Abstract
The ESCRT machinery functions in several important eukaryotic cellular processes. The AAA-ATPase Vps4 catalyzes disassembly of the ESCRT-III complex and may regulate membrane deformation and vesicle scission as well. Ist1 was proposed to be a regulator of Vps4, but its mechanism of action was unclear. The crystal structure of the N-terminal domain of Ist1 (Ist1NTD) reveals an ESCRT-III subunit-like fold, implicating Ist1 as a divergent ESCRT-III family member. Ist1NTD specifically binds to the ESCRT-III subunit Did2, and cocrystallization of Ist1NTD with a Did2 fragment shows that Ist1 interacts with the Did2 C-terminal MIM1 (MIT-interacting motif 1) via a novel MIM-binding structural motif. This arrangement indicates a mechanism for intermolecular ESCRT-III subunit association and may also suggest one form of ESCRT-III subunit autoinhibition via intramolecular interaction.
INTRODUCTION
The ESCRT (endosomal sorting complexes required for transport) machinery has been implicated in a number of physiological and pathological processes in eukaryotic cells, including MVB (multivesicular body) formation, HIV budding, and abscission during cytokinesis (Piper and Katzmann, 2007; Saksena et al., 2007). The process of MVB sorting involves the invagination of the endosomal membrane into the lumen of the compartment, giving rise to the intralumenal vesicles characteristic of the MVB. The ESCRTs were initially characterized in the yeast Saccharomyces cerevisiae through studies of this MVB-sorting process and are conserved throughout higher eukaryotes (Babst, 2005). MVB sorting sequesters transmembrane endocytic cargoes away from the cytoplasm. This process can contribute to attenuation of growth factor signaling both through segregating activated receptors from the cytoplasmic signal transduction machinery and by facilitating delivery of receptors to the lysosomal lumen for degradation (Katzmann et al., 2002). The proper function of the ESCRT machinery is thus critical for maintaining cellular homeostasis.
Major components of the ESCRT machinery include the three ESCRT complexes (ESCRT-I, -II, and -III), the Vps27—Hse1 complex, the AAA ATPase Vps4 and associated regulators of these proteins (Williams and Urbe, 2007; Hurley, 2008). ESCRT-III appears distinct from the other complexes in that it dynamically assembles during MVB sorting, whereas ESCRT-I and -II appear more stable. ESCRT-III also appears to be more complex because there are numerous, structurally similar subunits (Vps20, Snf7, Vps24, Vps2, Did2, and Vps60) that assemble into the oligomer through specific interactions (Babst et al., 2002; Saksena et al., 2009). Moreover, ESCRT-III subunits have been suggested to adopt closed and open conformations associated with the monomeric or oligomeric states (Muziol et al., 2006), with the C-terminus suggested to maintain the autoinhibited closed conformation (Zamborlini et al., 2006; Shim et al., 2007; Lata et al., 2008a). However, the precise natures of these conformations and ESCRT-III assembly remain unclear. ESCRT-III and Vps4 appear to be responsible for generating membrane curvature and vesicle budding. Although the exact molecular mechanism by which vesiculation occurs is unknown, it is generally believed that the ESCRT-III subunits form membrane-bound higher-order oligomers either in the shape of helical filaments or tubular structures (Ghazi-Tabatabai et al., 2008; Hanson et al., 2008; Lata et al., 2008b), which promote change in the membrane structure (Saksena et al., 2009; Wollert et al., 2009). Vps4 interacts with the ESCRT-III proteins and catalyzes remodeling of the ESCRT-III structures using energy derived from ATP hydrolysis (Babst et al., 1998; Xiao et al., 2007). The action of Vps4 is required to complete the vesicle budding process and recycle the system so that protein trafficking through the MVB pathway can be sustained. Given that Vps4 catalyzes the only known energy-consuming step in the MVB pathway, it is not surprising that the activity of Vps4 is closely regulated in vivo by a number of proteins, including Vps2, Did2, Vps60, Vta1, and Ist1(Azmi et al., 2006; Azmi et al., 2008; Dimaano et al., 2008; Xiao et al., 2008).
Although much progress has been made on the molecular mechanism by which Vps2, Did2, Vps60, and Vta1 function in the cell (Azmi et al., 2006, 2008; Dimaano et al., 2008; Xiao et al., 2008), less is known about Ist1. Ist1 was originally identified as an evolutionarily conserved protein that interacts with both Vps4 and Did2 (Dimaano et al., 2008; Rue et al., 2008). Binding of Ist1 to Vps4 interferes with the ATP-dependent oligomerization of Vps4 and inhibits the ATPase activity (Dimaano et al., 2008). Furthermore, overexpression of Ist1 in yeast leads to an MVB-sorting defect similar to that observed in cells depleted of Vps4, suggesting that Ist1 acts as a negative regulator of the MVB pathway (Dimaano et al., 2008). Ist1 binds to the N-terminal MIT (microtubule interacting and transport) domain of Vps4 through its C-terminal region, with its last 15 amino acids bearing strong homology to a conserved sequence motif called MIM1 (MIT interacting motif 1; Agromayor et al., 2009; Bajorek et al., 2009). Deletion of IST1 by itself has no obvious phenotype in the yeast, but simultaneous deletion of IST1 with either vta1Δ or vps60Δ gives rise to a strong MVB-sorting defect not seen in cells devoid of either Vta1 or Vps60 (Dimaano et al., 2008; Rue et al., 2008). Interestingly, deletion of DID2 is also synthetic with vta1Δ and vps60Δ, but not with ist1Δ. These results suggest that the in vivo function of Ist1 extends beyond its inhibitory activity toward Vps4 and that it may function together with Did2 and in parallel to Vta1/Vps60 in the MVB pathway.
To provide insights into the mechanism of Ist1 function, we sought to characterize the molecular structure of Ist1. To this end, we have determined the crystal structure of the N-terminal domain of Ist1 (Ist1NTD) and examined its interaction with Did2. Ist1NTD contains a fold similar to the ESCRT-III subunit Vps24/CHMP3 despite a low level of sequence identity, indicating that Ist1 is a divergent member of the ESCRT-III subunit family. Ist1NTD specifically interacted with the MIM1 element within Did2 via a novel MIM1-binding site. Disruption of Ist1–Did2 interaction through mutagenesis led to a synthetic MVB-sorting defect in vta1Δ yeast as well as a defect in abscission during mammalian cell division, validating the relevance of the Ist1–Did2 interaction observed. This characterization of the specific interaction between two ESCRT-III subunits, Did2 and Ist1, illustrates for the first time a potential mechanism by which ordered assembly of ESCRT-III is achieved via intermolecular association. Moreover, this complex may also suggest a conformation of an ESCRT-III subunit in which intramolecular interaction leads to an autoinhibited state.
MATERIALS AND METHODS
Cloning, Expression, and Purification
Detailed cloning, expression and purification information are described in Supplementary Materials. Briefly, DNA fragments encoding yeast Ist1 and Did2 were amplified from the S. cerevisiae genomic DNA. Ist1NTD and His6-tagged SUMO-Did2-MIM1 were coexpressed in Escherichia coli BL21(DE3). Ist1NTD/His6-tagged SUMO-Did2–MIM1 complex was initially purified on a Ni2+-NTA affinity column (Qiagen, Valencia, CA) followed by ULP1 protease cleavage to remove the His6-SUMO tag. The resulting protein mixture was passed over a second Ni2+-NTA column and was further purified by gel filtration chromatography on a Superdex-75 column (GE Healthcare, Piscataway, NJ). Purified protein complex was concentrated to 10–15 mg ml−1 and flash-frozen in liquid nitrogen. Selenomethionyl proteins were expressed in E. coli B834(DE3) using a minimal medium where methionines were replaced with selenomethionines and purified in a similar way.
Crystallization and Structure Determination
Crystals of both Ist1NTD and Ist1NTD/Did2-MIM1 were grown out of the purified Ist1NTD/Did2–MIM1 complex. Native crystals of Ist1NTD were grown by the hanging-drop method at 4°C against the reservoir solution of 6–10% (wt/vol) PEG 3000, 100 mM Na acetate (pH 5.0), 200 mM MgCl2, and 100 mM glycine. For data collection, crystals were transferred into a cryo solution of 16% (wt/vol) PEG 3000, 20% (vol/vol) ethylene glycol, 100 mM Na acetate (pH 5.0), 200 mM MgCl2, and 100 mM glycine in a stepwise manner before being flash-frozen under liquid nitrogen. The Ist1NTD crystal belongs to a space group of C2 with two molecules in the asymmetric unit (Table 1). Selenomethionyl crystals of Ist1NTD were grown under similar conditions but initially belonged to a space group of P212121. Overnight dialysis in the cryo solution transformed them into a C2 space group different from that of the native crystal and rendered them more resistant to radiation decay (Table 1). For preparation of the heavy atom derivative, native Ist1NTD crystals were soaked in a stabilizing solution containing 0.5 mM mercury acetate at 4°C for 2 h. The crystals were then back-soaked and stepwise transferred into the cryo solution. Diffraction data for both the native and the selenomethionyl crystals were collected at the Advanced Photon Source beam line 21-ID-D (Argonne National Laboratory). Diffraction data for the mercury derivative were collected with a Rigaku rotating anode generator and an R-AXIS IV image-plate detector system (The Woodlands, TX). The mercury derivative crystals are isomorphous to the cryo-soaked selenomethionyl crystals.
Table 1.
Crystallographic data and refinement statistics
| Ist1 alone |
Complex Native | |||||
|---|---|---|---|---|---|---|
| Native | Se (peak) | Se (inflection) | Se (remote) | Hg derivative | ||
| Wavelength (Å) | 0.9793 | 0.9793 | 0.9795 | 0.9643 | 1.5418 | 0.9793 |
| Space group | C2 | C2 | C2 | P4222 | ||
| Unit Cell (Å) | a = 144.84 | a = 145.63 | a = 145.26 | a = b = 165.93 | ||
| b = 44.17 | b = 44.31 | b = 44.33 | c = 121.56 | |||
| c = 66.95 | c = 138.34 | c = 136.93 | ||||
| β = 110.49° | β = 112.75° | β = 112.47° | ||||
| Resolution (Å) (last shell) | 1.70 (1.73–1.70) | 2.85 (2.90–2.85) | 2.80 (2.85–2.80) | 2.60 (2.64–2.60) | 2.90 (2.95–2.90) | 3.8 (3.87–3.8) |
| Completeness (%) (last shell) | 98.9 (95.1) | 96.9 (98.9) | 96.5 (87.6) | 97.5 (98.9) | 99.1 (99.9) | 99.6 (100.0) |
| I/σ (last shell) | 34.3 (5.9) | 26.5 (7.5) | 26.4 (5.8) | 30.8 (8.3) | 10.3 (3.0) | 16.6 (2.7) |
| Rmerge (%) (last shell) | 4.3 (20.4) | 6.1 (17.7) | 5.8 (20.7) | 6.1 (23.2) | 13.7 (42.5) | 9.1 (65.8) |
| Redundancy | 3.7 | 6.3 | 6.3 | 6.3 | 3.3 | 6.0 |
| Unique reflections | 43548 | 19282 | 19951 | 24749 | 18245 | 17219 |
| Refinement statistics | ||||||
| Rwork/Rfree (%) | 22.6/24.9 | 29.8/30.7 | ||||
| Rmsd, bonds (Å) | 0.004 | 0.011 | ||||
| Rmsd, angles (°) | 0.971 | 1.652 | ||||
| Mean B (Å2) | 23.8 | 147.4 | ||||
| Ramachandran (%) | ||||||
| Core | 92.9 | 83.3 | ||||
| Allowed | 7.1 | 15.3 | ||||
| Generously allowed | 0.0 | 1.4 | ||||
| Disallowed | 0.0 | 0.0 | ||||
| Molecules/ASUa | 2 | 4 complexes | ||||
a ASU: asymmetric unit
Ist1NTD/Did2–MIM1 complex crystals were grown by the sitting drop method at 20°C against the reservoir solution of 1.1–1.15 M Na citrate, 100 mM Na citrate (pH 5.5), and 10 mM urea. For data collection, crystals were transferred into a cryo solution of 1.26 M Na citrate, 10 mM urea, and 10% (vol/vol) glycerol before being flash-frozen under liquid nitrogen. Diffraction data were collected at the Advanced Photon Source beam line 21-ID-D. The complex crystals belong to the space group of P4222 with four complex molecules in the asymmetric unit (Table 1).
The structure of Ist1NTD was determined by a combination of multiwavelength anomalous dispersion (MAD) and single isomorphous replacement (SIR) using the data collected from the selenomethionyl crystal and the mercury derivative crystal. The structure of Ist1NTD/Did2–MIM1 complex was determined by the molecular replacement method with the refined Ist1NTD structure as a search model. More detailed description of the structure determination process can be found in Supplementary Materials (Supplementary Figure S5 and Table 1).
Biochemical Interactions
Glutathione S-transferase (GST) pulldown and immunoprecipitation experiments were performed following standard procedures (Chen et al., 2007) and are described in detail in Supplementary Materials.
Surface Plasmon Resonance
Binding of Ist1 proteins to the Did2-MIM1 fragment was analyzed using a BIACORE 3000 instrument (GE Healthcare) and an NTA sensor chip at room temperature. Did2-MIM1 was purified as a His10-GST fusion protein, diluted to 10 μM and immobilized by passage over the NTA sensor chip that had been preactivated with 0.5 mM nickel chloride. The His10-GST protein was also immobilized separately to serve as a control. Binding studies were performed by passing 0.05 to 40 μM wild-type or mutant Ist1 proteins over the immobilized proteins. The association and dissociation times were 120 and 180 s, respectively. Binding to the His10-GST was subtracted from that of the His10-GST-Did2-MIM1, and the data were fit using a one-site specific binding model.
Yeast Cell Biology
pRS415 HA-Ist1 was generated by first amplifying the Ist1 promoter from genomic DNA along with coding sequences for an N-terminal HA epitope and then cloning this fragment into pRS415. Ist1 coding sequences were then subcloned from the bacterial expression constructs (wild-type or mutants) to generate pRS415 HA-Ist1, pRS415 HA-Ist1L168A/Y172A, and pRS415 HA-Ist1E175R plasmids. Standard techniques were used to introduce the pRS415 HA-Ist1 constructs into ist1Δvta1Δ yeast (MCY2; Dimaano et al., 2008) along with a MVB reporter construct expressing GFP-CPS (pGO45; Odorizzi et al., 1998). Microscopy was performed on living cells using a Nikon fluorescence microscope (Melville, NY) fitted with filters optimized for EGFP detection and a Coolsnap HQ digital camera (Photometrics, Tucson, AZ). Images were analyzed using Delta Vision software (Applied Precision, Issaquah, WA).
Immunofluorescence and Confocal Microscopy
Immunofluorescence experiments were carried out as described previously (Chen et al., 2006). Hela cells grown on coverslips were fixed with 10% (vol/vol) Formalin for 10 min before being permeabilized with 0.5% (vol/vol) Triton X-100 in phosphate-buffered saline (PBS, GIBCO, Rockville, MD) for 5 min. Additional Formalin was quenched with 100 mM glycine in PBS. Cells were blocked with 1% (wt/vol) BSA plus 1% (wt/vol) chicken albumin in PBS and stained with rabbit anti-HA (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:500, mouse anti-α-tubulin (clone DM1A, Sigma, St. Louis, MO) at 1:500. Coverslips were mounted in Vectashield mounting media (Vector Laboratories, Burlingame, CA). Confocal microscopy was performed using Olympus FluoView 500 laser scanning confocal microscope (Melville, NY); images were taken under a 60× water immersion lens.
Figure Preparation
Multiple sequence alignment was performed with ClustalW (Larkin et al., 2007). Alignment figures with secondary structures labeled were prepared using ESPript (Gouet et al., 1999). Amino acid conservation scores are calculated using the ConSurf server (Landau et al., 2005). Structure figures were generated using Pymol (DeLano Scientific, South San Francisco, CA).
Accession Codes
Coordinates and structure factors for structures described in the study have been deposited into the Protein Data Bank with the accession codes of 3GGY (Ist1NTD) and 3GGZ (Ist1NTD-Did2).
RESULTS
Specific Interaction between Ist1 and Did2-MIM1
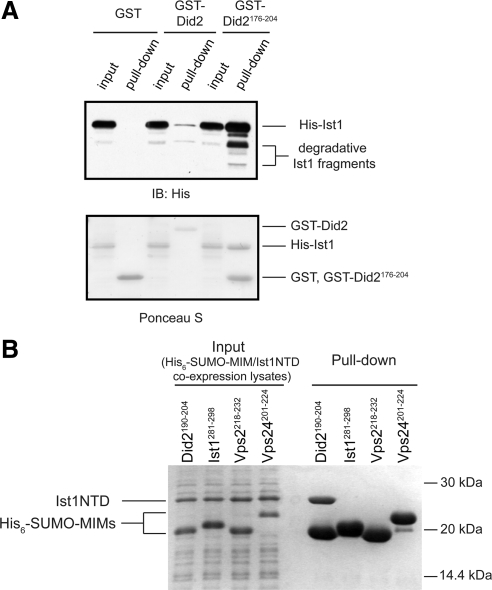
S. cerevisiae Ist1 has been shown to bind to Did2. The C-terminal region of Did2 is important for mediating this interaction (Dimaano et al., 2008; Rue et al., 2008). Indeed, GST-tagged Did2176-204, but not GST-tagged Did2104-175, interacts with the full-length Ist1 (Figures 1A and 3C). Interestingly, deletion of the N-terminal region of Did2 significantly strengthens the interaction between the two proteins (Figure 1A). This is consistent with the notion that Did2 adopts an autoinhibitory conformation at its resting state where its C-terminal region is masked, as has been previously observed in its interaction with Vps4 (Zamborlini et al., 2006; Shim et al., 2007; Azmi et al., 2008).
Figure 1.
Analysis of interaction between Ist1 and Did2. (A) Ist1 binds to the C-terminal region of Did2. GST, GST-Did2, or GST-Did2176-204 was used to pull down N-terminally His-tagged Ist1 as indicated. Proteins retained on the beads were analyzed by SDS-PAGE and visualized by anti-His immunoblotting (top) and Ponceau S staining (bottom). (B) Ist1NTD specifically interacts with Did2-MIM1. His-SUMO-tagged MIM1 fragment from Did2 (Did2190-204), Vps2 (Vps2218-232), Vps24 (Vps24201-224), or Ist1 (Ist1281-298) was coexpressed with Ist1NTD in E. coli. Proteins retained on the Ni2+-NTA beads were analyzed by SDS-PAGE and visualized by Coomassie staining.
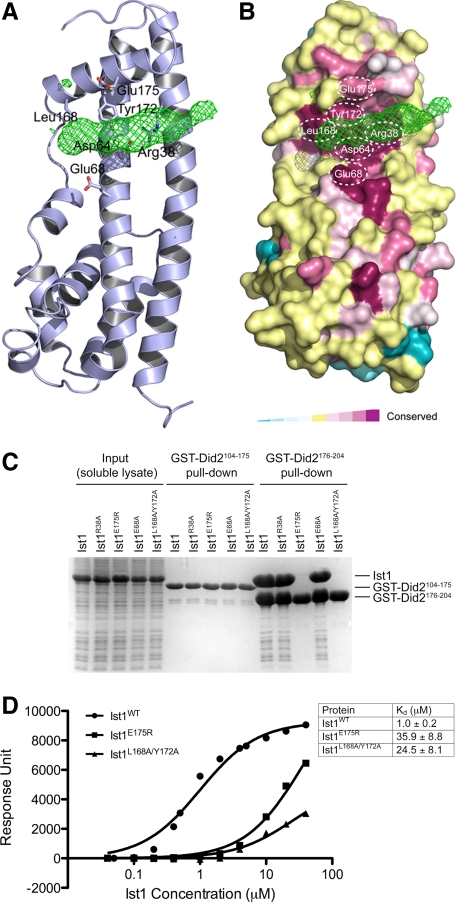
Figure 3.
Did2-MIM1 binds in a conserved surface groove of Ist1NTD. (A) Fo-Fc difference map (contoured at 3σ) showing the location of Did2-MIM1 helix in the Ist1NTD/Did2-MIM1 structure. Conserved residues of Ist1 at the interface are also shown. (B) Did2-MIM1 binds to Ist1NTD in a highly conserved surface groove. The molecular surface of Ist1NTD is colored based on conservation scores. The most conserved residues are colored in magenta and the least conserved in cyan. (C) Contribution of the conserved residues to the binding of Did2. GST-tagged Did2 fragments were used to pull down wild-type or mutant Ist1 as indicated. Proteins retained on the beads were analyzed by SDS-PAGE and visualized by Coomassie staining. (D) SPR analysis of wild-type and mutant Ist1 proteins binding to Did2176-204. Data represent the average of three independent measurements for each group. A simple one-to-one binding model was used to fit the data.
To identify the Did2-binding site on Ist1, we subjected purified Ist1 to limited proteolysis by subtilisin. Digestion of Ist1 generated two stable fragments (Supplementary Figure S1) with molecular weights of 22,576 and 8741 Da, respectively, as determined by mass spectrometry. We deduced that the larger fragment corresponds to the N-terminal amino acids 1-193 (Ist1NTD) and the smaller fragment corresponds to the C-terminal amino acids 226-298. When expressed by itself, Ist1NTD is not stable and precipitates rapidly in solution (data not shown). However, when Ist1NTD and Did2176-204 are coexpressed in E. coli, they can be purified as a stable complex (see Materials and Methods; data not shown), suggesting that Ist1NTD contains the Did2-binding site. By contrast, the smaller, C-terminal fragment of Ist1 does not bind to Did2 (data not shown).
The last 15 amino acids of Did2 have been previously characterized as a MIM1 motif because of its ability to interact with the MIT domain of Vps4. It is known to adopt a helical conformation (Stuchell-Brereton et al., 2007; Yang et al., 2008). This stretch of sequence is highly conserved, raising the question as to whether it also mediates binding to Ist1. Indeed, Did2-MIM1 (Did2190-204) can be purified in a stable complex with Ist1NTD (Figure 1B). Therefore, the minimal Ist1-interacting sequence of Did2 appears to be Did2-MIM1.
Several other proteins, including Vps2, Vps24, and Ist1 itself, contain C-terminal MIM1 or MIM1-like sequences that bind to Vps4 MIT domain (Obita et al., 2007; Stuchell-Brereton et al., 2007; Agromayor et al., 2009; Bajorek et al., 2009; Supplementary Figure S2). To test whether these sequences can interact with Ist1, we coexpressed His6-SUMO-tagged C-terminal fragments from Vps2 (Vps2218-232), Vps24 (Vps24201-224), and Ist1 (Ist1281-298) with Ist1NTD. We used Ni2+-NTA affinity chromatography to examine complex formation between these respective protein pairs. Interestingly, formation of protein complexes can only be observed between Ist1NTD and Did2-MIM1 under the assay conditions utilized (Figure 1B), suggesting that the interaction between Ist1NTD and Did2-MIM1 is highly specific.
The Crystal Structure of Ist1NTD
To understand the structural basis of Ist1 function, we used x-ray crystallography to study the structure of Ist1 and its molecular interaction with Did2. Given the perceived structural flexibility within the full-length protein and the aggregation problem of Ist1NTD experienced during protein preparation, we focused our structural study on the Ist1NTD/Did2–MIM1 complex. Two crystal forms were obtained from the purified complex. One was grown in the presence of high concentration of sodium citrate and diffracted to 3.8 Å resolution. It belongs to the space group of P4222 and contains both protein fragments. The other was grown under polyethylene glycol conditions and diffracted to 1.7-Å resolution. It belongs to the space group of C2 but contains only Ist1NTD. Because the C2 crystal form diffracts significantly better than the P4222 form, we first determined the structure of Ist1NTD (Table 1).
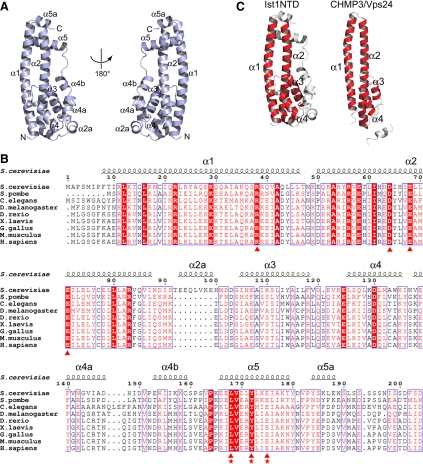
Ist1NTD has an all α fold with nine α-helices (Figure 2A). Helices α1 and α2 fold into a 60-Å long helical hairpin. Helix α2 takes a sharp turn at residues Arg83 and Val84 and is connected to helix α3 via a short, nonconserved helix α2a (Figure 2, A and B). Helices α3, α4, and α4a form a three-helix bundle, which packs against the bottom half of the α1/α2 hairpin to form the core of the structure. A short helix α4b and a flexible linker extending from the core subsequently direct the rest of the sequence to the other side of the molecule, which folds into two helices α5 and α5a and packs onto the top half of the α1/α2 hairpin. There are two Ist1NTD molecules in the asymmetric unit, related by a noncrystallographic two-fold symmetry. They are nearly superimposable with an rmsd (root mean square deviation) of 0.5 Å for the Cα atoms. The only major difference between the two molecules is located at the linker between helix α4b and α5 (residues 160-163), suggesting that this region is highly dynamic.
Figure 2.
Crystal structure of Ist1NTD. (A) Ribbon representation of Ist1NTD structure shown in two orientations. Secondary structural elements and the N- and C-termini of the structure are labeled. (B) Structure-based sequence alignments of Ist1NTD from various organisms. Secondary structural elements of S. cerevisiae Ist1NTD are shown above the sequence block. Residues involved in forming the conserved Did2-binding groove are highlighted with triangles at the bottom. Residues essential for Did2 binding are also noted with stars. (C) Ist1NTD has an ESCRT-III like fold. Helices present in both structures of Ist1NTD and the ESCRT-III protein CHMP3 are labeled and highlighted.
Ist1NTD shows remarkable structural similarity to the ESCRT-III subunit CHMP3/Vps24 (PDB ID: 2GD5; Muziol et al., 2006; Figure 2C). Pairwise comparison of the two structures by DaliLite (Holm et al., 2008) gives an rmsd of 2.6 Å for 103 aligned Cα atoms at 10% sequence identity. In particular, the α1/α2 helical hairpins from the two molecules have nearly identical structures, with helices α3 and α4 packing against the hairpin in a similar manner (Figure 2C). Because all known ESCRT-III subunits are believed to have a similar structure (Babst et al., 2002; Muziol et al., 2006), the fold represented by helices α1–α4 is likely shared by all ESCRT-III subunits and therefore can be generally referred to as the ESCRT-III fold. The presence of this fold in Ist1 suggests that this protein can be defined as a divergent ESCRT-III subunit.
The Crystal Structure of Ist1NTD in Complex with Did2-MIM1
The cocrystal structure of the Ist1NTD/Did2-MIM1 (Did2190-204) complex was determined at 3.8-Å resolution by molecular replacement using the refined Ist1NTD structure as an initial search model. A total of four Ist1NTD molecules were found in the asymmetric unit. Each Ist1NTD is bound with a Did2-MIM1 based on the electron density feature in the difference map (Figure 3A). Did2-MIM1 binds to a highly conserved surface groove on Ist1, formed by helix α5 and the α1/α2 helical hairpin (Figure 3, A and B). In particular, side chains of Arg38, Asp64, Glu68, Leu168, Tyr172, and Glu175 of Ist1 are all pointing toward the Did2-MIM1 electron density and therefore are likely involved in binding to the Did2-MIM1 peptide.
To examine the functional importance of these residues in contributing to Did2 binding, we introduced point mutations into the full-length Ist1 and examined their effects on Ist1–Did2 interaction. Mutating Arg38 to alanine (R38A) or Glu68 to alanine (E68A) had no obvious effect on binding to Did2 (Figure 3C), nor did mutating Leu168 or Tyr172 individually to alanine. However, mutating Glu175 to arginine (E175R) or mutating Leu168 and Tyr172 simultaneously to alanines (L168A/Y172A) compromised interaction between the two proteins (Figure 3C). Furthermore, although the Did2-MIM1 peptide bound to wild-type Ist1 with a Kd of 1 μM, both E175R and L168A/Y172A displayed significantly decreased binding affinities between the peptide and the protein (Figure 3D). These results suggest that Ist1 residues Glu175, Leu168, and Tyr172 are involved in direct binding to Did2.
The resolution limit of the electron-density map complicated our ability to register the sequence of Did2-MIM1 into the density. However, results from the above biochemical analysis put significant constraints on the potential mode of interaction between Ist1 and Did2. Given the functional importance of Ist1 Glu175, it is likely that a positively charged residue of Did2-MIM1 is in close proximity to form a salt bridge interaction with Ist1. Moreover, this positively charged residue is likely located in the midsection of the electron density, as dictated by the position of Glu175 (Figure 3A). The MIM1 sequence in human CHMP1A (human Did2 ortholog) has been previously shown to form an amphipathic α-helix (Stuchell-Brereton et al., 2007; Yang et al., 2008). By modeling yeast Did2-MIM1 into a similar helix based on the sequence conservation in this region, we reasoned that the absolutely conserved Arg198 in Did2-MIM1 is most likely close to Glu175 of Ist1. This provides us a marker to fit the entire 15 residues of Did2-MIM1 into the electron density map (Figure 4A). The working and free R factors for the complex structure are 29.8 and 30.7%, respectively, which is reasonable at the 3.8-Å resolution (Table 1). Additional analyses to evaluate the validity of our model are described in Supplementary Materials.
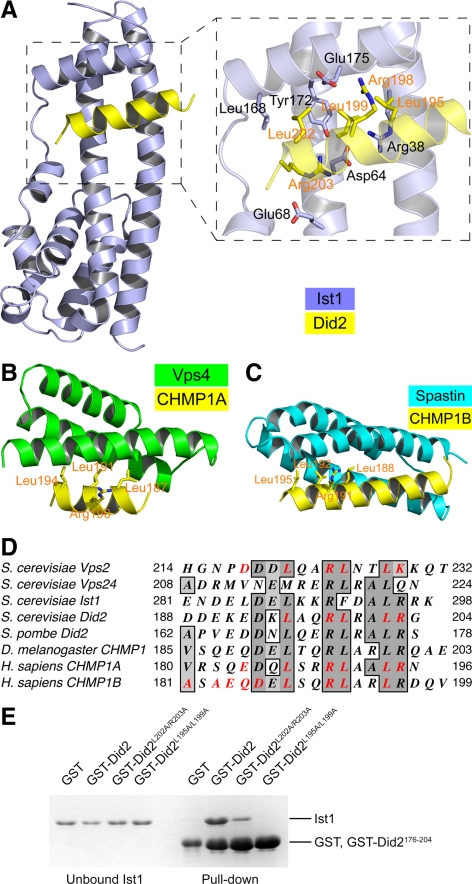
Figure 4.
A conserved surface of Did2 mediates interaction with Ist1NTD. (A) The structure of the Ist1NTD/Did2—MIM1 complex. Side chains of residues at the interface from the two proteins are shown in an enlarged view. The MIM1 helices in B and C are shown in the same general orientation. (B) The structure of the Vps4-MIT/CHMP1A–MIM1 complex (PDB ID: 2JQ9). (C) The structure of the spastin-MIT/CHMP1B–MIM1 complex (PDB ID: 3EAB). (D) MIM1 or MIM1-like sequences in Did2, Vps2, Ist1, and Vps24. Conserved sequence features are highlighted. Residues that have been experimentally demonstrated to be important for protein–protein interaction are shown in red. (E) Conserved residues in Did2 are important for Ist1 binding. GST-tagged wild-type or mutant Did2176-204 proteins were used to pull down purified Ist1 as indicated. Proteins retained on the beads were analyzed by SDS-PAGE and visualized by Coomassie staining.
Ist1 Displays a Novel Mode of Binding to Did2-MIM1
The structure of the MIM1 region of mammalian Did2 homolog CHMP1 has been previously characterized in complex with the MIT domains of mammalian Vps4 (CHMP1A) and spastin (CHMP1B), respectively (Stuchell-Brereton et al., 2007; Yang et al., 2008). The MIT domain has a three-helix bundle structure. When in complex with Did2/CHMP1-MIM1, it can use either a groove between α2 and α3 (in Vps4) or a groove between α1 and α3 (in spastin; Figure 4, B and C). The mode of interaction between Did2-MIM1 and Ist1NTD shows distinctive features unseen in the previous two cases. Unlike the MIT domain that binds to MIM1 with two parallel or antiparallel helices, the MIM1-binding site in Ist1NTD consists of a groove formed by three perpendicular helices (Figure 4A). Our structure shows that one helix (α5) forms the floor of the binding site, whereas the other two (α1 and α2) run perpendicular, forming the sidewall.
A number of residues are conserved among Did2 proteins, including Asp193, Leu195, Arg198, Leu199, Leu202, and Arg203 (Figure 4D). The three leucine residues as well as Arg198 participate in binding to the MIT domains of Vps4 and spastin (Figure 4, B and C; Stuchell-Brereton et al., 2007; Yang et al., 2008). In the best structural model we can derive for the Ist1–Did2 complex, the side chains for five of the six conserved residues are located at the interface of the peptide–protein complex and may contribute to binding. To evaluate their roles, point mutations were introduced into the Did2-MIM1 peptide and their effects on binding were evaluated. Although none of the single mutations had a measurable binding defect, double mutation of L195A/L199A or L202A/R203A displayed a reduced binding to Ist1 in comparison with the wild-type Did2 peptide (Figure 4E). These results suggest that the molecular surface of the Did2-MIM1 helix that mediates Vps4 MIT and spastin MIT associations also participates in binding to Ist1 despite structural differences of these binding partners.
Ist1–Did2 Interaction Is Important for MVB Sorting
Previous studies have suggested that Ist1 functions together with Did2 in a process parallel to the Vta1/Vps60 complex (Rue et al., 2008; Dimaano et al., 2008). Simultaneous deletions of IST1 or DID2 with VTA1 gives rise to a strong MVB-sorting defect in yeast. To assess whether a functional Ist1–Did2 complex is important in the MVB pathway, we examined the sorting of the model yeast MVB cargo, carboxypeptidase S (CPS), in response to the disruption of Ist1–Did2 interaction. In wild-type cells the green fluorescent protein (GFP)-CPS fusion protein was observed exclusively in the lumen of the vacuole as a result of proper vacuolar sorting (Figure 5A). Consistent with previous results, ist1Δ cells or vta1Δ cells have no obvious or only modest effect on GFP-CPS transport, whereas ist1Δvta1Δ cells display a dramatic mislocalization of GFP-CPS to the limiting membrane of the vacuole and the aberrant class E compartment (Figure 5A). To evaluate the role of Ist1–Did2 interaction in MVB sorting, the subcellular localizations of GFP-CPS in ist1Δvta1Δ yeast cells supplemented with plasmids expressing either wild-type IST1 or Did2-interaction–deficient ist1 mutants were examined. As expected, transforming wild-type IST1 into ist1Δvta1Δ cells partially restored delivery of GFP-CPS into the vacuole to a level similar to that observed in vta1Δ cells. However, neither of the Did2-interaction defective ist1 mutants were capable of alleviating the MVB-sorting defect observed in ist1Δvta1Δ cells. These results highlight the importance of Ist1–Did2 interaction for the in vivo function of Ist1, suggesting that a stable Ist1–Did2 complex is required for the proper function of the ESCRT machinery in the yeast MVB pathway.
Figure 5.
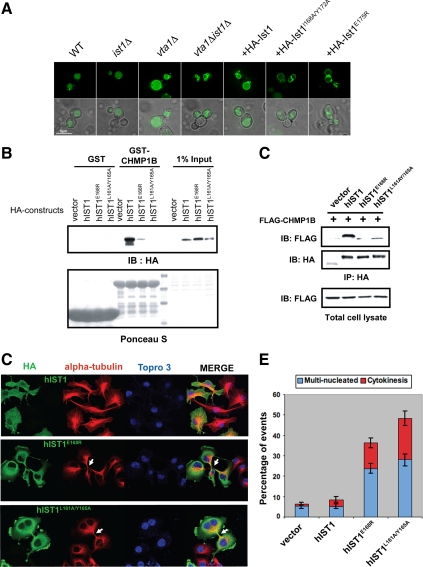
Ist1 residues involved in Did2 binding are important for both MVB sorting in yeast and mammalian cytokinesis. (A) The yeast MVB cargo GFP-CPS was visualized by fluorescent and brightfield microscopy in living cells corresponding to wild-type, ist1Δ, vta1Δ, ist1Δvta1Δ, or ist1Δvta1Δ cells expressing wild-type or mutant Ist1 proteins as indicated. The absence of lumenal GFP fluorescence in ist1Δvta1Δ cells expressing HA-Ist1L168A/Y172A or HA-Ist1E175R indicated that the Ist1–Did2 interaction contributes to Ist1 function in vivo. (B) In vitro interaction between hIST1 and CHMP1B. Cos-1 cells expressing HA-tagged hIST1 proteins were lysed and subjected to pulldown by GST or GST-CHMP1B. Proteins retained on the beads were analyzed by SDS-PAGE and visualized by anti-HA blotting (top) and Ponceau S staining (bottom). (C) In vivo interaction between hIST1 and CHMP1B. Cos-1 cells expressing FLAG-tagged CHMP1B and HA-tagged hIST1 constructs were lysed and subjected to immunoprecipitation with anti-HA antibody. CHMP1B proteins associated with hIST1 were detected by anti-FLAG blotting. (D) Disruption of hIST1-CHMP1 interaction leads to cytokinesis defects. Hela cells expressing wild-type or mutant HA-hIST1 proteins were fixed and subjected to immunofluorescence with anti-HA (green) and anti-α-tubulin (red). Nuclei were visualized with TORPO 3 (blue). (E) Quantification of the cytokinesis defects shown in C. Multinucleated cells (with two or more nuclei) or cells displaying apparent cytokinesis defect (with clear cytoplasmic bridge between the daughter cells as visualized by α-tubulin staining) were scored. Experiments were repeated four times, with 100 cells counted for each sample.
Ist1–Did2 Interaction Is Important for Efficient Abscission
Ist1 is evolutionarily conserved in eukaryotic cells. As the structure of the Ist1NTD/Did2–MIM1 complex illustrates, the Did2-binding site of Ist1 involves one of the most conserved regions on the Ist1NTD surface (Figure 3B). Furthermore, residues in Ist1 and Did2/CHMP1 that are important for the complex formation are evolutionarily conserved (Figures 2B and 4D). Mutating the conserved residues in yeast Ist1 significantly weakens its interaction with Did2 and disrupts its biological activity in the cell (Figures 3C and 5A). To examine whether the structure and function of Ist1–Did2 interaction is conserved in other organisms, we introduced corresponding mutations into the human Ist1 (hIST1) protein and tested the binding between hIST1 and one of the human Did2 homologues, CHMP1B.
As shown in Figure 5B, wild-type hIST1, but neither of the two mutants (hIST1E168R and hIST1L161A/Y165A), efficiently interacts with GST-CHMP1B. We further tested the in vivo interaction between these two proteins using coimmunoprecipitation (Figure 5C). Consistent with the pulldown experiment, wild-type hIST1 formed a stable complex with CHMP1B. In contrast, neither mutant of hIST1 interacted with CHMP1B to a level that can be detected under these experimental conditions, suggesting that the interaction mechanism between Ist1 and Did2 is evolutionarily conserved.
The ESCRT machinery has been recently demonstrated to be important in the abscission step during cytokinesis (Spitzer et al., 2006; Carlton and Martin-Serrano, 2007; Morita et al., 2007; Agromayor et al., 2009; Bajorek et al., 2009). To determine whether hIST1-CHMP1 interaction also plays a role in this process, we introduced wild-type and mutant hIST1 into Hela cells and examined whether the CHMP1-binding deficient mutants would compromise cytokinesis. Cells expressing wild-type hIST1 showed little alteration in cell cycle progression when compared with nontransfected cells. However, expressing either hIST1E168R or hIST1L161A/Y165A mutants led to a significant accumulation of cells arrested in a late phase of cytokinesis. This is evidenced by cells with four or more nuclei arrested in late cytokinesis stage that possess a cytoplasmic bridge (Figure 5, D and E). These cells were able to proceed through the nuclear division (mitosis) but failed to complete their cytoplasmic division (abscission/cytokinesis). Taken together, these data suggested that hIST1-CHMP1 interaction impacts cell abscission during cytokinesis, a process that requires the function of the ESCRT machinery.
DISCUSSION
The assembly of ESCRT-III subunits into a polymer has been implicated in deforming the endosomal membrane to generate the intralumenal vesicles of the MVB. The previously identified ESCRT-III subunits are similar in size (∼220 amino acids) and appear to adopt the CHMP3/Vps24-fold based on extensive conservation of secondary structure. Although these ESCRT-III subunits are all similar, in vivo analyses have demonstrated that these factors are not functionally redundant, suggesting that specific interactions within the ESCRT-III polymer and with ESCRT-III effectors are critical for MVB sorting. However, it is not clear how the specificity of subunit assembly is achieved to generate a functional ESCRT-III. Ist1 has previously been identified as an ESCRT-III effector via binding to Did2 and as a modulator of ESCRT-III function through regulating Vps4 activity; however, the mechanism of Ist1 function was unclear. Here we demonstrate that Ist1 possesses the conserved ESCRT-III subunit fold, identifying Ist1 as a divergent ESCRT-III subunit. Moreover, cocrystallization of Ist1NTD with the Did2 C-terminal MIM1 sequence identifies a novel MIM-binding structural motif in Ist1. The specificity of the Ist1–Did2 MIM1 association implicates this binding pocket in contributing to the order of Ist1 recruitment and suggests that similar structural motifs in other ESCRT-III subunits may facilitate the specific subunit associations required for ESCRT-III assembly/function. Additionally, this binding pocket may participate in an intramolecular interaction of the carboxyl-terminus, forming the closed or autoinhibited state of the ESCRT-III subunits.
A New Mode of MIM Binding
A common feature of the protein complexes within the ESCRT machinery is that they contain domains that bind to phosphoinositides, ubiquitin, or specific proteins. The MIT domain is a protein–protein interaction domain identified in a number of components of the ESCRT machinery, including Vps4, Vta1, AMSH, and UBPY (Hurley and Yang, 2008). The MIT domain recognizes sequence motifs present primarily within the ESCRT-III subunits. These sequences are collectively called the MIMs. Two types of MIM have been identified, MIM1 and MIM2 (Obita et al., 2007; Stuchell-Brereton et al., 2007; Kieffer et al., 2008; Samson et al., 2008). Interaction between MIT and MIM has been implicated in regulating the disassembly of ESCRT-III as well as targeting specific proteins to the site of ESCRT function.
Our biochemical and structural analyses of the Ist1/Did2–MIM1 complex reveal that MIM1 can interact with a structure other than the MIT domain. The increased MIM-binding structural repertoire reveals a previously unrecognized complexity within the protein–protein interaction network of the ESCRT machinery. It is interesting to note that when binding to Vps4-MIT and Ist1NTD, Did2 appears to utilize a similar binding surface (Figure 4, A and B; Stuchell-Brereton et al., 2007). If our complex structure model is accurate, this surface involves Glu192, Leu195, Ala196, Arg198, Leu199, Leu202, and Arg203 of Did2. As a result, superimposing the MIM1 motifs in these two structures results in significant clashes between Ist1NTD and Vps4-MIT (Supplementary Figure S3), consistent with the observation that Did2-MIM1 cannot bind to Ist1 and Vps4 simultaneously in vitro (Supplementary Figure S4). How these two interactions are coordinated in the cells is intriguing. It is unclear whether Vps4 and Ist1 bind to Did2 in a sequential and competing manner or if they simultaneously bind to different Did2 molecules in an ESCRT-III oligomer.
It is interesting to note that binding of MIM1 to Ist1NTD is more specific than binding to Vps4-MIT. Although the Vps4-MIT domain binds to MIM1s derived from different proteins with similar strength, Ist1NTD appears to selectively bind to Did2 under the conditions utilized (compare Figure 1B and Supplementary Figure S2). The apparent Kd for Ist1–Did2 MIM1 interaction is 1 μM (Figure 3D), whereas we have observed that the affinity of the Ist1NTD for Did2 MIM1 is stronger (data not shown). By contrast, the Vps4 MIT-Did2 MIM1 apparent Kd is ∼35 μM (Obita et al., 2007; Stuchell-Brereton et al., 2007). These distinctions suggest that Ist1 makes more extensive contacts with the Did2 MIM1 than does the Vps4 MIT and may explain the differential specificities exhibited by the Ist1NTD and Vps4 MIT; however, we have not been able to resolve the determinants of this specificity at the structural level. Recent studies have suggested that specific pairs of ESCRT-III subunit interactions facilitate the ordered assembly of ESCRT-III on the endosomal membrane (Babst et al., 2002; Saksena et al., 2009). The basis of specificity for inter-ESCRT-III subunit association remains unclear, but the structure of the Ist1NTD-Did2 MIM complex suggests one possible mechanism. The positioning of helix α5 perpendicular to helices α1 and α2 within one ESCRT-III subunit creates a pocket that may permit specific binding of the C-terminus of its partner subunit(s); this interaction may then facilitate formation of additional contacts between subunits, via α1-α4, that stabilize ESCRT-III subunit oligomerization. Higher resolution structural studies are required to fully evaluate the contributions of interface residues to the energetics and specificity of the Ist1–Did2 and other ESCRT-III subunit-subunit interactions.
Implication for ESCRT-III Activation
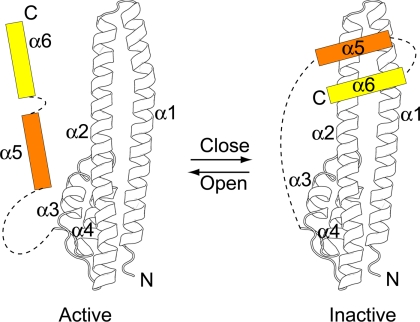
The Ist1NTD-Did2 MIM1 structure may also be informative for the mechanism of ESCRT-III subunit autoinhibition and activation via regulation of intramolecular association. The ESCRT-III subunits are meta-stable molecules that exist in two distinct conformations: a membrane-bound active form and a cytosolic inactive form. Transition between the two conformations is controlled by the C-terminal autoinhibitory region, which binds to the N-terminal ESCRT-III fold in the inactive form to prevent oligomerization (Zamborlini et al., 2006; Shim et al., 2007). The autoinhibitory region consists of helix α5 and additional C-terminal residues including the MIM. In the previous study of CHMP3, the fragment used in the structure determination lacks the C-terminal residues. Thus the conformation present in the crystal structure may represent an open and active form of the molecule (Muziol et al., 2006). In this form, helix α5 appears to assume an arbitrary conformation stabilized by lattice contact and makes little contact with the core of the structure (Figure 2C).
The surprising discovery that the MIM1-binding Ist1NTD contains an ESCRT-III-like fold provides a possible structural model for the elusive inactive or closed conformation of the ESCRT-III subunits. In the Ist1NTD structure, helix α5 is linked to helix α4 by a similar length connection (30 amino acids; Figure 2B). It is conceivable that the position of helix α5 in the Ist1NTD structure corresponds to the position of helix α5 in the ESCRT-III subunits when it folds back and interacts with the ESCRT-III fold (Figure 6). On the basis of the Ist1NTD/Did2–MIM1 complex structure, we hypothesize that in the inactive state of the ESCRT-III subunits, the C-terminus binds in a site formed by helices α1, α2, and α5 (Figure 6). Consistent with this proposal, it is interesting to note that in the crystal lattice of CHMP3 (Muziol et al., 2006), there is a neighboring helix α5 in what appears to be the same position proposed here. Thus, it is unclear whether the positioning of helix α5 in the CHMP3 structure represents the open conformation (as discussed above) or results from “domain swapping” of the α5 helix (Liu and Eisenberg, 2002).
Figure 6.
Model for the functional cycle of the ESCRT-III proteins. Left panel, the open, active conformation of the ESCRT-III proteins. Helices α1–α4 are adapted from the structure of human CHMP3. Helix α5 and the C-terminal helix α6 are shown as arbitrarily placed cartoons and the linkers as dash lines. Right panel, the speculative closed, inactive conformation of the ESCRT-III proteins. The positions of helices α5–α6 are inspired by the structure of the Ist1NTD/Did2–MIM1 complex.
During the course of ESCRT-III subunit activation, weakening of intramolecular interaction leads to the release of the C-terminus as well as ESCRT-III membrane recruitment/polymerization. In this model, transition to the active state may simply represent replacement of a weak intramolecular C-terminus interaction with a more stable intermolecular association via the same MIM-binding pocket. Three observations support this model of a transition between intra- and intermolecular interactions. First, Ist1 contains a MIM1 in its C-terminus, but the Ist1NTD-Ist1 MIM1 interaction is not detected in pulldown experiments (Figure 1B). Second, the Ist1NTD-Did2 MIM1 association appears more robust than the association between full-length Ist1 and Did2 MIM1, suggesting that the Ist1CTD partially inhibits Ist1NTD-Did2 MIM1 association. Third, purification of Ist1NTD requires coexpression of the Did2 MIM1, suggesting that MIM binding pocket within full length Ist1 is occupied as Ist1NTD is unstable in the absence of the C-terminal sequence. These observations collectively suggest that the Ist1 MIM1 binds weakly to the pocket formed by α5 to maintain the closed state in the absence of Did2. Whether helix α5 is first released and later refolded or repositioned during activation, as suggested by the CHMP3 structure (Muziol et al., 2006), is not presently clear.
Mechanism of Ist1 Function In Vivo
Previous studies have suggested a number of possible roles Ist1 plays with the ESCRT machinery. Binding of Ist1 to Vps4 inhibits oligomerization of the ATPase in vitro, and overexpression of Ist1 in yeast leads to an MVB sorting defect similar to loss of Vps4 function (Dimaano et al., 2008). Our study confirms the previous observation that Ist1 binds to Vps4 via its C-terminal MIM1 sequence. It should be noted that the C-terminal sequence alone is not sufficient for inhibition, which also requires Ist1NTD (Dimaano et al., 2008). It is intriguing to speculate that the Ist1NTD portion disfavors Vps4 oligomerization to inhibit ATPase activity, although how this might occur remains unclear.
It would appear that the inhibitory function toward Vps4 is not the only role of Ist1 in the cell. Instead, accumulating evidence has suggested that Ist1 functions through its interaction with Did2 (Dimaano et al., 2008; Rue et al., 2008; Agromayor et al., 2009; Bajorek et al., 2009). We have demonstrated that Ist1 binds to the C-terminal MIM1 sequence of Did2 via its N-terminal domain (Ist1NTD), which adopts an ESCRT-III-like fold. Disruption of this conserved interaction through point mutations within Ist1NTD leads to defects in both yeast MVB sorting and mammalian cytokinesis. Our studies also suggest that the Ist1–Did2 interaction is highly specific despite the existence of other homologous MIM1 or MIM1-like sequences in the ESCRT machinery. Furthermore, it is unlikely that Ist1 and Did2 exist as a preformed complex in the cytosol as the affinity between full-length Ist1 and Did2 is rather weak. Consequently, we suggest that interaction between Ist1 and Did2 is coupled to Did2 assembly into ESCRT-III on the membrane.
Did2 appears to be recruited to the endosomal membrane via the Vps24 and Vps2 subunits (Nickerson et al., 2006), which can also recruit Vps4. Did2 membrane localization appears to be correlated with a conformational change leading to exposure of its Ist1-binding MIM1 sequence. Binding of Ist1 to Did2 may regulate the ESCRT machinery in several ways. Ist1 may stabilize the active conformation of Did2, allowing additional regulators of MVB sorting to bind. Alternatively, Did2-association may reduce the inhibitory effect of Ist1 on MVB sorting through altering Ist1-Vps4 interactions. Binding of Ist1 to Did2 may also release the Ist1 C-terminal MIM1, which could potentially interact with multiple partners in the ESCRT machinery. The order and interconnection of these interactions need to be further delineated to better understand their biological consequences.
In summary, we have presented structural and biochemical analyses of the N-terminal domain of Ist1 that binds the Did2 C-terminus. Our results reveal not only that Ist1 is a divergent member of the ESCRT-III family but also that Ist1 interacts with the Did2 MIM1 via a novel MIM-binding structural motif. This information coupled with cell biology experiments provide important insights into the molecular mechanism of Ist1 function and the biological role of the Ist1–Did2 interaction. Perhaps of greater interest, these results suggest a conserved molecular mechanism pertaining to the interaction between the C-terminal amphipathic helices of the ESCRT-III proteins and their binding partners. This interaction may be utilized in both the inhibition (intramolecular) and the assembly (intermolecular) of the ESCRT-III subunits. Clearly, the molecular details that govern the specificity of these interactions deserve further exploration in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to staffs at the Center for Structural Biology at the University of Michigan Life Sciences Institute for maintaining the x-ray facility and staffs at the Advanced Photon Source beam line 21-ID-D of Argonne National Laboratory for access and help with data collection. We thank S. Patury and J. Gestwicki for access and help with BIACORE instrument and data analysis and J. Tesmer, J. Smith, R. Fuller, and D. Klionsky for critically reading the manuscript. This work was supported by Grants RO1 DK65980 (Z.X.) and RO1 GM73024 (D.J.K.) from the National Institutes of Health.
Abbreviations used:
- ESCRT
endosomal sorting complexes required for transport
- MVB
multivesicular body
- MIM
MIT-interacting motif
- MIT
microtubule interacting and transport.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-05-0403) on May 28, 2009.
REFERENCES
- Agromayor M., Carlton J. G., Phelan J. P., Matthews D. R., Carlin L. M., Ameer-Beg S., Bowers K., Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Morita E., Skalicky J. J., Morham S. G., Babst M., Sundquist W. I. Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell. 2009;20:1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J. G., Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Chen X. W., Inoue M., Hsu S. C., Saltiel A. R. RalA-exocyst-dependent recycling endosome trafficking is required for the completion of cytokinesis. J. Biol. Chem. 2006;281:38609–38616. doi: 10.1074/jbc.M512847200. [DOI] [PubMed] [Google Scholar]
- Chen X. W., Leto D., Chiang S. H., Wang Q., Saltiel A. R. Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev. Cell. 2007;13:391–404. doi: 10.1016/j.devcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Dimaano C., Jones C. B., Hanono A., Curtiss M., Babst M. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell. 2008;19:465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S., Saksena S., Short J. M., Pobbati A. V., Veprintsev D. B., Crowther R. A., Emr S. D., Egelman E. H., Williams R. L. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D. I., Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Roth R., Lin Y., Heuser J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Kaariainen S., Rosenstrom P., Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H., Yang D. MIT domainia. Dev. Cell. 2008;14:6–8. doi: 10.1016/j.devcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., Sundquist W. I. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. ConSurf 2005, the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lata S., Roessle M., Solomons J., Jamin M., Gottlinger H. G., Svergun D. I., Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 2008a;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gottlinger H. G., Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008b;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Gottlinger H., Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., West M., Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S. D. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Piper R. C., Katzmann D. J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue S. M., Mattei S., Saksena S., Emr S. D. Novel Ist1-did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell. 2008;19:475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Sun J., Chu T., Emr S. D. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;31:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R. Y., Obita T., Freund S. M., Williams R. L., Bell S. D. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S., Kimpler L. A., Hanson P. I. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Spitzer C., Schellmann S., Sabovljevic A., Shahriari M., Keshavaiah C., Bechtold N., Herzog M., Muller S., Hanisch F. G., Hulskamp M. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Williams R. L., Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell. Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Xia H., Yoshino-Koh K., Zhou J., Xu Z. Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J. Mol. Biol. 2007;374:655–670. doi: 10.1016/j.jmb.2007.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Xia H., Zhou J., Azmi I. F., Davies B. A., Katzmann D. J., Xu Z. Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev. Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Rismanchi N., Renvoise B., Lippincott-Schwartz J., Blackstone C., Hurley J. H. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A., Usami Y., Radoshitzky S. R., Popova E., Palu G., Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl. Acad. Sci. USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.