Abstract
Amyloidogenic proteins and polypeptides can be divided into two structural classes, namely those which are flexible and are intrinsically disordered in their unaggregated state and those which form a compact globular structure with a well-defined tertiary fold in their normally soluble state. This review article is focused on amyloid formation by natively disordered polypeptides. Important examples of this class include islet amyloid polypeptide (IAPP, amylin), pro-IAPP processing intermediates, α-synuclein, the Aβ peptide, atrial natriuretic factor, calcitonin, pro-calcitonin, the medin polypeptide, as well as a range of de novo designed peptides. Amyloid formation is a complex process consisting of a lag phase during which no detectable fibril material is formed, followed by a rapid growth phase that leads to amyloid fibrils. A critical analysis of the literature suggests that a subset of intrinsically disordered polypeptides populate a helical intermediate during the lag phase. In this scenario, early formation of multimeric species is promoted by helix–helix association involving one region of the polypeptide chain which leads to a high effective concentration of an amyloidogenic sequence located in a different region of the chain. Helical intermediates appear to be particularly important in membrane-catalyzed amyloid formation and have been implicated in glycosaminoglycan mediated amyloid formation as well. There is suggestive evidence that targeting helix–helix interactions can be a viable strategy to inhibit amyloid formation. The characterization of transient helical intermediates is challenging, but new methods are being developed that offer the prospect of providing residue-specific information in real time.
Keywords: Aβ, amyloid, helical intermediate, IAPP, natively unfolded
Introduction
Amyloid formation plays an important role in a diverse range of human disorders including the spongiform encephalopathy, Alzheimer's disease, Parkinson's disease and type 2 diabetes (Sipe, 1994; Selkoe, 2003; Chiti and Dobson, 2006). An even larger number of polypeptides that show no evidence of amyloid formation in vivo do so in vitro, leading to the notion that the ability to form amyloid is a general feature of protein chains (Dobson, 1999; Vendruscolo et al., 2003). A specific primary sequence is not required to form amyloid, but all amyloid fibrils share a number of common structural features. Amyloid fibrils are long, unbranched and the ordered regions adopt the classic cross-β-structure, in which the β-sheet hydrogen bonds are oriented parallel to the fibril axis, with the β-strands running perpendicular to the long axis of the fibril (Tycko, 2004; Nelson et al., 2005). The strands are parallel in the in vitro amyloid formed by the Aβ peptide of Alzheimer's disease and by islet amyloid polypeptide (IAPP, also known as amylin), the two polypeptides for which the most detailed structural information is available. It has also been demonstrated, however, that fibrils can also be made up of anti-parallel strands (Petkova et al., 2002; Luca et al., 2007). The super molecular organization of amyloid fibrils can be complex, and fibrils are usually composed of 2–9 protofilaments per fibril (Goldsbury et al., 2000). Structure determination of amyloid fibrils is an active and exciting area of research, and promising results have been obtained for IAPP and Aβ by solid-state NMR and for a range of short seven-residue peptides by X-ray diffraction (Sawaya et al., 2007). The emerging hypothesis that intermediates in amyloid formation are the toxic species has lead to a renewed focus on the nature of intermediate species (Kirkitadze et al., 2002; Caughey and Lansbury, 2003; Kayed et al., 2003; Walsh and Selkoe, 2004; Lansbury and Lashuel, 2006). The time course of amyloid formation is complex and the details of fibril formation are not well understood, despite considerable effort. Amyloid formation involves a lag phase during which a critical nucleus is formed and during which no appreciable amyloid is detectable. The lag phase is followed by a rapid growth phase which leads to the production of amyloid fibrils (Oosawa and Asakura, 1975; Padrick and Miranker, 2002; Cannon et al., 2004; Ferrone, 2006; Wetzel, 2006). A detailed understanding of the structure and energetics of amyloidogenic intermediates should provide critical information for the design of inhibitors.
Amyloidogenic proteins and polypeptides can be divided into two distinct structural classes: those which are flexible, and are intrinsically disordered or only marginally structured in their unaggregated state (‘natively disordered’) and those which form a compact globular structure with a well-defined tertiary fold in their normally soluble state. Globular proteins normally have to unfold, or at least have to partially unfold, to form amyloid. Natively unfolded sequences usually have lower intrinsic propensity to form amyloid than do sequences derived from globular proteins, but there are still many important examples of unstructured polypeptides which form amyloid in vivo (Table I) (Linding et al., 2004; Monsellier et al., 2008). This review article focuses on amyloid formation by natively unfolded polypeptides. Important examples of ‘natively disordered polypeptides’ which form amyloid include: IAPP, which is the protein component of diabetes-associated islet amyloid; pro-IAPP processing intermediates; the Aβ peptide of Alzheimer's disease; atrial natriuretic factor (ANF), which forms atrial amyloid; calcitonin and pro-calcitonin, the protein component of thyroid amyloid; medin polypeptide which is involved in medial amyloidosis; α-synuclein, responsible for Lewy body formation in Parkinson's disease; as well as a range of de novo designed peptides. The importance of Parkinson's disease, type 2 diabetes and Alzheimer's disease has stimulated interest in the mechanism of amyloid formation by natively unfolded polypeptides. A critical analysis of the literature argues that some natively unfolded polypeptides populate a helical intermediate during amyloid formation in solution.
Table I.
Natively unfolded polypeptides which form amyloid in vivo
| Polypeptide | Disease |
|---|---|
| Aβ1–40 and Aβ1–42 | Alzheimer's disease |
| Atrial natriuretic factor | Artial amyloid |
| Glucagon | Glucagon amyloid-like fibrils: non-toxic |
| Pro-calcitonin | Medullary carcinoma of the thyroid |
| Islet amyloid polypeptide (IAPP, amylin) | Type 2 diabetes |
| Pro-IAPP processing intermediatesa | Type 2 diabetes |
| Medin | Amyloid deposits in the medial layer of arteries |
| α-Synuclein | Parkinson's disease |
aPro-IAPP processing intermediates are thought to play a role in the in vivo generation of pancreatic amyloid and there is immunological evidence that they are found in islet amyloid deposits together with mature IAPP.
The mechanism of amyloid formation in vivo may be quite different from that observed in dilute aqueous solution. Amyloid formation in vivo takes place in a heterogeneous environment with the potential for interactions with the extracellular matrix and with membranes (Snow and Wight, 1989; Sipe, 1994; Inoue, 2001; Ancsin, 2003; Knight and Miranker, 2004; Quist et al., 2005; Lashuel and Lansbury, 2006). Thus, mechanistic studies of amyloid formation in heterogeneous environments are an important goal. Biophysical studies with components of the extracellular matrix and with well-defined model membranes have shown that a variety of amyloidogenic polypeptides readily adopt helical structures when bound to surfaces, and the helical intermediates generated appear to play an important role in amyloid formation.
Here, we review the evidence for the population of helical intermediates during amyloid formation by natively unfolded polypeptides in homogenous solution, and in the presence of membranes, and components of the extracellular matrix. We describe how the targeting of putative helical intermediates may lead to inhibitors of amyloid formation which act early in the self-assembly process. We point out suggestive recent studies which indicate that some inhibitors might actually target helical structures. We conclude with a brief discussion of the techniques that can be applied to probe the secondary structure of amyloid intermediates. Characterization of amyloidogenic intermediates is a major technical challenge, since many of the spectroscopic techniques which are routinely applied to soluble proteins are either not applicable to amyloid assembly or are much harder to employ and interpret. Mutational approaches which have proven to be powerful in the analysis of protein folding may not prove so effective when applied to amyloid formation. Fortunately, several newly developed non-invasive methods hold considerable promise.
Results
Aβ and IAPP appear to populate helical intermediates during amyloid formation in solution
Probably, the first evidence for the involvement of helical intermediates in amyloid formation came from the work of the Teplow lab on Aβ (Kirkitadze et al., 2001; Teplow et al., 2006). A number of lines of experimental evidence argue that the formation of a helical intermediate is important in amyloid formation by Aβ (Kirkitadze et al., 2001). A transient increase in helicity was observed via CD immediately before the appearance of β-structure, suggesting that the helical intermediate is a precursor to β-sheet formation. Mutational studies involving 18 separate variants of Aβ demonstrated that the intermediate is populated in each case, and showed that the kinetics of its formation are correlated with the time course of the appearance of fibrils (Kirkitadze et al., 2001; Teplow et al., 2006). Solvent perturbation experiments which made use of fluorinated alcohols such as hexafluoroisopropanol (HFIP) and trifluoroethanol (TFE) are also highly suggestive. TFE and HFIP are well known to promote helix formation in peptides, and HFIP is more effective on a per volume basis. The effect of these co-solvents on the kinetics of Aβ aggregation is consistent with formation of a helical intermediate. The mutational studies, the solvent perturbation analysis and the time-resolved CD studies of wild-type Aβ are all consistent with the intermediate being on-pathway. It is very important to point out that it is extremely difficult to prove that an intermediate is on- or off-pathway. This is widely appreciated in studies of the folding of soluble proteins where it is often impossible to determine if a kinetic intermediate is on- or off-pathway, owing to the fact that experimental data can usually be equally well fit to the two competing models depicted below.
![]()
This inherent ambiguity is less well appreciated in the field of amyloid formation, but recent works on the kinetics of membrane-catalyzed amyloid formation by IAPP clearly show that the problems which plague protein folding also come into play in studies of amyloid formation. Zanni and coworkers used newly developed two-dimensional infrared (2DIR) methods to follow the formation of amyloid by IAPP in the presence of model membranes in real time (Ling et al., 2009). A rise and then a decay of a spectroscopic signal which was associated with the intermediate was detected; however, straightforward analysis showed that the data could be equally well fit to a model in which the intermediate was on-pathway, or to a model in which it was off-pathway. The ambiguity is not a consequence of the use of IR methods, but rather is a general feature of reaction kinetics and is a problem for time-resolved CD and fluorescence studies as well.
IAPP offers a second example of a system in which helical intermediates appear to play a role; although the evidence, at least for studies in homogenous solution, is a bit more indirect. IAPP does not adopt a well-ordered structure in its monomeric state, but it does have a tendency to transiently populate partial helical structure as judged by studies of rat IAPP, and more robust helical structure can be induced in a variety of ways. Rat IAPP is not amyloidogenic, yet differs from the highly amyloidogenic human peptide at only 6 of 37 positions. Thus, rat IAPP offers the opportunity to study a soluble monomeric analog of IAPP. Analysis of the rat IAPP 1H and 13C NMR secondary shifts indicates that the segment encompassing residues 5 through 20 or residues 8–20 transiently samples helical regions of the ϕ,ψ map, whereas 1H NMR of solubilized human IAPP indicate that it also has some propensity to transiently sample helical states (Williamson and Miranker, 2007; Yonemoto et al., 2008; Wei et al., 2009).
IAPP can be induced to form more robust helical structure in a variety of ways. Early studies show that the polypeptide forms well-ordered helical structure in mixed HFIP/H2O solvents (Cort et al., 1994). In 25% HFIP, rat IAPP forms a single helix from residues 5–20, as does the human peptide. An additional helical region encompassing residues 23–29 is observed in the human peptide in 25% HFIP. Eisenberg and coworkers have recently solved the crystal structure of a fusion protein consisting of human IAPP fused to maltose-binding protein (MBP). They found that human IAPP adopts α-helical structure from residues 8 to 18 and from residues 22 to 27 (Wiltzius et al., 2009). Two molecules are observed in the asymmetric unit, and the pair of IAPP molecules was found to interact via the 8–18 helical domains. The observed structure and IAPP–IAPP interactions are likely stabilized by interactions with MBP. Nonetheless, it is strikingly similar to the structure observed in the mixed HFIP/H2O solvent system. In addition, helical structure within the 8–18 region is observed in a fragment of IAPP bound to model membranes, and NMR studies of full-length IAPP bound to SDS micelles reveal helical structure between residues 5 and 28 with a kink near the 18–22 segments (Namga et al., 2008; Patil et al., 2009). Thus, although IAPP is dynamic and does not adopt rigid helical structure in aqueous solution in its monomeric state, it can clearly be induced to do so in a wide variety of ways. The fact that very similar helical structure is observed in the X-ray structure of the fusion protein, in a mixed HFIP/H2O solvent system, and when IAPP is bound to membranes or SDS micelles is highly suggestive.
IAPP has not been subjected to the same sort of analysis that the Teplow group applied to Aβ, but examination of the literature suggests that IAPP also populates an early helical intermediate. For example, the rate of amyloid formation by IAPP is strongly dependent upon co-solvent concentration and increases as the volume fraction of the helix stabilizing solvent HFIP increases, at least up to a point, whereas the rate decreases in the presence of small amounts of helix destabilizing co-solvents such as DMSO (Padrick and Miranker, 2002; Abedini and Raleigh, 2005). The MBP–IAPP structure suggests that IAPP may dimerize early in the self-assembly pathway and implies that those interactions could stabilize the helices. This hypothesis is supported by studies utilizing cross-linking methods (Wiltzius et al., 2009).
A number of other polypeptides appear to populate helical intermediates during amyloid formation
There is also some, albeit not conclusive, evidence that helical intermediates may be involved in amyloid formation by insulin under certain conditions. Helical intermediates have also been shown to play a role in the aberrant aggregation of the natively unfolded tau protein, while the assembly of silk into its cross-β-structure in vivo is known to proceed via a helical intermediate (van Beek et al., 2000; Kunjithapatham et al., 2005). Helical intermediates have been observed during amyloid formation by several de novo designed polypeptides, and in peptide fragments derived from globular proteins (Mihara and Takahashi, 1997; Fezoui et al., 2000; Liu et al., 2004; D'Auria et al., 2009). These studies, together with the more extensive work on Aβ and IAPP, strongly suggest that helical intermediates can play a role in amyloid formation by natively unfolded polypeptides in solution.
Helical intermediates might promote amyloid formation by generating a high local concentration of an amyloidogenic sequence
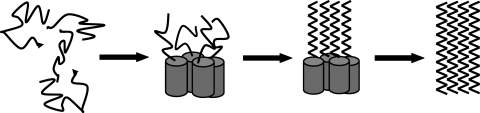
How might the formation of helical structure promote the conversion from a flexible unstructured ensemble of conformations to β-sheet-rich amyloid structures? A number of amyloidogenic polypeptides have a propensity to transiently populate helical structure, which could be further stabilized by peptide–peptide or peptide–membrane interactions. Helix formation and self-association are thermodynamically linked in many systems; classic examples include peptides with a tendency to form amphiphillic helices, coiled coils and other oligomerization domains, and numerous designed systems. In this model, the initial formation of oligomers would be driven by the linkage between helix formation and association (Fig. 1). It is unlikely that helical structure would extend throughout the molecule, and there is good evidence that it does not do so in the systems studied to date. In this case, helix-mediated association will lead to a high local concentration of an aggregation prone sequence, and this in turn could promote intermolecular β-sheet formation. A highly schematic representation of this scenario is depicted in Fig. 1. It is important to appreciate that helical intermediates do not always have to lead to an increase in the rate of amyloid formation. If the helical intermediate were too stable, it could actually decrease the rate of amyloid formation since it would represent a kinetic trap. Such effects appear to be operative in IAPP membrane systems when the content of anionic lipids is high, and may potentially be important during the aggregation of pro-IAPP processing intermediates in the presence of HSPGs (Jayasinghe and Langen, 2005; Meng et al., 2007; Apostolidou et al., 2008; Yonemoto et al., 2008).
Fig. 1.
A schematic diagram of how an α-helical intermediate might promote amyloid formation. α-Helices are depicted as cylinders, β-strands as zigzagged lines. Initial oligomerization is driven by the thermodynamic linkage between self-association and helix formation (step 1). This in turn generates a high local concentration of a region of the protein chain which has a high propensity to adopt β-structure. Propagation of β-structure leads to the formation of β-sheet-rich assemblies. The diagram is schematic and is not meant to imply a specific pathway of assembly. A tetramer is shown here for the purposes of illustration, but a range of oligomeric species could be formed. The diagram implies a sequential zipping of the β-strands and unwinding of the helices, but this is simply meant to be illustrative and a diversity of pathways is likely.
Unstructured amyloidogenic polypeptides readily adopt helical structure when bound to model membranes and other surfaces
A wide range of studies have demonstrated that natively unfolded amyloidogenic polypeptides can adopt helical structure when bound to surfaces (Perrin et al., 2000; Eliezer et al., 2001; Lee et al., 2002; Zhu et al., 2003; Jayasinghe and Langen, 2005; Knight et al., 2006; Jayasinghe and Langen, 2007; Meng et al., 2007; Olofsson et al., 2007; Apostolidou et al., 2008). The two most extensively characterized examples are α-synuclein and IAPP, whose interactions with model membranes have been investigated by a number of laboratories (Perrin et al., 2000; Eliezer et al., 2001; Lee et al., 2002; Zhu et al., 2003; Knight and Miranker, 2004; Jayasinghe and Langen, 2005; Knight et al., 2006; Jayasinghe and Langen, 2007; Munishkina and Fink, 2007; Apostolidou et al., 2008). The rate of amyloid formation by IAPP is enhanced by peptide–membrane interactions. IAPP undergoes a transition from its natively disordered monomeric state to an α-helical structure when it binds to membranes; and the ability to adopt membrane-induced α-helical structure is conserved in IAPP molecules from different species (Knight et al., 2006). Experiments have been conducted with a range of lipid compositions and efficient peptide–membrane interactions require the presence of anionic lipids. This is not surprising since IAPP has a net positive charge at neutral pH. Some of the studies of IAPP–membrane interactions used mole fractions of anionic lipids that were likely not within the normal physiological range, but the basic principle that interactions with membranes induce helical structure in IAPP is robust.
NMR and EPR studies have provided important information about the conformation of IAPP on the membrane. The NMR studies outlined in the proceeding section reveal helical structure which is remarkably consistent with that observed in the crystal structure of the MBP–IAPP fusion protein. EPR is a more perturbing technique than NMR, since it involves the use of analogs of IAPP which lack the native Cys-2 to Cys-7 disulfide bond, but contain a spin-label attached to a non-native Cys (Apostolidou et al., 2008). Nonetheless, the results are in good general agreement with the NMR work and lead to a model in which residues 9 through 22 of membrane-bound IAPP adopt helical structure, whereas the N- and C-termini are much less ordered. The structural models are attractive because the region which becomes helical on the membrane is similar to the region which has the highest propensity to sample helical structure in the monomeric state, and because they also provide a plausible frame work for envisaging how a membrane-bound helical intermediate promotes the development of β-sheet structure. A number of factors come into play; association with the membrane leads to a high local concentration of peptide, and membrane-bound IAPP monomers collide via diffusion in two, rather than in three dimensions. In these models, the C-terminal region of IAPP is not interacting with membranes and is much more flexible. This portion of IAPP is highly amyloidogenic, and thus the association of the helical regions could lead to a high local concentration of a very amyloidogenic region of the molecule (Nilsson and Raleigh, 1999). The model predicts that variants of IAPP which contain substitutions that inhibit β-sheet formation by the C-terminal region would lead to a membrane-bound helical species that will not progress to amyloid. Along these lines, comparative 2DIR studies of rat and human IAPP show that rat IAPP forms a membrane-bound intermediate which appears to be very similar to the intermediate detected with human IAPP (Ling et al., 2009). The time scale for formation of the rat and human IAPP membrane-bound intermediates are very similar; however, the rat peptide is trapped in the intermediate non-amyloid state. Five of the six amino acid differences between the rat and the human sequence lie within the region extending from the C-terminus to the major helical region; these include several prolines and other residues expected to significantly decrease amyloidogenicity. The model also predicts that formation of helical structure could inhibit amyloid formation if the helices were overly stabilized, since the system would be trapped in a non-amyloid conformation. There is experimental evidence that such effects can be observed in IAPP–membrane systems (Jayasinghe and Langen, 2005; Apostolidou et al., 2008).
A large body of work indicates that α-synuclein adopts partial helical structure when bound to model membranes, but the effects of protein–membrane interactions on the rate of aggregation are not clear, and there are conflicting reports in the literature. Some workers have proposed that membrane–protein interactions inhibit amyloid formation, whereas others present evidence that they enhance it (Lee et al., 2002; Zhu et al., 2003). The different results may reflect differences in the stability of the membrane-bound helical structure under the different experimental conditions. Medin, another natively unfolded protein, has also been shown to populate helical structure when it interacts with membranes, and formation of the helical intermediate appears to promote amyloid formation (Olofsson et al., 2007).
Helical intermediates and amyloid formation are not limited to membrane–peptide interactions. For example, recent work has probed the potential role of pro-IAPP processing intermediates in amyloid formation in type 2 diabetes (Park and Verchere, 2001; Abedini et al., 2006). The studies were motivated by the fact that partially processed pro-IAPP is found in amyloid deposits in vivo. One theory postulates that interactions between the pro-IAPP processing intermediates and sulfated proteoglycans of the extracellular matrix lead to amyloid formation (Park and Verchere, 2001). In vitro biophysical studies have shown that these interactions can efficiently promote amyloid formation by pro-IAPP processing intermediates, and the work offers evidence that helical intermediates are involved (Meng et al., 2007).
Targeting helical intermediates may provide a route to inhibiting amyloid formation
Preventing the formation of a helical intermediate will inhibit amyloid formation if the intermediate is on-pathway, and trapping amyloidogenic peptides in the intermediate helical state would prevent amyloid formation whether the intermediate was on- or off-pathway. Insulin is among the most potent inhibitors of IAPP aggregation, and its proposed mode of action is consistent with it targeting a helical intermediate. There are a number of lines of evidence in support of this assertion. NMR studies have shown that residual helical structure in human IAPP appears to be important for its interactions with insulin. Independent peptide mapping studies and docking studies using the MBP–IAPP structure all argue that insulin interacts with IAPP via the putative helical region of IAPP (Gilead et al., 2006; Wei et al., 2009; Wiltzius et al., 2009).
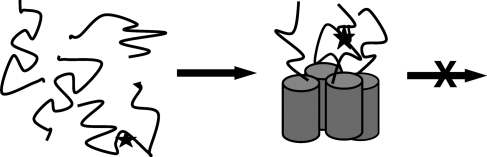
One could envisage inhibitors that combine a recognition motif that targets helical oligomeric species with a unit which prevents the conversion of the helical oligomers into β-sheet-rich structures. Studies with IAPP point mutants are highly suggestive that this strategy is viable. For example, an I26P point mutant converts human IAPP from a highly amyloidogenic molecule into a potent inhibitor of amyloid formation by wild-type IAPP (Abedini et al., 2007). Note that the mutation lies outside of the major putative helical domain, and thus the molecule should be capable of interacting with wild-type IAPP to form a heterogeneous helical intermediate, in which one or more of the peptide chains contain a mutation that prevents the conversation to β-sheet structure (Fig. 2). This proposed mechanism is similar to the mode of action of certain dominant negative inhibitors of transcription factors.
Fig. 2.
A schematic diagram of how a hypothetical inhibitor which combines a helix recognition motif with a β-sheet breaker could act as an inhibitor. The diagram is meant to represent a mutant of an amyloidogenic peptide in which the mutation (depicted as an X) is located in a region outside of the helix recognition motif, but within a region critical for conversion to β-structure. Some IAPP point mutants may act via this mechanism.
New methods are needed to define helical intermediates with residue-specific resolution
The method of choice for obtaining residue-specific information about the folding of soluble proteins is to analyze the effect of point mutations on the rate of folding, and on the stability of the protein. If certain conditions are met, the ratio of the change in the activation free energy to the change in the equilibrium free energy of folding, the ϕ-value, can be interpreted to reflect the development of structure in the transition state for folding. Although the mutational analysis of amyloid formation is powerful, it is unlikely that it can be reliably applied in the same fashion that ϕ-values are used in protein folding. The mechanism of amyloid formation is complex and may involve multiple pathways and the production of heterogeneous species during the lag phase. Thus, mutations may alter the mechanism of fibril formation or redirect flux through alternative pathways. In addition, amyloid fibrils can be polymorphic and mutations could alter the final structure of the fibril, either at the level of influencing which regions adopt cross-β-structure, or at the supramolecular level. All of these considerations argue that mutational studies need to be analyzed with care. Unfortunately, standard spectroscopic methods lack the resolution to provide residue-specific information. Studies of helical intermediates have largely relied upon the use of CD to follow secondary structure formation. CD spectroscopy is very sensitive to the presence of helical structure, but it can be difficult to quantitatively deconvolve CD spectra to deduce relative populations of secondary structure, particularly if light scattering becomes a problem. In a perfect world, one would like to define the regions of the polypeptide which are sampling helical structure in both homogenous and heterogeneous systems, but this is not possible with CD, and the same problem applies to standard IR studies. A third complicating factor is that the CD signal of an α-helix is sensitive to the length of the helical segment, and the rotational strength (intensity) of the helical bands is much weaker for short helices. Consequently, the CD spectrum of a polypeptide which transiently samples a set of short helical segments could appear to arise from a system which is largely unfolded, even though it has a significant tendency to sample helical structure. NMR does not suffer from this difficulty, and it is interesting to note that there are examples of unfolded soluble proteins which appear to have more helical structure as judged by NMR then by CD. Unfortunately, high-resolution NMR studies of aggregating systems are extraordinarily difficult, and aggregation is usually too fast to allow modern multi-dimensional NMR to be applied. Fortunately, recent advances in IR spectroscopy and specific isotopic labeling offer the exciting prospect of being able to obtain site-specific structural information with high intrinsic time resolution in a non-perturbing manner (Zanni and Hochstrasser, 2001; Mukherjee et al., 2006; Shim et al., 2007; Kim et al., 2008, Ling et al., 2009). 2DIR with 13C18O isotopic labeling of specific carbonyls appears to be the method of choice (Shim et al., 2009). The method offers good spectral resolution while retaining structural information. The labeling shifts the frequency of the amide-I mode of interest well away from the broad natural abundance background, whereas the 2D experiment allows the weak coupling between the perturbed mode and the unlabelled oscillators to be detected, and thus, provides the structural information. Advances in instrumentation now allow one to record 2DIR spectra ‘on-the-fly’, opening the door to high-resolution kinetic studies (Shim et al., 2007, 2009). 2DIR studies of amyloid formation are still relatively novel, but real-time kinetic investigations of amyloid formation in homogenous solution and in membrane-bound systems have already been reported; multiple labeling schemes will allow the delineation of helical regions.
Conclusions
The generality of helical intermediates and their exact role, i.e. on- or off-pathway, remains an open question, but the literature reviewed above, and careful consideration of data published on Aβ and IAPP, argues that they play a role in amyloid formation in those two systems in vitro. The issue of whether or not other natively unfolded polypeptides form helical intermediates during amyloid formation is less clear, although the studies reviewed here are strongly suggestive. The role of helical intermediates in amyloid formation in vivo is an open question. The case for helical intermediates appears to be even stronger for membrane-catalyzed amyloid formation in particular, and for surface mediated amyloid formation in general. Initial observations suggest that targeting helical oligomeric intermediates offers a route to the rational design of amyloid inhibitors. Mechanistic studies of amyloid formation in real time have suffered from low structural resolution and/or limited time resolution; however, recent methodological developments hold considerable promise. We hope this review will help to stimulate further investigations of the potential role of helical intermediates in amyloid formation, and will also help to motivate the application of new methodologies to a range of amyloidogenic systems.
Funding
This work was supported by NIH grant GM078114.
Acknowledgements
We thank Professor Martin Zanni and members of the Raleigh group for helpful discussions.
Footnotes
Edited by Valerie Dagget
References
- Abedini A., Raleigh D. Biochemistry. 2005;44:16284–16291. doi: 10.1021/bi051432v. [DOI] [PubMed] [Google Scholar]
- Abedini A., Tracz S.M., Cho J., Raleigh D. Biochemistry. 2006;45:9228–9237. doi: 10.1021/bi0510936. [DOI] [PubMed] [Google Scholar]
- Abedini A., Meng F., Raleigh D. J. Am. Chem. Soc. 2007;129:11300–11301. doi: 10.1021/ja072157y. [DOI] [PubMed] [Google Scholar]
- Ancsin J.B. Amyloid: J. Protein Folding Disord. 2003;10:67–79. doi: 10.3109/13506120309041728. [DOI] [PubMed] [Google Scholar]
- Apostolidou M., Jayasinghe S.A., Langen R. J. Biol. Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M.J., Williams A.D., Wetzel R., Myszka D.G. Anal. Biochem. 2004;328:67–75. doi: 10.1016/j.ab.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Caughey B., Lansbury P.T., Jr Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Cort J., Liu Z., Lee G., Harria S.M., Prickett K.S., Gaeta L.S.L., Andersen N.H. Biochem. Biophys. Res. Commun. 1994;204:1088–1095. doi: 10.1006/bbrc.1994.2574. [DOI] [PubMed] [Google Scholar]
- D'Auria G., Vacatello M., Falcigo L., Paduano L., Mangiapia G., Calvanese L., Gambaretto R., Dettin M., Paolillo L. J. Pept. Sci. 2009;15:210–219. doi: 10.1002/psc.1083. [DOI] [PubMed] [Google Scholar]
- Dobson C.M. Trends Biochem. Soc. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- Eliezer D., Kutluay E., Bussell R., Jr, Brown G. J. Mol. Biol. 2001;307:799–807. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- Ferrone F. Methods Enzymol. 2006;412:285–299. doi: 10.1016/S0076-6879(06)12017-0. [DOI] [PubMed] [Google Scholar]
- Fezoui Y., Hartley D.M., Walsh D.M., Selkoe D., Osterhout J.J., Teplow D.B. Nat. Struct. Biol. 2000;7:1095–1099. doi: 10.1038/81937. [DOI] [PubMed] [Google Scholar]
- Gilead S., Wolfenson H., Gazit E. Angew. Chem. Int. Ed. 2006;45:6476–6480. doi: 10.1002/anie.200602034. [DOI] [PubMed] [Google Scholar]
- Goldsbury C., Goldie K., Pellaud J., Seelig J., Frey P., Muller S.A., Kistler J., Cooper G.J.S., Aebi U. J. Struct. Biol. 2000;130:352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- Inoue S. Int. Rev. Cytol. 2001;210:121–161. doi: 10.1016/s0074-7696(01)10005-7. [DOI] [PubMed] [Google Scholar]
- Jayasinghe S.A., Langen R. Biochemistry. 2005;44:12113–12119. doi: 10.1021/bi050840w. [DOI] [PubMed] [Google Scholar]
- Jayasinghe S.A., Langen R. BBCA Biomembr. 2007;1768:2002–2009. doi: 10.1016/j.bbamem.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Kayed R., Head E., Thompson J.L., McIntire T.M., Milton S.C., Cotman C.W., Glable C.G. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Liu L., Axelsen P.H., Hochstrasser R.M. Proc. Natl Acad. Sci. USA. 2008;105:7720–7725. doi: 10.1073/pnas.0802993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkitadze M.D., Condron M.M., Teplow D.B. J. Mol. Biol. 2001;312:1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- Kirkitadze M.D., Bitan G., Teplow D.B. J. Neurosci. Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- Knight J.D., Miranker A.D. J. Mol. Biol. 2004;341:1175–1187. doi: 10.1016/j.jmb.2004.06.086. [DOI] [PubMed] [Google Scholar]
- Knight J.D., Hebda J.A., Miranker A.D. Biochemistry. 2006;45:9496–9508. doi: 10.1021/bi060579z. [DOI] [PubMed] [Google Scholar]
- Kunjithapatham R., Oliva F.Y., Doshi U., Pérez M., Àvila J., Munoz V. Biochemistry. 2005;44:149–156. doi: 10.1021/bi048564t. [DOI] [PubMed] [Google Scholar]
- Lansbury P.T., Lashuel H.A. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- Lashuel H.A., Lansbury P.T., Jr Q. Rev. Biophys. 2006;39:167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. J. Mol. Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- Ling Y.L., Strasfeld D.B., Shim S.-H., Raleigh D.P., Zanni M.T. J. Phys. Chem. B. 2009;113:2498–2505. doi: 10.1021/jp810261x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Prausnitz J.M., Blanch H.W. Biomacromolecules. 2004;5:1818–1823. doi: 10.1021/bm049841e. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Choi C., Lee S.J. J. Biol. Chem. 2002;277:671–678. doi: 10.1074/jbc.M107045200. [DOI] [PubMed] [Google Scholar]
- Luca S., Yau W-M., Leapman R., Tycko R. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Abedini A., Raleigh D.P. Biochemistry. 2007;46:12091–12099. doi: 10.1021/bi7004834. [DOI] [PubMed] [Google Scholar]
- Mihara H., Takahashi Y. Curr. Opin. Struct. Biol. 1997;7:501–508. doi: 10.1016/s0959-440x(97)80113-3. [DOI] [PubMed] [Google Scholar]
- Monsellier E., Ramazzo M., Taddei N., Chiti F. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000199. e1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P., Kass I., Arkin I., Zanni M.T. Proc. Natl Acad. Sci. USA. 2006;103:3528–3533. doi: 10.1073/pnas.0508833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munishkina L.A., Fink A.L. BBCA Biomembr. 2007;1768:1862–1885. doi: 10.1016/j.bbamem.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Namga R.P.R., Brender J.R., Xu J.D., Veglia G., Ramamoorthy A. Biochemistry. 2008;47:12689–12697. doi: 10.1021/bi8014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R., Sawaya M.R., Balbirnie M., Madsen A.O., Riekel C., Grothe R., Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M.R., Raleigh D.P. J. Mol. Biol. 1999;294:1375–1385. doi: 10.1006/jmbi.1999.3286. [DOI] [PubMed] [Google Scholar]
- Olofsson A., Borowik T., Grobner G., Sauer-Eriksson A.E. J. Mol. Biol. 2007;374:186–194. doi: 10.1016/j.jmb.2007.08.064. [DOI] [PubMed] [Google Scholar]
- Oosawa F., Asakura S. Thermodynamics of the Polymerization of Protein. New York: Academic Press; 1975. [Google Scholar]
- Padrick S.B., Miranker A.D. Biochemistry. 2002;41:4694–4703. doi: 10.1021/bi0160462. [DOI] [PubMed] [Google Scholar]
- Park K., Verchere C.B. J. Biol. Chem. 2001;276:16611–16616. doi: 10.1074/jbc.M008423200. [DOI] [PubMed] [Google Scholar]
- Patil S.M., Xu S.H., Sheftic S.R., Alexandrescu A.T. J. Biol. Chem. 2009;284:11982–11991. doi: 10.1074/jbc.M809085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R.J., Woods W.S., Clayton D.F., George J.M. J. Biol. Chem. 2000;275:34393–34398. doi: 10.1074/jbc.M004851200. [DOI] [PubMed] [Google Scholar]
- Petkova A.T., Ishii Y., Balbach J.J., Antzutkin O.N., Leapman R.D., Delaglio F., Tycko R. Proc. Natl Acad. Sci. USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quist A., Doudevski I., Lin H., Azimova R., Ng D., Frangione B., Kagan B., Ghiso J., Lal R. Proc. Natl Acad. Sci. USA. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya M.R., et al. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Selkoe D. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- Shim S.-H., Strasfeld D.B., Ling Y.L., Zanni M.T. Proc. Natl Acad. Sci. USA. 2007;104:14197–14202. doi: 10.1073/pnas.0700804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S.-H., Gupta R., Ling Y.L., Strasfeld D.B., Raleigh D.P., Zanni M.T. Proc. Natl Acad. Sci. USA. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe J.D. Crit. Rev. Clin. Lab. Sci. 1994;31:325–354. doi: 10.3109/10408369409084679. [DOI] [PubMed] [Google Scholar]
- Snow A.D., Wight T.N. Neurobiol. Aging. 1989;10:481–497. doi: 10.1016/0197-4580(89)90108-5. [DOI] [PubMed] [Google Scholar]
- Teplow D.B., et al. Acc. Chem. Res. 2006;39:635–645. doi: 10.1021/ar050063s. [DOI] [PubMed] [Google Scholar]
- Tycko R. Curr. Opin. Struct. Biol. 2004;14:96–103. doi: 10.1016/j.sbi.2003.12.002. [DOI] [PubMed] [Google Scholar]
- van Beek J.D., Beaulieu L., Schafer H., Demura M., Aaskura T., Meier B.H. Nature. 2000;405:1077–1079. doi: 10.1038/35016625. [DOI] [PubMed] [Google Scholar]
- Vendruscolo M., MacPhee C.E., Dobson C.M. Philos. Trans. R. Soc. Series A. 2003;361:1205–1222. doi: 10.1098/rsta.2003.1194. [DOI] [PubMed] [Google Scholar]
- Walsh D.M., Selkoe D.J. Protein Pept. Lett. 2004;11:213–228. doi: 10.2174/0929866043407174. [DOI] [PubMed] [Google Scholar]
- Wei L., Jiang P., Yau Y.H., Summer H., Shocat S.G., Pervushin K. Biochemistry. 2009;48:2368–2376. doi: 10.1021/bi802097b. [DOI] [PubMed] [Google Scholar]
- Wetzel R. Acc. Chem. Res. 2006;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- Williamson J., Miranker A. Protein Sci. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltzius J.J.W., Sievers S.A., Sawaya M.R., Eisenberg D. Protein Sci. 2009 doi: 10.1002/pro.145. in press, doi:10.1002/pro.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemoto I.T., Kroon G.J.A., Dyson H.J., Blach W.E., Kelly J.W. Biochemistry. 2008;47:9900–9910. doi: 10.1021/bi800828u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanni M.T., Hochstrasser R.M. Curr. Opin. Struct. Biol. 2001;11:516–522. doi: 10.1016/s0959-440x(00)00243-8. [DOI] [PubMed] [Google Scholar]
- Zhu M., Li J., Fink A.L. J. Biol. Chem. 2003;278:16873–16877. doi: 10.1074/jbc.M210136200. [DOI] [PubMed] [Google Scholar]