Abstract
Mitochondrial fusion requires the coordinated fusion of the outer and inner membranes. Three large GTPases—OPA1 and the mitofusins Mfn1 and Mfn2—are essential for the fusion of mammalian mitochondria. OPA1 is mutated in dominant optic atrophy, a neurodegenerative disease of the optic nerve. In yeast, the OPA1 ortholog Mgm1 is required for inner membrane fusion in vitro; nevertheless, yeast lacking Mgm1 show neither outer nor inner membrane fusion in vivo, because of the tight coupling between these two processes. We find that outer membrane fusion can be readily visualized in OPA1-null mouse cells in vivo, but these events do not progress to inner membrane fusion. Similar defects are found in cells lacking prohibitins, which are required for proper OPA1 processing. In contrast, double Mfn-null cells show neither outer nor inner membrane fusion. Mitochondria in OPA1-null cells often contain multiple matrix compartments bounded together by a single outer membrane, consistent with uncoupling of outer versus inner membrane fusion. In addition, unlike mitofusins and yeast Mgm1, OPA1 is not required on adjacent mitochondria to mediate membrane fusion. These results indicate that mammalian mitofusins and OPA1 mediate distinct sequential fusion steps that are readily uncoupled, in contrast to the situation in yeast.
INTRODUCTION
Mitochondria are dynamic organelles that continually exchange contents through membrane fusion. Mitochondrial fusion controls the morphology of the organelle and is critically important for maintaining the function of the mitochondrial network. Loss of fusion has been linked to reduced respiratory activity, embryonic lethality, apoptosis, and neurodegeneration (Okamoto and Shaw, 2005; Detmer and Chan, 2007; Suen et al., 2008). In addition, mutations in Mfn2 and OPA1, two genes involved in mitochondrial fusion, cause the human neurodegenerative diseases Charcot-Marie-Tooth type 2A (CMT2A) (Zuchner et al., 2004) and dominant optic atrophy (DOA) (Alexander et al., 2000; Delettre et al., 2000), respectively.
Mitochondrial fusion is a multi-step process requiring the coordinated fusion of both the outer and inner membranes, ultimately resulting in mixing of matrix contents. The coordination of outer and inner membrane fusion has been studied in budding yeast (Sesaki et al., 2003; Wong et al., 2003; Meeusen et al., 2006; Hoppins et al., 2007). In yeast, the outer membrane proteins Fzo1p and Ugo1p and the inner membrane protein Mgm1p are essential for mitochondrial fusion. In an in vitro mitochondrial fusion assay, Mgm1p is required for fusion of the inner but not outer mitochondrial membranes (Meeusen et al., 2006). However, in vivo, yeast lacking Mgm1 have no outer membrane fusion (Sesaki et al., 2003). Ugo1p forms a complex with both Fzo1p and Mgm1p and may coordinate the activity of both proteins (Wong et al., 2003; Sesaki and Jensen, 2004).
In mammals, mitochondrial fusion requires the action of three large GTPases: the mitofusins (Mfns) Mfn1 and Mfn2, which are orthologues of Fzo1p, and the dynamin-related protein OPA1, which is the ortholog of Mgm1p (Chen et al., 2003; Cipolat et al., 2004; Chen et al., 2005; Chan, 2006). The coordination of outer membrane versus inner membrane fusion in mammalian mitochondria can be disrupted by pharmacological treatments (Malka et al., 2005), but the molecular basis of this uncoupling has not been characterized. To address whether these large GTPases act at distinct steps during mitochondrial fusion, we have developed assays to analyze mitochondrial outer membrane fusion in mouse embryonic fibroblasts (MEFs) containing null alleles of OPA1 or both mitofusins. Our results provide clear evidence that mitofusins act early and are essential for outer membrane fusion, whereas OPA1 is required for only inner membrane fusion. In addition, we find several differences in the properties of mitochondrial fusion between yeast and mammals.
MATERIALS AND METHODS
Cell Culture and Retroviral Expression
All cell lines were grown in Dulbecco's Modified Eagle's Medium supplemented with 10% FBS, 1 mM L-glutamine, penicillin, and streptomycin. Cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C.
PA-GFP-OMP25 was constructed by inserting a segment encoding the last 37 amino acids of OMP25 in frame with PA-GFP. The terminal residues of OMP25 have been shown to be sufficient to direct localization to the mitochondrial outer membrane (Nemoto and De Camilli, 1999). To construct mCherry-OMP25, mCherry was amplified by PCR and cloned in-frame to the last 37 amino acids of OMP25. Mito-PA-GFP (provided by Richard Youle, National Institutes of Health), PA-GFP-OMP25, and mCherry-OMP25 were subcloned into the Moloney-based retroviral vector pCLβW. Retroviral stocks were produced by cotransfection of 293T cells with retroviral constructs and the ecotropic retroviral packaging vector pCL-Eco using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according the manufacturer's protocol.
A retroviral shRNAi construct for Phb1 was made as previously described (Chen et al., 2005) using the following primers:
5′ gatccccGACTTGCAGAAtGTCAATAttcaagagaTATTGACGTTCTGCAAGTCtttttggaaa 3′
5′ agcttttccaaaaaGACTTGCAGAACGTCAATAtctcttgaaTATTGACaTTCTGCAAGTCggg 3′. The retroviral construct targets the sequence gacttgcagaacgtcaata in mouse Phb1.
PEG Fusion Assay
MEFs expressing mitochondrially targeted DsRed or mCherry were coplated with MEFs expressing mitochondrially targeted enhanced green fluorescent protein (EGFP). The next morning, cells were incubated with PEG 1500 (Roche, Indianapolis, IN) for exactly 1 min at room temperature and then washed with medium 3 times. The cells were then cultured in medium containing 33 μg/ml cyclohexmide for 1 or 7 h. The cultures were fixed in 10% Formalin, and the coverslips were then mounted onto microscope slides with GelMount (Biomeda, Foster City, CA).
Confocal Microscopy and Image Processing
Confocal microscopy was performed with a Zeiss LSM 510 Meta microscope with a 100× oil-immersion objective. Live cells were maintained at 37°C under mineral oil. In cells expressing mito-PA-GFP or PA-GFP-OMP25, mitochondria within a region of interest were photoactivated with a two-photon laser set at 750 nm. With the multi-track scanning mode, mt-DsRed was imaged by excitation with a 1-mW 543-nm helium/neon laser, with emission recorded through an LP560-nm filter. EGFP or photoactivated PA-GFP was imaged by excitation with a 25-mW 488-nm argon laser, with emission recorded through a BP 500–560-nm filter.
Image processing and analysis were performed with ImageJ software. Individual mitochondria were selected manually in each still frame, and its fluorescence intensity, corrected for background fluorescence, was recorded. Mitochondria were randomly selected for quantitative analysis.
Electron Microscopy
Cultured cells were prepared for electron microscopy as previously described (Perkins and McCaffery, 2007). Briefly, cells were fixed at room temperature (3.0% formaldehyde, 1.5% glutaraldehyde, 0.1 M sodium cacodylate, 5 mM Ca2+, 2.5% sucrose pH 7.4 buffer) and stored at 4°C. Cells were washed 3 times in ice-cold 0.1 M sodium cacodylate containing 2.5% sucrose and then postfixed in Palade's OsO4 for 1 h on ice in the dark. After washing 3 times with 0.06 M veronol acetate, the fixed cell pellet was stained and stabilized en bloc with Kellenberger's uranyl acetate at room temperature in the dark. After one rinse with 50% ethanol (4°C) the cells were dehydrated at 4°C through a graded series of ethanol (70%, 95%, 100%) for 5 min each and treated 3 times for 5 min each in fresh 100% acetone at room temperature. Cells were then infiltrated in a well-mixed Epon-acetone resin series of 33% for 3 h, 66% for 6 h followed by 100% at least overnight with agitation at room temperature, after which the samples were allowed to polymerize in 100% Epon blocks at 60°C for 48 h. For conventional electron microscopy, sections of ≈80 nm were cut using a diamond knife and poststained with 2% uranyl acetate and lead citrate. For electron tomography, sections of ≈300 nm were cut and poststained with 2% uranyl acetate and lead citrate. Colloidal gold particles were applied (10 nm) for use as fiducial marks for alignment before recording single axis tilt series at 2° from −60° to + 60° on an FEI Tecnai 12 transmission electron microscope operated at 120 kV with images recorded on a Tietz 214 CCD camera. Tomograms were calculated from the tilt series, segmented, and displayed as described previously (Sun et al., 2007).
RESULTS
OPA1 Is Not Required on Adjacent Mitochondria for Fusion
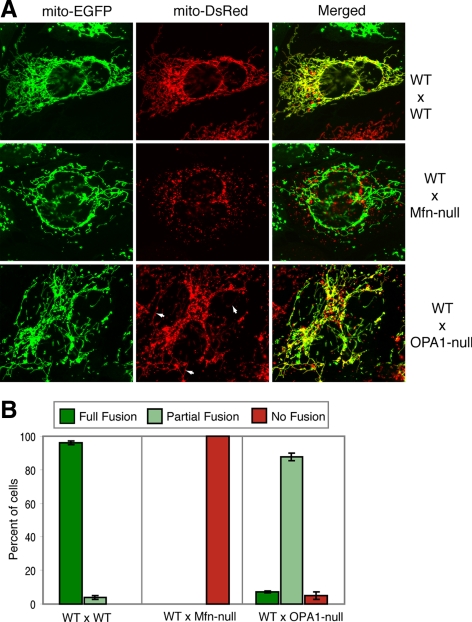
We previously showed that both Mfn-null cells (double null cells lacking Mfn1 and Mfn2) and OPA1-null cells lack any detectable mitochondrial fusion activity (Koshiba et al., 2004; Chen et al., 2005; Song et al., 2007), as measured in fusion assays that monitor mixing of fluorescent proteins targeted to the mitochondrial matrix. One of the key properties of mitofusins is that they are required on adjacent mitochondria during fusion, supporting their proposed role in tethering the outer mitochondrial membranes (Koshiba et al., 2004). To test whether there is a similar requirement for OPA1 function, we performed PEG cell hybrid assays in which OPA1-null cells containing DsRed in the mitochondrial matrix were fused against wild-type cells containing EGFP in the mitochondrial matrix. In 95% of the cell hybrids, we detected substantial colocalization of DsRed and EGFP, indicating fusion of both the outer and inner membranes to yield merging of the contents of the mitochondrial matrix (Figure 1). In 7% of the cell hybrids, all the mitochondria showed colocalization of DsRed and EGFP (Figure 1B; scored as “full fusion”). In the other 88% of cell hybrids, clear colocalization of DsRed and EGFP was found in a fraction of the mitochondrial population (scored as “partial fusion”). Many of the DsRed-containing mitochondria in these OPA1-null/wild-type cell hybrids were elongated tubules, whereas mitochondria of the OPA1-null parental cells were completely fragmented. This observation is consistent with the conclusion that substantial mitochondrial fusion has occurred. In contrast, cell hybrids between two OPA1-null cells show no mitochondrial fusion (Song et al., 2007). In agreement with previous studies, no mitochondrial fusion was detected when Mfn-null cells were fused with wild-type cells.
Figure 1.
Distinct requirements for mitofusins and OPA1 in mitochondrial fusion. (A) Representative images of three PEG cell hybrid fusion assays done by fusing the parental cells indicated on the right. In the bottom two panels, the mutant cells (Mfn-null or OPA1-null) express mito-DsRed, whereas the wild-type cell (WT) expresses mito-EGFP. In the bottom panel, a cell hybrid between a wild-type and an OPA1-null cell shows extensive mixing of the green (left panel) and red (middle panel) fluorophores. Some elongated mitochondrial tubules containing DsRed in the cell hybrid are indicated by arrows. (B) Quantitation of the PEG cell hybrid experiments. Error bars indicate standard deviations from 3 independent experiments in which 100 cell hybrids were scored. The cell hybrids were scored by estimating the percentage of mitochondria showing colocalization of fluorophores. Full fusion indicates that all the mitochondria in the cell hybrid contained both fluorophores. Cell hybrids with a combination with fused and unfused mitochondria were scored as partial fusion. Hybrids with no doubly stained mitochondria were scored as no fusion.
The results above indicate that OPA1 is needed on only one mitochondrion to facilitate fusion between a pair. It should be noted, though, that multiple OPA1 isoforms arise through differential RNA splicing and protein processing. Long isoforms of OPA1 are anchored to the inner membrane, whereas short isoforms have no apparent membrane anchor. A combination of long and short isoforms is necessary for fusion activity (Song et al., 2007). One potential concern is that short isoforms of OPA1 can diffuse in the intermembrane space if outer membrane fusion occurs, complicating the interpretation of the PEG cell hybrid results. To address this concern, we used the recent observation that an OPA1 long isoform alone can promote fusion in the presence of cycloheximide, in contrast to the requirement for a combination of long and short forms during normal cell growth (Tondera et al., 2009). Consistent with this study, we found that whereas OPA1-null cells expressing isoform 1ΔS1, a noncleavable long form of OPA1 isoform 1 (Song et al., 2007), have fragmented mitochondria, incubation with cycloheximide leads to formation of more elongated mitochondria (Figure S1, A and B). Western blot analysis showed that the elongation of mitochondria with cycloheximide treatment was not attributable to production of a short form of OPA1 (Figure S1C). In contrast, cells expressing isoform 3, which produces only a short form, showed fragmented mitochondria under both conditions.
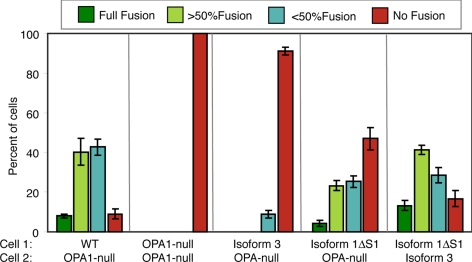
Because cycloheximide is used to inhibit protein synthesis during the PEG fusion assay, we asked whether the membrane-bound long form of isoform 1 could mediate mitochondrial fusion when present on only one of the parental cells. When OPA1-null cells expressing 1ΔS1 were fused with OPA1-null cells, we found that ≈50% of the cell hybrids contained substantial mitochondrial fusion (Figure 2). This observation further supports the idea that OPA1, unlike mitofusins, is not needed on adjacent mitochondria promote fusion.
Figure 2.
Mitochondrial fusion in hybrids of parental cells with different OPA1 isoforms. Mitochondrial fusion was measured in cell hybrids between wild-type cells, OPA1-null cells, or OPA1-null cells expressing the indicated OPA1 isoforms. Error bars indicate SD from 3 experiments.
Outer Membrane Fusion in OPA1-Null Cells in the PEG Fusion Assay
To study the function of mitofusins and OPA1 in mitochondrial outer membrane fusion, we used the PEG cell hybrid assay with one of the parental cell lines expressing mCherry fused to the membrane targeting sequence of OMP25 (Nemoto and De Camilli, 1999), a mitochondrial outer membrane protein. At the standard 7-h time point used for PEG cell fusion experiments, we found that OPA1-null cell hybrids showed extensive mixing of mCherry-OMP25 throughout the mitochondrial population (Figure S2). Under these conditions, however, Mfn-null cells also showed mixing of the outer membrane marker, although less extensively (Figure S2). We were concerned that the extended time used in a standard PEG cell hybrid experiment might lead to artifactual results with the outer membrane marker. When we repeated these experiments using shorter incubation times, we found a marked difference in the extent of mCherry-OMP25 mixing in OPA1-null cells versus Mfn-null cells (Figure 3). At 1 h after cell fusion, Mfn-null hybrids show little evidence for outer membrane fusion, whereas more than 95% of OPA1-null hybrids show clear partial fusion (Figure 3B). These results suggest that outer membrane fusion occurs in OPA1-null cells. However, our results also suggest that the PEG cell hybrid assay is not an ideal system to measure mitochondrial outer membrane fusion. It is possible that the residual mCherry-OMP25 mixing in Mfn-null cells reflects an artifact of the PEG fusion assay. Because of this concern, we decided to develop an assay to directly visualize outer membrane fusion events in real-time.
Figure 3.
Outer membrane fusion in OPA1-null cells. (A) Representative images of three PEG cell hybrid fusion assays done by fusing the parental cells indicated on the right and analyzing cells 1 h after cell fusion. Mitochondrial outer membrane mixing was monitored with mCherry-OMP25. In the bottom panel, an OPA1-null cell hybrid shows partial mixing of the green (left panel) and red (middle panel) fluorophores. The large inset is a magnified image of the small box. Arrows highlight individual mitochondria within the inset. (B) Quantitation of the PEG cell hybrid experiments. Scoring and quantitation was done as for Figure 1B, except that because cells only showed partial mitochondrial fusion at 1 h, the partial fusion category was further separated into cells in which greater or <50% of the mitochondria showed fluorophore mixing. Error bars indicate SD from 3 experiments.
Outer Membrane Fusion Observed in OPA1-Null Cells With a New Outer Membrane Fusion Assay
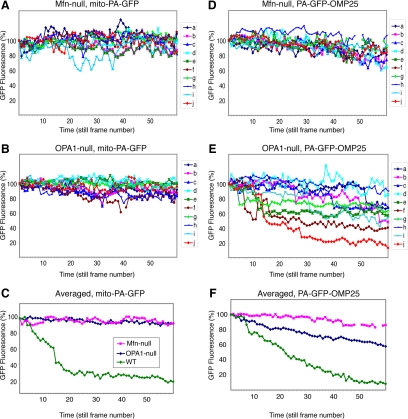
We adopted an assay (Karbowski et al., 2004; Haigh et al., 2007) for mitochondrial fusion that uses photoactivatible GFP (PA-GFP), a GFP derivative whose fluorescence increases 100-fold upon photoactivation (Patterson and Lippincott-Schwartz, 2002). In this assay, cells express both mito-PA-GFP and mito-DsRed, variants that are targeted to the mitochondrial matrix. Within a single cell, a small subset of mito-PA-GFP–labeled mitochondria is photoactivated by 2-photon excitation and then tracked by time-lapse microscopy. In wild-type cells, mitochondrial fusion causes the PA-GFP in photoactivated mitochondria to transfer to nonactivated mitochondria. Because mitochondrial fusion results in content exchange, such events are characterized by sudden stepwise changes in PA-GFP and DsRed fluorescence (Figure S3). In cells where mitochondrial fusion activity is high, the PA-GFP fluorescence intensity in the activated mitochondria is steadily diluted because of cycles of fusion with surrounding mitochondria. In cells with little or no mitochondrial fusion, however, the PA-GFP fluorescence in the activated mitochondria remains stable, apart from fading attributable to photobleaching. As expected, both OPA1-null and Mfn-null cells behave similarly in this assay, with activated mito-PA-GFP showing only modest declines in fluorescence over a 20 min recording session (Figure 4,A–C). In addition, inspection of individual mitochondria in the time-lapse movies revealed no evidence for matrix mixing. In both cell lines, we tabulated the fluorescence of 10 individual photoactivated mitochondria (Figure 3, A and B). Over the 20-min recording session, the overall fluorescence intensity for the photoactivated mitochondria in both cell lines declined by only 7% (Figure 3C). This gradual reduction is attributed to photobleaching, because examination of the time-lapse movies reveal that individual mitochondria undergo gradual reductions in fluorescence intensity without fluorescence exchange with neighboring mitochondria. In contrast, the photoactivated mitochondria in wild-type cells lose most of their fluorescence within 10 min (Figure 4C).
Figure 4.
Behavior of matrix and outer membrane markers in Mfn-null and OPA1-null cells. Full fusion and outer membrane fusion were monitored by measuring the dilution of the matrix marker mito-PA-GFP (A–C) or the outer membrane marker PA-GFP-OMP25 (D–F), respectively, in Mfn-null (A and D) and OPA1-null (B and E) cells. Panels A, B, D, and E show individual normalized fluorescence traces of 10 individual mitochondria every 20 s throughout the entire 20-min recording session. In a few traces, discontinuities are attributable to brief defocusing during recording. The data in A and B are averaged to yield the traces in C. The data in D and E are averaged to yield the traces in F. In E and F, mitochondria from OPA1-null cells demonstrate a much greater decline in PA-GFP-OMP25 fluorescence compared with Mfn-null cells. Data from 6 mitochondria in wild-type cells were used to obtain the wild-type averages in C; data from 5 mitochondria were used to obtain the wild-type averages in F. The legend to F is shown in C.
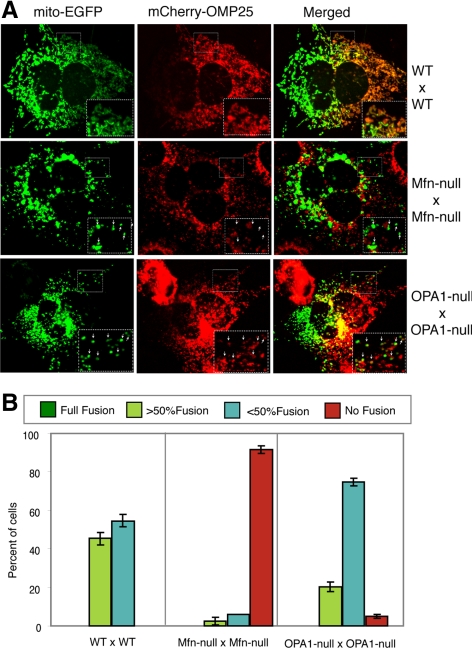
To examine the issue of mitochondrial outer membrane fusion, we modified the photoactivation assay by localizing PA-GFP to the mitochondrial outer membrane through fusion with the targeting sequences of OMP25. We expressed both mito-DsRed and PA-GFP-OMP25 in cells and used two-photon excitation to photoactivate a small group of mitochondria in a region of interest. After photoactivation, cells were monitored by time-lapse microscopy every 20 s for 20 min. In wild-type cells, mitochondrial fusion results in tightly coupled outer and inner membrane fusion (Figure S4). Such events result in stepwise changes in the fluorescence intensity of both fluorescent markers. When a PA-GFP-OMP25 labeled mitochondrion fuses with a nonactivated mitochondrion, its GFP fluorescence shows a stepwise decline as its DsRed fluorescence suddenly increases.
In Mfn-null cells, we found that PA-GFP-OMP25 behaved similarly to mito-PA-GFP, and we never detected evidence for outer membrane fusion (Figure 4, D and F). From the time-lapse images of Mfn-null cells, we tracked and quantified the fluorescence profiles of individual photoactivated mitochondria as a function of time. These plots indicate that the fluorescence intensities were stable (Figure 4D). The averaged fluorescence of the photoactivated mitochondria declined only 14% after 20 min of continual recording (Figure 4F). The slightly higher level of photobleaching in this assay is attributable to the higher laser power necessary to image PA-GFP-OMP25, which has lower fluorescence than mito-PA-GFP.
In OPA1-null cells, however, we detected frequent outer membrane fusion events. The average fluorescence intensity of the photoactivated mitochondria declined by 42% after 20 min (Figure 4F). When the PA-GFP-OMP25 fluorescence intensities of individual photoactivated mitochondria were analyzed, most mitochondria (7 of 10) show stochastic stepwise drops in intensity that are highly suggestive of outer mitochondrial fusion (Figures 4E and 5). Although frequent outer membrane fusion events are observed (see below) in OPA1-null cells, these events happen less frequently than in wild-type cells (Figure 4F). Consistent with this idea, OPA1-null cells have fragmented mitochondrial morphology even when analyzed with outer membrane markers.
Figure 5.
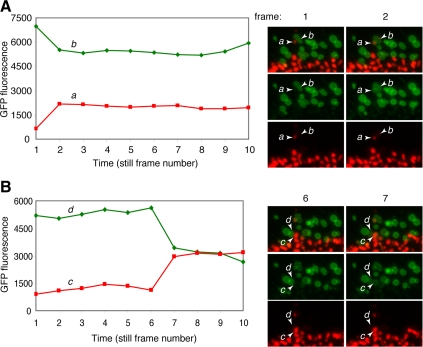
Visualization of outer membrane fusion events in OPA1-null cells. (A) PA-GFP fluorescence traces in a pair of mitochondria in an OPA1-null cell (expressing PA-GFP-OMP25 and mito-DsRed). Mitochondrion b was photoactivated and initially contained high levels of PA-GFP fluorescence. Mitochondrion a was not photoactivated and therefore initially did not show PA-GFP fluorescence. In frame 2, a stepwise drop in the PA-GFP fluorescence of b is matched by an increase the fluorescence of a. Still frames corresponding to these changes are shown on the right. The frames before and after the fluorescence change are shown as a merged (top), green (middle), or red image (bottom). The relevant mitochondria involved in each outer membrane fusion event are labeled. Although exchange of PA-GFP fluorescence occurs, there is no transfer of DsRed. Under our PA-GFP photoactivation conditions, DsRed is photobleached. Therefore, photoactivated mitochondria are easily visualized by high GFP fluorescence and no DsRed fluorescence. (B) Another example similar to A. The still frames shown are from Movies S1 and S2.
Examination of the time-lapse movies revealed unambiguous outer membrane fusion events (Supplemental Movies 1 and 2). Figure 5 shows fluorescence traces of mitochondria from a time-lapse movie of OPA1-null cells, along with selected still frames. In the two cases shown, the fluorescence traces show a stepwise drop in the PA-GFP fluorescence of one mitochondrion that is correlated with a stepwise increase in the PA-GFP fluorescence of an adjacent mitochondrion. Examination of the corresponding still frames (Figure 5) indicates that PA-GFP fluorescence has been transferred from the first mitochondrion to the adjacent mitochondrion, which was not photoactivated and therefore initially contained only mito-DsRed fluorescence. Importantly, there was no transfer of mito-DsRed fluorescence, indicating that outer membrane exchange has occurred in the absence of matrix transfer. The stepwise nature of the fluorescence change implies that once outer membrane mixing has initiated, it is completed within the 20-s recording interval.
Ultrastructure of OPA1-Null Cells
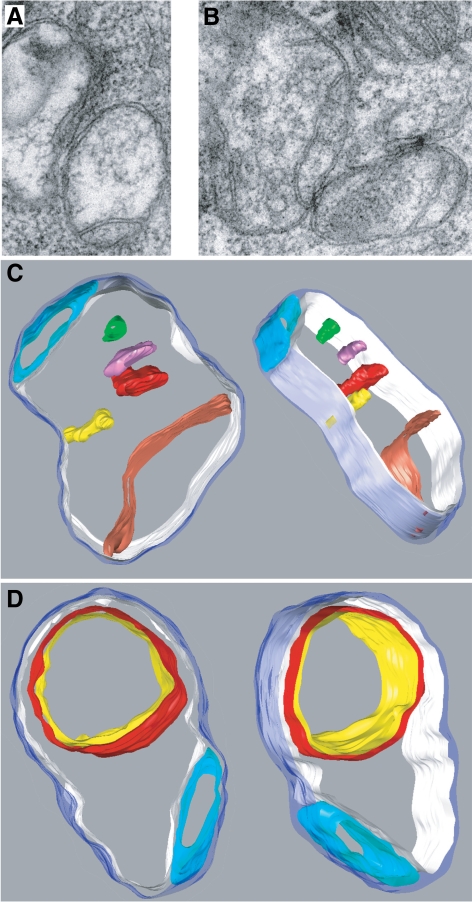
These results indicate that whereas mitofusins are essential for outer membrane fusion and consequently also for inner membrane fusion, OPA1 functions in only inner membrane fusion. In OPA1-null cells, frequent mitochondrial outer membrane events occur but are always uncoupled from inner membrane fusion (Figure 5). Such uncoupled membrane fusion events would be expected to lead to mitochondria containing multiple matrix compartments in OPA1-null cells. To test this prediction, we scored mitochondrial profiles in OPA1-null cells by thin-section electron microscopy. Consistent with previous reports using RNAi knockdown (Olichon et al., 2003; Griparic et al., 2004; Frezza et al., 2006), we found that OPA1-null cells had severe ultrastructural defects in the mitochondria, including swollen mitochondria and defects in cristae organization, such as concentric inner membranes. Such changes have been attributed to the role of OPA1 in cristae maintenance (Olichon et al., 2003; Griparic et al., 2004; Frezza et al., 2006; Meeusen et al., 2006), which appears to be distinct from its role in membrane fusion. In addition, however, we found many mitochondria with multiple matrices. Of 280 mitochondria scored, 83 (30%) had apparently multiple matrix compartments bounded by a single outer membrane (Figure 6,A and B). Such mitochondrial profiles were not observed in wild-type or Mfn-null cells (Figure S5). We calculated three-dimensional electron tomograms from tilt series of a number of mitochondria to confirm the nature of the inner membrane defects, and views of two three-dimensional models derived from segmented tomograms are shown in Figures 6C and 6D. In both, a single outer membrane encloses two separate matrix compartments. Mitochondria with multiple matrix compartments could, in principle, arise from inner membrane fission uncoupled from outer membrane fission or from outer membrane fusion uncoupled from ensuing inner membrane fusion. The fluorescence imaging experiments above indicate that it is attributable to the latter process.
Figure 6.
EM tomography showing multiple matrices in OPA1-null mitochondria. (A and B) Electron micrographs of conventional thin sections showing mitochondria that appear to contain multiple matrix compartments in the mitochondrion at lower right in A and in the mitochondria on the left and lower right in B. (C and D) Views of three-dimensional models calculated from segmented electron tomograms. Both show mitochondria with at least two separate matrix compartments. The outer membrane is rendered in translucent blue. The mitochondrion in (C) contains a large matrix compartment bounded by an inner boundary membrane rendered in white with one large crista rendered in red, a smaller crista in yellow, and three separate membrane compartments in red, magenta, and green that do not connect to the inner boundary membrane within this section; a smaller, separate matrix compartment is rendered in turquoise at the top. The mitochondrion in D contains a smaller matrix compartment rendered in turquoise at the bottom and a larger matrix compartment rendered in white. The latter contains another large compartment bounded by a double membrane rendered in red and yellow.
Similar Defects in Cells Lacking Prohibitins
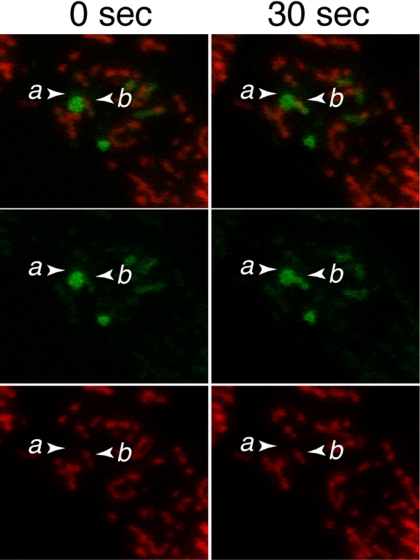
Prohibitin 1 (Phb1) and Prohibitin 2 (Phb2) are related large protein complexes residing on the mitochondrial inner membrane that are essential for proper proteolytic processing of OPA1 (Merkwirth et al., 2008). In the absence of prohibitins, OPA1 is fully processed into short isoforms only, leading to severe defects in mitochondrial function. Our results above would predict that cells lacking prohibtins might also show defects in inner membrane fusion. To test this prediction, we used short hairpin RNAi to knockdown the expression of Phb1. As expected, this knockdown led to reduced levels of Phb1 (Figure S6). The levels of Phb2 are similarly reduced, consistent with previous results that the expression of the two proteins is interdependent (Merkwirth et al., 2008). Knockdown of Phb1 also led to increased processing of OPA1 predominantly into the short isoforms (Figure S6). Using time-lapse imaging, we readily found examples of outer membrane fusion events uncoupled from inner membrane fusion (Figure 7).
Figure 7.
Uncoupled outer membrane fusion in Phb1 knockdown cells. Mito-DsRed and PA-GFP-OMP25 were expressed in wild-type MEFs containing shRNAi for Phb1. In the left panel, mitochondrion a, containing high levels of PA-GFP fluorescence, is adjacent to mitochondrion b, which contains DsRed fluorescence but little PA-GFP fluorescence. In the right panel, 30 s later, PA-GFP fluorescence from a has transferred into b without exchange of DsRed.
DISCUSSION
Similarities and Differences Between Yeast and Mammalian Mitochondrial Fusion
Our results indicate that OPA1 and mitofusins act at distinct steps during mitochondrial fusion. Most importantly, we find that, in vivo, deletion of OPA1 in mammalian cells does not abolish mitochondrial outer membrane fusion. Our work agrees with previous in vitro work in yeast showing that Mgm1 is required for inner membrane fusion (Meeusen et al., 2006). However, it should be noted that yeast lacking Mgm1 show no outer membrane fusion in vivo (Sesaki et al., 2003). This difference suggests that fusion of the outer and inner membranes is not as tightly coupled in mammalian cells as in yeast cells. The reason for this difference remains to be determined. However, it is worth noting that the adaptor protein Ugo1 in yeast physically binds to both Fzo1 and Mgm1 and is thought to coordinate fusion of the outer and inner membranes (Wong et al., 2003; Sesaki and Jensen, 2004; Hoppins et al., 2009). So far, no mammalian ortholog of Ugo1 has been found.
We also find that OPA1, unlike mitofusins (Koshiba et al., 2004), is not required on adjacent mitochondria for fusion to occurs. This striking distinction implies a difference in molecular mechanism for these two types of GTPases during mitochondrial fusion. With in vitro fusion assays with yeast mitochondria, Mgm1p is required on both mitochondria for efficient fusion (Meeusen et al., 2006). It is unclear whether this discrepancy is attributable to the different fusion assays used or whether it reflects another difference between mammalian and yeast mitochondrial fusion.
Implications for Neurodegenerative Disease
Although the neurodegenerative diseases CMT2A and DOA are both thought to arise from reduced mitochondrial fusion, our results suggest that they impact distinct steps in the mitochondrial fusion pathway. This insight may help to understand the pathogenic basis for these diseases. CMT2A, caused by mutations in Mfn2, is a disease characterized by degeneration of the longest peripheral nerves (Zuchner et al., 2004). In contrast, DOA is a disease of retinal ganglion cell degeneration that leads to atrophy of the optic nerve (Alexander et al., 2000; Delettre et al., 2000). In both diseases, a subset of patients show broader and overlapping tissue involvement, indicating that mitochondrial fusion is important to a wider range of cell types (Zuchner and Vance, 2006; Hudson et al., 2008; Zanna et al., 2008; Zeviani, 2008). The basis for the tissue-specific differences between these two diseases is unknown. It will be worth exploring whether they arise, in part, because of the distinct roles of Mfn2 and OPA1 in mitochondrial fusion.
Mutations in OPA1 cause autosomal dominant DOA, and some cases are caused by haploinsufficiency of OPA1 (Delettre et al., 2002). In addition, mice with a 50% reduction of OPA1 protein have an age-dependent degeneration of retinal ganglion cells (Alavi et al., 2007; Davies et al., 2007). In contrast, our OPA1-null cells lack any OPA1 protein and would be expected to have a much more severe loss of mitochondrial fusion compared with cells from patients. Nevertheless, the preservation of outer membrane fusion even in OPA1-null cells implies that DOA patients likely have intact outer membrane fusion.
Every mitochondrial fusion event mediates a dramatic remodeling between two mitochondria. Fusion of the four lipid bilayers results in not only mixing of membrane lipids and proteins but also the merging of mitochondrial matrices. At a cellular level, loss of mitofusins or OPA1 results in severe consequences for mitochondrial function, including heterogeneity in membrane potential, heterogeneity in mtDNA nucleoids, and greatly reduced respiratory capacity (Chen et al., 2005; Chen et al., 2007). It is possible that the role of OPA1 in maintenance of cristae structure (Olichon et al., 2003; Griparic et al., 2004; Frezza et al., 2006; Meeusen et al., 2006) may contribute in part to these functional defects, although it should be noted that yeast Mgm1p is not necessary for cristae structure when mitochondrial fission is blocked (Sesaki et al., 2003). Instead, we favor the model that the defect in mitochondrial inner membrane fusion underlies the cellular defects in OPA1-null cells. These observations suggest that inner membrane fusion, perhaps because of its effect in merging mitochondrial matrices, has an important physiological function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Steven Barlow for assistance with electron microscopy. We are grateful to Dr. Hsiuchen Chen (California Institute of Technology) for early work on this study and to Drs. Chen and Christiane Alexander (Max Delbrück Center) for use of the OPA1-null cells. This work was supported by NIH grant GM062967 to D.C.C., and a Blasker Science and Technology Grant from the San Diego Foundation to T.G.F. Z.S. was supported by an Elizabeth Ross Fellowship.
Abbreviations used:
- CMT
Charcot-Marie-Tooth
- DOA
dominant optic atropy
- Mfn
mitofusins.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-03-0252) on May 28, 2009.
REFERENCES
- Alavi M. V., et al. A splice site mutation in the murine Opa1 gene features pathology of autosomal dominant optic atrophy. Brain. 2007;130:1029–1042. doi: 10.1093/brain/awm005. [DOI] [PubMed] [Google Scholar]
- Alexander C., et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Chan D. C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- Chen H., Chomyn A., Chan D. C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E., Chan D. C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., McCaffery J. M., Chan D. C. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Cipolat S., Martins de Brito O., Dal Zilio B., Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies V. J., Hollins A. J., Piechota M. J., Yip W., Davies J. R., White K. E., Nicols P. P., Boulton M. E., Votruba M. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum. Mol. Genet. 2007;16:1307–1318. doi: 10.1093/hmg/ddm079. [DOI] [PubMed] [Google Scholar]
- Delettre C., et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Delettre C., Lenaers G., Pelloquin L., Belenguer P., Hamel C. P. OPA1 (Kjer type) dominant optic atrophy: a novel mitochondrial disease. Mol. Genet. Metab. 2002;75:97–107. doi: 10.1006/mgme.2001.3278. [DOI] [PubMed] [Google Scholar]
- Detmer S. A., Chan D. C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Frezza C., et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Griparic L., van der Wel N. N., Orozco I. J., Peters P. J., van der Bliek A. M. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J. Biol. Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- Haigh S. E., Twig G., Molina A. A., Wikstrom J. D., Deutsch M., Shirihai O. S. PA-GFP: a window into the subcellular adventures of the individual mitochondrion. Novartis Found Symp. 2007;287:21–36. doi: 10.1002/9780470725207.ch3. discussion 36–46. [DOI] [PubMed] [Google Scholar]
- Hoppins S., Horner J., Song C., McCaffery J. M., Nunnari J. Mitochondrial outer and inner membrane fusion requires a modified carrier protein. J. Cell Biol. 2009;184:569–581. doi: 10.1083/jcb.200809099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Hudson G., et al. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain. 2008;131:329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- Karbowski M., Arnoult D., Chen H., Chan D. C., Smith C. L., Youle R. J. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J. Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T., Detmer S. A., Kaiser J. T., Chen H., McCaffery J. M., Chan D. C. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Malka F., Guillery O., Cifuentes-Diaz C., Guillou E., Belenguer P., Lombes A., Rojo M. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep. 2005;6:853–859. doi: 10.1038/sj.embor.7400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J. M., Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Lower B., Wunderlich F. T., von Kleist-Retzow J. C., Waisman A., Westermann B., Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22:476–488. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y., De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Patterson G. H., Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Perkins E. M., McCaffery J. M. Conventional and immunoelectron microscopy of mitochondria. Methods Mol. Biol. 2007;372:467–483. doi: 10.1007/978-1-59745-365-3_33. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J. Biol. Chem. 2004;279:28298–28303. doi: 10.1074/jbc.M401363200. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S. M., Yaffe M. P., Jensen R. E. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell. 2003;14:2342–2356. doi: 10.1091/mbc.E02-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Chen H., Fiket M., Alexander C., Chan D. C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen D. F., Norris K. L., Youle R. J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M. G., Williams J., Munoz-Pinedo C., Perkins G. A., Brown J. M., Ellisman M. H., Green D. R., Frey T. G. Correlated three-dimensional light and electron microscopy reveals transformation of mitochondria during apoptosis. Nat. Cell Biol. 2007;9:1057–1065. doi: 10.1038/ncb1630. [DOI] [PubMed] [Google Scholar]
- Tondera D., et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. D., Wagner J. A., Scott S. V., Okreglak V., Holewinske T. J., Cassidy-Stone A., Nunnari J. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 2003;160:303–311. doi: 10.1083/jcb.200209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanna C., et al. OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain. 2008;131:352–367. doi: 10.1093/brain/awm335. [DOI] [PubMed] [Google Scholar]
- Zeviani M. OPA1 mutations and mitochondrial DNA damage: keeping the magic circle in shape. Brain. 2008;131:314–317. doi: 10.1093/brain/awm339. [DOI] [PubMed] [Google Scholar]
- Zuchner S., et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- Zuchner S., Vance J. M. Molecular genetics of autosomal-dominant axonal Charcot-Marie-Tooth disease. Neuromolecular Med. 2006;8:63–74. doi: 10.1385/nmm:8:1-2:63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.