Abstract
During tumor development, cells acquire multiple phenotypic changes upon misregulation of oncoproteins and tumor suppressor proteins. Hakai was originally identified as an E3 ubiquitin-ligase for the E-cadherin complex that regulates cell–cell contacts. Here, we present evidence that Hakai plays a crucial role in various cellular processes and tumorigenesis. Overexpression of Hakai affects not only cell–cell contacts but also proliferation in both epithelial and fibroblast cells. Furthermore, the knockdown of Hakai significantly suppresses proliferation of transformed epithelial cells. Expression of Hakai is correlated to the proliferation rate in human tissues and is highly up-regulated in human colon and gastric adenocarcinomas. Moreover, we identify PTB-associated splicing factor (PSF), an RNA-binding protein, as a novel Hakai-interacting protein. By using cDNA arrays, we have determined various specific PSF-associated mRNAs encoding proteins that are involved in several cancer-related processes. Hakai affects the ability of PSF to bind these mRNAs, and expression of PSF short hairpin RNA or a dominant-negative PSF mutant significantly suppresses proliferation of Hakai-overexpressing cells. Collectively, these results suggest that Hakai is an important regulator of cell proliferation and that Hakai may be an oncoprotein and a potential molecular target for cancer treatment.
INTRODUCTION
Carcinoma results from a series of transformations in epithelial cells (Hanahan and Weinberg, 2000). During the transformations, cells acquire several characteristic phenotypes, including increased cell proliferation, enhanced cell motility and invasion, and altered cell–cell and cell–substratum adhesions. Disruption of cell–cell contacts is associated with the transition from adenoma to carcinoma, and loss of E-cadherin, a crucial membrane protein for the formation of adherens junctions (Perez-Moreno et al., 2003; Gumbiner, 2005), has been shown to be involved in this process (Birchmeier and Behrens, 1994; Cavallaro and Christofori, 2004).
Hakai was originally identified as an E3 ubiquitin-ligase for the E-cadherin complex (Fujita et al., 2002). Hakai contains Src homology (SH)2, RING-finger, and proline-rich domains, and it is structurally and functionally related to c-Cbl, a RING-finger type E3 ubiquitin-ligase for receptor tyrosine kinases (Joazeiro et al., 1999; Levkowitz et al., 1999). Tyrosine phosphorylation of the E-cadherin complex by receptor or nonreceptor tyrosine kinases promotes the binding to the SH2 domain of Hakai. Subsequently, Hakai acts as an E3-ligase for the E-cadherin complex and mediates its ubiquitination, endocytosis, and degradation in the lysosomes (Fujita et al., 2002; Palacios et al., 2005; Shen et al., 2007; Bonazzi et al., 2008), causing the perturbation of cell–cell adhesions. At present, this is the only reported function of Hakai.
However, there are several lines of evidence that suggest additional roles for Hakai. First, Northern blot analysis revealed that Hakai is ubiquitously expressed in tissues (Fujita et al., 2002). The Hakai mRNA was detected in spleen and skeletal muscle where E-cadherin is absent, suggesting that Hakai may have substrates other than E-cadherin. Second, many E3-ligases have multiple substrates for ubiquitination (Maniatis, 1999; Shenoy et al., 2001; Mantovani and Banks, 2003; Li et al., 2004). By ubiquitinating multiple proteins, these E3-ligases may coordinate and interconnect apparently distinct cellular processes. Likewise it is plausible that Hakai also has multiple substrates. Finally, as shown below, Hakai localizes in the nucleus as well as in the cytoplasm, indicating possible functional roles at multiple cellular locations.
In the present study, we show that Hakai is involved in the regulation of cell proliferation in addition to cell–cell contacts. Hakai can regulate cell proliferation in an E-cadherin down-regulation–independent manner. We also demonstrate that Hakai interacts with PTB-associated splicing factor (PSF), a multifunctional RNA-binding protein. Hakai regulates the ability of PSF to bind transcripts, and the expression of PSF affects proliferation of Hakai-overexpressing cells. Furthermore, we demonstrate that expression of Hakai is up-regulated in colon and gastric cancer and present data suggesting that Hakai is a potential drug target in cancer treatment.
MATERIALS AND METHODS
Antibodies and Materials
The rabbit polyclonal anti-Hakai antibody (Hakai-2498) was generated by using peptides (GPHHPDQTRYRPYYQ and DHTDNELQGTNSSG) as antigen (Eurogentec, Seraing, Belgium). Antibody to the cytoplasmic portion of E-cadherin (BD Biosciences Transduction Labs, Lexington, KY) was used for Western blotting. Antibodies to the extracellular portion of E-cadherin, HECD-1 and ECCD-2 (Zymed Laboratories, South San Francisco, CA), were used for immunofluorescence of human and canine cells, respectively. Anti-β-catenin antibody was from BD Biosceiences Pharmingen. Anti-zona occludens (ZO)-1 antibody was from Zymed Laboratories. Anti-hemagglutinin (HA) was from Roche Diagnostics (Mannheim, Germany). Anti-FLAG (M2) and anti-PSF antibodies were from Sigma-Aldrich (St. Louis, MO). Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was from Chemicon International (Hampshire, United Kingdom). Anti-α-tubulin antibody was from Immunologicals Direct (Oxford Biotechnology, United Kingdom). Anti-5-bromo-2′-deoxyuridine (BrdU) antibody was from Calbiochem (Darmstadt, Germany). Anti-Ki67 antibody (MM1) was from Novocastra Laboratories (Newcastle upon Tyne, United Kingdom). Anti-cdk2, anti-cdk4, anti-cyclin D1, anti-cyclin E, anti-p21, and anti-HuR (3A2) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-cdk6 antibody was from Neomarkers (Fremont, CA). Anti-ubiquitin antibody was from BAbCO (Richmond, CA). All antibodies were used at dilutions of 1:1000 for Western blotting and 1:100 for immunofluorescence, except for the following antibodies: anti-cdk2 and anti-cdk4 (1:2000), anti-cyclin D1 (1:5000), anti-cyclin E (1:200), and anti-p21 (1:200) antibodies for Western blotting. pcDNA-HA-Hakai and pGEX-Hakai (N terminus: amino acids 20-216) were kindly provided by Walter Birchmeier (Max-Delbrück-Center, Berlin, Germany) (Fujita et al., 2002). pEGFP-PSF (full length), -PSF-N (amino acids 1-340), -PSF-C (amino acids 338-707), and -PSF-ΔRRM2 (RNA recognition motif) were kindly provided by James G. Patton (Vanderbilt University, Nashville, TN) (Dye and Patton, 2001). To construct the Hakai mutant (Hakai-ΔRING: amino acids 1-108 and 147-491) lacking the RING-finger domain, cDNAs of the Hakai N- and C-terminal domains were amplified by polymerase chain reaction (PCR) using the following primers: for the N terminus, 5′-GGA ATT CGG CCG GAT CAC ACT GAC AAT GAG TTA CAA GG-3′ and 5′-GGA ATT CGC GGC CGC GAA ATG TAC TGG TGT ATC GTC CTT TTC-3′; for the C terminus, 5′-GGA ATT CGG CCG GAT GTA GTG ATC CTG TGC AGC GGA TTG-3′ and 5′-GGA ATT CGC GGC CGC TCA CTG GTA ATA CGG TCT GTA TCT TGT TTG ATC-3′. The cDNA of the Hakai N terminus was cleaved by EagI and cloned into a NotI site of pcDNA-HA. Then, the cDNA of the Hakai C terminus was cleaved by EagI and cloned into a NotI site of pcDNA-HA-Hakai-N-terminus. The hepatocyte growth factor (HGF)/scatter factor was purchased from Chemicon International.
Cell Culture, Western Blotting, Immunoprecipitation, Immunofluorescence, Time-Lapse Analysis, and Affinity Protein Purification
Human embryonic kidney (HEK)293, Madin-Darby canine kidney (MDCK), MCF-7, MDA-MB231, NIH3T3, and A431 cells were cultured in DMEM containing penicillin/streptomycin and 10% fetal calf serum (FCS) at 37°C and ambient air supplemented with 5% CO2. MCF-10A cells were cultured in DMEM/F-12 containing penicillin/streptomycin, 5% horse serum, 20 ng/ml epidermal growth factor (Millipore, Billerica, MA), 0.5 μg/ml hydrocortisone (Calbiochem), 100 ng/ml cholera toxin (Sigma-Aldrich), and 10 μg/ml Insulin (Invitrogen, Paisley, United Kingdom) at 37°C and ambient air supplemented with 5% CO2. To establish cell lines stably expressing Hakai, MDCK cells were transfected with pcDNA-HA-Hakai using Lipofectamine Plus reagent (Invitrogen) and were selected in a medium containing 800 μg ml−1 G418 (Calbiochem). More than 10 stable clones were obtained from two independent transfections. All clones represented comparable phenotypes and results using clone 4 were presented if not indicated. Western blotting, immunoprecipitation, and immunofluorescence were performed as described previously (Hogan et al., 2004). For Western blotting, cell lysates (30 μg of proteins) were loaded in SDS-polyacrylamide gel electrophoresis (PAGE) if not indicated. For immunoprecipitation, we used 1% Triton X-100 lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, and 1% Triton X-100) containing 5 μg ml−1 leupeptin, 50 mM phenylmethylsulfonyl fluoride, and 7.2 trypsin inhibitor units of aprotinin. For Supplemental Figure S8, 10 mM N-ethylmaleimide (Sigma-Aldrich) was added in lysis buffer. Immunofluorescence images were analyzed by epifluorescence microscopy, if not indicated. For Supplemental Figure S1, human fibronectin-coated coverslips (BD Biosciences, Erembodegem, Belgium) were used for immunofluorescence. To visualize nuclei, we used Hoechst 33342 (Invitrogen). Cell death was measured by using LIVE/DEAD Viability/Cytotoxicity kit (Invitrogen) according to manufacturer's instructions. To obtain epifluorescence images, we used an Axioskop 1 (Carl Zeiss, Jena, Germany) with a CoolSNAP camera (Roper Scientific, Trenton, NJ). To obtain phase contrast images, we used a DMIRB microscope (Leica, Wetzlar, Germany) with a C4742-95 Orca camera (Hamamatsu, Bridgewater, NJ). For time-lapse experiments, cells were cultured in Leibovitz's medium containing 10% FCS during 24 h at 37°C. To obtain time-lapse images we used an Axiovert 100M microscope (Carl Zeiss) with a Biopoint Controller (Ludl Electronic Products, Hawthrorne, NY) and a C4742–95 Orca camera (Hamamatsu). Time-lapse images were captured and analyzed using Openlab software (Improvision, Coventry, United Kingdom). For Figures 1B and 7C, confocal images were captured using a TCS SPE confocal microscope and Leica Application Suite (Lecia) and were analyzed using ImageJ 1.36b (National Institutes of Health, Bethesda, MD). For affinity protein purification, HEK293 cells from four 15-cm plates (Figure 7A) or transfected HEK293 cells in a 9-cm dish (Supplemental Figure S6) were lysed in Triton X-100 lysis buffer, and the cell lysates were subjected to glutathione beads coupled to 20 μg of glutathione transferase (GST) or GST-Hakai (N terminus: amino acids 20-216), followed by SDS-PAGE, and Coomassie protein staining or Western blotting. Hakai-binding proteins were excised from the gel. The amino acid sequences were determined by liquid chromatography-tandem mass spectrometry.
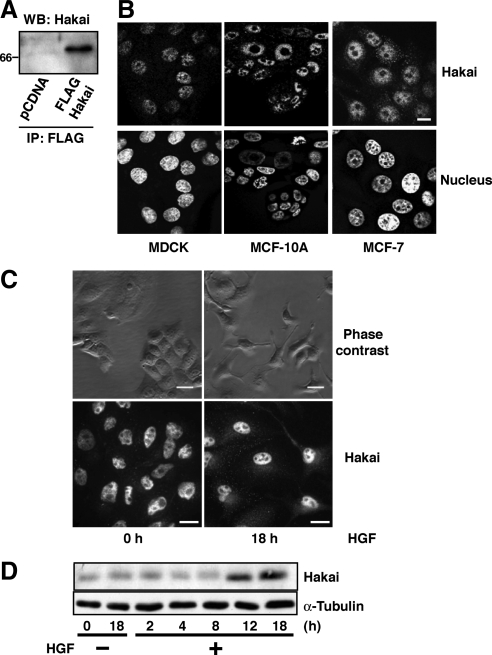
Figure 1.
Subcellular localization of Hakai in different cell lines. (A) Detection of exogenous Hakai protein by anti-Hakai antibody. Cell lysates of HEK293 cells expressing FLAG-tagged Hakai were immunoprecipitated with anti-FLAG antibody, followed by Western blotting with anti-Hakai antibody. (B) Immunofluorescence analyses of MDCK, MCF-10A, and MCF-7 epithelial cells using anti-Hakai antibody. Nuclei were stained with Hoechst dye. Images were analyzed by confocal microscopy. (C and D) Effect of HGF on the localization and expression of Hakai in MDCK cells. Cells were incubated in the presence or absence of HGF (40 ng/ml) for the indicated times. (C) Cells were examined by phase contrast and immunofluorescence with anti-Hakai antibody. (D) Cell lysates were examined by Western blotting with anti-Hakai and anti-α-tubulin antibodies. Bars (B and C), 10 μm.
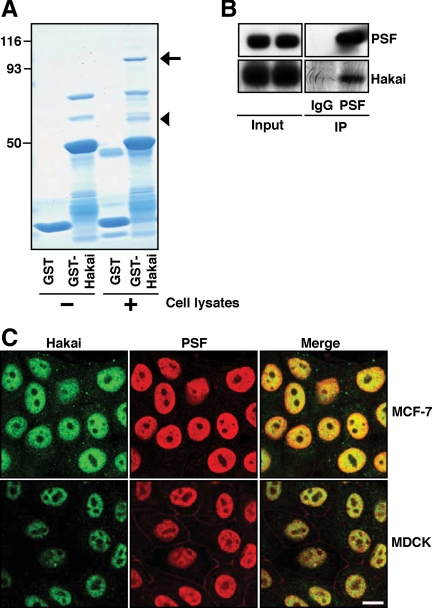
Figure 7.
Interaction between Hakai and PSF. (A) Identification of PSF as a novel Hakai-binding protein by affinity purification using HEK293 cells lysates. Proteins bound to GST or GST-Hakai (N terminus: amino acids 20-216) were examined by Coomassie protein staining. Arrow and arrowhead indicate the position of PSF and p54nrb, respectively. (B) Interaction between endogenous Hakai and PSF proteins. HEK293 cell lysates were immunoprecipitated with control IgG or anti-PSF antibody, followed by Western blotting with anti-PSF and anti-Hakai antibodies. (C) Colocalization of Hakai and PSF in the nucleus. The subcellular localization of Hakai and PSF was examined in MCF-7 and MDCK cells using anti-Hakai and anti-PSF antibodies. Bar, 10 μm.
Proliferation Assay and Flow Cytometry Analysis
For quantification of the cell number, 2.5 × 104 cells were plated in a six-well plate, and the cell number was counted during 5 d using hemocytometer. For colorimetric analysis, 1 × 104 cells were plated in a 96-well plate, and after 24 h they were treated with 10 μM BrdU for 3 h. BrdU incorporation into newly synthesized DNA was measured using a cell proliferation colorimetric immunoassay kit (Calbiochem) according to manufacturer's instructions. For the immunofluorescence assay, 5 × 105 cells were plated in a six-well plate. After 24 h, cells were transfected if indicated, and they were treated with 10 μM BrdU for 5 h for MDCK cells or 3 h for other cell lines. They were fixed with 3.7% paraformaldehyde/phosphate-buffered saline (PBS) for 15 min and incubated with 2 N HCl and 0.5% Triton X-100 for 30 min before the primary antibody incubation. For the flow cytometry analysis, cells were trypsinized, washed with PBS containing 1 mM EDTA, and fixed with chilled 70% ethanol for 30 min at 4°C, followed by incubation in PBS containing 100 μg/ml RNase and 50 μg/ml propidium iodide (Sigma-Aldrich). Stained DNA was analyzed by FACSscan flow cytometer (Clontech, Mountain View, CA). Data were acquired and analyzed by Cell Quest Program (BD Biosciences, Cowley, United Kingdom).
RNA Interference
Oligonucleotides (oligos) used for Hakai siRNA were Hakai-1 (CTCGATCGGTCAGTCAGGAAA) and Hakai-2 (CACCGCGAACTCAAAGAACTA). The small interfering RNA (siRNA) oligos were transfected into MCF-7, MDA-MB231, or HEK293 cells using HiPerFect reagent (QIAGEN, Dorking, Surrey, United Kingdom) according to manufacturer's instructions. Cells (5 × 104) were plated in a 24-well plate, and 1.5 μg of siRNA was transfected with 9 μl of HiPerFect reagent per well. As a negative control, we used nonsilencing control siRNA (AF 488) from QIAGEN. At 96 h after transfection, cells were lysed in Triton X-100 lysis buffer and examined by Western blotting. Hakai-overexpressing MDCK cells stably expressing PSF short hairpin RNA (shRNA) were produced as follows: PSF shRNA oligonucleotides (5′-CACCGCAAAGGATTCGGGCTTATTACGAATAATAAGCCCGAATCCTTTGC-3′ and 5′-AAAAGCAAAGGATTCGGGCTTATTATTCGTAATAAGCCCGAATCCTTTGC-3′) were cloned into pENTR/H1/TO using BLOCK-iT Inducible H1 RNAi Entry Vector kit (Invitrogen) according to manufacturer's instructions. MDCK HA-Hakai cells (clone 4) were transfected with pENTR/H1/TO PSF shRNA using Lipofectamine 2000 (Invitrogen), followed by selection in medium containing 400 μg ml−1 of Zeocin (Invitrogen) and 800 μg ml−1 G418. To ensure equal loading, protein concentration of lysates was quantified using the DC Protein Assay reagent (Bio-Rad Laboratories, Hercules, CA) and measured by a VERSAmax microplate reader (Molecular Devices, Sunnyvale, CA).
Immunohistochemistry
Tissue samples for immunohistochemistry were fixed overnight in neutral-buffered Formalin and embedded in paraffin wax. Immunohistochemistry was performed on paraffin wax sections as described previously (Toda et al., 1999). Antigen retrieval was performed by microwave method for anti-Hakai and anti-E-cadherin antibodies or autoclave method for anti-Ki67 antibody. For the former, the slides were microwaved in 10 mM citrate buffer, pH 6.0, for 10 min in a pressure proof cooker (Rakuchin Gozen, Daiya Industries, Tokyo, Japan), and then placed on a metal rack and boiled for 20 min, followed by placing into distilled water. For the latter, a Coplin jar containing glass slides in citrate buffer was covered with a loose fitting cap and heated in a prestige stainless steel pressure cooker for 5 min at 121°C. The pressure cooker was then removed from heat source, and the glass slides were rinsed in distilled water. Endogenous peroxidase activity was then blocked by 0.3% H2O2 in methyl alcohol for 30 min, followed by washing with PBS. Nonspecific binding was blocked by incubating the sections in 1% goat serum (Sigma-Aldrich) for 30 min. Polyclonal primary antibodies were diluted at 1:2000 in PBS/1% bovine serum albumin (BSA) and incubated overnight at 4°C, followed by incubation for 30 min with biotinylated secondary antibody (Vector Laboratories, Burlingame, CA) diluted at 1:300 in PBS. After five washes with PBS, the avidin-biotin-peroxidase complex (ABC-Elite; Vector Laboratories) was applied for 50 min at a 1:100 dilution in PBS/1% BSA. After washing with PBS, coloring reaction was detected with 0.3 mg/ml diaminobenzidine and 0.003% H2O2 in 50 mM Tris-HCl, pH 7.6. Hematoxylin was used as a nuclear counterstain. Informed consent was obtained from each patient.
Soft Agar Anchorage-Independent Growth Assay
To examine anchorage-independent cell growth, we used a colony formation assay in soft agar. First, 3 ml of DMEM/10% FCS containing 0.5% low-melting agarose was placed in six-well dishes as a bottom support layer. Then, 1 × 103 parental or Hakai-overexpressing MDCK cells were suspended in DMEM/10%FCS containing 0.3% low-melting agarose (Lonza Rockland, Rockland, ME) and overlaid as a top layer. After 12 d of incubation at 37°C, colonies were stained with 100 μl of 10 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (Sigma-Aldrich) for 1 h at 37°C.
Analyses of PSF–mRNA Complexes
Immunoprecipitation of PSF-mRNA complexes was performed as described previously (Mazan-Mamczarz et al., 2006), except that cell lysates were precleared for 30 min at 4°C using 30 μg of mouse immunoglobulin G (IgG) (Sigma-Aldrich) coupled to 50 μl of protein A-Sepharose 4B beads (GE Healthcare, Chalfont St. Giles, United Kingdom), which had been swollen in NT2 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, and 0.05% Nonidet P-40) supplemented with 5% bovine serum albumin. From the immunoprecipitates, RNAs were extracted and used either for hybridization of cDNA arrays or for verification of individual PSF target transcripts. The array analysis was performed as reported previously (Mazan-Mamczarz et al., 2006). In brief, the RNAs extracted from immunoprecipitates with anti-PSF antibody or control IgG were reverse transcribed in the presence of [α-33P]dCTP, and the radiolabeled product was used to hybridize cDNA arrays (Mammalian Gene Collection [MGC] arrays, containing ∼6000 individual human genes) as described previously (Tenenbaum et al., 2002). All of the data were analyzed using the Array-Pro software (Media Cybernetics, Carlsbad, CA), and then normalized by Z score transformation and used to calculate differences in signal intensities. Significant values were tested using a two-tailed Z-test and p values <0.01 (n = 3). It should be noted that human cDNA arrays were hybridized with cDNAs from canine MDCK cells; therefore, it is plausible that some of transcripts might have been missed under this experimental condition. For the analysis of individual transcripts, the RNAs from the immunoprecipitation (IP) material were used in reverse transcription. Reverse transcription (RT)-PCR was carried out using SuperScript II reverse transcriptase with random hexamers (Invitrogen). To determine the mRNA level coding β-actin, we used human β-actin primers (Invitrogen). Specific primers were designed to correspond to the target mRNA sequences of both dog and human: for PAI-RBP1 (5′-CCTGGGCACTTACAGGAAGG-3′ and 5′-CACCCTTTTCTTCAAGTGGC-3′); for NF2 (5′-GATGCGGTCTGAGGAGACAGC-3′ and 5′-CTGACTCCTCAGCCATCTTCAG-3′); for LKB1 (5′-GGACACGTTCATCCACCGCATC-3′ and 5′-CTTGAGCTTCTTCTTCTTGAG-3′); for α-catenin (5′-GCCATGTCCCTGATCCAGGC-3′ and 5′-GCACCGGGTTCACATGCTTCTTC-3′); and for tubulin α-6 (5′-CAAGTGACAAGACCATTGG-3′ and 5′-CTACCATGAAGGCACAATCAG-3′). PCR products were visualized after electrophoresis in 2% agarose gels stained with ethidium bromide to verify that a single band was amplified in each reaction. HuR–mRNA complexes were analyzed in the same way as described above, except that anti-HuR antibody was used for IP.
Statistical Analysis
Student's t test assuming unequal variance was performed for statistical analysis.
RESULTS
Subcellular Localization of Hakai in Various Cell Lines
In the previous study, we showed the role of Hakai as an E3 ubiquitin-ligase for the E-cadherin complex (Fujita et al., 2002). However, the subcellular localization of Hakai still remains unknown. To determine the localization of endogenous Hakai protein, we newly generated polyclonal antibody that specifically recognizes Hakai. Expression of endogenous Hakai protein was detected by Western blotting with this antibody in different cell lines such as MDCK, MCF-7, and HEK293 cells (see Figure 1D and Supplemental Figure 3A). The antibody also recognized exogenously expressed FLAG-tagged Hakai in HEK293 cells (Figure 1A). Using this antibody, we analyzed the localization of Hakai in various epithelial cell lines by immunofluorescence. In nontransformed epithelial cell lines, MDCK and MCF-10A cells, we observed enriched nuclear localization of Hakai (Figure 1B). In transformed epithelial cell lines, MCF-7 and A431 cells, punctuate cytoplasmic localization of Hakai was observed together with the nuclear distribution (Figure 1B; data not shown).
Next, we examined whether the localization of Hakai was affected by addition of growth factor. We analyzed the effect of HGF, which enhances proliferation, dissociation of cell–cell contacts and cell motility in epithelial cells (Birchmeier et al., 2003). After 18 h of HGF treatment, MDCK cells lost epithelial morphology with disrupted intercellular adhesions and became more fibroblastic (Figure 1C, top). Interestingly, during this process, the increased cytoplasmic localization of Hakai was observed (Figure 1C, bottom). We also found that the expression level of Hakai protein was enhanced by HGF treatment (Figure 1D). These data suggest that the localization of Hakai is dynamically regulated under different conditions.
Overexpression of Hakai Induces Attenuation of Cell–Cell Adhesions and Enhances Cell Proliferation
To further understand the functional role of Hakai, we newly established MDCK cell lines stably overexpressing Hakai. In these cells, the expression of exogenous Hakai was four- to sixfold higher than that of endogenous Hakai (Supplemental Figure S1C), and twofold higher than that of the previously established clones (Fujita et al., 2002). Nontransfected parental MDCK cells showed a typical epithelial morphology with tight cell–cell contacts (Supplemental Figure S1A, left). In contrast, all of the Hakai-overexpressing MDCK cell lines represented fibroblastic morphology with decreased cell–cell contacts and increased protrusion formation (Supplemental Figure S1A, right). Time-lapse microscopic analyses revealed that, compared with parental cells (Supplemental Movie 1), Hakai-overexpressing cells frequently produced spiky protrusions that were dynamically extended and retracted (Supplemental Movie 2). Exogenously expressed Hakai was localized in the nucleus as well as in the cytoplasm (Supplemental Figure S1B). In the cytoplasm, Hakai showed a diffuse and some punctuate localization and was also enriched at the tip of protrusions (Supplemental Figure S1B, arrows), suggesting a role in the protrusion dynamics.
Using these stable cell lines, we examined the effect of overexpression of Hakai on cell–cell adhesions. In Hakai-overexpressing cells, the expression of E-cadherin was substantially reduced (Supplemental Figure S1C) and E-cadherin disappeared from cell–cell contacts (Supplemental Figure S1E). This effect on E-cadherin was greater than that observed in the previous report (Fujita et al., 2002), probably because of the higher expression level of Hakai in the newly established clones. The expression of β-catenin (another adherence junction protein) and ZO-1 (tight junction protein) was not altered in Hakai-overexpressing cells (Supplemental Figure S1D), and they remained at cell–cell contact sites (Supplemental Figure S1E). This is consistent with a previous report showing that reducing E-cadherin expression alone has little effect on the localization of β-catenin and ZO-1 in MDCK cells (Capaldo and Macara, 2007). Our data indicate the specific effect of Hakai on E-cadherin at cell–cell contacts.
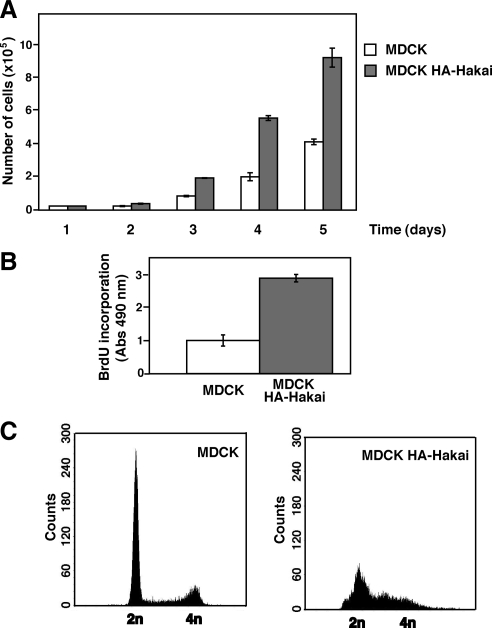
While culturing Hakai-overexpressing MDCK cells, we also found that they underwent cell division much faster than parental MDCK cells. We quantified this observation by counting the number of cells during 5 d of incubation. As shown in Figure 2A, we found the increased cell number in Hakai-overexpressing cells. BrdU is a synthetic nucleoside that is an analogue of thymidine. It is incorporated into newly synthesized DNA during S phase of the cell cycle and substitutes for thymidine during DNA replication; thus, it is commonly used to detect proliferating cells in living tissues. Using this method, we observed a significant increase in the proliferation rate of Hakai-overexpressing MDCK cells compared with parental MDCK cells (Figure 2B). As shown in Supplemental Figure S2, cell death was not decreased in Hakai-overexpressing cells, indicating that the increased cell number in Hakai-overexpressing cells is not due to decreased cell death. By using flow cytometry, we also examined the effect of overexpression of Hakai on cell cycle. The percentage of sub-G0/G1 cell population in parental and Hakai-overexpressing MDCK cells is 70 and 44%, respectively (Figure 2C), indicating that overexpression of Hakai promotes the transition from G0/G1 to S phase. Indeed, in Hakai-overexpressing MDCK cells, the protein expression level of cell cycle regulators controlling transition from G0/G1 to S phase was substantially affected (Supplemental Figure S3A). Overexpression of Hakai also significantly enhanced cell proliferation in NIH3T3 fibroblast cells that do not express E-cadherin (Supplemental Figure S3, B and D), suggesting that the effect of Hakai occurs in an E-cadherin-independent manner. Collectively, these data indicate that overexpression of Hakai induces a higher proliferation rate.
Figure 2.
Effect of overexpression of Hakai on cell proliferation in MDCK cells. (A and B) Increased proliferation rate in Hakai-overexpressing MDCK cells. (A) The cell number of parental and Hakai-overexpressing MDCK cells was monitored during 5 d. (B) Parental and Hakai-overexpressing MDCK cells were labeled with BrdU, followed by colorimetric assay. Statistical analyses from nine independent experiments indicate the significantly increased BrdU incorporation in Hakai-overexpressing MDCK cells (p < 0.005). (C) Effect of overexpression of Hakai on cell-cycle regulation. The DNA content of parental and Hakai-overexpressing MDCK cells was analyzed by flow cytometry. (A and B) Data are represented as mean ± SD.
Next, we examined whether E3 ubiquitin-ligase activity of Hakai is involved in regulation of proliferation. It has been reported that the RING-finger domain is required for the E3 ligase activity of Hakai (Fujita et al., 2002). Thus, we analyzed the effect of expression of the Hakai mutant (Hakai-ΔRING) lacking the RING-finger domain (Supplemental Figure S4, A–C) on cell proliferation in MDCK cells. We found that expression of Hakai-ΔRING strongly suppressed cell proliferation, whereas expression of wild-type Hakai enhanced it (Supplemental Figure S4D). This result suggests that E3 ubiquitin-ligase activity of Hakai, at least partially, plays a role in regulation of cell proliferation.
Knockdown of Hakai Decreases the Proliferative Rate of Transformed Epithelial Cells
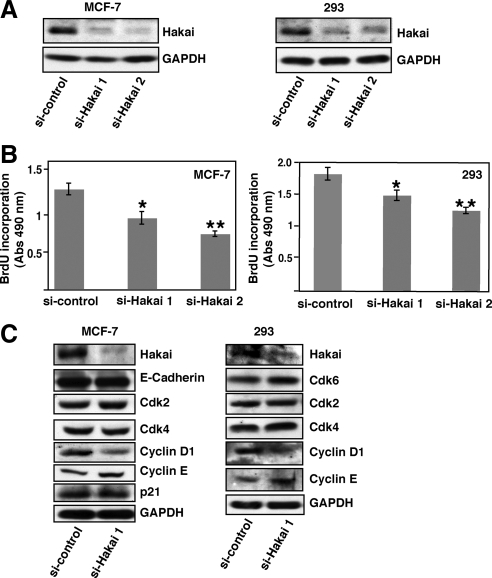
Next, we examined whether the knockdown of Hakai in transformed epithelial MCF-7 and HEK293 cells affects their proliferation rate. The expression of Hakai was strongly reduced by transiently transfecting either of two different Hakai-specific siRNA oligos (Figure 3A and Supplemental Figure S5). Under these experimental conditions, MCF-7 and HEK293 cells transfected with Hakai siRNA had a significantly decreased BrdU incorporation as determined by colorimetric assay (Figure 3B). We observed 35–45% reduction of the BrdU incorporation in MCF-7 cells transfected with Hakai siRNA, compared with those with control siRNA (Figure 3B). Furthermore, the effect of Hakai knockdown on the expression of cell cycle regulators was examined. We found that the protein expression level of cyclin D1 was substantially decreased in Hakai-knockdown MCF-7 and HEK293 cells (Figure 3C). The expression of other cell cycle regulators or E-cadherin was not affected (Figure 3C), suggesting the specific effect of Hakai knockdown on cyclin D1. Comparable effect of Hakai knockdown on cyclin D1 was also observed in MDA-MB231 cells where E-cadherin is not expressed (Supplemental Figure S3, C and D). Together, we conclude that Hakai plays a crucial role in the regulation of cell proliferation.
Figure 3.
Effect of the knockdown of Hakai on cell proliferation in MCF-7– and HEK293–transformed epithelial cells. (A) Effect of Hakai siRNA in MCF-7 and HEK293 cells. MCF-7 or HEK293 cells were transiently transfected with either of two different Hakai siRNA oligos or a control nonsilencing oligo. After 72 h of transfection, cell lysates were examined by Western blotting with anti-Hakai and anti-GAPDH antibodies. (B) BrdU incorporation determined by colorimetric assay. Transfected MCF-7 or HEK293 cells were labeled with BrdU, followed by colorimetric assay. Statistical analyses indicate the significantly decreased BrdU incorporation in Hakai-knockdown MCF-7 cells (*p < 0.01, **p < 0.005; n = 9) or HEK293 cells (*p < 0.01, **p < 0.00002; n = 3). Data are represented as mean ± SD. (C) Effect of Hakai-knockdown on cell cycle regulators. MCF-7 or HEK293 cells were transiently transfected with a Hakai siRNA or control nonsilencing oligo. After 72 h of transfection, cell lysates were examined by Western blotting with antibodies against various cell cycle regulators.
Enhanced Expression of Hakai in Proliferative Tissues
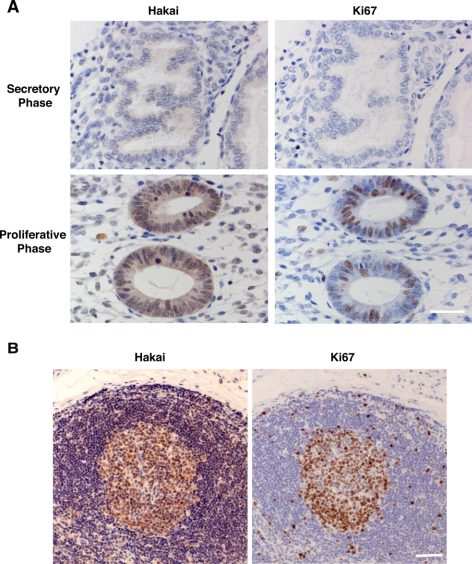
As shown in Figures 2 and 3, our results indicated a role of Hakai in cell proliferation in cell culture systems. To ascertain whether Hakai is indeed involved in a proliferation process in vivo, we investigated the expression and localization of Hakai in different human tissues. First, we analyzed Hakai expression in endometrium of the uterus. Throughout the reproductive age, every woman undergoes a hormonal cycle lasting for an average of 28 d. In this ovulation cycle, there are two phases in the uterus endometrium: proliferative and secretory phases. During the proliferative phase, cells lining the uterine cavity begin to proliferate under the effect of estrogen. In contrast, during the secretory phase, cells stop proliferating and increase their secretory activity under the effect of progesterone. Thus, the endometrium is an excellent model system to analyze the involvement of proteins in cell proliferation. By immunohistochemistry we observed very low expression of Hakai in the endometrial cells in the secretory phase (Figure 4A, top left). This correlated with low expression of Ki67, a proliferation marker (Figure 4A, top right). In contrast, enhanced expression of Hakai was detected in the proliferative phase (Figure 4A, bottom left), together with increased Ki67 expression (Figure 4A, bottom right). Enhanced expression of Hakai was observed both in the nucleus and cytoplasm. Expression of E-cadherin remained at a low level during both secretory and proliferative phases (Supplemental Figure S6). These data indicate that the expression of Hakai is highly up-regulated in the proliferative phase of the human endometrium.
Figure 4.
Hakai expression in human tissues. (A) Sections of secretory and proliferative phases of the uterus endometrium. Immunohistological analyses were performed with anti-Hakai and anti-Ki67 antibodies (brown) using samples from a secretory phase (top) and a proliferative phase (bottom) of the endometrium. (B) Sections of cervical lymph node. The germinal center in the lymph node shows enhanced Hakai and Ki67 immunoreactivity (brown). (A and B) Tissues were also stained with hematoxylin (blue). Bars, 50 μm.
We also analyzed the expression level of Hakai in the lymph node that contains germinal centers where lymphocytes are actively proliferating. There was a marked increase in Hakai and Ki67 expression in the germinal center (Figure 4B). These results suggest that, at least in the endometrium and lymph node, the expression of Hakai is up-regulated where cells proliferate actively, further supporting a role for Hakai in the proliferation process.
Involvement of Hakai in Oncogenesis
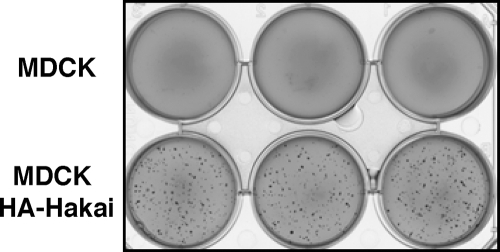
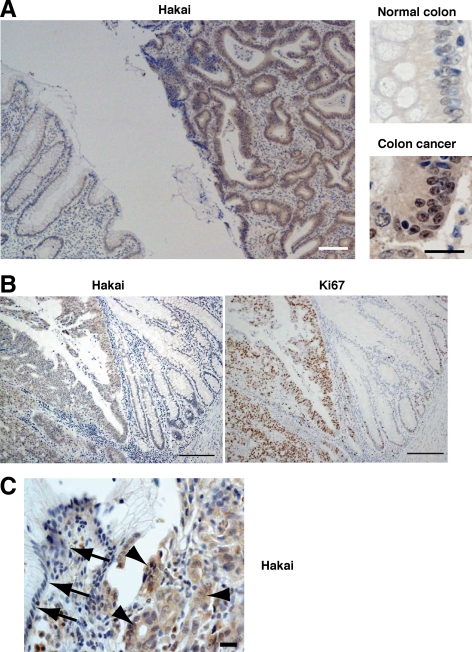
The effects observed in Hakai-overexpressing and Hakai-knockdown epithelial cells suggested a potential role for Hakai in tumor progression. By using soft agar assays, we first examined the colony formation driven by Hakai-overexpressing cells compared with parental MDCK cells. As shown in Figure 5, Hakai-overexpressing cells formed colony foci in soft agars, whereas parental MDCK cells did not. These data indicate that overexpression of Hakai induces anchorage-independent cell growth; thus, Hakai exhibits an oncogenic potential. Next, by immunohistochemistry, we examined the expression and localization of Hakai in human colon cancer tissues. The section shown in the left panel of Figure 6A includes normal colon epithelium (left half of the image) and colon adenocarcinoma (right half of the image) in the same slide. Immunostaining with anti-Hakai antibody clearly demonstrated the increased expression of Hakai in colon adenocarcinoma compared with normal colon epithelium. The magnified images revealed that the expression of Hakai was enhanced both in the nucleus and the cytoplasm in colon cancer cells (Figure 6A, right). In Figure 6B, we showed a section from another patient that also includes normal colon (right half of the image) and colon adenocarcinoma (left half of the image) in the same slide. Immunohistochemical analyses with anti-Hakai and anti-Ki67 antibodies showed that both Hakai and Ki67 were highly expressed in colon adenocarcinoma cells. We have observed comparable up-regulation of Hakai in all of the four colon cancer tissues thus far analyzed. The expression of Hakai was further examined in gastric adenocarcinoma. The section shown in Figure 6C includes normal stomach epithelium (arrows) and gastric adenocarcinoma (arrowheads) in the same slide. Immunostaining with anti-Hakai antibody showed the increased expression of Hakai in gastric adenocarcinoma. The comparable up-regulation of Hakai was observed in 60% of the independent gastric adenocarcinoma samples (n = 5). These data suggest that expression of Hakai is often enhanced in colon and gastric cancer and is correlated with enhanced proliferation of cancer cells.
Figure 5.
Anchorage independent-growth of Hakai-overexpressing MDCK cells. Cells were suspended in soft agars, and colony formation was examined after 12 d. The result shown is a representative of three independent experiments.
Figure 6.
Enhanced Hakai expression in colon and gastric cancer tissues. (A) A section of normal colon epithelium (left half of the image) and grade I colon adenocarcinoma (right half of the image). A section was examined by immunohistochemistry using anti-Hakai antibody. Bar, 100 μm. Magnified images are shown in the right panels. Bar, 20 μm. (B) Sections of normal colon epithelium (right half of the image) and grade II colon adenocarcinoma (left half of the image). Sections were examined by immunohistochemistry using anti-Hakai and anti-Ki67 antibodies. Bars, 200 μm. (C) A section of normal stomach epithelium (arrows) and grade II gastric adenocarcinoma (arrowheads). A section was examined by immunohistochemistry using anti-Hakai antibody. Bar, 20 μm. (A–C) Tissues also were stained with hematoxylin (blue).
Hakai Interacts with PSF and Affects Its RNA-Binding Ability
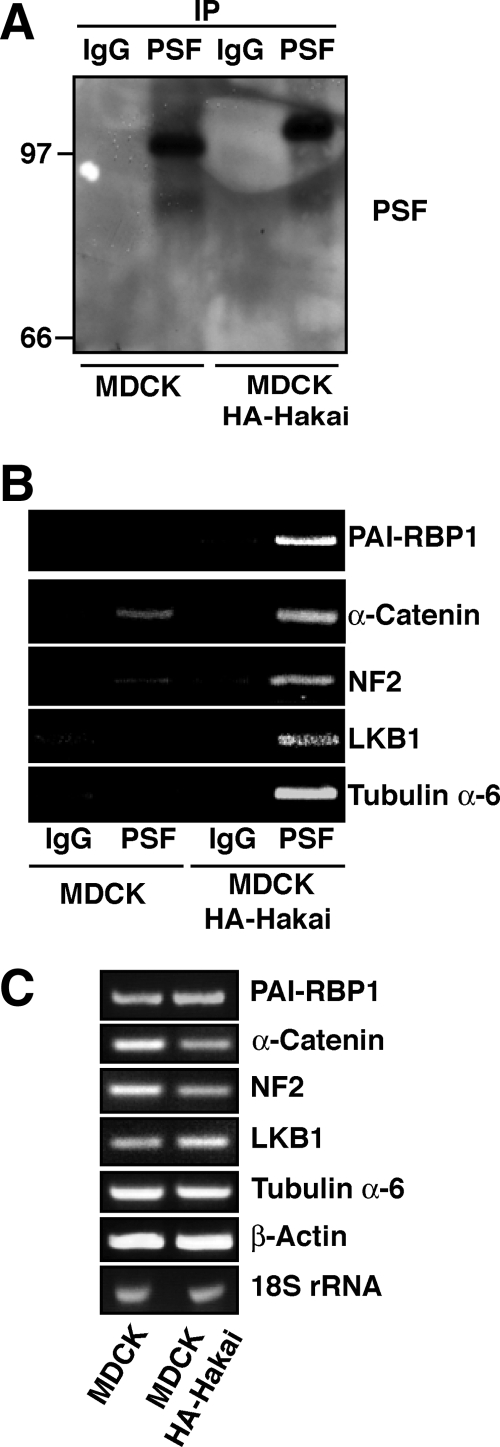
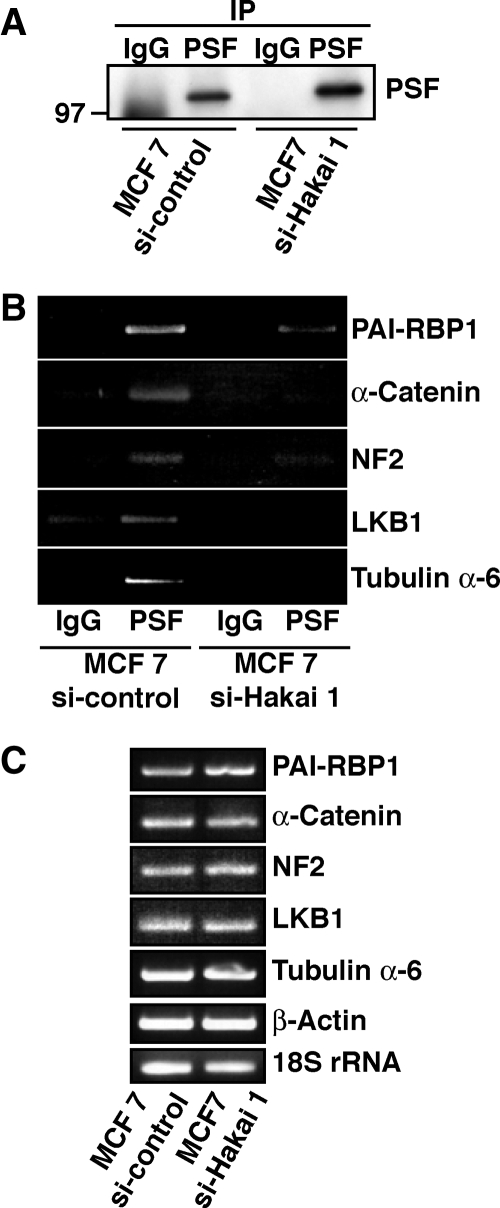
We have shown that Hakai induces various phenotypic changes in epithelial cells. However, not all these changes can be solely attributed to the decreased expression of E-cadherin. We have also revealed that Hakai is localized in the nucleus. Therefore, we further investigated a possible role of Hakai in the nucleus. To identify novel Hakai-interacting proteins, we performed affinity protein purification using HEK293 cell lysates with glutathione beads coupled to GST or GST-Hakai. Two proteins were recovered in GST-Hakai beads but were not present in GST beads (Figure 7A, arrow and arrowhead). Mass spectrometric analysis revealed that the novel Hakai-interacting proteins were PSF and its binding partner p54nrb. We also confirmed the interaction between endogenous Hakai and PSF proteins in HEK293 cells by immunoprecipitation (Figure 7B). Using several truncation mutants, we showed that the N terminus of PSF is, at least partially, responsible for the interaction with Hakai (Supplemental Figure S7). PSF is a RNA-binding protein that was initially termed as a splicing factor (Patton et al., 1993), but recently it has been shown to regulate several cellular processes, including transcription, pre-mRNA processing, nuclear retention of edited RNA, and DNA relaxation (Shav-Tal and Zipori, 2002). It contains two consensus RNA-binding domains; however, only a few mRNAs have been reported to bind PSF (Zolotukhin et al., 2003; Buxade et al., 2008; Cobbold et al., 2008). To investigate the physiological significance of the interaction between Hakai and PSF, their subcellular localization was first analyzed in MCF-7 and MDCK cells by immunofluorescence. We found that Hakai and PSF are colocalized in the nucleus in both cell lines (Figure 7C). Overexpression of Hakai did not induce ubiquitination of PSF as determined by Western blotting with anti-ubiquitin antibody after immunoprecipitation of PSF (Supplemental Figure S8), suggesting that PSF is not a substrate of E3 ubiquitin-ligase Hakai. Next, we examined whether overexpression of Hakai affects the ability of PSF to bind RNA. To identify the mRNAs bound to PSF, cell lysates of parental and Hakai-overexpressing MDCK cells were immunoprecipitated with control IgG or anti-PSF antibody, followed by extraction of mRNAs. The RNAs from the immunoprecipitates were then reverse transcribed, and the resulting products were hybridized to human cDNA arrays (http://www.grc.nia.nih.gov/branches/rrb/dna/dna.htm#, MGC arrays). It is interesting that in parental MDCK cells, we could not detect significant binding of any transcripts to PSF. In contrast, in Hakai-overexpressing cells, we found that 176 array spots (∼2.93% of the total spot on the array) had -fold increase of >1.20 when comparing the signals in PSF immunoprecipitate arrays with those in control IgG immunoprecipitate arrays, and they were considered to represent specific PSF-associated transcripts.
In Supplemental Table 1, we show a subset of selected transcripts from the experimental data, including those encoding proteins involved in tumor progression and angiogenesis (PAI-RBP1), tumor suppression (NF2, LKB1), cell–cell adhesion (α-catenin), and cytoskeleton dynamics (tubulin α-6). To further validate these results, we performed RT-PCR using PSF immunoprecipitates from parental and Hakai-overexpressing MDCK cells (Figure 8A). In Hakai-overexpressing cells, the amount of PSF-bound transcripts was increased compared with that in parental cells (Figure 8B), indicating a positive regulatory role of Hakai in the RNA-binding capacity of PSF. Total amount of mRNAs were not affected by overexpression of Hakai (Figure 8C). The RNA-binding ability of another RNA-binding protein HuR was not affected by overexpression of Hakai (Supplemental Figure S9), suggesting the specific effect of Hakai on PSF. To ascertain this result, the role of Hakai in the RNA-binding ability of PSF was examined in transformed epithelial cell lines, MCF-7 cells (Figure 9A). Interestingly, we found that the considerable amount of mRNAs bound to PSF in MCF-7 cells (Figure 9B). Thus, the effect of knockdown of Hakai on the interaction between PSF and mRNAs was examined. Total amounts of mRNAs were not affected by knockdown of Hakai (Figure 9C). On knockdown of Hakai, PSF could no longer firmly interact with mRNAs (Figure 9B), confirming that Hakai plays a crucial role in the RNA-binding ability of PSF.
Figure 8.
Enhanced RNA-binding ability of PSF in Hakai-overexpressing MDCK cells. (A) Cell lysates of parental and Hakai-overexpressing MDCK cells were immunoprecipitated with control IgG or anti-PSF antibody, followed by Western blotting with anti-PSF antibody. It should be noted that PSF in Hakai-overexpressing cells migrated more slowly in SDS-PAGE than that in parental cells, but the identity of this modification is currently unknown. (B) Interaction of PSF with mRNAs encoding PAI-RBP1, α-catenin, NF2, LKB1, and tubulin α-6 was examined by RT-PCR after immunoprecipitation with control IgG or anti-PSF antibody using parental and Hakai-overexpressing MDCK cells. (C) Total RNA levels of the indicated mRNAs in parental and Hakai-overexpressing MDCK cells. The amount of mRNAs encoding PAI-RBP1, α-catenin, NF2, LKB1, tubulin α-6, and β-actin was examined by RT-PCR. 18S rRNA was used for control RNA loading.
Figure 9.
Reduced RNA-binding ability of PSF in Hakai-knockdown MCF-7 cells. (A) Cell lysates of MCF-7 cells transfected with a Hakai siRNA or control nonsilencing oligo were immunoprecipitated with control IgG or anti-PSF antibody, followed by Western blotting with anti-PSF antibody. (B) Interaction of PSF with mRNAs encoding PAI-RBP1, α-catenin, NF2, LKB1, and tubulin α-6 was examined by RT-PCR after immunoprecipitation with control IgG or anti-PSF antibody using MCF-7 cells transfected with a Hakai siRNA or control nonsilencing oligo. (C) Total RNA levels of the indicated mRNAs in MCF-7 cells transfected with a Hakai siRNA or control nonsilencing oligo. The amount of mRNAs encoding PAI-RBP1, α-catenin, NF2, LKB1, tubulin α-6, and β-actin was examined by RT-PCR. 18S rRNA was used for control RNA loading.
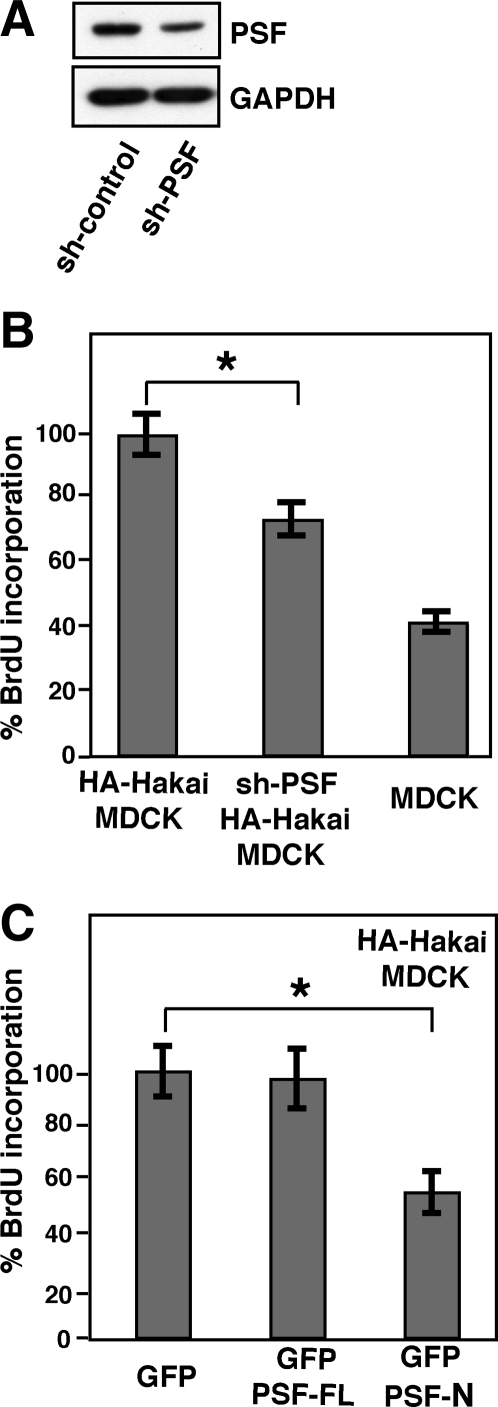
To examine the functional relation between Hakai and PSF, we established Hakai-overexpressing MDCK cell lines that stably express PSF shRNA. In these cells, expression of PSF was knocked down by ∼60% (Figure 10A). BrdU incorporation analyses showed that the knockdown of PSF partially suppressed the effect of Hakai overexpression on cell proliferation (Figure 10B). PSF knockdown did not affect the expression of E-cadherin in Hakai-overexpressing MDCK cells (unpublished observation). Furthermore, transient expression of the N terminus of PSF that binds to Hakai but lacks RNA-binding sites (Supplemental Figure S7) significantly suppressed cell proliferation in Hakai-overexpressing MDCK cells (Figure 10C). These data suggest an involvement of PSF in the regulation of cell proliferation.
Figure 10.
PSF is involved in the proliferative control in Hakai-overexpressing MDCK cells. (A) Effect of PSF shRNA on PSF expression in Hakai-overexpressing MDCK cells. Cell lysates of Hakai-overexpressing MDCK cells stably transfected with control shRNA or PSF shRNA were examined by Western blotting with anti-PSF and anti-GAPDH antibodies. (B) Effect of PSF shRNA on cell proliferation in Hakai-overexpressing MDCK cells. Parental, Hakai-overexpressing MDCK cells, or Hakai-overexpressing MDCK cells stably transfected with PSF shRNA were labeled with BrdU and analyzed by immunofluorescence with anti-BrdU antibody. Statistical analyses indicate the significantly decreased BrdU incorporation in PSF shRNA Hakai-overexpressing cells compared with Hakai-overexpressing cells (*p < 0.01; n = 3). Results are expressed as a percentage relative to Hakai-overexpressing cells, and are represented as mean ± SD. (C) Effect of expression of the N terminus of PSF on cell proliferation in Hakai-overexpressing MDCK cells. GFP, GFP-PSF (full length), or GFP-PSF-N (amino acids 1-340) was transiently expressed in Hakai-overexpressing MDCK cells. Cells were labeled with BrdU and analyzed by immunofluorescence with anti-BrdU antibody. Statistical analyses indicate the significantly decreased BrdU incorporation in Hakai-overexpressing MDCK cells that express GFP-PSF-N (*p < 0.001; n = 3). Results are expressed as a percentage relative to GFP-expressing cells and are represented as mean ± SD.
DISCUSSION
In this article, we show that Hakai is involved in the regulation of not only cell–cell contacts but also cell proliferation. Overexpression of Hakai in epithelial cells induces dramatic morphological effects that are reminiscent of the epithelial–mesenchymal transition. The expression level of E-cadherin is specifically reduced and the strength of cell–cell contacts is strongly decreased. The loss of cell–cell contacts is accompanied by increased protrusion formation. However, one of the most obvious phenotypic changes is observed in cell proliferation. On overexpression of Hakai, cells proliferate two- to threefold faster. In contrast, there is a significant decrease in the proliferation rate in cells expressing Hakai siRNA. Close correlation between Hakai expression and proliferation is also observed in two different human tissues. Our data suggest that there are two proteins through which Hakai may regulate cell proliferation: PSF and cyclin D1. Expression of PSF shRNA or a dominant-negative PSF mutant significantly suppresses proliferation of Hakai-overexpressing MDCK cells (Figure 10, B and C). The knockdown of Hakai specifically affects the protein expression level of cyclin D1 in MCF-7, HEK293, and MDA-MB231 cells (Figure 3C and Supplemental Figure S3C). These two proteins may function independently, because overexpression of Hakai does not affect the ability of PSF to interact with the mRNA that encodes cyclin D1 (unpublished observation). We also show that the expression of the Hakai mutant lacking the RING-finger domain suppresses cell proliferation, suggesting that E3 ubiquitin-ligase activity of Hakai is, at least partially, involved in cell proliferation (Supplemental Figure S4, A–D). This Hakai mutant is able to bind PSF (Supplemental Figure S4E), indicating that the interaction with PSF itself cannot promote cell proliferation. We have obtained no evidence showing that Hakai induces ubiquitination of PSF. Thus, it is plausible that there are unidentified substrate protein(s) that are involved in Hakai/PSF-mediated proliferative control. It remains to be elucidated whether and how PSF, cyclin D1, and E3 ubiquitin-ligase activity of Hakai are functionally correlated.
There are several lines of evidence indicating that Hakai can affect multiple cellular phenotypes in an E-cadherin–independent manner. For example, MDCK cells expressing E-cadherin shRNA do not extend spiky protrusions that are seen in Hakai-overexpressing cells (unpublished observation). Overexpression and knockdown of Hakai affects cell proliferation in NIH3T3 and HEK293 cells, respectively, where no or little expression of E-cadherin is detected (Figure 3B and Supplemental Figure S3B). Thus, it is plausible that Hakai fulfills its functions by interacting with other proteins than E-cadherin. Here, we identify PSF as a novel Hakai-binding protein. PSF has been recently shown to affect multiple cellular processes (Shav-Tal and Zipori, 2002; Kaneko et al., 2007; Cobbold et al., 2008); however, it is still not clearly known which RNAs specifically interact with PSF. By using cDNA arrays, we have determined several transcripts that bind to PSF. Furthermore, we show that Hakai regulates the interaction between PSF and its target mRNAs. In the future, it needs to be further investigated how the functions of PSF targets are affected by Hakai and attributed to Hakai-mediated cellular processes.
Another observation is that Hakai localizes in the nucleus as well as in the cytoplasm in cultured cell lines and human tissues. On addition of HGF, cytoplasmic localization of Hakai increases, indicating that subcellular localization of Hakai is dynamically regulated; however, the molecular mechanisms that regulate Hakai localization are not clear. It is interesting that this enriched nuclear localization also was observed in β-transducin repeat-containing protein (β-TrCP) (Davis et al., 2002). β-TrCP is an F-box-type E3 ubiquitin-ligase that ubiquitinates many cytoplasmic proteins such as β-catenin and inhibitor of nuclear factor-κBα (Maniatis, 1999; Polakis et al., 1999; Karin and Ben-Neriah, 2000). Despite its important roles in the cytoplasm, β-TrCP localizes predominantly in the nucleus. In the nucleus it binds to a pseudosubstrate hnRNP-U, and this interaction is responsible for the nuclear localization of β-TrCP (Davis et al., 2002), although its nuclear role is not clearly understood. In a similar way, Hakai may also have nuclear binding protein(s) that regulate localization of Hakai.
By using immunohistochemistry, we have analyzed the expression and localization of Hakai in several human cancer tissues. In colon cancer, the expression of Hakai is highly enhanced both in the nucleus and cytoplasm. This enhanced Hakai expression is also observed in benign colon adenoma (unpublished observation), suggesting that Hakai may be involved in the early stages of tumorgenesis by up-regulating cell proliferation. Similar up-regulation of Hakai is also found in gastric carcinoma, whereas we have not observed increased expression of Hakai in melanoma and glioblastoma cells (unpublished observation), indicating that up-regulation of Hakai in cancer is cell type specific. In addition, we have shown that the knockdown of Hakai in transformed epithelial cells significantly reduces the proliferation rate. Together, these observations suggest that Hakai can be a potential drug target in cancer treatment. Further investigations of the physiological and pathological functional roles of Hakai would lead us to a novel therapeutic treatment for cancer.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Marsh for critical reading of the manuscript. We thank Cristina Azevedo for helpful discussions. We also thank Fiona Watt and Hector Garcia-Palmer for technical advice for immunohistochemistry, Alison Lloyd for providing antibodies against various cell cycle regulators, James G. Patton for providing PSF constructs, and Walter Birchmeier for providing Hakai constructs. A.F. was supported by a postdoctoral fellowship from Ministerio de Educacion, Cultura y Deporte of Spain. This work is supported by Medical Research Council funding to the Cell Biology Unit.
Abbreviations used:
- IP
immunoprecipitation
- MDCK
Madin-Darby canine kidney
- PSF
PTB-associated splicing factor.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0845) on June 17, 2009.
REFERENCES
- Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G. F. Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Birchmeier W., Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim. Biophys. Acta. 1994;1198:11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Bonazzi M., Veiga E., Cerda J. P., Cossart P. Successive post-translational modifications of E-cadherin are required for InlA-mediated internalisation of Listeria monocytogenes. Cell Microbiol. 2008;10:2208–2222. doi: 10.1111/j.1462-5822.2008.01200.x. [DOI] [PubMed] [Google Scholar]
- Buxade M., Morrice N., Krebs D. L., Proud C. G. The PSF.p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor alpha. J. Biol. Chem. 2008;283:57–65. doi: 10.1074/jbc.M705286200. [DOI] [PubMed] [Google Scholar]
- Capaldo C. T., Macara I. G. Depletion of E-cadherin disrupts establishment but not maintenance of cell junctions in Madin-Darby canine kidney epithelial cells. Mol. Biol. Cell. 2007;18:189–200. doi: 10.1091/mbc.E06-05-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro U., Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- Cobbold L. C., Spriggs K. A., Haines S. J., Dobbyn H. C., Hayes C., de Moor C. H., Lilley K. S., Bushell M., Willis A. E. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 2008;28:40–49. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M., Hatzubai A., Andersen J. S., Ben-Shushan E., Fisher G. Z., Yaron A., Bauskin A., Mercurio F., Mann M., Ben-Neriah Y. Pseudosubstrate regulation of the SCF(beta-TrCP) ubiquitin ligase by hnRNP-U. Genes Dev. 2002;16:439–451. doi: 10.1101/gad.218702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B. T., Patton J. G. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp. Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Krause G., Scheffner M., Zechner D., Leddy H. E., Behrens J., Sommer T., Birchmeier W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002;4:222–231. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- Gumbiner B. M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hogan C., Serpente N., Cogram P., Hosking C. R., Bialucha C. U., Feller S. M., Braga V. M., Birchmeier W., Fujita Y. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 2004;24:6690–6700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C. A., Wing S. S., Huang H., Leverson J. D., Hunter T., Liu Y. C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- Kaneko S., Rozenblatt-Rosen O., Meyerson M., Manley J. L. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Levkowitz G., et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Li Y., Kumar K. G., Tang W., Spiegelman V. S., Fuchs S. Y. Negative regulation of prolactin receptor stability and signaling mediated by SCF(beta-TrCP) E3 ubiquitin ligase. Mol. Cell. Biol. 2004;24:4038–4048. doi: 10.1128/MCB.24.9.4038-4048.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T. A ubiquitin ligase complex essential for the NF-kappaB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- Mantovani F., Banks L. Regulation of the discs large tumor suppressor by a phosphorylation-dependent interaction with the beta-TrCP ubiquitin ligase receptor. J. Biol. Chem. 2003;278:42477–42486. doi: 10.1074/jbc.M302799200. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K., Lal A., Martindale J. L., Kawai T., Gorospe M. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios F., Tushir J. S., Fujita Y., D'Souza-Schorey C. Lysosomal targeting of E-cadherin: a unique mechanism for the down-regulation of cell-cell adhesion during epithelial to mesenchymal transitions. Mol. Cell. Biol. 2005;25:389–402. doi: 10.1128/MCB.25.1.389-402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J. G., Porro E. B., Galceran J., Tempst P., Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M., Jamora C., Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. doi: 10.1016/s0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- Polakis P., Hart M., Rubinfeld B. Defects in the regulation of beta-catenin in colorectal cancer. Adv. Exp. Med. Biol. 1999;470:23–32. doi: 10.1007/978-1-4615-4149-3_3. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y., Zipori D. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Shen Y., Hirsch D. S., Sasiela C. A., Wu W. J. CDC42 regulates E-cadherin ubiquitination and degradation through an EGF receptor to SRC-mediated pathway. J. Biol. Chem. 2007;283:5127–5137. doi: 10.1074/jbc.M703300200. [DOI] [PubMed] [Google Scholar]
- Shenoy S. K., McDonald P. H., Kohout T. A., Lefkowitz R. J. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- Tenenbaum S. A., Lager P. J., Carson C. C., Keene J. D. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- Toda Y., Kono K., Abiru H., Kokuryo K., Endo M., Yaegashi H., Fukumoto M. Application of tyramide signal amplification system to immunohistochemistry: a potent method to localize antigens that are not detectable by ordinary method. Pathol. Int. 1999;49:479–483. doi: 10.1046/j.1440-1827.1999.00875.x. [DOI] [PubMed] [Google Scholar]
- Zolotukhin A. S., Michalowski D., Bear J., Smulevitch S. V., Traish A. M., Peng R., Patton J., Shatsky I. N., Felber B. K. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell. Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.