Abstract
Eukaryotic cells have evolved quality control mechanisms to degrade aberrant mRNA molecules and prevent the synthesis of defective proteins that could be deleterious for the cell. The exosome, a protein complex with ribonuclease activity, is a key player in quality control. An early quality checkpoint takes place cotranscriptionally but little is known about the molecular mechanisms by which the exosome is recruited to the transcribed genes. Here we study the core exosome subunit Rrp4 in two insect model systems, Chironomus and Drosophila. We show that a significant fraction of Rrp4 is associated with the nascent pre-mRNPs and that a specific mRNA-binding protein, Hrp59/hnRNP M, interacts in vivo with multiple exosome subunits. Depletion of Hrp59 by RNA interference reduces the levels of Rrp4 at transcription sites, which suggests that Hrp59 is needed for the exosome to stably interact with nascent pre-mRNPs. Our results lead to a revised mechanistic model for cotranscriptional quality control in which the exosome is constantly recruited to newly synthesized RNAs through direct interactions with specific hnRNP proteins.
INTRODUCTION
Expression of protein-coding genes is a complex multistep process. Precursor messenger RNAs (pre-mRNAs) are synthesized by RNA polymerase II (Pol-II) and assembled into ribonucleoprotein (RNP) complexes during transcription. The pre-mRNPs must be processed to become mature mRNPs, which are exported to the cytoplasm and translated into protein. Mutations in the DNA or errors in the transcription and processing reactions can lead to the synthesis of aberrant mRNPs. Eukaryotic cells have evolved quality control mechanisms that identify aberrant mRNPs and prevent their expression into protein. In the yeast Saccharomyces cerevisiae there are at least three nuclear steps of mRNP biogenesis that are under surveillance: the cleavage and polyadenylation of the 3′ end (Hilleren et al., 2001), the assembly of the mRNA–protein complexes (Zenklusen et al., 2002; Luna et al., 2005), and the removal of introns (Galy et al., 2004). In all cases, mRNPs with assembly and/or processing defects are detected by nuclear surveillance mechanisms, retained in the nucleus, and degraded (reviewed by Vinciguerra and Stutz, 2004; Sommer and Nehrbass, 2005; Saguez et al., 2005). Genetic studies in S. cerevisiae have identified the nuclear exosome as a key player in the recognition and retention of defective transcripts (reviewed by Jensen et al., 2003; Houseley et al., 2006; Vanacova and Stefl, 2007; Schmid and Jensen, 2008).
The exosome is a protein complex with ribonuclease activity (Mitchell et al., 1997; Allmang et al., 1999; Mitchell and Tollervey, 2000). The core of the exosome has a barrel-like architecture (reviewed by Lorentzen et al., 2008). The barrel is made of nine protein subunits organized into two rings, a hexameric ring and a trimeric ring, and the structure of the barrel is conserved throughout evolution (Lorentzen et al., 2005; Liu et al., 2006; Wang et al., 2007). Two additional proteins, Dis3/Rrp44 and Rrp6, are associated with the barrel and provide the ribonucleolytic activity (Liu et al., 2006; Dziembowski et al., 2007; Schneider et al., 2007).
The exosome has several roles. It is involved in the surveillance of mRNA biogenesis, the cytoplasmic turnover of mRNA, and the degradation of unstable transcripts derived from intergenic regions in the genome of the S. cerevisiae (Wyers et al., 2005; Preker et al., 2008). The exosome also plays an essential role in the processing of noncoding RNA precursors, including pre-rRNA, pre-snRNA, pre-snoRNA, and pre-tRNA (reviewed by Butler, 2002). The specificity and the regulation of the activity of the exosome require cofactors or auxiliary proteins. For cytoplasmic mRNA turnover, the exosome requires Ski7 and the Ski complex (van Hoof et al., 2002). At least three auxiliary factors are involved in nuclear surveillance in yeast. Rrp6 is required for cotranscriptional surveillance of mRNA biogenesis and for many of the functions of the exosome in RNA processing (see, for example, Hilleren et al., 2001; Zenklusen et al., 2002; Galy et al., 2004). The nucleic acid–binding protein Rrp47 is necessary for the processing of rRNA, snoRNA, and snRNA precursors (Mitchell et al., 2003). The TRAMP polyadenylation complex, furthermore, acts as a cofactor for the degradation of cryptic transcripts and structured RNA substrates (LaCava et al., 2005; Vanacova et al., 2005), and for mRNA surveillance (Rougemaille et al., 2007).
Much of our knowledge about mRNA surveillance comes from studies in yeast. The composition and structure of the exosome are highly conserved from yeast to humans, which suggests that the functions of the exosome are also conserved in complex eukaryotes. However, little is known about the mechanisms by which the exosome is recruited to the nascent pre-mRNP in metazoans. In Drosophila melanogaster, the exosome interacts physically with the transcription elongation factors Spt5/6, which provides a means for cotranscriptional recruitment of the exosome to transcribed genes (Andrulis et al., 2002). To get further insight into the mechanisms of recruitment of the nuclear exosome to protein-coding genes, we have studied the core exosome subunit Rrp4 in Drosophila and Chironomus, two insect model systems that offer different advantages for studies of gene expression. We first studied the association of Rrp4 with the Balbiani ring (BR) genes in the polytene chromosomes of Chironomus tentans. The BR genes code for large secretory proteins of the larval salivary glands, and the nucleotide sequences of the BR genes have been determined (reviewed by Wieslander, 1994). Large chromosomal puffs known as “BR puffs” form in the polytene chromosomes when the BR genes are transcribed. The BR pre-mRNAs have all the features of typical protein-coding transcripts and undergo normal pre-mRNA processing (reviewed by Wieslander et al., 1996). The BR genes can be easily identified in polytene chromosome preparations, and the cotranscriptional association of specific proteins with the newly synthesized BR pre-mRNAs can be studied by immunoelectron microscopy (immuno-EM). Thanks to these features, the BR genes are a useful experimental system for in situ studies of mRNA biogenesis (reviewed by Daneholt, 2001). We have also analyzed the association of Rrp4 with several genes of D. melanogaster by chromatin immunoprecipitation (ChIP) and we present evidence that, both in Drosophila and Chironomus, a fraction of Rrp4 interacts with the nascent pre-mRNP. We also show that this interaction can be mediated by the heterogeneous nuclear ribonucleoprotein (hnRNP) Hrp59 and that Hrp59 interacts not only with Rrp4 but also with other exosome subunits. Our results suggest that hnRNP proteins play an important role in mediating the interactions between the surveillance machinery and nascent transcripts.
MATERIALS AND METHODS
Culturing Conditions
C. tentans was cultured as described by Meyer et al. (1983). Salivary glands were isolated from fourth instar larvae. C. tentans tissue culture cells were cultivated at 24°C as previously described (Wyss, 1982). D. melanogaster S2 cells were cultured at 28°C following the Drosophila Expression System manual from Invitrogen (Carlsbad, CA).
Amplification and Cloning of Rrp4 cDNA
Primers were designed to match conserved amino acid regions in the Rrp4 sequence. Sequence conservation was based on alignment of the D. melanogaster Rrp4 (CG3931) and Anopheles gambiae Rrp4 (BWB5561) amino acid sequences using the Align program at http://workbench.sdsc.edu.
C. tentans total RNA was isolated, reverse-transcribed, and used for PCR amplification following standard procedures. The PCR product was ligated into the TOPO vector (Invitrogen), and the resulting plasmid was called TOPO-Rrp4-471. Detailed procedures and primer sequences are provided as Supplemental Data.
SDS-PAGE and Western Blotting
Proteins were separated by SDS-PAGE using the Mini-Protean II system (Bio-Rad, Richmond, CA) and transferred to polyvinylidenefluoride (PVDF) membranes (Millipore, Bedford, MA) in Tris-glycine buffer with 0.02% SDS and 4 M urea using a semidry electrophoretic transfer cell (Bio-Rad). The membranes were probed with antibodies following standard procedures.
Antibodies
A synthetic peptide spanning amino acids 143 to156 (LIKRRKNHFHNLPC) of Ct-Rrp4 was conjugated to keyhole limpet hemocyanin and used to immunize rabbits following standard procedures. The antibody was purified by affinity chromatography. The anti-V5 antibody was from Invitrogen. The anti Pol-II CTD antibody 4H8 (ab5408) was from Abcam (Cambridge, MA), and the negative control anti-IgG antibody (Z0456) was from DakoCytomation (Carpenteria, CA). The following antibodies were used for Western blotting: polyclonal anti-Hrp59 (Kiesler et al., 2005), polyclonal against Rrp6 (kindly provided by E. Andrulis, Case Western Reserve University School of Medicine, Cleveland, OH), mAb 10:3G1 against Chironomus Hrp36/sqd (Kiseleva et al., 1997), mAb Bj6 against NonA (kindly provided by H. Saumweber, Humboldt University, Berlin, Germany), and mAb against PEP (kindly provided by S. Amero, Loyola University Chicago, Maywood, IL). The mAb Y12 was used for the detection of snRNP proteins (Lerner et al., 1981). Gold-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA).
Immunofluorescence
C. tentans polytene chromosomes were isolated from the salivary glands of fourth instar larvae and fixed essentially as described by Björkroth et al. (1988). The isolated chromosomes were incubated with primary antibody, anti-Rrp4, mAb Y12, or mAb 4H8, in 0.5% BSA in TKM buffer (10 mM triethanolamine-HCl, 100 mM KCl, and 1 mM MgCl2) at 4°C overnight. The working concentrations of the antibodies were 5–10 μg/ml. The chromosomes were washed, incubated with fluorescent secondary antibodies, mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA), and examined in an LSM 510 laser confocal microscope (Carl Zeiss, Thornwood, NY). For 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) treatments, fourth instar larvae were treated with 0.4 mM DRB for 45 min before dissection of the glands and chromosome isolation. For experiments of RNA digestion, the chromosomes were incubated with 100 μg/ml RNase A for 30 min and washed with TKM before being fixed and processed as above. Immunofluorescent staining of salivary glands was carried out as described by Kiesler et al. (2005).
Immuno-EM
Salivary glands were prefixed for 30–60 s with 2% paraformaldehyde in TKM buffer, washed with TKM, and subsequently permeabilized and fixed as described by Björkroth et al. (1988). The isolated chromosomes were blocked in 2% BSA in TKM for 30 min and incubated with 10 μg/ml purified anti-Rrp4 antibody diluted in TKM containing 0.5% BSA. The secondary antibody was an anti-rabbit IgG conjugated to 6-nm gold particles. The chromosomes were postfixed with 2% glutaraldehyde and embedded in Agar 100. Thin sections were examined and photographed in an FEI 120-kV Tecnai electron microscope (Eindhoven, The Netherlands) at 80 kV using a Gatan US 1000P CCD camera (Pleasanton, CA). Quantitative analysis was carried out using images recorded from random fields. The numbers of gold particles in the proximal, middle, and distal regions of the BR genes were counted and presented as the concentration of immunogold labeling in each region relative to the proximal region. A more detailed description of the experimental procedure is provided as Supplemental Data. For double-labeling experiments, the isolated chromosomes were incubated with a mixture of anti-Rrp4 and anti-Pol-II (mAb 4H8) antibodies. Secondary antibodies conjugated to 6- and 12-nm colloidal gold were used to detect the binding of anti-Pol-II and anti-Rrp4 antibodies, respectively.
Stably Transfected S2-Rrp4, S2-Rrp6, and S2-Ski6 Cells
cDNAs comprising the complete open reading frames (ORFs) of the Rrp4, Rrp6, and Ski6 genes, respectively, were amplified using specific primers (Supplemental Table S1) and cloned into the pMT/V5-His B plasmid (Invitrogen). The pMT-Rrp4, pMT-Rrp6, and pMT-Ski6 plasmids were sequenced to confirm the accuracy of the cloning. Stable cells were generated by cotransfecting S2 cells with pCoHygro (Invitrogen) and either pMT-Rrp4, pMT-Rrp6, or pMT-Ski6 using the Calcium Phosphate Transfection Kit (Invitrogen). Stable mock-transfected cells were generated by transfection with the pCoHygro plasmid alone. Stable transfectants were selected in the presence of 300 μg/ml hygromycin B following the procedure recommended by Invitrogen. The details of the cloning are provided as Supplemental Data. Expression of the V5-tagged proteins was induced with 400 μM CuSO4 for 24 h.
Fractionation of S2 Cells
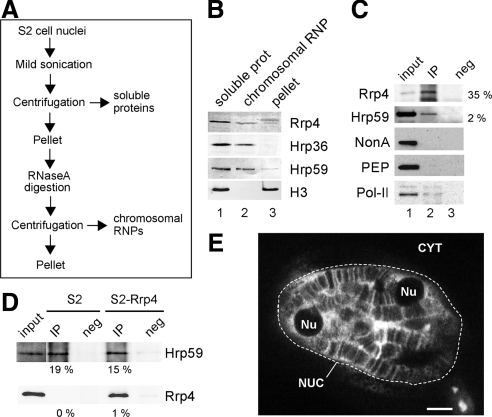
The cells were washed with PBS, resuspended in lysis buffer (PBS, 2 mM MgCl2, 0.2% Nonidet P-40, and Complete protease inhibitors; Roche, Indianapolis, IN), and homogenized using a glass homogenizer (tight pestle). The homogenate was centrifuged at 4000 × g for 10 min at 4°C. The pellet containing the nuclei was resuspended in PBS containing 2 mM MgCl2 and Complete protease inhibitors, sonicated three times for 4–5 s each time, and centrifuged at 16,000 × g for 10 min at 4°C. The resulting supernatant was the soluble nuclear extract referred to as soluble proteins in Figure 6. The pellet was resuspended in PBS, digested with 0.1 mg/ml RNase A at room temperature for 20 min, and centrifuged as above. The supernatant contained chromosomal RNA-binding proteins (chromosomal RNP in Figure 6).
Figure 6.
Rrp4 interacts with Hrp59 on the chromosomes. (A) The flow chart summarizes the fractionation scheme used to prepare two types of nuclear protein extracts containing nuclear soluble proteins and proteins associated with the chromosomes through RNA-mediated interactions. (B) Soluble (lane 1) and chromosomal (lane 2) protein extracts were prepared from S2 cells. The proteins in each fraction were analyzed by Western blot with the anti-Rrp4 antibody. A significant fraction of Rrp4 was found in the chromosomal RNP fraction. Antibodies against the two mRNA-binding proteins, Hrp36 and Hrp59, and against histone H3 were used in parallel to assess the specific composition of the fractions. (C) Chromosomal RNP extracts prepared from S2 cells expressing Rrp4-V5 and used for coimmunoprecipitation experiments. The proteins immunoprecipitated by the anti-V5 antibody (ip, lane 2) were probed by Western blot with antibodies against Rrp4 and Hrp59, as indicated. The percentage of protein immunoprecipitated in each case is given to the right. NonA and PEP, two mRNA-binding proteins abundant in the chromosomal RNP extract, were not coimmunoprecipitated, which supports the specificity of the Rrp4-Hrp59 interaction. Negative control immunoprecipitations were carried out in parallel (neg, lane 3). The input was also analyzed by Western blot (inp, lane 1). Pol-II, detected by mAb 4H8 against the CTD, was coimmunoprecipitated with Rrp4-V5 from soluble extracts. (D) Reciprocal immunoprecipitation. Chromosomal RNP extracts were prepared from S2-Rrp4 cells and from mock-transfected S2 cells, as indicated. The extracts were used for immunoprecipitation using an anti-Hrp59 antibody. The percentage of protein immunoprecipitated in each case is given under each lane. Hrp59 was immunoprecipitated from both S2 and S2-Rrp4 cells, as expected. In S2-Rrp4 cells, a significant fraction of Rrp4-V5 was coimmunoprecipitated. (E) The distribution of Hrp59 was analyzed by immunofluorescence on whole-mount salivary gland preparations. The glands were fixed, permeabilized, and immunostained with an anti-Hrp59 antibody. The image is a confocal section showing the distribution of Hrp59 in the nucleus of a typical salivary gland cell. NUC, nucleus; CYT, cytoplasm; Nu, nucleolus. Scale bar, 10 μm.
ChIP
ChIP was performed essentially as described by Takahashi et al. (2000). Approximately 1 × 107 cells were used for each immunoprecipitation. Expression of Rrp4-V5 in S2-Rrp4 cells was induced with 400 μM CuSO4 for 24 h. The cells were fixed at room temperature by the addition of a fixing solution containing formaldehyde to give a final concentration of 2%. The chromatin was sheared by sonication to give a DNA size of 250–900 base pairs. Immunoprecipitation was performed for 90 min at 4°C using 10 μg/ml primary antibody. A mixture of protein A and protein G coupled to Sepharose (Sigma, St. Louis, MO) was added, and the incubation was continued for an additional 60 min. The specimens were washed with RIPA buffer. The immunoprecipitated DNA was purified and amplified by quantitative real-time PCR in an ABI7000 system using SYBR Green (Applied Biosystems, Foster City, CA). The sequences of primer sets for the different parts of the five genes analyzed are provided as Supplemental Data. At least two independent ChIP experiments were quantified for each gene region analyzed, and each experiment was quantified in triplicate. The relative amounts of coimmunoprecipitated sequences for each PCR product were determined on the basis of the threshold cycle (Ct). Within each experiment, the values obtained were expressed relative to the input, and the values obtained with the negative control antibody were subtracted. The results presented in the histograms are averages relative to the input after subtraction of the negative control values. Error bars show the SDs. ChIP experiments performed with Chironomus salivary glands were carried out as described in Botelho et al. (2008) using the anti-Rrp4 antibody.
Coimmunoprecipitation Assays
The cells were washed with PBS, resuspended in lysis buffer (PBS, 2 mM MgCl2, 0.2% Nonidet P40, and Complete protease inhibitors; Roche), and homogenized using a glass homogenizer (tight pestle). The homogenate was centrifuged at 4000 × g for 10 min at 4°C. The supernatant was the cytoplasmic fraction. The pellet containing the nuclei was resuspended in PBS containing 2 mM MgCl2 and Complete protease inhibitors, sonicated three times for 4–5 s each, and centrifuged. The resulting supernatant was the soluble nuclear extract referred to as soluble RNPs in Figure 4. The pellet was resuspended in PBS, digested with 0.1 mg/ml RNase A, and centrifuged. The supernatant contained chromosomal RNA-binding proteins. These three fractions were precleared and used for coimmunoprecipitation assays using the anti-V5 (Invitrogen) antibody at 5 μg/ml and a mixture of protein A and protein G coupled to Sepharose (Sigma). The bound proteins were eluted with 1% SDS and analyzed by SDS-PAGE and Western blotting.
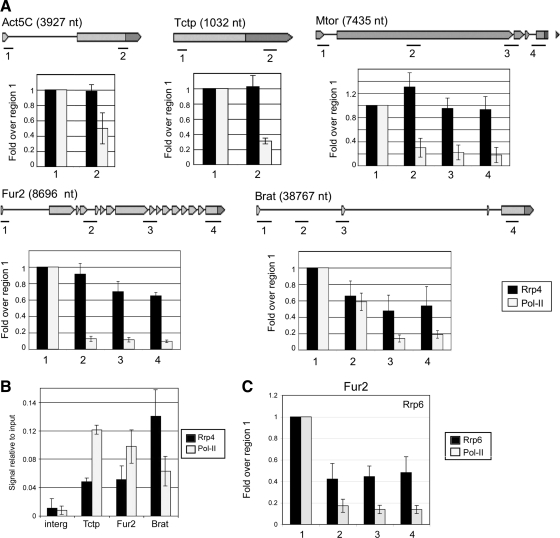
Figure 4.
ChIP analysis of Rrp4 distribution in genes of D. melanogaster. The expression of V5-tagged Rrp4 in S2 cells was induced, and the cells were fixed and processed for ChIP analysis using the anti-V5 antibody (■). Parallel experiments were carried out with antibody 4H8 against Pol-II (□). (A) The histograms summarize the ChIP results obtained for five different genes: Act5C, Tctp, Mtor, Fur2, and Brat, as indicated. The lengths of the genes and their exon–intron structure are depicted (data from http://flybase.bio.indiana.edu). For each gene, the regions analyzed are indicated as short black bars and are numbered 1–4. The relative levels of Rrp4 in each region are calculated relative to the input and expressed as a fold change to region 1 in order to reveal variations along the genes. The values are averages from six PCR reactions from at least two different experiments. Error bars, SDs. (B) The ChIP experiment was as above, but the histogram shows the signals obtained for Rrp4 and Pol-II in region 1 relative to the input, in order to compare the levels of Rrp4 and Pol-II among the different genes. An intergenic sequence (interg) that is devoid of annotated genes according to FlyBase was also analyzed for comparison. (C) The histogram shows the distribution of Rrp6 along the Fur2 gene. The ChIP was carried out as above using S2 cells that express V5-tagged Rrp6.
Immunoprecipitation and High-Performance Liquid Chromatography/Tandem Mass Spectrometry
S2 cells expressing V5-tagged Rrp4, Rrp6, or Ski6 were homogenized in a lysis buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.1% Nonidet P-40, 10% glycerol, and protease inhibitors. Mock-transfected S2 cells were used in parallel as a negative control. The homogenates were centrifuged at 7100 × g for 10 min at 4°C. The nuclear pellets were resuspended in lysis buffer without detergent, sonicated, supplemented with Nonidet P-40 to a final concentration of 0.1%, and digested with 0.1 mg/ml RNase A. The RNase-treated samples were centrifuged at 15,000 × g for 20 min at 4°C and used for immunoprecipitation as described above. The immunoprecipitated proteins were resolved by SDS-PAGE, and each lane was excised and digested with sequence-grade trypsin (Roche). The peptides were extracted and purified essentially as described by Shevchenko et al. (2006).
High-performance liquid chromatography/tandem mass spectrometry (HPLC/MS/MS) was carried out as described by Zubarev et al. (2008). Database searches were carried out using Mascot (version 2.1.3, Matrix Science, London, United Kingdom), and the peptides were identified by searching against the NCBInr (National Center for Biotechnology Information nonredundant) database. The Mascot output files were used as input files for quantification using an in-house written program package (C++) as described in Supplemental Materials and Methods. Three immunoprecipitation experiments followed by HPLC/MS/MS analysis were carried out for each exosome subunit (Rrp4, Rrp6, and Ski6). Protein abundances were normalized by the abundance of the immunoglobulin heavy-chain variable region [Mus musculus] (gi 27728890).
RNA Interference in Drosophila S2 Cells
RNA interference (RNAi) was carried out essentially as described by Hase et al. (2006). Double-strand RNAs (dsRNAs) against Hrp59 and green fluorescent protein (GFP) were prepared by in vitro transcription from PCR products with T7 promoters on both ends of the amplimers, using the Megascript RNAi kit (Ambion, Austin, TX). The PCR primers are described in the Supplemental Data. dsRNA, 20 μg, was applied to S2 cells every 48 h, and the cells were harvested after 96 h.
RESULTS
Ct-Rrp4 Is Associated with Transcribed Genes
The sequences of the exosome proteins of C. tentans were not known when we started the work described here. We cloned the cDNA for C. tentans Rrp4 (referred to as Ct-Rrp4). We aligned the amino acid sequences of the Rrp4 proteins from D. melanogaster and A. gambiae, and we designed degenerate PCR primers corresponding to three highly conserved regions. The primers were used in nested PCR reactions using cDNA prepared from C. tentans as a template. A PCR product of length 471 nt was obtained and sequenced. A BLAST analysis showed that the sequence of the PCR product was homologous to the central part of Rrp4. The flanking 5′ and 3′ sequences of the Ct-Rrp4 cDNA were obtained by RACE.
The full-length cDNA for Ct-Rrp4 contains an ORF of 291 amino acids that encodes a protein of 32.8 kDa. BLAST comparisons showed that Ct-Rrp4 was 61 and 63% identical to the D. melanogaster and A. gambiae orthologues, respectively.
A search for conserved protein domains revealed the existence of two types of RNA-binding domains in Ct-Rrp4. A conserved S1 domain was found between amino acids 81 and 156 (Supplemental Figure S1). A putative KH domain was found downstream of the S1 domain but was not as highly conserved. No obvious nuclear localization signals were identified.
An anti-peptide antibody against Ct-Rrp4 was raised in rabbit and purified by affinity chromatography. The specificity of the purified anti-Rrp4 antibody was analyzed by Western blotting on a total protein extract of C. tentans and on recombinant Rrp4 expressed in E. coli. After affinity purification, the anti-Rrp4 antibody was monospecific (Supplemental Figure S2).
We used the anti-Rrp4 antibody for immunocytochemical studies. We first analyzed the distribution of Rrp4 in the polytene chromosomes of C. tentans by immunofluorescence (Figure 1A). Ct-Rrp4 was detected at numerous loci in all four chromosomes, including the BR puffs in chromosome IV. The nucleolus was intensely stained, as expected from the important role that the exosome plays in pre-rRNA maturation.
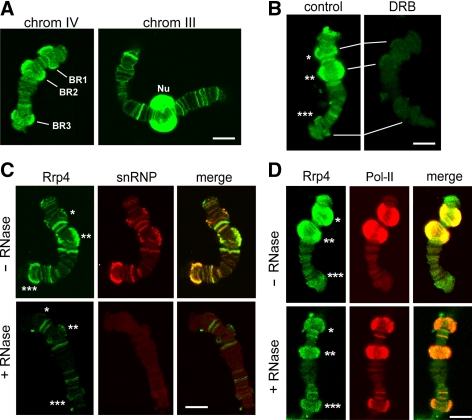
Figure 1.
The association of Ct-Rrp4 with the polytene chromosomes studied by immunofluorescence. (A) Rrp4 is associated with transcribed genes in the polytene chromosomes. Confocal sections of isolated polytene chromosomes stained with the purified anti-Rrp4 antibody. Multiple loci were intensely stained in the chromosomes, including the Balbiani ring puffs BR1, BR2, and BR3 in chromosome IV. The nucleolus (Nu) was also intensely stained. (B) The association of Rrp4 with the chromosomes requires ongoing transcription. Polytene chromosomes from control larvae and from larvae treated with the transcription inhibitor DRB stained for Rrp4 as above. (C) The association of Rrp4 with the chromosomes is RNA-dependent. Polytene chromosomes were isolated and treated with 100 μg/ml RNase A before fixation and immunostaining (+RNase). Control chromosomes were incubated in parallel without RNase A (−RNase). The chromosomes were double-stained with purified anti-Rrp4 antibody (green) and mAb Y12 against snRNP proteins (red). BR1, BR2 and BR3 are indicated with one, two and three asterisks, respectively. (D) The fraction of Rrp4 that is resistant to RNase colocalizes with Pol-II. Polytene chromosomes were isolated and treated with RNase A as in C. The chromosomes were double-stained with purified anti-Rrp4 antibody (green) and mAb 4H8 against the CTD of Pol-II (red). BR1, BR2, and BR3 are indicated with one, two, and three asterisks, respectively. Scale bars, 10 μm.
We next asked whether Rrp4 was associated with the chromosomes only during ongoing transcription (Figure 1B). We treated the larvae with the transcription inhibitor DRB, and we isolated polytene chromosomes for immunostaining. The effect of the DRB treatment was assessed by the reduction of the diameter of the BR puffs (see for instance the reduced size of BR2 in the DRB-treated sample shown in Figure 1B). Figure 1B shows that the immunofluorescent staining was nearly abolished by the DRB treatment.
Preparations of polytene chromosomes were digested with RNase A before immunostaining to determine whether the association of Rrp4 with the chromosomes was mediated by RNA (Figure 1C). The chromosomes were costained with Y12, a mAb against core snRNP proteins, to monitor the effect of the RNase treatment. Control chromosomes were incubated in parallel in the absence of RNase A. The snRNP staining (red in Figure 1C) was abolished by the RNase treatment, as expected. The intensity of the Rrp4 staining (green in Figure 1C) was significantly reduced, and this reduction was very pronounced at the BR puffs (asterisks in Figure 1C). However, a part of the Rrp4 staining was resistant to the RNase treatment. Double-staining of polytene chromosomes with anti-Rrp4 (green in Figure 1D) and anti-Pol-II (red in Figure 1D) antibodies revealed that the RNase-resistant fraction of Rrp4 overlaps with Pol-II. These results suggest that there are two modes of interaction of Rrp4 with the chromosomes. One mode is independent of RNA and may be explained by a direct association of the exosome with the transcription machinery, as reported by Andrulis and coworkers (2002). The other mode is predominant in highly transcribed genes such as the BR genes, and this mode requires RNA.
Ct-Rrp4 Is Present along the Entire BR Transcription Unit
We analyzed the distribution of Ct-Rrp4 along the BR genes in the polytene chromosomes of C. tentans by immuno-EM in order to test the hypothesis that the exosome travels with the transcription machinery along transcribed genes. The exon–intron structure of the BR genes is known (Wieslander and Paulsson, 1992; Figure 2A), and the morphology of the active BR genes and BR mRNPs has been characterized in detail (Skoglund et al., 1983). The active BR genes are loaded with multiple RNA polymerases and, at any given time, each BR gene is being transcribed simultaneously by many RNA polymerases that have reached different elongation stages. This gives the BR genes a distinct polarity and the different regions of the gene show specific morphological features due to the progressive growth of the nascent BR pre-mRNPs (Figure 2B). Full-length genes are not available in the sections used for transmission electron microscopy, but partial gene segments are observed (Figure 2C). The gene segments can be classified into proximal, middle, and distal segments, based on the morphology of the nascent pre-mRNPs. The pre-mRNPs are seen as fibers of increasing length in the proximal portion of the BR gene. In the middle and distal portions, the growing pre-mRNPs appear as stalked granules of increasing diameter (Figure 2, B and C).
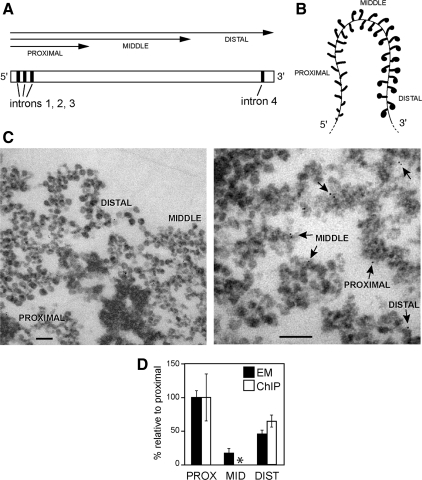
Figure 2.
The association of Ct-Rrp4 with the BR1 and BR2 genes analyzed by immuno-EM. (A) Schematic representation of the BR1 and BR2 genes. The exon–intron organization is shown. The arrows indicate the sequences included in the transcripts that are associated with each portion of the gene. (B) Scheme of the active BR transcription unit illustrating the distinct morphology of the growing pre-mRNPs in the proximal, middle, and distal portions of the BR gene. (C) Immunoelectron microscopic analysis of the Rrp4 distribution in the active BR1 and BR2 transcription units. Polytene chromosomes were immunostained with the purified anti-Rrp4 antibody and the labeling was visualized with a secondary antibody conjugated to colloidal gold markers (6 nm). The left panel shows a section of a chromosome where the proximal, middle, and distal portions of the BR genes can be easily recognized. Right panel, examples of immunogold labeling (arrows). Scale bars, 100 nm. (D) The number of immunogold markers located in each portion of the BR genes was counted, and the relative concentrations of Rrp4 in each portion were calculated by dividing the percentage of immunogold in each portion by the fraction that the portion represents of the total length of the gene (proximal 20%, middle 60%, distal 20%). The histogram shows a compilation of data from the BR1 and BR2 puffs and gives labeling densities relative to the proximal region (■). Gold markers analyzed: n = 681. Error bars, SDs from three independent experiments. The densities of Rrp4 in the proximal and distal regions of the BR1 gene were also analyzed by ChIP (□). The middle region (asterisk) could not be quantified by ChIP because its sequence is highly repetitive. For the proximal and distal regions, the histogram shows average ChIP signals from three independent experiments.
For immuno-EM experiments, polytene chromosomes were manually isolated and immunostained with the purified anti-Rrp4 antibody. The binding sites were revealed with a gold-conjugated secondary antibody. Negative control samples were processed in parallel without primary antibody (not shown). After the immunolabeling, the chromosomes were embedded in plastic and sectioned for EM analysis. Examples of immunolabeling are shown in Figure 2C. Samples from three independent experiments were used to quantify the distribution of labeling in the different parts of the BR gene. For quantification, each gold marker was classified according to its association with the proximal, middle, or distal part of the BR gene. The distribution of the labeling was not uniform along the gene, but significant levels of Rrp4 were detected in all the parts of the BR gene (Figure 2D).
The association of Rrp4 with initial and distal segments of the BR gene was confirmed by ChIP. The association of Rrp4 with the middle region could not be quantified by ChIP because the sequence of the middle region is highly repetitive.
In summary, the results reported above show that Rrp4 is present along the entire length of the BR gene.
Ct-Rrp4 Is Associated with BR Transcripts
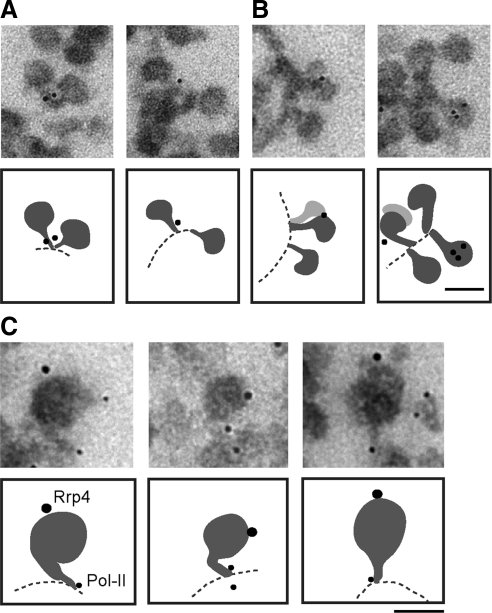
We next analyzed the precise location of Rrp4 in the active BR genes. The BR pre-mRNPs in the distal part of the gene are large enough to allow us to discriminate between labeling near the transcription machinery and labeling to the RNP. We selected immuno-EM images in which the relative positions of the chromatin axis and the nascent pre-mRNPs could be seen, and for each image we determined whether the gold markers were close to the chromatin (within 25 nm from the axis) or distant from the chromatin (more than 25 nm from the axis). Given the dimensions of the antibodies, this latter group contains only gold markers associated with BR pre-mRNP. The markers close to the chromatin axis, in contrast, may label Rrp4 molecules bound to the stalk of the growing pre-mRNP particles, bound to the chromatin, or bound to the transcription machinery. The gold markers were close to the axis in 9 of 19 selected images (Figure 3A), whereas the labeling was far from the axis in 10 of 19 cases (Figure 3B). We conclude that a significant fraction of Rrp4 is associated with the nascent BR pre-mRNPs without being in contact with the transcription machinery. This conclusion was further strengthened by double-labeling experiments in which Pol-II and Rrp4 were detected simultaneously. As shown in Figure 3C, Rrp4 (large gold markers) is located far from the Pol-II (small gold markers) on the nascent pre-mRNP.
Figure 3.
The association of Ct-Rrp4 with nascent BR pre-mRNP particles. The figure shows examples of immunolabeled BR pre-mRNP particles at high magnification. Schematic interpretations of the images are provided under each micrograph. Isolated polytene chromosomes were stained with the anti-Rrp4 antibody (A and B) or with anti-Rrp4 and Pol-II simultaneously (C). (A) Two examples of immunolabeling with anti-Rrp4 antibody showing the association of immunogold markers with the stalk of the nascent BR pre-mRNP. (B) Two examples of immunolabeling showing the association of Rrp4 with the globular domain of the nascent BR pre-mRNP. (C) Double-labeling of Rrp4 and Pol-II. Rrp4 and Pol-II were detected with 12- and 6-nm gold-conjugated secondary antibodies, respectively. Scale bars, 50 nm.
The Distribution of Rrp4 along Transcribed Genes Is Independent of Exon–Intron Organization
The concentration of Rrp4 along the BR gene was not constant (Figure 2D). This observation suggests that the levels of Rrp4 that are present at a given gene region may be determined by sequence-specific features, such as the presence of splice sites. This prompted us to analyze the distribution of the exosome along other genes with different structural features. It is possible to map proteins along only the large BR genes by immuno-EM, and we were therefore compelled to use ChIP instead. ChIP can detect proteins that are bound to the DNA as well as proteins that interact with the nascent transcript (see for example, Görnemann et al., 2005). We used D. melanogaster for these experiments, because the genomic sequence information is available. We constructed stable S2 cells that expressed a V5-tagged version of the Drosophila Rrp4 protein (Rrp4-V5) under the control of an inducible promoter. This cell line was called S2-Rrp4. The expressed Rrp4-V5 was present in both the nucleus and cytoplasm, as expected (Supplemental Figure S4A). The anti-V5 antibody specifically immunoprecipitated Rrp4-V5 (Supplemental Figure S4B and Supplemental Table S2), and the exosome cofactor Rrp6 was coimmunoprecipitated (Supplemental Figure S4C), which indicated that Rrp4-V5 was able to engage in functional interactions.
We used the S2-Rrp4 cells to study the association of Rrp4 with genes of D. melanogaster by ChIP. We arbitrarily selected five genes that differ in length and exon-intron organization: Act5C, Tctp, Mtor, Fur2, and Brat. The expression of these five selected genes in S2 cells was documented in the FLIGHT database (http://flight.licr.org/) and was validated by RT-PCR (Supplemental Figure S5). The expression of Rrp4-V5 was induced for 24 h, chromatin was prepared, and immunoprecipitation was carried out using the anti-V5 antibody. Quantification of the immunoprecipitated DNA was based on real-time PCR assays using primer-pairs specific for different regions of each gene. Negative control ChIP reactions were carried out in parallel using an unrelated antibody. For each gene, the background values obtained in the negative control were subtracted, and the relative density of Rrp4 was expressed relative to the input. To illustrate variations along each gene, the relative abundance of Rrp4 in each region was expressed relative to the most upstream region situated near the transcription start (Figure 4A, ■). Rrp4 was present in all the regions of all the genes analyzed. For Act5C, Tctp, and Mtor, the relative abundance of Rrp4 was relatively constant along the genes. For Fur2 and Brat, the levels of Rrp4 were significantly higher in the 5′ region than at the 3′ end, as immuno-EM had shown for the BR genes.
We could not establish any obvious correlation between the levels of Rrp4 associated with the genes and the presence of splice sites. The patterns of Rrp4 distribution were similar for intronless and intron-containing genes and, within a given gene, the distribution of Rrp4 could not be correlated with intron–exon features. For instance, region 2 of Brat, located in the middle of a very long intron, had levels of Rrp4 similar to those in regions 3 and 4, located in exonic sequences (Figure 4A).
The specificity of the ChIP results was tested using the parental S2 cells that did not express V5-tagged Rrp4. No significant signals were obtained for any of the analyzed genes (data not shown).
We also determined the distribution of Pol-II along the same genes (Figure 4, □). The rationale of this experiment was that if the exosome was tethered to transcribed genes through interactions with the transcription machinery, the levels of Rrp4 would parallel those of Pol-II. However, in all the genes analyzed, the levels of Pol-II decreased in a very pronounced manner toward the 3′ end, and the Pol-II levels in the downstream region of the genes were much lower than those obtained for Rrp4. Similar profiles have been reported for Pol-II in mammalian cells (see, for example, Listerman et al., 2006), and the decrease can be attributed to the fact that many of the Pol-II complexes that are recruited to the gene fail to proceed to productive elongation. In summary, we have shown that the pattern of distribution of Rrp4 does not parallel that of Pol-II. The difference observed between the Rrp4 and Pol-II distributions could be explained if multiple Rrp4 molecules become associated with each transcript during transcription (see Discussion).
Finally, we asked whether the levels of Rrp4 recruited to the upstream region of the genes were similar for all the genes analyzed and proportional to the density of Pol-II. For this purpose, we compared the ChIP signals obtained in the upstream regions of each gene (regions 1). Figure 4B shows that the levels of Rrp4 were different from gene to gene, and did not correlate with the levels of Pol-II in the same region.
We conclude that the amount of Rrp4 and its distribution are to some extent determined by gene-specific features, but are not directly correlated with the exon–intron structure of the gene nor with the levels of Pol-II. The density of Rrp4 decreased in the 5′ to 3′ direction along long genes and this decrease was much less pronounced than that observed for Pol-II, which can be explained if each growing mRNP associates with multiple Rrp4 molecules. The presence of multiple copies of Rrp4 associated with the nascent transcripts in a given region of the gene would result in increased ChIP signals for that particular region.
An important question was whether the results obtained for Rrp4 reflect the behavior of the entire exosome. To address this point, we carried out ChIP experiments using S2 cells that express a V5-tagged version of Rrp6, one of the catalytic subunits of the exosome, and we analyzed the distribution of Rrp6 along the Fur2 gene. As shown in Figure 4C, the density of Rrp6 also decreased in the 5′-to-3′ direction but remained higher than that of Pol-II, as observed for Rrp4.
The Exosome Interacts Physically with the hnRNP Protein Hrp59
The immuno-EM and ChIP results reported above suggest that additional interactions contribute to the localization of the exosome to transcribed genes, in addition to the binding of the exosome to the transcription machinery. We have examined whether Rrp4 interacts with proteins that are associated with the nascent pre-mRNA using a coimmunoprecipitation assay, and we have identified coimmunoprecipitated proteins by mass spectrometry. We prepared nuclear protein extracts from mock-transfected S2 cells and from S2 cells expressing the V5-tagged Rrp4, and we treated the extracts with RNase A to disrupt RNA-mediated interactions. These RNase-treated nuclear extracts were used in immunoprecipitation experiments using an anti-V5 antibody (lane 2 in Figure 5A). Negative control immunoprecipitations were run in parallel using the mock-transfected S2 cells (lane 3 in Figure 5A). The immunoprecipitated proteins were resolved by SDS-PAGE, digested in-gel with trypsin, and analyzed by HPLC/MS/MS. Biological triplicates were carried out for statistical purposes (not shown). In all the experiments, Mascot searches identified Hrp59, the product of the CG9373 gene, as a protein significantly enriched in the immunoprecipitates (Figure 5B, Supplemental Tables S3 and S4), which indicates that Hrp59 and Rrp4 interact physically in vivo. Hrp59, also known as Rumpelstiltskin, is the hnRNP M protein of D. melanogaster (Kiesler et al., 2005; Hase et al., 2006; Jain and Gavis, 2008).
Figure 5.
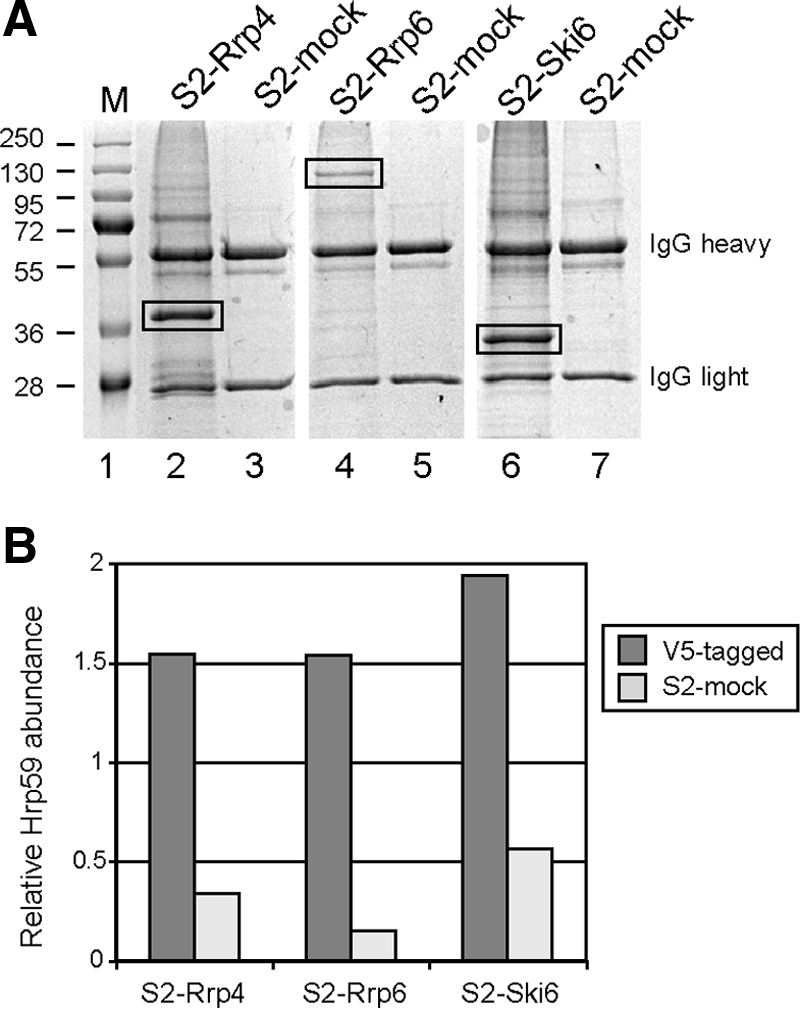
Rrp4, Rrp6 and Ski6 interact with the hnRNP protein Hrp59. Nuclear protein extracts were prepared from S2 cells expressing V5-tagged Rrp4, Rrp6 or Ski6. Mock-transfected S2 cells (S2-mock) were also processed in parallel. The nuclear protein extracts were treated with RNase A and used for immunoprecipitation using the V5 antibody. The immunoprecipitated proteins were resolved by SDS-PAGE and stained with Coomassie brilliant blue. (A) Example of the bound proteins stained with Coomassie. The heavy and light chains of the V5 antibody are indicated to the right. The mobility of prestained molecular mass markers (M) is shown to the left in kDa. (B) The proteins in each lane were processed for HPLC/MS/MS analysis. The mRNA-binding protein Hrp59 was found significantly enriched in the immunoprecipitates of S2-Rrp4, S2-Rrp6, and S2-Ski6 cells. The histogram shows the normalized relative Hrp59 amounts in one immunoprecipitation experiment for each of the cell lines, in arbitrary units. The IgG heavy chain was used for normalization.
We also carried out coimmunoprecipitation experiments using S2 cells that express either Rrp6-V5 or Ski6/Rrp41-V5 (lanes 4 and 6, respectively, in Figure 5A). In both cases, Hrp59 was found significantly enriched in the immunoprecipitates (Figure 5B). These results suggest that Hrp59 is not associated with the individual Rrp4 subunit but with the exosome complex.
The exosome had previously been shown to interact with Pol-II and with the transcription elongation factors Spt5 and Spt6 (Andrulis et al., 2002). These interactions were also detected in our HPLC/MS/MS study (Supplemental Table S4).
We used cell fractionation and immunoprecipitation methods to further characterize the interaction between Rrp4 and Hrp59 and to determine whether this interaction takes place cotranscriptionally. We isolated nuclei from S2 cells and prepared two types of protein extracts (Figure 6A). One of the extracts contained soluble nuclear proteins and RNPs (soluble proteins), and the other extract contained proteins bound to the chromosomes via RNA (chromosomal RNPs). These proteins were released by RNase A digestion. Rrp4 was present in the soluble nuclear fraction, in the chromosomal RNP fraction, and in the pellet (Figure 6B). Antibodies against Hrp36, an abundant hnRNP protein of the A/B type, and against histone H3 were used as controls to assess the quality of the fractions.
We used a chromosomal RNP extract prepared from cells expressing Rrp4-V5 in immunoprecipitation experiments using the V5 antibody. Rrp4 itself was efficiently precipitated as expected (Figure 6C). A significant fraction of Hrp59 (2%) was also detected in the immunoprecipitate (lane 2 in Figure 6C). Other abundant mRNA-binding proteins such as NonA and PEP were analyzed in parallel but were not detected in the immunoprecipitate. Pol-II, which is known to interact with the exosome (Andrulis et al., 2002), was coimmunoprecipitated with Rrp4 (Figure 6C).
We also carried out the reciprocal immunoprecipitation: we immunoprecipitated Hrp59 from S2 cells expressing Rrp4-V5, and we detected coimmunoprecipitated Rrp4 by Western blot using the anti-V5 antibody (Figure 6D). Mock-transfected S2 cells were used in parallel as negative control. It is worth noticing that the immunoprecipitations were carried out using a chromosomal RNP extract that was digested with RNase A (Figure 6A). This suggests that the association of Rrp4 with Hrp59 takes place on the chromosome and that the Rrp4-Hrp59 interaction is not mediated by RNA. From these experiments we conclude that Rrp4 interacts, directly or indirectly, with Hrp59 in vivo.
Hrp59 associates cotranscriptionally with transcripts from many different genes, as shown by the fact that an anti-Hrp59 antibody stains many bands in the polytene chromosomes (Figure 6E and Kiesler et al., 2005). The nucleoli appear as dark spots in the Hrp59-stained cells (Nu in Figure 6E), which indicates that Hrp59 is not present in the nucleoli.
The results presented above indicate that Rrp4 interacts with the hnRNP M protein of Drosophila, Hrp59 and suggest that this interaction does not take place in the nucleolus but on the chromosomes. The HPLC/MS/MS data also suggest that the exosome complex, not Rrp4 alone, is associated with Hrp59. Which of the exosome subunits interacts directly with Hrp59 remains to be elucidated.
Hrp59 Depletion Inhibits the Association of Rrp4 with Transcribed Genes
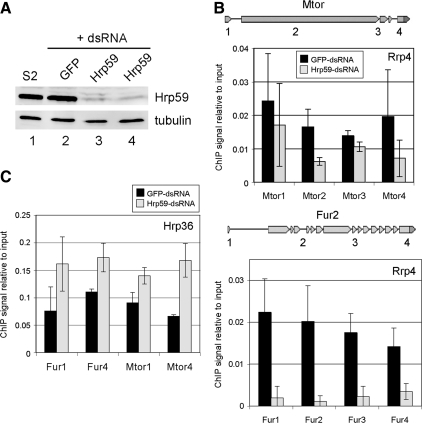
We next explored the functional significance of the interaction between the exosome and Hrp59. If Hrp59 had a role in the recruitment and/or tethering of the exosome to nascent transcripts, depletion of Hrp59 by RNAi would lead to reduced levels of Rrp4 at the transcription sites. We silenced the expression of Hrp59 by RNAi in S2 cells expressing Rrp4-V5, and we analyzed the effects of the silencing on the association of Rrp4 with transcribed genes by ChIP. The efficiency of the RNAi was checked by Western blotting (Figure 7A). Control cells were treated in parallel with GFP-dsRNA. The association of Rrp4 with two genes, Mtor and Fur2, was analyzed by ChIP (Figure 7B). For both genes, depletion of Hrp59 resulted in decreased levels of Rrp4. We asked whether this decrease was a general effect due to limited accessibility of proteins to the transcribed genes in the absence of Hrp59, and we analyzed the association of Hrp36, an abundant hnRNP protein, with the Mtor and Fur2 genes (regions 1 and 4 for each gene). Hrp36 signals were found not significant in an intergenic region (data not shown). As shown in Figure 7C and Supplemental Figure S6, the levels of Hrp36 associated with Mtor and Fur2 increased in response to Hrp59 depletion. This observation rules out a general concealing of the RNP structure and can be explained by the fact that hnRNP proteins have overlapping binding sites on the mRNAs (Dreyfuss et al., 2002). Thus in the absence of Hrp59, other abundant hnRNP proteins are likely to gain access to the transcripts.
Figure 7.
Depletion of Hrp59 by RNAi in S2-Rrp4 cells inhibits with the association of Rrp4 with the transcribed genes. (A) S2-Rrp4 cells were treated with dsRNA for either GFP (lane 2) or Hrp59 (lanes 3 and 4, two independent experiments). The efficiency of the silencing was analyzed by Western blot with and anti-Hrp59 antibody. Tubulin was analyzed in parallel. Untreated S2 cells were analyzed in parallel (lane 1). (B) Chromatin was prepared from S2-Rrp4 cells treated with dsRNA for either Hrp59 or GFP as a control. The association of Rrp4-V5 with the Mtor and Fur2 genes was analyzed by ChIP using the anti-V5 antibody, as in Figure 4. The histograms show the average quantitative PCR (qPCR) signals from two independent experiments relative to input. Each qPCR measurement was done in triplicate. Error bars, SDs. (C) ChIP as in B using an antibody against the mRNA-binding protein Hrp36. Regions 1 and 4 for Mtor and Fur2 were analyzed. The histogram shows three qPCR reactions from one experiment. Another experiment of the same type is shown in Supplemental Figure S6. Note that the Hrp36 signal was increased in Hrp59-depleted cells.
In summary, our results show that depletion of Hrp59 interferes with the association of Rrp4 with the transcribed genes, which suggests that Hrp59 is involved in the interaction between the exosome and the pre-mRNP. Our results also suggest that other hnRNP proteins bind to the nascent pre-mRNA in the absence of Hrp59, and that the resulting mRNP complexes have a reduced ability to interact with the surveillance machinery.
DISCUSSION
We have studied the association of Rrp4 with protein-coding genes, and we have shown that Rrp4 is recruited to transcriptionally active loci in the polytene chromosomes of C. tentans. This agrees with previous observations in D. melanogaster, in which the exosome is associated with the RNA polymerase II and with the transcription elongation factors Spt5 and Spt6 (Andrulis et al., 2002). These interactions have led to the proposal that the transcription machinery is responsible for recruiting the exosome to sites of pre-mRNA synthesis and that the exosome travels along the transcribed genes bound to the transcription elongation complex (Andrulis et al., 2002). The results of our work on the BR genes of C. tentans are compatible with the hypothesis that the transcription machinery is involved in the recruitment and tethering of the exosome to transcribed genes. However, our work has also revealed the existence of an additional mechanism that operates in the cotranscriptional surveillance of nascent pre-mRNPs. This additional mechanism involves an interaction between the exosome and the nascent pre-mRNP and requires the hnRNP M protein Hrp59.
We have determined the precise location of Rrp4 in the BR1 and BR2 genes by immuno-EM, and we have shown that a fraction of Rrp4 is directly associated with the nascent BR pre-mRNP without any contact with the transcription machinery. Our results from cell fractionation experiments and from experiments in which the chromosomes were digested with RNase A before they were immunostained also point to the existence of a fraction of Rrp4 that is bound to the chromosomes via RNA. We propose that a fraction of Rrp4 binds directly to the nascent pre-mRNP, whereas another fraction is associated with the chromosomes independently of RNA (Figure 8). This latter fraction is likely to be associated with the transcription elongation factors Spt5 and Spt6, as reported by Andrulis et al. (2002). Given that the number of Pol-II molecules present in a given gene region equals the number of transcripts, the finding that the relative density of Rrp4 along the genes remains higher than the density of Pol-II suggests that multiple Rrp4 molecules bind to each nascent transcript during transcription. This would be difficult to explain by interactions between the exosome and the transcription machinery, but is in full agreement with the finding that at least three exosome subunits can also interact with hnRNP proteins, which are usually associated with the pre-mRNA in multiple copies.
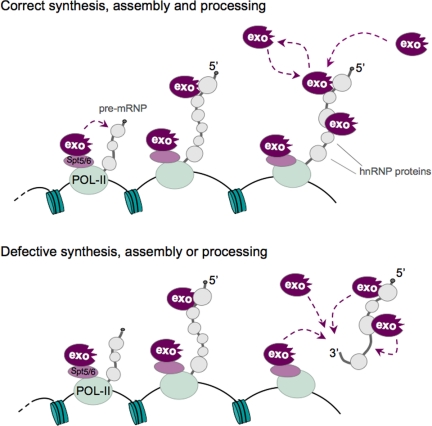
Figure 8.
A model for the cotranscriptional surveillance of mRNA biogenesis. The recruitment of the exosome takes place continuously during transcription and is based on two types of interaction that are independent of gene expression errors. On the one hand, the exosome has been shown previously to bind to the transcription machinery. On the other hand, our data demonstrates that the exosome interacts with the nascent pre-mRNPs and that this interaction is mediated by specific hnRNP proteins, such as Hrp59. The exosomes associated with the pre-mRNP could either be recruited by the transcription machinery and transferred to the transcript during elongation or recruited directly to the pre-mRNP. The interactions of the exosome with the transcription machinery and with the nascent pre-mRNP would be sufficient to establish an increased local concentration of surveillance factors around the transcript that could ensure that any RNA substrates that arise from transcription and/or processing errors are efficiently degraded.
We have shown that three different exosome subunits can be coimmunoprecipitated with the mRNA-binding protein Hrp59 in a RNA-independent manner. Given that Hrp59 is an hnRNP protein that binds to nascent mRNPs cotranscriptionally (Kiesler et al., 2005) and given that Hrp59 interacts with Rrp4, Ski6/Rrp41, and Rrp6, we propose that Hrp59 mediates the association of the exosome to the nascent pre-mRNPs. Several exosome subunits, including Rrp4, contain RNA-binding domains and can, in principle, bind RNA directly. However, the pronounced effect that Hrp59 depletion has on the association of Rrp4 with transcribed genes indicates that direct RNA binding does not play a major role in the association of the exosome with nascent transcripts. Alternatively, Hrp59 could act as a cofactor necessary for the direct binding of Rrp4—or the exosome—to RNA.
Hrp59 is evolutionarily conserved. The closest orthologues of Hrp59 in S. cerevisiae are Gbp2 and Hrb1, two shuttling mRNA-binding proteins that are recruited to the pre-mRNA cotranscriptionally (Hurt et al., 2004), and a genetic interaction between Gbp2 and Rrp6 has been reported (Wilmes et al., 2008). Further research is needed to determine whether Gbp2 and/or Hrb1 are involved in the recruitment of the exosome to active genes.
It is unclear how the exosome discriminates between correct and defective transcripts. We have found Rrp4 associated with many transcriptionally active loci under normal growth conditions in C. tentans, which suggests that the discrimination between correct and defective transcripts is not based on the selective recruitment of the exosome to defective RNAs. Our results suggest that the exosome instead is recruited to expressed genes by molecular interactions that are independent of expression defects. It has been proposed that the packaging of the mRNA into an mRNP complex plays a crucial role in the definition of aberrant transcripts. According to this proposal, the correct assembly of the nascent mRNA into a mRNP complex protects the transcript from degradation by the exosome. Instead, aberrant transcripts become assembled into abnormal mRNP structures that render the mRNA unprotected and susceptible to degradation (Jensen et al., 2003).
Two different strategies can be envisioned for the cotranscriptional recruitment of surveillance factors to sites of mRNA synthesis. The exosome could be specifically recruited to nascent transcripts as specific processing reactions take place. For example, the exosome could be recruited to pre-mRNA splice sites by the spliceosome for surveillance of intron removal. Alternatively, the recruitment of the exosome could be constitutive and independent of specific processing events. For instance, the degradation of cryptic unstable transcripts in S. cerevisiae is closely coupled to the Nrd1p-Nab3p–dependent transcription termination pathway (Arigo et al., 2006; Thiebaut et al., 2006). Our results suggest, in contrast, that protein-coding genes in insects are monitored by a constitutive mechanism. Our ChIP analysis shows that the exosome is present along the complete length of protein-coding genes, even of very long exons and in intronless transcripts, and the density of Rrp4 along the genes does not correlate with their exon–intron structure. The interactions of the exosome with the Pol-II machinery and with the hnRNP protein Hrp59 provide two general mechanisms for the constitutive recruitment of the exosome to protein-coding transcripts.
Hrp59 is an abundant hnRNP protein that belongs to the hnRNP M family. Hrp59 associates cotranscriptionally with transcripts from many different genes, as shown by immunostaining of polytene chromosomes, and it is probable that Hrp59 has a general role in the packaging of mRNPs (Kiesler et al., 2005). Hrp59 has also a specific role in the regulating the alternative splicing of some transcripts (Hase et al., 2006). We have now shown that pre-mRNAs synthesized in the absence of Hrp59 are defective in the recruitment of the surveillance machinery. On the basis of the fact that Hrp59 is associated with the nascent mRNA and with Rrp4, we propose that Hrp59 is directly involved in the recruitment of the exosome to expressed genes and works as an adaptor for mRNA surveillance.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to B. Björkroth (CMB, Karolinska Institutet) for technical support and George Farrants for language editing. We are also grateful to E. Andrulis, H. Saumweber, and S. Amero for the generous gift of antibodies against Rrp6, NonA, and PEP, respectively. Alexander R. Zubarev wrote the code for protein quantification. This work was supported by the Swedish Research Council (Grants 2006-5398 to N.V. and 621-2007-4410 to R.Z.), the Carl Trygger Foundation, the Swedish Cancer Society, the Knut and Alice Wallenberg Foundation (instrumental grant to R.Z.), and the European Science Foundation (RNAQuality Programme to N.V.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0079) on June 10, 2009.
REFERENCES
- Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E., Werner J., Nazarian A., Erdjument-Bromage H., Tempst P., Lis J. T. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Arigo J. T., Eyler D. E., Carroll K. L., Corden J. L. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Björkroth B., Ericsson C., Lamb M. M., Daneholt B. Structure of the chromatin axis during transcription. Chromosoma. 1988;96:333–340. [Google Scholar]
- Botelho S. C., Tyagi A., Hessle V., Farrants A. K., Visa N. The association of Brahma with the Balbiani ring 1 gene of Chironomus tentans studied by immunoelectron microscopy and chromatin immunoprecipitation. Insect Mol. Biol. 2008;17:505–513. doi: 10.1111/j.1365-2583.2008.00825.x. [DOI] [PubMed] [Google Scholar]
- Butler J. S. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- Daneholt B. Assembly and transport of a pre-messenger RNP particle. Proc. Natl. Acad. Sci. USA. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Narry K. V., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Dziembowski A., Lorentzen E., Conti E., Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Galy V., Gadal O., Fromont-Racine M., Romano A., Jaquier A., Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Görnemann J., Kotovic K. M., Hujer K., Neugebauer K. M. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Hase M. E., Yalamanchili P., Visa N. The Drosophila heterogeneous nuclear ribonucleoprotein M protein, HRP59, regulates alternative splicing and controls the production of its own mRNA. J. Biol. Chem. 2006;281:39135–39141. doi: 10.1074/jbc.M604235200. [DOI] [PubMed] [Google Scholar]
- Hilleren P., McCarthy T., Rosbash M., Parker R., Jensen T. H. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Hurt E., Luo M. J., Röther S., Reed R., Strässer K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. USA. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. A., Gavis E. R. The Drosophila hnRNP M homolog Rumpelstiltskin regulates nanos mRNA localization. Dev. 2008;135:973–982. doi: 10.1242/dev.015438. [DOI] [PubMed] [Google Scholar]
- Jensen T. H., Dower K., Libri D., Rosbash M. Early formation of mRNP: license for export or quality control? Mol. Cell. 2003;11:1129–1133. doi: 10.1016/s1097-2765(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Kiesler E., Hase M. E., Brodin D., Visa N. Hrp59, an hnRNP M protein in Chironomus and Drosophila, binds to exonic splicing enhancers and is required for expression of a subset of mRNAs. J. Cell Biol. 2005;168:1013–1025. doi: 10.1083/jcb.200407173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E., Visa N., Wurtz T., Daneholt B. Immunocytochemical evidence for a stepwise assembly of Balbiani ring premessenger ribonucleoprotein particles. Eur. J. Cell Biol. 1997;74:407–416. [PubMed] [Google Scholar]
- LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lerner E. A., Lerner M. R., Janeway C. A., Jr., Steitz J. A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E., Basquin J., Conti E. structural organization of the RNA-degrading exosome. Curr. Opin. Cell Biol. 2008;18:709–713. doi: 10.1016/j.sbi.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Liu Q., Greimann J. C., Lima C. D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [erratum: Cell 131, 188–189] [DOI] [PubMed] [Google Scholar]
- Listerman I., Sapra A. K., Neugebauer K. M. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- Lorentzen E., Walter P., Fribourg S., Evguenieva-Hackenberg E., Klug G., Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- Luna R., Jimeno S., Martin M., Huertas P., Garcia-Rubio M., Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Meyer B., Mähr R., Eppenberger H. M., Lezzi M. The activity of Balbiani rings 1 and 2 in salivary glands of Chironomus tentans larvae under different modes of development and after pilocarpine treatment. Dev. Biol. 1983;98:265–277. doi: 10.1016/0012-1606(83)90357-3. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Tollervey D. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 2000;7:843–846. doi: 10.1038/82817. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Houalla R., Podtelejnikov A., Mann M., Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M. S., Mapendano C. K., Schierup M. H., Jensen T. H. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008 doi: 10.1126/science.1164096. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Rougemaille M., Gudipati R. K., Olesen J. R., Thomsen R., Seraphin B., Libri D., Jensen T. H. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguez C., Olesen J. R., Jensen T. H. Formation of export-competent mRNP: escaping nuclear destruction. Curr. Opin. Cell Biol. 2005;17:287–293. doi: 10.1016/j.ceb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Schmid M., Jensen T. H. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Schneider C., Anderson J. T., Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Skoglund U., Andersson K., Björkroth B., Lamb M. M., Daneholt B. Visualization of the formation and transport of a specific hnRNP particle. Cell. 1983;34:847–855. doi: 10.1016/0092-8674(83)90542-1. [DOI] [PubMed] [Google Scholar]
- Sommer P., Nehrbass U. Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr. Opin. Cell Biol. 2005;17:294–301. doi: 10.1016/j.ceb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Rayman J. B., Dynlacht B. D. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Thiebaut M., Kisseleva-Romanova E., Rougemaille M., Boulay J., Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S., Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Staples R. R., Baker R. E., Parker R. Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol. 2002;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P., Stutz F. mRNA export: an assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Wang H. W., Wang J., Ding F., Callahan K., Bratkowski M. A., Butler J. S., Nogales E., Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc. Natl. Acad. Sci. USA. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L. The Balbiani ring multigene family: coding repetitive sequences and evolution of a tissue-specific cell function. Prog. Nucleic Acid Res. Mol. Biol. 1994;48:275–313. doi: 10.1016/s0079-6603(08)60858-2. [DOI] [PubMed] [Google Scholar]
- Wieslander L., Bauren G., Bernholm K., Jiang W. Q., Wetterberg I. Processing of pre-mRNA in polytene nuclei of Chironomus tentans salivary gland cells. Exp. Cell Res. 1996;229:240–246. doi: 10.1006/excr.1996.0366. [DOI] [PubMed] [Google Scholar]
- Wieslander L., Paulsson G. Sequence organization of the Balbiani ring 2.1 gene in Chironomus tentans. Proc. Natl. Acad. Sci. USA. 1992;89:4578–4582. doi: 10.1073/pnas.89.10.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes G. M., et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F., et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Wyss C. Chironomus tentans epithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp. Cell Res. 1982;139:297–307. doi: 10.1016/0014-4827(82)90255-5. [DOI] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra P., Wyss J. C., Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev R. A., Nielsen M. L., Fung E. M., Savitski M. M., Kel-Margoulis O., Winbender E., Kel A. Identification of dominant signaling pathways from proteomics expression data. J. Proteomics. 2008;71:89–96. doi: 10.1016/j.jprot.2008.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.