121:9879–9880. [Google Scholar]
- Andersen N.H., Brodsky Y., Neidigh J.W., Prickett K. Bioorg. Med. Chem. 2002;10:79–85. doi: 10.1016/s0968-0896(01)00263-2. [DOI] [PubMed] [Google Scholar]
- Andersen N.H., Olsen K.A., Fesinmeyer R.M., Tan X., Hudson F.M., Eidenschink L.A., Farazi S.R. J. Am. Chem. Soc. 2006;128:6101–6110. doi: 10.1021/ja054971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai J., Nakazato M., Kangawa K., Matsukura S., Matsuo H. Biochem. Biophys. Res. Commun. 1989;164:400–405. doi: 10.1016/0006-291x(89)91733-6. [DOI] [PubMed] [Google Scholar]
- Ashburn T.T., Lansbury P.T., Jr J. Am. Chem. Soc. 1993;115:11012–11013. [Google Scholar]
- Bai Y., Milne J.S., Mayne L., Englander S.W. Proteins Struct. Funct. Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger Y., Khiat A., Larocque A., Fournier A., St Pierre S. Int. J. Pept. Protein Res. 1996;47:477–483. doi: 10.1111/j.1399-3011.1996.tb01098.x. [DOI] [PubMed] [Google Scholar]
- Breeze A.L., Harvey T.S., Bazzo R., Campbell I.D. Biochemistry. 1991;30:575–582. doi: 10.1021/bi00216a036. [DOI] [PubMed] [Google Scholar]
- Brender J.R., Lee E.L., Cavitt M.A., Gafni A., Steel D.G., Ramamoothy A. J. Am. Chem. Soc. 2008;a 130:6424–6429. doi: 10.1021/ja710484d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brender J.R., Hartman K., Reid K.R., Kennedy R.T., Ramamoothy A. Biochemistry. 2008;b 47:12680–12688. doi: 10.1021/bi801427c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruch M.D., Dhingra M.M., Gierasch L.M. Proteins Struct. Funct. Genet. 1991;10:130–139. doi: 10.1002/prot.340100206. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. X-PLOR Version 3.1, a System for X-Ray Crystallography and NMR. New Haven, CT: Yale University Press; 1992. [Google Scholar]
- Brushweiler R., Blackledge M., Ernst R.R. J. Biomol. NMR. 1991;1:3–11. doi: 10.1007/BF01874565. [DOI] [PubMed] [Google Scholar]
- Bundi A., Grathwohl C., Hochmann J., Keller R.G., Wagner G., Wüthrich K. J. Magn. Reson. 1975;18:191–198. [Google Scholar]
- Butler A.E., Jang J., Gurlo T., Carty M.D., Soeller W.C., Butler P.C. Diabetes. 2004;53:1509–1516. doi: 10.2337/diabetes.53.6.1509. [DOI] [PubMed] [Google Scholar]
- Carpenter K.A., Schmidt R., vonMentzer B., Haglund U., Robert E., Walpole C. Biochemistry. 2001;40:8317–8325. doi: 10.1021/bi0102860. [DOI] [PubMed] [Google Scholar]
- Cooper G.J.S., Willis A.C., Clark A., Turner R.C., Sim R.B., Reid K.B.M. Proc. Natl Acad. Sci. USA. 1987;84:8628–8632. doi: 10.1073/pnas.84.23.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cort J., Andersen N.H. Biochem. Biophys. Res. Commun. 1997;233:687–691. doi: 10.1006/bbrc.1997.6524. [DOI] [PubMed] [Google Scholar]
- Cort J., Liu Z., Lee G., Harris S.M., Prickett K.S., Gaeta L.S.L., Andersen N.H. Biochem. Biophys. Res. Commun. 1994;204:1088–1095. doi: 10.1006/bbrc.1994.2574. [DOI] [PubMed] [Google Scholar]
- Deftos L.J. Endocrinology. 1997;138:519–520. doi: 10.1210/endo.138.2.4987. [DOI] [PubMed] [Google Scholar]
- Driscoll P.C., Clore G.M., Beress L., Gronenborn A.M. Biochemistry. 1989;28:2178–2187. doi: 10.1021/bi00431a032. [DOI] [PubMed] [Google Scholar]
- Dyson H.J., Wright P.E. Annu. Rev. Biophys. Biophys. Chem. 1991;20:519–538. doi: 10.1146/annurev.bb.20.060191.002511. [DOI] [PubMed] [Google Scholar]
- Eidenschink L.A., Kier B.L., Huggins K., Andersen N.H. Proteins Struct. Funct. Genomics. 2009;75:308–322. doi: 10.1002/prot.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesinmeyer R.M., Hudson F.M., Andersen N.H. J. Am. Chem. Soc. 2004;126:7238–7243. doi: 10.1021/ja0379520. [DOI] [PubMed] [Google Scholar]
- Fesinmeyer R.M., Hudson F.M., Olsen K.A., White G.W.N., Euser A., Andersen N.H. J. Biomol. NMR. 2005;33:213–231. doi: 10.1007/s10858-005-3731-7. [DOI] [PubMed] [Google Scholar]
- Gebre-Medhin S., Olofsson C., Mulder H. Diabetologia. 2000;43:687–695. doi: 10.1007/s001250051364. [DOI] [PubMed] [Google Scholar]
- Glenner G.G., Eanes D., Wiley C. Biochem. Biophys. Res. Commun. 1988;155:608–614. doi: 10.1016/s0006-291x(88)80538-2. [DOI] [PubMed] [Google Scholar]
- Goldsbury C., Goldie K., Pellaud J., Seelig J., Frey P., Muller S.A., Kistler J., Cooper G.J., Aebi U. J. Struct. Biol. 2000;130:352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- Green J., Goldsbury C., Min T., Sunderji S., Frey P., Kistler J., Cooper G., Aebi U. J. Mol. Biol. 2003;326:1147–1156. doi: 10.1016/s0022-2836(02)01377-3. [DOI] [PubMed] [Google Scholar]
- Haataja L., Gurlo T., Huang C.J., Butler P.C. Endocr. Rev. 2008;29:303–316. doi: 10.1210/er.2007-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt S.G., Kilk K., Wang Y., Smith P.M., Langel U., Poyner D.R. Br. J. Pharmacol. 2003;138:325–332. doi: 10.1038/sj.bjp.0705040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard J.A.M., Martin S.R., Chaplin L.C., Bose C., Kelly S.M., Price N.C. Biochem. J. 1991;275:785–788. doi: 10.1042/bj2750785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson F.M., Andersen N.H. Biopolymers. 2004;76:298–308. doi: 10.1002/bip.20126. [DOI] [PubMed] [Google Scholar]
- Johnson W.C., Jr Proteins Struct. Funct. Genet. 1991;7:205–214. [Google Scholar]
- Kajava A.V., Aebi U., Steven A.C. J. Mol. Biol. 2005;348:247–252. doi: 10.1016/j.jmb.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Kapurniotu A., Schmauder A., Tenidis K. J. Mol. Biol. 2002;315:339–350. doi: 10.1006/jmbi.2001.5244. [DOI] [PubMed] [Google Scholar]
- Knight J.D., Hebda J.A., Miranker A.D. Biochemistry. 2006;45:9496–9508. doi: 10.1021/bi060579z. [DOI] [PubMed] [Google Scholar]
- Knight J.D., Williamson J.A., Miranker A.D. Protein Sci. 2008;17:1850–1856. doi: 10.1110/ps.036350.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B.W., Miranker A.D. Protein Sci. 2005;14:231–239. doi: 10.1110/ps.041051205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B.W., Hebda J.A., Miranker A.D. Protein Eng. Des. Sel. 2008;21:147–154. doi: 10.1093/protein/gzm076. [DOI] [PubMed] [Google Scholar]
- Kudva Y.C., Mueske C., Butler P.C., Eberhardt N.L. Biochem. J. 1998;331:809–813. doi: 10.1042/bj3310809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X., Chen C., Andersen N.H. J. Magn. Reson. 1993;101B:271–288. [Google Scholar]
- Lee G.M., Chen C., Marschner T.M., Andersen N.H. FEBS Lett. 1994;355:140–146. doi: 10.1016/0014-5793(94)01153-2. [DOI] [PubMed] [Google Scholar]
- Luca S., Yau W.-M., Leapman R., Tycko R. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine J., Jack E., Stockley P.G., Radford S.E., Serpell L.C., Middleton D.A. J. Am. Chem. Soc. 2008;130:14990–15001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- Mascioni A., Porcelli F., Ilangovan U., Ramamoorthy A., Veglia G. Biopolymers. 2003;69:29–41. doi: 10.1002/bip.10305. [DOI] [PubMed] [Google Scholar]
- Matsuura J., Manning M.C. J. Pharm. Biomed. Anal. 1993;11:89–93. doi: 10.1016/0731-7085(93)80128-n. [DOI] [PubMed] [Google Scholar]
- Mazor Y., Gilead S., Benhar I., Gazit E. J. Mol. Biol. 2002;322:1013–1024. doi: 10.1016/s0022-2836(02)00887-2. [DOI] [PubMed] [Google Scholar]
- McLean L.R., Balasubramaniam A. Biochim. Biophys. Acta. 1992;1122:317–320. doi: 10.1016/0167-4838(92)90411-6. [DOI] [PubMed] [Google Scholar]
- Meadows R.P., Nikonowicz E.P., Jones C.R., Bastian J.W., Gorenstein D.G. Biochemistry. 1991;30:1247–1254. doi: 10.1021/bi00219a012. [DOI] [PubMed] [Google Scholar]
- Meyers J.-P., Pelton J.T., Hoflack J., Saudek V. Biopolymers. 1991;31:233–241. doi: 10.1002/bip.360310210. [DOI] [PubMed] [Google Scholar]
- Mihalyi E. J. Chem. Eng. Data. 1969;13:179–182. [Google Scholar]
- Nanga R.P.R., Brender J.R., Xu J., Veglia G., Ramamoothy A. Biochemistry. 2008;47:12689–12697. doi: 10.1021/bi8014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidigh J.W., Fesinmeyer R.M., Prickett K.S., Andersen N.H. Biochemistry. 2001;40:13188–13200. doi: 10.1021/bi010902s. [DOI] [PubMed] [Google Scholar]
- Nishi M., Bell G.I., Steiner D.F. Diabetologia. 1990;33:628–630. doi: 10.1007/BF00400208. [DOI] [PubMed] [Google Scholar]
- Olsen K.A., Fesinmeyer R.M., Stewart J., Andersen N.H. Proc. Natl Acad. Sci. USA. 2005;102:15483–15487. doi: 10.1073/pnas.0504392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick S.B., Miranker A.D. J. Mol. Biol. 2001;308:783–796. doi: 10.1006/jmbi.2001.4608. [DOI] [PubMed] [Google Scholar]
- Padrick S.B., Miranker A.D. Biochemistry. 2002;41:4694–4703. doi: 10.1021/bi0160462. [DOI] [PubMed] [Google Scholar]
- Patil S.M., Xu S., Sheftic S.R., Alexandrescu A.T. J. Biol. Chem. 2009;284:11982–11991. doi: 10.1074/jbc.M809085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotto M., Saudek V., Sklenar V. J. Biomol. NMR. 1992;2:661–666. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Porat Y., Mazor Y., Efrat S., Gazit E. Biochemistry. 2004;43:14454–14462. doi: 10.1021/bi048582a. [DOI] [PubMed] [Google Scholar]
- Poyner D.R., Sexton P.M., Marshall I., Smith D.M., Quirion R., Born W., Muff R., Fischer J.A., Foord S.M. Pharmacol. Rev. 2002;54:233–246. doi: 10.1124/pr.54.2.233. [DOI] [PubMed] [Google Scholar]
- Pratum T.K., Moore B.S. J. Magn. Reson. 1993;102B:91–97. [Google Scholar]
- Ruschak A.M., Mirander A.D. Proc. Natl Acad. Sci. USA. 2007;104:12341–12346. doi: 10.1073/pnas.0703306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha J., Mahadevan D. Protein Eng. 1991;4:539–544. doi: 10.1093/protein/4.5.539. [DOI] [PubMed] [Google Scholar]
- Schwarzinger S., Kroon G.J.A., Foss T.R., Wright P.E., Dyson H.J. J. Biomol. NMR. 2000;18:43–48. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- Schwarzinger S., Kroon G.J.A., Foss T.R., Chung J., Wright P.E., Dyson H.J. J. Am. Chem. Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- Scrocchi L.A., Ha K., Chen Y., Wu L., Wang F., Fraser P.E. J. Struct. Biol. 2003;141:218–227. doi: 10.1016/s1047-8477(02)00630-5. [DOI] [PubMed] [Google Scholar]
- Sklenar V., Piotto M., Leppik R., Saudek V. J. Magn. Reson. 1993;102A:241–245. [Google Scholar]
- Wei L., Jiang P., Yau Y.H., Summer H., Shochat S.G., Mu Y., Pervushin K. Biochemistry. 2009;48:2368–2376. doi: 10.1021/bi802097b. [DOI] [PubMed] [Google Scholar]
- Westermark P., Wilander E. Diabetologia. 1978;15:417. doi: 10.1007/BF01219652. [DOI] [PubMed] [Google Scholar]
- Westermark P., Wernstedt C., Wilander E., Sletten K. Biochem. Biophys. Res. Commun. 1986;140:827–831. doi: 10.1016/0006-291x(86)90708-4. [DOI] [PubMed] [Google Scholar]
- Westermark P., Wernstedt C., Wilander E., Hayden D., O'Brian T.D., Johnson K.H. Proc. Natl Acad. Sci. USA. 1987;a 84:3881–3885. doi: 10.1073/pnas.84.11.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark P., Engstrom U., Johnson K.H., Westermark G.T., Betsholtz C. Proc. Natl Acad. Sci. USA. 1990;87:5036–5040. doi: 10.1073/pnas.87.13.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark P., Liu Z.-C., Westermark G.T., Leckström A., Steiner D.F. FEBS Lett. 1996;379:203–206. doi: 10.1016/0014-5793(95)01512-4. [DOI] [PubMed] [Google Scholar]
- Williamson J.A., Miranker A.D. Protein Sci. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Sykes B.D. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Sykes B.D., Richards F.M. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- Yan L-M., Tararek-Nossol M., Velkova A., Kazantzis A., Kapumiotu A. Proc. Natl Acad. Sci. USA. 2006;103:2046–2051. doi: 10.1073/pnas.0507471103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.T., Wu C.-S.C., Martinez H.M. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- Yonemoto I.T., Kroon G.J.A., Dyson H.J., Balch W.E., Kelly J.W. Biochemistry. 2008;47:9900–9910. doi: 10.1021/bi800828u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorski M.G. J. Magn. Reson. 1990;89:608–614. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.