Abstract
Several well-studied promoters in yeast lose nucleosomes upon transcriptional activation and gain them upon repression, an observation that has prompted the model that transcriptional activation and repression requires nucleosome remodeling of regulated promoters. We have examined global nucleosome positioning before and after glucose-induced transcriptional reprogramming, a condition under which more than half of all yeast genes significantly change expression. The majority of induced and repressed genes exhibit no change in promoter nucleosome arrangement, although promoters that do undergo nucleosome remodeling tend to contain a TATA box. Rather, we found multiple examples where the pre-existing accessibility of putative transcription factor binding sites before glucose addition determined whether the corresponding gene would change expression in response to glucose addition. These results suggest that selection of appropriate transcription factor binding sites may be dictated to a large extent by nucleosome prepositioning but that regulation of expression through these sites is dictated not by nucleosome repositioning but by changes in transcription factor activity.
INTRODUCTION
Chromatin, whose basic unit is a nucleosome that consists of 147 base pairs of DNA wrapped twice around a histone octamer, plays a central role in the regulation of genes. Several experiments in yeast have shown that nucleosome depletion causes derepression of PHO5, GAL1, CUP1, SUC2, and HIS3 in the absence of their transcriptional activators (Han and Grunstein, 1988; Han et al., 1988; Durrin et al., 1992; Hirschhorn et al., 1992). These observations suggested that nucleosomes in promoters can function as nonspecific repressors, consistent with the concept that DNA sequences incorporated into nucleosomes are less accessible to DNA binding proteins such as transcription factors (Morse, 2003, 2007). Subsequent observations have demonstrated that chromatin structure can participate actively in gene regulation through repositioning of nucleosomes so as to block access of a transcription factor to its cognate binding sites or to remove a barrier to such access. Consistent with this notion, several examples have been reported in which gene induction involves recruitment to a promoter of chromatin remodeling factors and histone modifying activities designed to remove or reposition a nucleosome to allow access to an otherwise masked transcription factor binding site (Li et al., 2007; Williams and Tyler, 2007). For example, remodeling factors act at the above-mentioned promoters for PHO5, GAL1, CUP1, and SUC2 to facilitate nucleosome loss on transcriptional activation and conversely to assemble or stabilize nucleosomes on the gene promoter during transcriptional repression (Almer et al., 1986; Fedor and Kornberg, 1989; Shen et al., 2001; Kim et al., 2006).
Chromatin can also play a more passive role in gene regulation by simply controlling which transcription factor binding sites are available for participation in gene regulation. Recent genome-wide studies of nucleosome positioning found that most yeast promoters contain an extended nucleosome-depleted region (NDR) that is enriched for transcription factor binding sites (Lee et al., 2004; Yuan et al., 2005; Lee et al., 2007; Mavrich et al., 2008; Shivaswamy et al., 2008). Moreover, sequences to which a transcription factor can bind are often more prevalent in the genome than the sites to which that factor is actually bound in vivo. For example, the yeast genome contains many more high-affinity binding motifs for Rap1 and Leu3 transcription factors than are bound by the cognate factor in vivo (Lieb et al., 2001; Liu et al., 2006). In addition, not all genes with a given transcription factor binding site show correlated expression patterns. These discrepancies may be explained by selective occlusion of transcription factor binding sites by positioned nucleosomes. For example, Leu3 sites to which Leu3 is not bound in vivo tend to lie under well positioned nucleosomes (Liu et al., 2006). Thus, as Morse has proposed, chromatin may be “instructive” for transcription factor binding (Morse, 2007).
To distinguish active versus passive roles of chromatin in regulating gene expression, Elgin and coworkers postulated two general categories of promoter architecture (Lu et al., 1994; Wallrath et al., 1994): “pre-set” promoters whose nucleosome structure does not significantly change with changes in transcription, such as the Drosophila hsp26 and hsp70 promoters; and “remodeling” promoters whose nucleosome structure changes significantly with gene expression changes, such as the mammalian mouse mammary tumor virus long terminal repeat promoter and the yeast PHO5 promoter. To assess whether this dichotomy applies to most yeast promoters and, if so, which yeast promoters fall into each category, we have explored the positions of nucleosomes across the entire yeast genome under conditions in which a majority of yeast genes undergo significant change in gene expression. Several recent studies have reported the precise nucleosome positions across the yeast genome (Yuan et al., 2005; Lee et al., 2007; Mavrich et al., 2008; Shivaswamy et al., 2008; Kaplan et al., 2009). Most studies of genome-wide nucleosome positioning have been conducted at steady states by using cells grown in rich media. However, the Segal laboratory observed that most nucleosome positions remain unchanged in several alternative carbon sources (Kaplan et al., 2009). Shivaswamy et al. (2008) examined nucleosome positioning before and after heat shock and observed discrete local nucleosome remodeling events at gene promoters but found no general relationship between transcriptional changes and nucleosome remodeling events. In contrast, Lee et al. (2004) also examined nucleosome occupancy changes during heat shock, by using lower resolution techniques, and concluded that promoter nucleosome occupancy changed inversely with transcriptional change.
We determined nucleosome occupancy before and after addition of glucose to cells grown in a poor carbon source, a nutrient upshift in which more than half of all yeast genes significantly change expression (Wang et al., 2004; Zaman et al., 2009). Accordingly, we have been able to correlate changes in nucleosome occupancy to changes in transcriptional activity in a large number of genes and relate those changes to the presence of previously predicted transcription factor binding motifs. We find that most genes that undergo significant expression changes do not exhibit any observable change in nucleosome positioning within their promoters, although those that do are significantly enriched for TATA boxes. Moreover, we find quite often that the accessibility of specific transcription factor binding motifs within a promoter before the nutrient upshift determines whether the corresponding gene will exhibit a change in expression. Thus, our results ratify Morse's postulate of a largely instructive role for chromatin in regulating gene expression.
MATERIALS AND METHODS
Nucleosomal DNA Isolation
We prepared nucleosomal DNA using an adaptation of previously described procedures (Kent et al., 1993; Yuan et al., 2005). From a 2-liter culture of strain Y2864 (Zaman et al., 2009) grown at 30° in SC + 3% glycerol to 2–3 × 106 cells/ml (A660 = 0.2–0.3), we removed and cross-linked (see below) 650 ml of culture as the zero time point and added glucose to 2% to the remaining culture. We then removed 650 ml of culture after 20 and 60 min and cross-linked cells by addition of formaldehyde to a concentration of 4% and incubation at room temperature for 30 min with gentle shaking; we found that spheroplasts from glycerol-grown cells are extremely fragile, necessitating increased cross-linking relative to previously published protocols. Cross-linking was terminated by addition of glycine to a final concentration of 250 mM and incubation for 5 min. Cross-linked cells were harvested, washed in 40 ml of 0°C phosphate-buffered saline, harvested, and the cell pellets snap-frozen in liquid nitrogen. We resuspended cells from the pellets in 10 ml of prespheroplasting buffer (100 mM Tris, pH 9.0, and 10 mM dithiothreitol [DTT] added freshly), incubated the suspension at 10 min at room temperature, harvested cells, and resuspended them in 20 ml spheroplasting buffer (50 mM KH2PO4/K2HPO4 pH 7.5, 1.0 M sorbitol, and 10 mM DTT added fresh) containing 0.25 mg/ml Zymolyase 100T (Seikagaku, Tokyo, Japan). Cells were incubated at 30°C until converted to >95% spheroplasts (∼30 min). We washed spheroplasts twice with 40 ml of 0°C spheroplasting buffer, gently resuspended them in 5 ml of NP-buffer (50 mM NaCl, 10 mM Tris, pH 7.4, 1 M sorbitol, 5 mM MgCl2, 1 mM CaCl2, with 1 mM β-mercaptoethanol and 500 μM spermidine added fresh), and then added Nonidet-P40 at a final concentration of 0.025%. We added micrococcal nuclease (10 U; Sigma-Aldrich, St. Louis, MO) and incubated the lysate for 45 min at 37°C, digestion conditions in which chromatin was converted predominantly to mononucleosomes (Supplemental Figure S1). We stopped digestions by addition of 1 ml of 5% SDS and 50 mM EDTA, followed by addition of 10 μl of 10 mg/ml RNaseA and incubation at 37°C for 30 min. We then added 5 μl of 10 mg/ml proteinase K and reversed the cross-links by incubation at 65°C for at least 4 h. DNA was purified using a polymerase chain reaction (PCR) clean-up kit (QIAGEN, Valencia, CA) and then analyzed by gel electrophoresis to assess the extent of digestion. We performed this experiment for four biological replicates, all of which yielded essentially identical results. Data for one of the replicates are presented in the Results.
Microarray Hybridization and Data Analysis
DNA (8 μg) was labeled and hybridized to 1.0R yeast tiling arrays (Affymetrix, Santa Clara, CA) as described previously (Gresham et al., 2006), except that fragmentation was accomplished by digestion with 0.05 U of DNaseI (Invitrogen, Carlsbad, CA) for 2 min at 37°C. Raw microarray data for individual time points was analyzed with Affymetrix Tiling Analysis Software, version 1.1 with settings as described previously (Lee et al., 2007). The microarray data are available for download from http://puma.princeton.edu/publications.html. Yeast mRNA expression data have been published previously (Zaman et al., 2009).
A Probabilistic Algorithm for Predicting Nucleosome Positions from Tiled Microarray Data
We developed a hidden Markov model (HMM) (Rabiner, 1989) to infer genome-wide nucleosome positions from hybridization data (Supplemental Figure S2). The model specifies nucleosome positions as peaks of probability that DNA will enter a nucleosome at a given base pair in the genome. The model also returns the overall nucleosome occupancy of any base pair in the genome as the probability that it is covered by any nucleosome. To compute the nucleosome occupancy for a base pair, we sum over posterior probabilities for all nucleosomes that initiate in the appropriate direction within 148 base pairs (length of 37 probes separated by 4 base pairs) of that site (Morozov et al., 2008). Hybridization values from the tiled Affymatrix arrays were used as input to the HMM, and the parameters of the model were fit by maximum likelihood. Because consecutive probes are not always equidistant on the chip, all log intensities were first placed on a regular four-base pair grid by linear interpolation. Gaps of 50 base pairs or longer were assigned zero hybridization values for the HMM but omitted from all subsequent bioinformatic analysis. The parameters of the HMM are fit on a set of 19 genes that show no appreciable changes in expression between 0 and 20 min (POL4, CDC20, RNR3, RAM2, RSM22, CDC73, ASC1, SCP160, ILV3, NNF1, AKR1, SKI2, MDL1, SAS4, SNF5, URE2, YFB3, SRL2, and MEC3), and their associated 5′ intergenic regions. The model uses a mixture of two Gaussians to represent both nucleosome state and linker state. The final set of fitting parameters includes means and widths of four one-dimensional Gaussians, two independent mixture coefficients, and 38 initial probabilities for the nucleosome and linker states. The topology of the Bayesian network (Supplemental Figure S2) reflects the fact that all 37 nucleosome states are forced to share the same mixture of two Gaussians. The HMM parameters inferred independently for all 19 loci were averaged and the resulting model was run genome-wide on each data set. We implemented the HMM in Matlab (The MathWorks, Natick, MA) as a dynamic Bayesian network using standard libraries from the Bayes Net Toolbox (BNT; http://www.cs.ubc.ca/∼murphyk/Software/BNT/bnt.html). Our Matlab code is available upon request.
RESULTS
Genome-wide Positions of Nucleosomes in Saccharomyces
To determine the changes in nucleosome positioning that accompany changes in gene expression, we determined nucleosome positions across the entire yeast genome before and after shifting cells from growth in glycerol to growth in glucose. This carbon source upshift significantly alters expression of more than half of all yeast genes (Wang et al., 2004; Zaman et al., 2009) and thereby provides numerous individual examples to examine the relationship between nucleosome positioning and gene expression. To perform this analysis, samples of glycerol grown cells were collected and formaldehyde cross-linked both before glucose addition and 20 and 60 min after addition. Chromatin isolated from these samples was digested with sufficient micrococcal nuclease to produce predominantly mononucleosomes (Supplemental Figure S1). These digested preparations were then heated to reverse the cross-links and release the nucleosomal-protected DNA, which was then hybridized to Affymetrix tiling microarrays that contain 25-mer probes offset by four to five base pairs across the entire unique sequence of the yeast genome.
To facilitate interpretation of the nucleosome positioning data, we developed a novel HMM to predict nucleosome occupancy from hybridization intensity data. Although HMMs have been previously used to predict nucleosome positions (Yuan et al., 2005; Lee et al., 2007), our approach has several important distinctions. First, all nucleosomes have a canonical length of 148 base pairs (37 probes separated by 4 base pairs). Second, the probability of starting a new nucleosome (PLN in Supplemental Figure S2) is not fit by maximum likelihood but rather is adjusted manually to reproduce physiologically relevant nucleosome occupancies in bulk chromatin (Wolffe, 1998). The manual adjustment reflects our lack of control over the zero intensity baseline, which can shift depending on the relative amounts of DNA used in the nucleosomal and control samples. Third, we model the probability of observing a nucleosome or linker state by a mixture of two Gaussians. We sought to account for the fact that a single Gaussian will cause the assignment of small nucleosome probabilities to high log intensity values. This problem is resolved by using a mixture model which can in principle fit functions of arbitrary shape, although in practice the number of Gaussian components has to be limited to two or three to reduce the total number of fitting parameters. In sum, our model avoids the need for post hoc assumptions of dubious biological accuracy, such as nucleosomes of noncanonical length, whereas, as described below, maintaining accurate predictions of experimentally determined in vivo nucleosome positions.
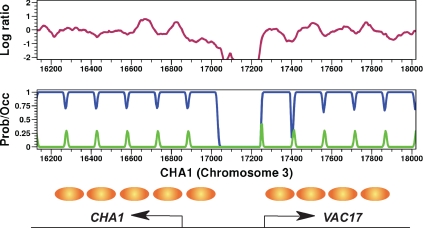
To test our HMM, we applied it to our hybridization data for a set of seven reference loci, independently of those used for training, with previously determined nucleosome positions. This stringent analysis indicated that interpretation of our data by this model places nucleosomes with statistically significant accuracy (Supplemental Figure S3) similar to that of a previously published study (Lee et al., 2007). We also applied the HMM to our hybridization data at the CHA1 gene, whose nucleosome organization has been previously determined both by indirect end labeling and by global nucleosome positioning studies (Moreira and Holmberg, 1998; Yuan et al., 2005; Lee et al., 2007). Our HMM places nucleosomes at locations determined by previous studies (Figure 1). CHA1 expression is glucose independent, and we find that our calculated nucleosome occupancy at this locus remains unchanged before glucose addition and 20 and 60 min post-glucose addition (Supplemental Figure S4). From these analyses, we conclude that our data sets, in conjunction with the HMM we developed to interpret those data, yield results highly consistent with prior nucleosome position determinations, and we conclude that both the model and the data are reliable.
Figure 1.
Nucleosome positioning at the CHA1 promoter. Top, log ratio of nucleosomal DNA to genomic DNA as determined by separate hybridizations to Affymetrix tiling arrays is plotted as a function of genomic position. Increasing values represent increasing MNase protection. Middle, nucleosome positions as predicted by the HMM used in this study. Blue track represents predicted nucleosome occupancy, from unoccupied (0) to fully occupied (1). Green track represents the probability of initiating a nucleosome at that location. Bottom, previously determined in vivo nucleosome positions (Moreira and Holmberg, 1998) are shown as brown ovals.
Certain genomic regions, such as the middle of longer open reading frames (ORFs), tend to yield hybridization patterns inconsistent with precisely positioned nucleosomes but rather present a pattern interpreted to indicate that different cells in the population contain nucleosomes at different positions (Mavrich et al., 2008). Unlike previously described HMMs, which account for these regions by invoking a “delocalized nucleosome state” of indefinite length (Yuan et al., 2005; Lee et al., 2007), our HMM deconstructs regions of delocalized nucleosomes as multiple overlapping arrays of nucleosomes with partial occurrence in the population. This modification improves the resolution of our nucleosome predictions in the regions of the genome not packaged in well-positioned nucleosomes. For example, glucose addition represses the ARP2 gene and induces a slight shift at 60 min post addition in the positioning of the nucleosome array in the 5′ of the ORF (Supplemental Figure S5). Interestingly, at the 20-min time point our HMM predicts the nucleosome positions to be delocalized relative to the 0- and 60-min time points, suggesting that different cells in the population may contain the nucleosome array in both the pre- and post-glucose addition conformation.
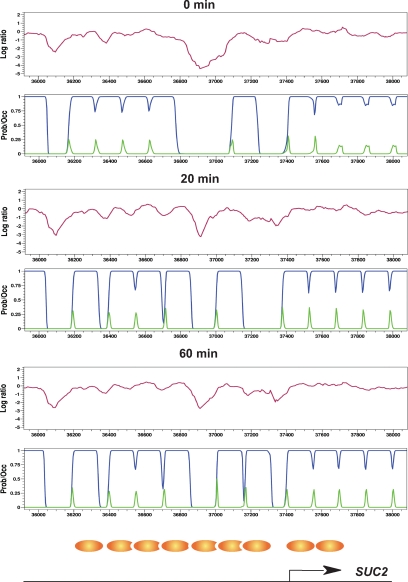
To further validate our data and methodology, we examined nucleosome occupancy at SUC2 and ADH2, two glucose-regulated genes that undergo well-characterized glucose-induced changes in nucleosome positioning. Glucose addition represses SUC2 expression by fivefold. Twenty minutes after glucose addition, we detect a shift in a promoter nucleosome to a site further upstream from the ORF. Sixty minutes after glucose addition, we detect addition of a new nucleosome adjacent to the shifted nucleosome (Figure 2), as has been reported previously (Perez-Ortin et al., 1987; Hirschhorn et al., 1992; Gavin and Simpson, 1997). Glucose addition represses ADH2 gene expression >50-fold. Our analysis indicates the addition of a well-positioned nucleosome to the ADH2 promoter 20 min after glucose addition (Supplemental Figure S6), as has been observed in previous studies (Verdone et al., 1996). In addition, the array of nucleosomes extending into the ADH2 ORF shifts toward the promoter upon repression. This repressed promoter structure is retained 60 min after glucose addition. For both ADH2 and SUC2, we compared our nucleosome positions with previously published positions determined for cells grown in glucose (Verdone et al., 1996; Gavin and Simpson, 1997). The correlation between the published positions and our post-glucose nucleosome occupancy is quite high, and clearly higher than for our pre-glucose positions (Supplemental Figure S7), indicating that our nucleosome position determinations are consistent with those reported previously.
Figure 2.
Glucose-induced nucleosome remodelling at the SUC2 promoter. Nucleosome protection data, represented as the log ratio of nucleosomal DNA to genomic DNA by tiling microarray (purple line), HMM prediction of the probability of nucleosome occupancy (blue line) and HMM prediction of the probability of initiating a nucleosome (green line) are shown for the SUC2 promoter, diagrammed at the bottom, at the indicated times before (0 min) or after (20 and 60 min) glucose addition. Previously determined in vivo nucleosome positions for the SUC2 promoter in cells grown in glucose (Gavin and Simpson, 1997) are shown as brown ovals.
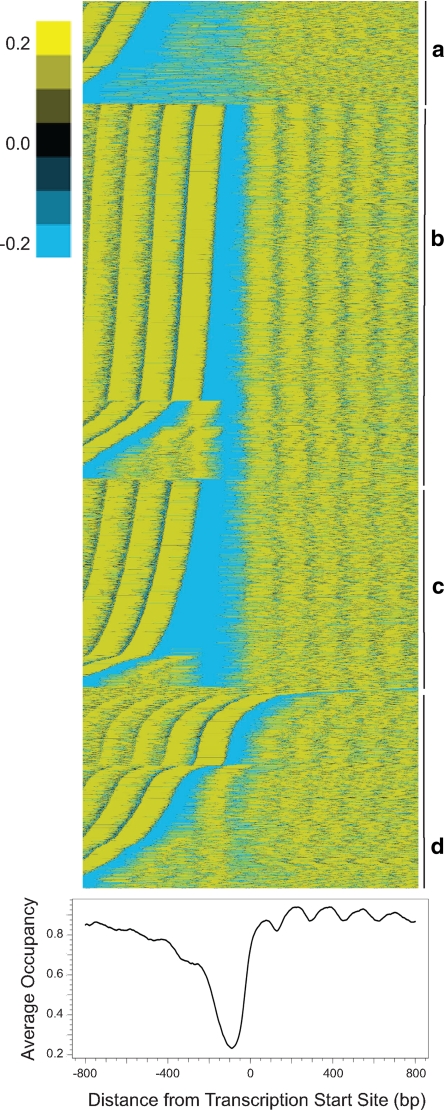
To investigate promoter nucleosome structure genome-wide, promoter nucleosome occupancy profiles before glucose addition were aligned by their experimentally determined transcriptional start sites (Nagalakshmi et al., 2008). Promoters with similar occupancy profiles were grouped by K-means clustering and then promoters within each cluster were sorted by the position of the least-occupied point (Figure 3). Similar to previous work (Lee et al., 2007), we found that clustering the promoter nucleosome occupancy profiles into more than four groups did not significantly alter our results (data not shown). Our analysis revealed features evident in previous studies of genome-wide nucleosome occupancy (Yuan et al., 2005; Lee et al., 2007; Mavrich et al., 2008; Shivaswamy et al., 2008). Most genes contain 1) a nucleosome-depleted region (NDR) of roughly 200 base pairs in their promoters; 2) a strongly positioned +1 nucleosome, which anchors an array of nucleosomes extending into the ORF; 3) a well positioned −1 nucleosome; 4) a nucleosome-depleted region at the 3′ end of the transcript (Supplemental Figure S8); and 5) a deeper NDR in the promoters of highly expressed genes than in those of poorly expressed genes (Supplemental Figure S9). Thus, our nucleosome occupancy data set recapitulates previously published results both at the level of individual promoters and on a genome-wide scale.
Figure 3.
Genome-wide promoter nucleosome structures. Top, nucleosome structure at individual promoters. Nucleosome occupancy for individual promoters aligned relative to the transcriptional start site (Nagalakshmi et al., 2008) was clustered by K-means into four groups (a–d) and then sorted sequentially within each group by the position of the minimum occupancy value. The mean nucleosome occupancy was subtracted from all values. Bottom, genome-wide average promoter nucleosome profile. The nucleosome occupancies for all promoters were aligned relative to the transcription start site, which is set as position 0, and then averaged at every nucleotide 800 base pairs upstream and 800 base pairs downstream over all genes to yield the average occupancy, which ranges from 0 (no nucleosome) to 1 (fully occupied).
Most Promoter Nucleosome Profiles Do Not Change with Transcriptional Change
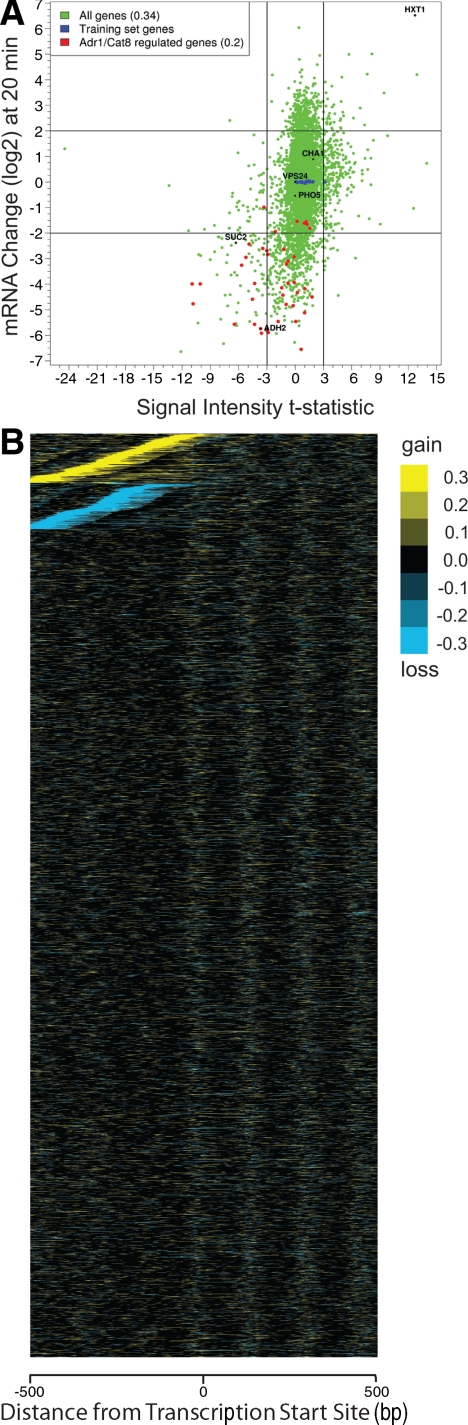
To address more completely the genome-wide relationship between changes in promoter nucleosome occupancy and gene transcription, we examined the correlation between a statistical measure of nucleosome occupancy differences and transcriptional change (Figure 4A). The scatter plot shown in Figure 4A correlates the change in gene expression of every gene, represented as the log base 2 change in mRNA level for that gene, with the t-statistic of the log ratio of MNase hybridization intensities at that gene promoter before and after glucose addition.
Figure 4.
Nucleosome remodelling occurs infrequently during transcriptional reprogramming. (A) Scatter plot showing the relationship between transcriptional change and nucleosome remodelling. Each point represents a single gene providing, on the x-axis, the t-statistic measure of the change in promoter nucleosome density 20 min after glucose addition relative to that before addition, plotted against the log2 change in mRNA levels 20 min after glucose addition. Promoters are defined as the region 800 base pairs upstream of the ORF, or until the next ORF. Correlation values (r) between transcriptional change and the nucleosome occupancy change are provided in the legend. (B) Representation of the differences in nucleosome occupancy at each nucleotide in individual promoters (vertical axis) 20 min after glucose addition relative to that before addition were calculated and plotted relative to the transcription start sites (horizontal axis, extending from −500 base pairs to +500 base pairs). These are clustered by K-means into three groups and then sorted within each group sequentially by maximum difference value (top cluster) or minimum difference value (middle cluster). The genes in the bottom cluster were not sorted.
Genes not regulated by glucose, such as CHA1, PHO5, and the genes used for training the HMM (highlighted in Figure 4A), tend not to undergo changes in promoter nucleosome occupancy. Both SUC2 and ADH2 undergo promoter nucleosome addition on gene repression (Figures 2 and Supplemental S6), and the position of these genes within Figure 4A accurately reflects our observations.
Of the genes that exhibit changes in promoter nucleosome occupancy, repressed genes tend to gain nucleosomes and induced genes tend to lose promoter nucleosomes. However, across all promoters the relationship between changes in nucleosome occupancy and transcription is surprisingly weak, with an overall correlation of 0.34. Similar results are obtained 60 min after glucose addition. Moreover, the same result is obtained by plotting changes in expression versus changes in nucleosome occupancy as predicted by our HMM rather than changes in hybridization intensity (Supplemental Figure S10). To visualize changes in nucleosome occupancy across all promoters, we calculated the difference in nucleosome occupancy between 0 and 20 min, relative to the transcription start site (TSS), at each promoter and then grouped these by K-means clustering into three groups. This clustering separated promoters into those that gained nucleosomes, those that lost nucleosomes, and those that remained unchanged (Figure 4B). Consistent with the results in Figure 4A, only 5% of all promoters gained nucleosomes and only 5% lost nucleosomes, despite that 50% of all genes changed expression by more than twofold. Moreover, for most of those promoters that gained or lost nucleosomes, only a single nucleosome was affected and most changes occurred between −550 and −100 base pairs from the transcriptional start site. In sum, 77 of 578 genes repressed by more than fourfold gained promoter nucleosomes, whereas 39 of 398 genes induced by more than fourfold lost promoter nucleosomes. However, the majority of promoters underwent no significant change in nucleosome occupancy profiles.
Our analysis of the relationship between changes in promoter nucleosome occupancy and gene expression measures the latter by mRNA microarrays. However, mRNA microarrays measure steady-state mRNA levels, which can be altered by changes in mRNA turnover as well as changes in transcription rate. To assess whether the dearth of nucleosome remodeling we observe on glucose upshift derives from a limited change in transcription rates of most genes, we performed chromatin immunoprecipitation (ChIP) against RNA polymerase II in ORFs (Supplemental Figure S11). At the SUC2 and HXT1 genes, changes in promoter nucleosome occupancy are associated with changes in mRNA levels (Figure 4A); after glucose upshift, we observe an increase in RNA polymerase II detected by ChIP at the HXT1 gene and a decrease at the SUC2 gene, which in both cases correlates with the changes in their mRNA levels. Examining several genes with glucose-regulated changes in mRNA levels but no changes in promoter nucleosome structure (HSP104, ALD3, REX4, and DHR2), we observed in every case a change in the occupancy of RNA polymerase II that correlated strongly with the change in mRNA level. These data indicate that genes with glucose-regulated changes in mRNA levels do undergo associated changes in transcription initiation rates, even in the absence of changes in promoter nucleosome occupancy.
Because we observed relatively few changes in nucleosome occupancy with transcriptional change, we considered the possibility that glucose-regulated promoters might alter their nucleosome positioning without altering their net nucleosome occupancy. To address this possibility, we determined the average nucleosome occupancy profiles across the promoters of genes whose transcription is either induced or repressed by glucose, before and after glucose addition (Supplemental Figure S12). The promoter nucleosome profiles of both glucose-induced and repressed genes showed little change, demonstrating that glucose-regulated genes do not undergo coherent nucleosome movements. Examination of nucleosome changes at individual promoters similarly failed to reveal examples of genes regulated by large-scale nucleosome sliding (Figure 4B). Because this level of analysis would not capture small shifts in nucleosome positioning that might alter the ability of transcription factors to bind, we specifically examined the relation between nucleosome position and putative transcription factor binding motifs. These results, described below, are consistent with the above observations that nucleosome repositioning plays a limited role in altering gene expression in response to glucose upshift.
We also examined changes in nucleosome occupancy at the 3′ end of transcripts. Difference maps of nucleosome occupancy at the 3′ end of all genes revealed a small number of genes that undergo nucleosome loss or addition at the 3′ end (Supplemental Figure S13A). Genes that exhibit nucleosome occupancy changes at their 3′ end are equally divided between those convergent with their downstream neighboring gene and those collinear with the downstream gene, indicating that the nucleosome remodeling at the 3′ end cannot be explained solely as remodeling of a downstream promoter. However, these changes in nucleosome occupancy at the 3′ end are not correlated with changes in gene expression (r = −0.01) (Supplemental Figure S13B). Thus, the function of 3′ NDR and changes to that structure remain unresolved.
Promoters Containing TATA Boxes Are More Likely to Undergo Nucleosome Remodeling
We observed that genes containing TATA boxes, which constitute ∼20% of all genes in yeast (Basehoar et al., 2004), were unusual in several respects. First, as noted previously (Ioshikhes et al., 2006; Albert et al., 2007; Mavrich et al., 2008), we found that, although the 5′ NDRs in non-TATA containing promoters were all 200 base pairs wide and stereotypically centered 100 base pairs upstream of the TSS, the NDRs in promoters with TATA boxes were of the same size as those at non-TATA promoters but were dispersed over a much wider region, with each gene containing an individually positioned NDR. This pattern is evident in the heat map of nucleosome positions in individual promoters, shown in Figure 3, with the TATA-containing genes significantly overrepresented in cluster a, and in plot of the average nucleosome density as a function of distance from the TSS (Supplemental Figure S14). Second, we observed that glucose-repressed genes were significantly enriched for TATA box-containing promoters (71% contain TATA boxes as opposed to a genome-wide average of 22%; binomial p = 1.3 × 10−36), whereas glucose-induced genes were significantly depleted for TATA box-containing promoters (15% contain TATA boxes; binomial p = 3.0 × 10−3). In sum, TATA box-containing genes behave differently from other genes both in nucleosome positioning and in expression changes after a glucose upshift.
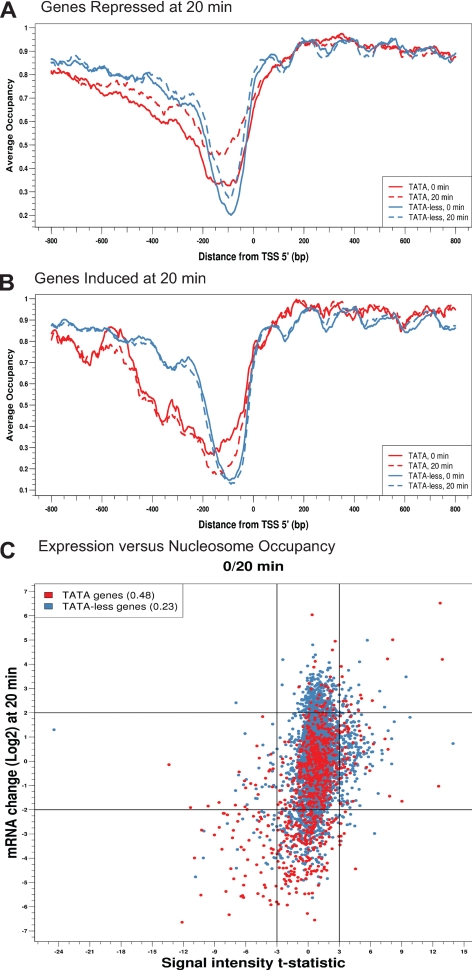
Because the asymmetry in the distribution of TATA box containing genes between glucose-induced and repressed genes mirrored the asymmetry in nucleosome remodeling in induced versus repressed genes, we directly examined if promoters containing a TATA box were more likely to undergo nucleosome remodeling. The promoter nucleosome profile for glucose repressed genes was subdivided between genes with or without a TATA box (Figure 5A). Strikingly, TATA box-containing promoters gained considerable nucleosome occupancy during gene repression, whereas promoters lacking TATA boxes gained little nucleosome occupancy during repression. We also subdivided the promoter nucleosome profile of glucose-induced genes between those with or without a TATA box (Figure 5B). Most glucose-induced genes lack TATA boxes and showed little difference in their promoter nucleosome profile before or after induction, whereas the minority of glucose-induced genes with TATA boxes lost nucleosome occupancy after induction. Thus, promoters of genes that contain TATA boxes tend to undergo changes in nucleosome occupancy upon changes in transcriptional activity, whereas those genes lacking TATA boxes tend to retain the same nucleosome profile upon changes in transcriptional activity.
Figure 5.
Promoters with TATA boxes are more likely to undergo nucleosome remodelling. (A) All genes repressed fourfold or more 20 min after glucose addition were subdivided into those containing TATA box in their promoters (blue lines) and those lacking TATA boxes (red lines) (Basehoar et al., 2004). For each subgroup, the average nucleosome occupancy at every nucleotide around the TSS was calculated before glucose addition (solid line) and 20 min after glucose addition (dashed line) and the values plotted relative to the position of the nucleotide from the TSS. (B) As in A, but for genes induced fourfold or more 20 min after glucose addition. (C) As in Figure 4A, but with all genes subdivided by the presence or absence of a TATA box within the promoter. Correlation values (r) are shown in the legend.
We calculated the relationships between transcriptional change and promoter nucleosome remodeling as a function of the presence or absence of TATA boxes (Figure 5C). The correlation between promoter log ratio MNase hybridization intensity change and transcriptional change for genes lacking a TATA box is weak (r = 0.23). However, the correlation between transcriptional change and promoter log ratio MNase hybridization intensity change for TATA-containing genes is much stronger (r = 0.48). Similar results were obtained both 20 and 60 min after glucose addition and with or without the application of the HMM nucleosome occupancy predictions (Supplemental Figure S15). These data demonstrate an association between TATA box-containing promoters and nucleosome remodeling during transcriptional change.
Functional Transcription Factor Binding Motifs Reside in Regions of Low Nucleosome Occupancy
Our data allowed us to examine the relationship among transcription factor binding motifs, nucleosome occupancy, and transcriptional changes. In particular, we explored the degree to which nucleosome occupancy over transcription factor binding motifs changed as a function of changes in expression or influenced their use in promoting transcriptional change. Genes regulated by the glucose-repressive transcription factors Adr1 and Cat8 (Tachibana et al., 2005) gain promoter nucleosomes during glucose repression (Verdone et al., 1996; Agricola et al., 2004). Consistent with these prior observations, we observed that many Adr1/Cat8-regulated genes gained promoter nucleosomes on transcriptional repression (Figure 4A). However, the overall correlation between changes in promoter nucleosomes and transcription for these genes is weak (r = 0.2), although this correlation is higher if alternative measures of nucleosome occupancy are used (Supplemental Figure S10).
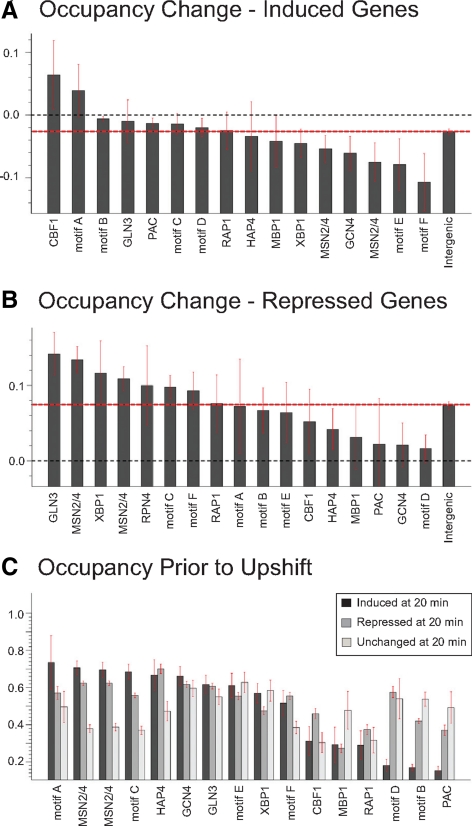
We extended our analysis to determine whether shifts in nucleosome positions expose or occlude transcription factor binding motifs in conjunction with changes in gene expression. We previously applied a mutual information based algorithm, FIRE (Elemento et al., 2007), to identify promoter-localized sequence motifs that mediate glucose-induced changes in gene expression, many of which corresponded to binding sites for transcription factors known to be involved in the glucose response, such as Msn2/4 and Hap4 (Zaman et al., 2009). Limiting our analysis to promoters, we measured differences in nucleosome occupancy in response to glucose addition directly over all instances of these individual motifs and observed little change in their nucleosome occupancy upon glucose addition (data not shown). To reduce the effect of spurious, nonfunctional representatives of these motifs, we calculated the difference in nucleosome occupancy over occurrences of each motif at 20 and 60 min after glucose addition only for promoters of those genes that exhibited transcriptional induction or repression (Figure 6, A and B). Most motifs undergo no significant change in average nucleosome occupancy. However, a small number of motifs, such as the Hap1-related CCG(N)5CC motif, become exposed on average during induction or, such as the Gln3 binding motif, become occluded during repression. This bias remains even after excluding those promoters that undergo overall changes in nucleosome occupancy during the upshift (data not shown). Examining the actual distribution of individual nucleosome occupancy changes over motifs, rather than the average changes, reveals that most motif occurrences undergo no significant changes in nucleosome occupancy, but for some motifs a subpopulation undergoes substantial changes in nucleosome occupancy (Supplemental Figure S16). Thus, at a limited number of promoters subtle shifts in nucleosome positioning can occasionally lead to the exposure or occlusion of transcription factor binding motifs.
Figure 6.
Nucleosomes are instructive for transcription factor regulation. For all genes induced (A) or repressed (B) fourfold or more 20 min after glucose addition, all occurrences of the listed motifs in promoters of those genes were noted and the average change in nucleosome occupancy directly over that motif determined. Values range from −1, which denotes a motif fully occupied at 0 min that becomes fully unoccupied 20 min after glucose addition, to 1, which denotes a motif fully unoccupied at 0 min that becomes fully occupied at 20 min. The bar labeled “intergenic” is the average change in nucleosome occupancy over all (A) glucose-induced or (B) repressed promoters. Error bars designate SE. (C) All instances of the listed motifs present in gene promoters were subdivided into those in genes whose expression was repressed following glucose addition, unchanged after glucose addition or induced after glucose addition. For each subset for each motif, the average nucleosome occupancy directly over that motif prior to glucose addition is plotted, with values ranging from completely unoccupied (0) to fully occupied (1). Error bars designate SE. Transcription factor motifs are as follows: Cbf1, [CGT]CA[CG]GTG[AG][AC]; Gln3, [ACT]GATAAG[ACG]; PAC, CTCATC[GT]C; Rap1, A[CT]CC.ACA[CT]; Hap4, [ACG]CCA[AC]TCA; Mbp1, T.[AT]CGCGT[ACT]; Xbp1, [CT][CT]TCG[AC]G[AG][CGT]; Msn2/4, [ACG][AG][ACT]. GGGG or CCCCT[AGT]; Gcn4, TGACT[ACT]A. Motif definitions are as follows: A, CGC[AG]C[CT]C[AT]; B, the RRPE motif [ACG]AAANTTTT; C, [AGT][AT][AT]AAGGG; D, GATCN3TGA[AG]; E, [CGT]TA[AT]ACGA.; F, [CGT]CCGN5CC[ACG].
In contrast to the weak correlation in changes in nucleosome occupancy over transcription factor binding motifs with changes in gene expression, we find in several cases a strong correlation between initial accessibility of transcription factor binding motifs and responses in gene expression. We examined each of the motifs involved in glucose transcriptional response and subdivided all the genes in which each motif appeared into those that were induced, repressed, or remained transcriptionally unchanged. We then measured the absolute nucleosome occupancy of the motif in each group before glucose addition (Figure 6C).
Promoters of the glucose-induced ribosomal biogenesis genes are highly enriched for RRPE and PAC motifs (Beer and Tavazoie, 2004; Jorgensen et al., 2004; Wade et al., 2006). Quite strikingly, in the promoters of those genes containing a PAC or RRPE motif (labeled Motif B in Figure 6) that were subsequently induced by glucose addition, the motifs were significantly more exposed than they were in those genes that were subsequently repressed or remained unchanged upon glucose addition, suggesting that likely physiologically relevant occurrences of RRPE/PAC motifs may be free of nucleosomes and accessible, whereas likely spurious instances of the RRPE/PAC motifs in promoters of repressed or unchanged genes are more likely to be contained within nucleosomes and perhaps inaccessible. The Msn2/4 transcription factors mediate portions of the general stress response which is alleviated by glucose addition (Gasch et al., 2000; Zaman et al., 2009), suggesting that physiologically relevant occurrences of the Msn2/4 motifs would be expected to occur in the promoters of glucose-repressed genes. We find that occurrences of the Msn2/4 motifs in promoters of glucose-repressed genes are significantly more exposed than in genes that were subsequently induced or transcriptionally unchanged. Several other motifs exhibited this dichotomy in occupancy, although many others did not. Nonetheless, these results suggest that accessibility dictated by nucleosome occupancy likely presages the functional activity of several transcription factor binding motifs.
DISCUSSION
We have investigated the relationship between gene expression and nucleosome positioning in yeast on a global scale. We focused on nucleosome position changes associated with transcriptional changes following glucose upshift, because more than half of all genes undergo significant change in gene expression, providing numerous individual examples for analysis. Our results demonstrate that yeast promoters fall into two clearly distinct categories: those that undergo changes in nucleosome occupancy upon change in expression, referred to as “remodeling promoters” by Elgin and colleagues, and those “preset promoters” whose nucleosome structure remains essentially unchanged during transcriptional alteration.
Remodeling Promoters
The promoters of only 10% of all genes gain or lose nucleosomes after glucose addition despite that >50% of all genes exhibit a change in mRNA levels of twofold or more. Those genes that gain promoter nucleosomes are predominantly those that undergo repression, whereas those that lose nucleosomes are predominantly unchanged in expression or, to a lesser extent, undergo induction. Those promoters that undergo nucleosome remodeling—both those that lose as well as those that gain nucleosomes—are highly enriched for predicted TATA boxes, with approximately half of all remodeled promoters containing a TATA box. As a consequence, TATA box-containing promoters exhibit a much higher correlation between transcriptional change and nucleosome remodeling than do promoters lacking TATA boxes. Pugh and coworkers noted previously that TATA boxes are enriched in the promoters of genes induced by starvation and other cell stresses and that these genes are particularly dependent on chromatin modification activities (Basehoar et al., 2004). We can now appreciate this dependency as a consequence of the significant proportion of TATA-containing promoters, relative to TATA-less promoters, that undergo nucleosome remodeling during transcriptional reprogramming. However, we still do not understand why TATA-containing boxes are associated with promoter remodeling. The TATA box itself does not seem to be required for nucleosome remodeling, as mutant PHO5, SUC2, and CUP1 promoters that lack TATA boxes still undergo wild type nucleosome remodeling (Hirschhorn et al., 1992; Fascher et al., 1993; Shen et al., 2001). Moreover, artificial recruitment of TBP to several remodeling promoters is not sufficient to initiate nucleosome remodeling (Ryan et al., 2000). Thus, although the correlation between TATA boxes and chromatin remodeling is clear, the mechanistic basis for that correlation is not.
In addition to remodeling at TATA-containing promoters, half the promoters that undergo remodeling under our conditions do not contain TATA boxes. What determines whether the promoter of a gene will undergo nucleosome remodeling during transcriptional reprogramming? We were unable to find any significant genome-wide correlation between binding motifs for particular transcription factors and nucleosome remodeling. Similarly, we were unable to detect specific motifs enriched at sites of nucleosome removal or addition (data not shown). One possibility is that remodeling is limited to those promoters in which the intrinsic energetics of nucleosome position, dictated by the ease at which segments of DNA can conform to the structure dictated by a nucleosome, does not strongly favor a single chromatin conformation. For many genes at which remodeling does not take place, the two nucleosomes bracketing the NDR (the +1 and −1 nucleosomes) assume their positions largely as a result of the intrinsic DNA sequence within the promoter (Ioshikhes et al., 2006; Segal et al., 2006). Such promoters may not be readily reconfigured, whereas other promoters in which multiple distinct sites are equally energetically favorable may be more easily remodeled. The availability from this study of an extensive list of remodeling versus nonremodeling promoters, in conjunction with improved computational methods to determine the free energy associated with nucleosome binding to any specific sequence of DNA, should allow us to directly test this hypothesis.
Preset Promoters
The majority of genes transcriptionally induced or repressed by glucose do not alter their promoter nucleosome structure. These preset promoters tend to be underrepresented for TATA boxes and tend to have a strong nucleosome-depleted region at a stereotypical position relative to the transcriptional start site, a promoter architecture that has been termed “type II” (Field et al., 2008). Previous work has characterized TATA-less promoters as enriched in constitutively expressed housekeeping genes (Basehoar et al., 2004), but the work reported here clearly demonstrates that individual TATA-less preset promoters are capable of dramatic gene expression changes.
The unaltered promoter nucleosome structure for most glucose-regulated genes implies the existence of constitutively accessible binding sites for the factors that control expression of these genes. In agreement with this inference, previous studies mapping genome-wide nucleosome positions at steady state have discovered a strong enrichment of predicted transcription factor binding sites in NDRs (Yuan et al., 2005; Lee et al., 2007; Morse, 2007; Field et al., 2008). Extending these studies, we found that gene promoters with an accessible transcription factor binding motif are significantly more likely to be regulated like known targets of that transcription factor than promoters where the motif is occluded by a nucleosome. This correlation was particularly noteworthy for induction through the RRPE/PAC elements and repression through Msn2/4 binding motifs. Because we did not directly measure transcription factor binding to these motifs, we cannot formally conclude that nucleosomes occlude transcription factor binding at any given motif; however, these results are consistent with previous work that found significant differences between the predicted binding sites and actual in vivo binding profile of the Leu3 and Rap1 transcription factors (Lieb et al., 2001; Liu et al., 2006). Given these observations, we would anticipate that coupling nucleosome position data with clustered gene expression data should improve computational approaches to predicting regulatory motifs and transcription factor binding sites associated with specific transcriptional responses (Beer and Tavazoie, 2004; Elemento et al., 2007). In sum, our results suggest that while nucleosome repositioning does not seem to be a major driving force in yeast for altering transcriptional activity, nucleosome positioning seems to play a significant role in determining which transcription factor binding sites are available for directing transcriptional changes.
Unexpected Promoter Nucleosome Stability in Yeast
Despite the massive transcriptional reprogramming induced by glucose addition we observed surprisingly little change in nucleosome positioning, particularly across coding regions. Moreover, a significant majority of yeast promoters show little or no change in nucleosome occupancy in association with significant change in expression. This stability was unanticipated given previous work on promoters for genes such as PHO5, ADH2, SUC2, and CHA1, where gene induction leads to substantial promoter nucleosome remodeling. We also observe the previously reported nucleosome remodeling in the promoters of the ADH2 and SUC2 genes, but our genome-wide analysis of promoter nucleosomes indicates that these remodeling promoters cannot be considered typical. Thus, the majority of yeast promoters fall into the category of promoters Elgin and colleagues described as preset.
Our analysis is limited to nucleosome positioning and does not examine many other changes that may occur at promoter nucleosomes, such as altered histone modifications, changing incorporation of histone variants into positioned nucleosomes, or even histone turnover. In addition, we have only examined nucleosome positioning changes in response to glucose addition, leaving the formal possibility that other perturbations might induce nucleosome remodeling to a greater degree. However, both our work and recent work of the Iyer laboratory, which examined global nucleosome positioning changes in response to heat shock (Shivaswamy et al., 2008), demonstrate remarkably stable nucleosome structure for most genes in yeast.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Donna Storton and Jessica Buckles for assistance with tiling microarrays, John Matese for management of the microarray data, Guo-Cheng Yuan for sharing HMM code with us, and Paul Schedl for valuable suggestions and discussions. This research was supported by National Institutes of Health grants GM-076562 and GM-045840 (to J.R.B.) and HG-004708 (to A.V.M.), and Center for Quantitative Biology/National Institutes of Health grant P50 GM-071508.
Abbreviations used:
- HMM
hidden Markov model
- NDR
nucleosome-depleted region
- ORF
open reading frame.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0111) on June 3, 2009.
REFERENCES
- Agricola E., Verdone L., Xella B., Di Mauro E., Caserta M. Common chromatin architecture, common chromatin remodeling, and common transcription kinetics of Adr1-dependent genes in Saccharomyces cerevisiae. Biochemistry. 2004;43:8878–8884. doi: 10.1021/bi049577+. [DOI] [PubMed] [Google Scholar]
- Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., Schuster S. C., Pugh B. F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Almer A., Rudolph H., Hinnen A., Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar A. D., Zanton S. J., Pugh B. F. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Beer M. A., Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Durrin L. K., Mann R. K., Grunstein M. Nucleosome loss activates CUP1 and HIS3 promoters to fully induced levels in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:1621–1629. doi: 10.1128/mcb.12.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O., Slonim N., Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fascher K. D., Schmitz J., Horz W. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J. Mol. Biol. 1993;231:658–667. doi: 10.1006/jmbi.1993.1317. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Kornberg R. D. Upstream activation sequence-dependent alteration of chromatin structure and transcription activation of the yeast GAL1-GAL10 genes. Mol. Cell. Biol. 1989;9:1721–1732. doi: 10.1128/mcb.9.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y., Kaplan N., Fondufe-Mittendorf Y., Moore I. K., Sharon E., Lubling Y., Widom J., Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin I. M., Simpson R. T. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and Swi-Snf and their effect on chromatin structure. EMBO J. 1997;16:6263–6271. doi: 10.1093/emboj/16.20.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D., Ruderfer D. M., Pratt S. C., Schacherer J., Dunham M. J., Botstein D., Kruglyak L. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–1936. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- Han M., Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Han M., Kim U. J., Kayne P., Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988;7:2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J. N., Brown S. A., Clark C. D., Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Ioshikhes I. P., Albert I., Zanton S. J., Pugh B. F. Nucleosome positions predicted through comparative genomics. Nat. Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J. R., Schneper L., Broach J. R., Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N., et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N. A., Bird L. E., Mellor J. Chromatin analysis in yeast using NP-40 permeabilised sphaeroplasts. Nucleic Acids Res. 1993;21:4653–4654. doi: 10.1093/nar/21.19.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., McLaughlin N., Lindstrom K., Tsukiyama T., Clark D. J. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol. Cell. Biol. 2006;26:8607–8622. doi: 10.1128/MCB.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. K., Shibata Y., Rao B., Strahl B. D., Lieb J. D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Lee W., Tillo D., Bray N., Morse R. H., Davis R. W., Hughes T. R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lieb J. D., Liu X., Botstein D., Brown P. O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Liu X., Lee C. K., Granek J. A., Clarke N. D., Lieb J. D. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 2006;16:1517–1528. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Wallrath L. L., Elgin S. C. Nucleosome positioning and gene regulation. J. Cell. Biochem. 1994;55:83–92. doi: 10.1002/jcb.240550110. [DOI] [PubMed] [Google Scholar]
- Mavrich T. N., Ioshikhes I. P., Venters B. J., Jiang C., Tomsho L. P., Qi J., Schuster S. C., Albert I., Pugh B. F. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira J. M., Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov A. V., Fortney K., Gaykalova D. A., Studitsky V. M., Widom J., Siggia E. D. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp475. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H. Getting into chromatin: how do transcription factors get past the histones? Biochem. Cell Biol. 2003;81:101–112. doi: 10.1139/o03-039. [DOI] [PubMed] [Google Scholar]
- Morse R. H. Transcription factor access to promoter elements. J. Cell. Biochem. 2007;102:560–570. doi: 10.1002/jcb.21493. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ortin J. E., Estruch F., Matallana E., Franco L. Fine analysis of the chromatin structure of the yeast SUC2 gene and of its changes upon derepression. Comparison between the chromosomal and plasmid-inserted genes. Nucleic Acids Res. 1987;15:6937–6956. doi: 10.1093/nar/15.17.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner L. A tutorial on Hidden Markov Models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- Ryan M. P., Stafford G. A., Yu L., Morse R. H. Artificially recruited TATA-binding protein fails to remodel chromatin and does not activate three promoters that require chromatin remodeling. Mol. Cell. Biol. 2000;20:5847–5857. doi: 10.1128/mcb.20.16.5847-5857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I. K., Wang J. P., Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. H., Leblanc B. P., Alfieri J. A., Clark D. J. Remodeling of yeast CUP1 chromatin involves activator-dependent repositioning of nucleosomes over the entire gene and flanking sequences. Mol. Cell. Biol. 2001;21:534–547. doi: 10.1128/MCB.21.2.534-547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S., Bhinge A., Zhao Y., Jones S., Hirst M., Iyer V. R. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C., Yoo J. Y., Tagne J. B., Kacherovsky N., Lee T. I., Young E. T. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol. Cell. Biol. 2005;25:2138–2146. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L., Camilloni G., Di Mauro E., Caserta M. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol. Cell. Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade C. H., Umbarger M. A., McAlear M. A. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast. 2006;23:293–306. doi: 10.1002/yea.1353. [DOI] [PubMed] [Google Scholar]
- Wallrath L. L., Lu Q., Granok H., Elgin S. C. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994;16:165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- Wang Y., Pierce M., Schneper L., Guldal C. G., Zhang X., Tavazoie S., Broach J. R. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2004;2:E128. doi: 10.1371/journal.pbio.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. K., Tyler J. K. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Wolffe A. Chromatin: Structure and Function. San Diego, CA: Academic Press; 1998. [Google Scholar]
- Yuan G. C., Liu Y. J., Dion M. F., Slack M. D., Wu L. F., Altschuler S. J., Rando O. J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Schneper L., Slonim N., Broach J. R. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 2009;5:245. doi: 10.1038/msb.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.