Abstract
Dynamin exhibits a high basal rate of GTP hydrolysis that is enhanced by self-assembly on a lipid template. Dynamin's GTPase effector domain (GED) is required for this stimulation, though its mechanism of action is poorly understood. Recent structural work has suggested that GED may physically dock with the GTPase domain to exert its stimulatory effects. To examine how these interactions activate dynamin, we engineered a minimal GTPase-GED fusion protein (GG) that reconstitutes dynamin's basal GTPase activity and utilized it to define the structural framework that mediates GED's association with the GTPase domain. Chemical cross-linking of GG and mutagenesis of full-length dynamin establishes that the GTPase-GED interface is comprised of the N- and C-terminal helices of the GTPase domain and the C-terminus of GED. We further show that this interface is essential for structural stability in full-length dynamin. Finally, we identify mutations in this interface that disrupt assembly-stimulated GTP hydrolysis and dynamin-catalyzed membrane fission in vitro and impair the late stages of clathrin-mediated endocytosis in vivo. These data suggest that the components of the GTPase-GED interface act as an intramolecular signaling module, which we term the bundle signaling element, that can modulate dynamin function in vitro and in vivo.
INTRODUCTION
Dynamin (Dyn) is a multidomain GTPase that plays an essential role in clathrin-mediated endocytosis (CME; Hinshaw, 2000; Conner and Schmid, 2003; Praefcke and McMahon, 2004). It belongs to a family of atypical GTPases (including MxA/Mx, Drp1/Dlp1/Dnm1, Vps1, dynamin A, and Mgm1) that share the common properties of low affinity for guanine nucleotides (>10–100 μM), high basal turnover (∼0.4–1 min−1), and the propensity for oligomerization (Song and Schmid, 2003). In solution, dynamin exists as a tetramer and its intrinsic GTPase activity can be enhanced >100-fold by self-assembly on a lipid template (Barylko et al., 1998, Stowell et al., 1999). The mechanisms mediating dynamin's assembly-stimulated GTPase activity remain unknown.
Evidence suggests that dynamin's GTPase effector domain (GED, residues 624–750) plays a role in its assembly-stimulated GTPase activity. Mutations in GED impair dynamin's assembly-stimulated GTPase activity (Sever et al., 1999) and its ability to self-assemble (Song et al., 2004). Interestingly, overexpression of some of these mutants (K694A and R725A) stimulate transferrin (Tfn) uptake by CME (Sever et al., 1999, 2000), whereas others (I690K and I697K) are inhibitory (Song et al., 2004). A third class of mutations in GED (A738T and T749I) was identified as suppressors of a Drosophila shibirets allele (Narayanan et al., 2005). These second site mutations rescued a defect in GTP binding, suggesting that GED can negatively regulate dynamin function in vivo. Finally, addition of purified GED to unassembled dynamin in vitro stimulates GTP hydrolysis in a cooperative manner (Sever et al., 1999). Although the mechanism remains unknown (Marks et al., 2001), these data suggest that GED plays an important role in regulating dynamin self-assembly, assembly-stimulated GTPase activities, and its in vivo function.
Many small GTPases modulate their catalytic activity through interactions with GTPase activating proteins (GAPs). GAPs bind and stabilize the conformationally flexible switch I and switch II regions of the active site and promote GTP hydrolysis either by directly contributing catalytic residues in trans or by facilitating conformational changes that reorient catalytic machinery in cis (Scheffzek et al., 1997; Rittinger et al., 1997a,b; Tesmer et al., 1997; Seewald et al., 2002). Although the functional consequences of perturbing GED activity resemble those expected for a GAP, this paradigm for stimulation is inconsistent with structural descriptions of assembled dynamin obtained from cryo-electron microscopy (EM) and computational modeling (Zhang and Hinshaw, 2001; Chen et al., 2004; Mears et al., 2007). These studies suggest that the helical dynamin polymer is organized such that the active sites of adjacent dimer subunits face each other and move into close proximity upon the nucleotide-dependent constriction of the assembled lattice (Mears et al., 2007). This arrangement precludes any association of GED with switch I and switch II, making dynamin stimulation via a classical GAP mechanism highly unlikely.
However, because of the low resolution of the cryo-EM maps, the absence of fiducial markers such as gold labels that could approximate GED's position and the lack of a high-resolution GED structure that could be utilized for docking, the location of GED within the assembled dynamin polymer remains unclear. Less is known about the exact structural nature of GED's interaction with the GTPase domain and the extent to which such an interaction can enhance dynamin GTP hydrolysis. Nonetheless, a model for GED's association with the GTPase domain has been suggested based on high-resolution structural studies of the GTPase domains from rat dynamin and Dictyostelium dynamin A (Niemann et al., 2001; Reubold et al., 2005). These proteins were crystallized as myosin fusions and in each structure a myosin helix packs into a hydrophobic groove that is formed by the N- and C-terminal helices of the GTPase domain. It was proposed that this region on the back of the GTPase domain constitutes the GED docking site. This model is supported by yeast two-hybrid analysis showing that the C-terminus of the GTPase domain is required for interactions with GED (Smirnova et al., 1999). Here we identify the GTPase-GED interface and examine its role in regulating dynamin's assembly-stimulated GTPase activity in vitro and dynamin function in vivo.
MATERIALS AND METHODS
Cloning and Mutagenesis of GTPase-GED Fusion Constructs
The GTPase-GED (GG) fusion proteins were constructed piecewise in two fragments. The first fragment constituted the GTPase portion and contained residues 6-320 of human dynamin-1; the second fragment comprised the C-terminal region of GED and contained residues 726-750. These fragments were amplified from cDNA by PCR and subcloned sequentially into a modified N-terminal MBP fusion vector (pMAL c2xP_5D) using EcoRI/XbaI and XbaI/HindIII restriction enzymes (New England Biolabs, Beverly, MA). The pMAL c2xP_5D plasmid was generated by removing the Factor Xa site from the parent pMAL c2x plasmid (New England Biolabs) with Blp1 and EcoRI restriction enzymes and replacing it with a new DNA fragment that included a PreScission protease site (LEVLFQGP) followed by an additional five aspartate residues. All point mutations were introduced by Quikchange mutagenesis (Stratagene, La Jolla, CA) and confirmed by sequencing.
Expression of GTPase-GED Constructs
GTPase-GED constructs were transformed into BL21(DE3) cells and grown at 37°C in Terrific broth (TB) to an OD600 of 0.6–0.8. Cultures were then induced with 0.3 mM IPTG for 3 h at 30°C. Cells were pelleted, washed with 40 ml of MBPHCBK200 (20 mM HEPES, pH 7.5, 200 mM KCl, 1 mM EDTA, and 1 mM DTT), and then pelleted a second time (buffer was discarded). At this stage, pellets were typically flash-frozen in liquid nitrogen and stored at −80°C for later use.
Purification of GTPase-GED Constructs
Frozen MBP-GG pellets from 250-ml cultures were thawed and resuspended in 20 ml of sonication buffer (MBPHCBK200, 10 mM PMSF, and a Roche Complete protease inhibitor cocktail [PI] tablet). Lysozyme was added to 1 mg/ml, and the mixture was incubated for 15 min at 4°C. Cells were disrupted by sonication for a total of 2 min, and the cell lysate was centrifuged at 10,000 rpm (7840 × g) for 30 min at 4°C to remove debris. The supernatant was diluted to 30 ml with dilution buffer (MBPHCBK200, 0.1 mg/ml AEBSF, and PI tablet) and loaded onto a 6-ml amylose column (New England Biolabs) equilibrated with MBPHCB200. The column was washed with 5 column volumes (CV) of MBPHCBK25 (20 mM HEPES, pH 7.5, 25 mM KCl, 1 mM EDTA, and 1 mM DTT), and the protein was then eluted with 10 mM maltose in MBPHCBK25. Peak elution fractions were visualized by SDS-PAGE and pooled.
The N-terminal MBP tag was removed by PreScission protease cleavage. PreScission (GST-fusion; GE Healthcare) was added to a final concentration of 0.5 U/mg fusion and the mixture was incubated for at least 3 h at 4°C.
PreScission-cleaved GTPase-GED constructs were further purified by anion exchange and size exclusion chromatography (SEC). The postcleavage reaction was loaded onto a 3.5 ml Q Sepharose column equilibrated with MBPHCBK25, and the column was washed with 2.5 CV of MBPHCBK25. The GG proteins were eluted with MBPHCBK100 (20 mM HEPES, pH 7.5, 100 mM KCl, 1 mM EDTA, and 1 mM DTT). Peak elution fractions were visualized by SDS-PAGE, pooled, concentrated, and passed over a Superdex 75 HR 10/30 column equilibrated with 20 mM HEPES, pH 7.5, 150 mM KCl, 4 mM MgCl2, 2 mM EGTA, and 1 mM DTT. GTPase-GED eluted as a single peak corresponding to the expected mass of a monomer (39.3 kDa). The peak SEC fractions were collected, concentrated to 100–200 μl, flash frozen in aliquots, and stored at −80°C.
Cloning and Mutagenesis of Dynamin Constructs
The open reading frame of dynamin1 was subcloned from pMIEG3-Dyn1 construct into pIEx6 vector using MfeI and NotI restriction sites. To correctly orient the dynamin1 reading frame with the N-terminal 6xHis tag of vector, the pIEx6-Dyn1 construct was cleaved with BamHI and MfeI. Linearized pIEx6-Dyn1 was purified, and 5′ overhangs were filled using PfuTurbo DNA polymerase. The plasmid was recircularized through ligation of blunted ends and transformed into DH5α competent cells. The correction of the frame was verified by sequencing. Interface mutations were generated by Quikchange mutagenesis and also confirmed by sequencing.
Expression and Purification of Dynamin Constructs
Dynamin constructs were expressed by transient transfection in Sf9 cells. Purified plasmid DNA (200 μg) and Insect Gene-juice transfection reagent (1 ml, Novagen, Madison, WI) were diluted individually with 10 ml of Sf9 cell media, mixed thoroughly, and then incubated at room temperature for 15 min. The DNA/transfection reagent mixture was then added to 100 ml of Sf9 cells at a concentration of 1 × 106 cells/ml (108 cells in total) and incubated at 27–28°C for 48 h. Cells were harvested by centrifugation, washed with a small amount of supernatant, pelleted again, and flash-frozen in liquid nitrogen. Pellets were typically stored at −80°C for later use. Expression was checked by Western blotting with Hudy-1 mAb. Dynamin was purified by affinity chromatography as described previously using glutathione-S-transferase (GST)-tagged amphiphysin-II SH3 domain as an affinity ligand (Stowell et al., 1999).
Intramolecular Cross-Linking of GTPase-GED Cysteine Mutants
Stocks, 20 mM, of each bifunctional methanethiosulfonate (MTS) cross-linker (Toronto Research Chemicals, North York, Canada) were prepared fresh in DMSO with the exception of MTS-1–MTS, which was prepared in acetone. Stocks were then diluted further to 200 μM in the reaction buffer (20 mM HEPES, pH 7.5, and 150 mM KCl). For cross-linking, 5 μl of a GTPase-GED double cysteine mutant (0.4 μg/μl) was mixed with 5 μl of cross-linker (200 μM) and incubated for 10 min at 4°C. Reactions were quenched with 2 μl 6× sample buffer with 60 mM N-ethylmaleimide. Samples were visualized by nonreducing SDS-PAGE and Coomassie staining.
Mass Spectrometry
Cross-linked GTPase-GED cysteine mutants were precipitated with cold acetone, resuspended, and digested overnight with trypsin. The resulting peptides were separated using a C18 column (500 μm ID) and analyzed by Fourier transform mass spectrometry. Uncrosslinked samples were similarly digested and analyzed as a control. The resulting total ion current chromatograms were compared, and a mass spectrum was taken for each unique peak that appeared in the cross-linked sample but was absent in the control. A deconvoluted mass spectrum was reconstructed from the observed charge states to obtain an accurate mass measurement for each cross-linked peptide. The measured average mass was used to identify the sequences of the cross-linked regions.
In Vitro Assays of Dynamin Function
Basal and liposome-stimulated GTP hydrolysis by dynamin was measured as a function of time using a colorimetric malachite green assay that detects the release of inorganic phosphate (Leonard et al., 2005). Liposomes containing a mixture of 85% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC; Avanti Polar Lipids, Alabaster, AL) and 15% porcine brain l-α-phosphatidylinositol-4,5-bisphosphate (PIP2, Avanti Polar Lipids) were prepared by extrusion through polycarbonate membranes (Whatman, Clifton, NJ) with a pore size of either 0.1 or 0.4 μm using an Avanti Mini-Extruder. Lipids were mixed, dried, rehydrated in buffer (20 mM HEPES, pH 7.5, and 100 mM NaCl) to a final concentration of 2.5 mM (∼2 mg/ml), and subjected to a series of freeze-thaw cycles before extrusion. Sedimentation assays were performed exactly as previously described (Ramachandran et al., 2007).
EM of Dynamin Interface Mutants Assembled In Vitro
Selected full-length dynamin constructs at 1 mg/ml in 20 mM HEPES, pH 7.5/100 mM NaCl were mixed 1:1 (vol/vol) with 0.4-μm PIP2-containing liposomes at 1 mg/ml in 20 mM HEPES, pH 7.5, and incubated at room temperature for 2 h. This mixture was then applied to glow-discharged, carbon-coated grids, washed with 20 mM HEPES, pH 7.5, and stained with 1% uranyl acetate. Samples were visualized in a Philips Technai F20 electron microscope (Mahwah, NJ) operating at 120 kV, and images were collected using Legion (Potter et al., 1999; Suloway et al., 2005) at 2.0-μm underfocus with a 4K × 4K Gatan CCD camera (Pleasanton, CA) at a nominal magnification of 50,000×, corresponding to a resolution of 2.24 Å per pixel.
Assay of Dynamin-Catalyzed Membrane Fission on SUPER Templates
Supported bilayers with excess reservoir, SUPER templates, were deposited on 5-μm silica beads as previously described (Pucadyil and Schmid, 2008). The lipid bilayers contained DOPC, 1,2-dioleoyl-sn-glycero-3-(phospho-l-serine) (DOPS), triammonium salt of porcine brain PIP2, and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(Lissamine Rhodamine B Sulfonyl) (RhPE; 79:15:5:1 mol%). All lipids were purchased from Avanti Polar Lipids. For fission assays, SUPER templates (typically 5 × 105) were suspended in 100 μl of 20 mM HEPES, pH 7.5, 150 mM KCl, 1 mM MgCl2 buffer ± dynamin ± nucleotides in a 0.5-ml polypropylene centrifuge tube for 30 min at 25°C. Tubes were spun at low speed (260 × g) at 25°C in a swinging bucket rotor. Seventy-five microliters of the supernatant was removed and mixed with 25 μl of 0.4% Triton X-100. Total membrane fluorescence on the beads (Total) was estimated in a separate reaction by adding templates to 0.1% Triton X-100. Fluorescence intensity of the supernatant was read in a 96-well plate reader (BioTek Instruments, Winooski, VT) at 25°C using 530/25-nm excitation and 590/25-nm emission filters.
Retroviral Transduction and Analysis of Dynamin-2 Mutants in Dyn2flox/− Fibroblasts
Dyn2flox/− fibroblasts were grown and maintained as described previously (Liu et al., 2008). For knockin experiments, dynamin-2 (Dyn2) constructs (wild type [WT], L12N, F20N, or A738N) were fused to green fluorescent protein (GFP) on their C-termini and used to generate retroviruses for infection of Dyn2flox/− cells as previously described (Liu et al., 2008). Cells were harvested 48 h after infection and GFP positive cells with low expression levels were isolated using fluorescence-activated cell sorting. The sorted cells were then infected with adenoviruses expressing Cre recombinase to remove endogenous Dyn2.
Clathrin-Mediated Endocytosis Assay
Tfn internalization was performed exactly as described (Sever et al., 2000) using biotinylated transferrin as the ligand and assessing its internalization into an avidin- or MesNa-inaccessible compartment.
Immunofluorescence and EM Analysis
Dyn2 knockout (KO) cells expressing different GFP-tagged Dyn2 constructs (WT, L12N, F20N, or A738N) were fixed and permeabilized simultaneously with 2% warm paraformaldehyde and 0.5% TX-100 for 2 min, to reduce cytosolic background staining, and then fixed with 4% paraformaldehyde for 40 min and stained with AP6 antibody (against α-adaptin). After immunofluorescence staining or mounting alone, cells were observed under wide-field epifluorescence microscopy using an inverted Olympus IX-70 microscope (Melville, NY).
For EM analysis, Dyn2 KO cells were grown in 35-mm culture dishes, fixed, and embedded in epon and prepared for thin-section EM as previously described (Yarar et al., 2005). The sections were stained with uranyl acetate followed by lead citrate. Before being examined, the grids were coded to conceal their precise construct identity. Grids were examined on a Philips CM100 electron microscope at 80 kV, and images were collected at 34,000× magnification using a Megaview III CCD camera. Pits were counted and scored for morphology without knowledge of the identity of each sample.
RESULTS
Design and Characterization of a Minimal GTPase-GED Fusion
The structures of the dynamin A and rat dynamin GTPase domains (Niemann et al., 2001; Reubold et al., 2005) each revealed that a helix derived from the myosin fusion protein associates with a hydrophobic groove on the GTPase domain formed by its N- and C-termini (NGTPase and CGTPase; Figure 1A). This interaction was suggested to mimic GED docking, but which region of GED might dock at this site had not been identified. The discovery that second site mutations located at the C-terminus of GED rescue the phenotype of a GTPase domain mutant of dynamin (Narayanan et al., 2005) attracted our attention to this portion of GED.
Figure 1.
Model of dynamin GG interface. (A) Crystal structure of rat dynamin GTPase domain (green) fused to myosin motor core (PDB: 2aka). A helix from the myosin (red) packs onto a hydrophobic groove comprised of the N- and C-termini (teal) of the GTPase domain. Hydrophobic side chains are shown in yellow. The myosin helix is predicted to mimic the GED docking arrangement. (B) Sequence alignments of the NGTPase, CGTPase, and CGED regions. Hs, Homo sapiens; Rn, Rattus norvegicus; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sc, S. cerevisiae; Dd, Dictyostelium discoideum; Dyn, dynamin. Black boxes indicate conserved hydrophobic residues. Green and cyan bars indicate the helical segments presumed to form the helical bundle that is modeled in C. Asterisk denotes the position of the kink in the CGTPase helix. (C) Helical wheel diagrams of the GTPase and GED helices. Yellow, hydrophobic residues; green, GTPase residues; cyan, GED residues. The plots are arranged in the putative docking arrangement, with the conserved residues in B forming the buried hydrophobic core between the three helices.
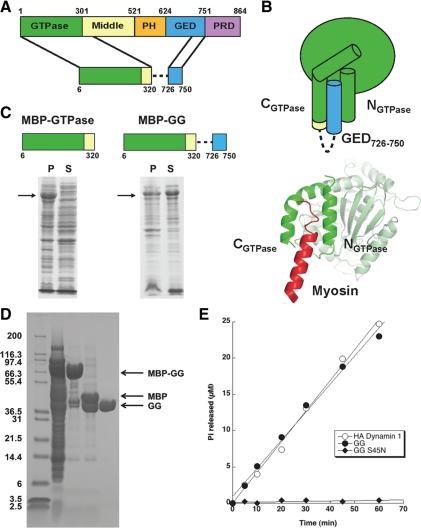
Secondary structure prediction algorithms model the C-terminus of GED (CGED) as a continuous α-helix (Chugh et al., 2006). A sequence alignment of dynamin family members revealed a number of highly conserved hydrophobic residues that map to one side of this putative CGED helix. (Figure 1, B and C). Given the similar amphipathic nature of the helices that constitute the predicted GED docking site, we speculated that the NGTPase, CGTPase, and CGED form a three-helix bundle (Figure 1C). We set out to test this hypothesis by designing a minimum GTPase-GED construct (Figure 2, A and B).
Figure 2.
Design and functional characterization of a minimal GTPase-GED fusion (GG). (A) Design of GG fusion. Dashed line represents flexible linker connecting the GTPase and GED fragments. (B) Diagram depicting the putative folding of GG. The crystal structure of the rat dynamin-myosin fusion (PDB: 2aka), which was used as a model to guide the design, is shown below for comparison. (C) Solubility of human dynamin1 GTPase domain in the absence (left) or presence (right) of the C-terminal GED helix. Proteins were expressed in E. coli as MBP fusions. Black arrows indicate bands corresponding to MBP-GTPase and MBP-GG, respectively. (D) Purification of GG from E. coli. GG was expressed as an MBP fusion and purified by a three-column procedure as described in Materials and Methods. Lane 1, soluble fraction following cell lysis; lane 2: pooled amylose column elutions; lane 3, sample from lane 2 after cleavage of MBP tag by PreScission protease; lane 4, purified GG after anion exchange and size exclusion chromatography to remove the MBP. Arrows denote the bands corresponding to MBP-GG fusion, MBP, and GG. (E) GTPase activities of purified GG and GG S45N. Proteins were assayed at a concentration of 2 μM in buffer containing 20 mM HEPES, pH 7.5, 150 mM KCl, 1 mM MgCl2 and 500 μM GTP. The activity of HA-tagged full-length dynamin-1 under the same reaction conditions is shown for comparison.
Dynamin's GTPase domain is insoluble when expressed in E. coli as an MBP fusion (Figure 2C). This instability could reflect exposure of the hydrophobic groove between NGTPase and CGTPase to solvent in the absence of the GED or myosin helix. Efforts to recover the GTPase domain by coexpression with GED failed (J. S. Chappie, unpublished data), suggesting that protein folding initially drives this interaction. If our hypothesis was correct, we reasoned that we could instead stabilize the GTPase domain by tethering the C-terminal portion of GED with a short flexible linker (Figure 2A). This design assumes that the incorporated GED fragment will form a helical peptide and dock with NGTPase and CGTPase in a manner analogous to the myosin helix (Figure 2B). As we predicted, expression of this GG fusion rescues the protein to the soluble fraction (Figure 2C), even after removal of the MBP fusion tag (Figure 2D). Purification yields a 39.3-kDa protein that is capable of hydrolyzing GTP at a rate comparable to the basal GTPase activity of full-length dynamin (Figure 2E). This activity can be inhibited by a P-loop mutation known to disrupt GTP binding in dynamin (S45N, Figure 2E). GG exists as a monomer in solution (data not shown), and we did not observe any detectable assembly at high concentrations of protein (>250 μM) or at low salt. Moreover, the observed hydrolysis activity is independent of protein concentration (data not shown). These data establish that the C-terminus of GED can provide structural stability through its interactions with the GTPase domain, allowing the latter to perform its basic chemical function outside the framework of the full-length protein. Monomeric GG and the full-length dynamin tetramer exhibit comparable GTPase activities, which strongly argues that the four GTPase domains within the tetramer function independently to hydrolyze GTP in the unassembled state. Although outside the scope of these studies, the minimal GG construct described here should also be a useful tool for future structure-function studies aimed at defining the mechanism of dynamin's basal GTPase activity.
Chemical Cross-Linking Confirms Direct Interaction of CGED with NGTPase
GG's activity implies that our design reconstitutes the minimal structural interactions required for dynamin's basal GTPase activity. However, this does not provide direct structural evidence that the GED interacts with the GTPase domain. In the absence of a high-resolution structure, we utilized chemical cross-linking to define this interaction further. Thiol-specific cross-linkers have been used extensively as molecular rulers to determine inter- and intramolecular distances, thereby providing constraints to guide structural modeling (Kenyon and Bruice, 1977; Loo and Clarke, 2001; Dalmas et al., 2005). In these cases, a bifunctional cross-linker is mixed with a protein containing two reactive cysteines, and cross-linked products are observed by nonreducing SDS-PAGE. The fixed position of the cysteines and the length of the cross-linker spacer arm each impose a distance constraint that determines the success of the cross-linking reaction. By varying the cross-linker length, the distance between the sulfhydryl side chains can be estimated. The extensive hydrophobic interface in the putative three-helix bundle would restrict the possible orientations of the amphipathic CGED helix in GG, making it an ideal template for this type of cross-link mapping.
To facilitate cross-linking, we generated a series of double cysteine mutants in GG and reacted them with a panel of bifunctional MTS compounds that ranged in length from 3.6 to 7.8 Å (Figure 3, A and B). Each cysteine pair is comprised of one substitution in the GTPase domain (R15C in the N-terminus or R297 in the C-terminus) and one substitution from an array of engineered cysteines in the GED (R730C, H733C, K736C, or S740C; Figure 3A). An additional mutation that removed a surface accessible reactive cysteine (C86S) was introduced into all constructs to limit nonspecific and intermolecular cross-linking. A third, partially buried cysteine (C169), thought to be critical for GTPase activity (Ramachandran and Schmid, 2008), was left unchanged. We directed our mutagenesis to the hydrophilic surfaces of the respective helices (Figure 1C) so as not to interfere with their hydrophobic packing and confirmed that the cysteine mutagenesis did not alter GG's GTPase activity (data not shown).
Figure 3.
Intramolecular cross-linking of GTPase and GED helices. (A) Residues selected for double cysteine mutagenesis in GG. Cysteine pairs included one substitution in the GTPase domain (R15C in NGTPase or R297C in CGTPase) and one substitution in the CGED region (R730C, H733C, K736C, or S740C). (B) Structure and estimated distances of bifunctional methanthiosulfonate (MTS) cross-linkers used for analyzing interactions between GTPase and GED helices in GG. (C) MTS cross-linking of GG double cysteine mutants. Cross-linkers were incubated with GG and analyzed by SDS-PAGE and Coomassie staining as described in Materials and Methods.
The cross-linking profile for each sample was visualized using nonreducing SDS-PAGE and Coomassie staining (Figure 3C). We detected efficient cross-linking between R15C in NGTPase and both R730C and H733C in CGED, as evidenced by a gel shift to a faster migrating species (Figure 3C, left panels). Mass spectrometry analysis of treated samples confirmed that the small gel-shift seen with the R15C/R730C construct corresponds to the presence of MTS-mediated covalent interactions between the engineered cysteines located in the NGTPase and CGED (Supplementary Figure S1). Although the efficiency increased with increasing length of the cross-linking reagent, we detected significant cross-linking even in the presence of the shortest reagent MTS-1–MTS, which approximates a distance of 3.6 Å between the cysteine pair. These findings confirm a close association of the NGTPase and CGED helix. We also detected slight gel shifts in the R15/K736C and R15C/S740C constructs, but only in the presence of longer cross-linking agents. However, the smaller degree of this gel shift makes interpretation more difficult. Overall, these data agree with our model for GED docking to the N-terminal helix of the GTPase domain.
In contrast, we were unable to detect cross-linking in the R297C GG mutants by SDS-PAGE. (Figure 3C, compare right panels with C86S control). This discrepancy may reflect an inherent difference in SDS-PAGE mobility for the two intramolecularly cross-linked species formed in GG. A covalent interaction between the NGTPase and CGED (R15C double mutants) connects the two termini of the GG construct, dramatically altering the shape of the unfolded protein; cross-linking between the CGTPase and CGED only modifies the structure of the extreme C-terminus, leaving the majority of the construct undisturbed. Thus, the shift in migration produced by the latter change may be so minor that it is not resolved by SDS-PAGE. Mass spectrometry analysis of the CGTPase-CGED cross-linked samples also proved ambiguous, as the corresponding CGTPase mutant peptide could not be detected, even in the non–cross-linked control digests. Thus, we cannot draw conclusions from these negative findings as to the positional relationship between CGTPase and CGED. However, the mutagenesis studies described below support an interaction between these two helices.
Perturbation of the GG Interface Disrupts Dynamin GTP Hydrolysis
To probe the functional significance of the GTPase-GED interface, we next generated a series of point mutations targeting the highly conserved hydrophobic residues within each of the three interface helices (Figure 1B). Mutation of a number of these side chains to alanine in GG, both individually and in pairs, produced no effect on GTPase activity (data not shown). Although it is possible that the alanine substitutions were maintaining rather than disrupting the conserved hydrophobic interface, we suspected that these interactions might be specifically involved in modulating dynamin's assembly-stimulated activity. Therefore, for subsequent analyses we decided to engineer mutations in the context of full-length dynamin. Initial mutagenesis of several hydrophobic residues in GED to alanine also yielded proteins whose activities were indistinguishable from wild type (data not shown). Therefore, we introduced asparagine residues and determined how each substitution affected basal and assembly-stimulated hydrolysis.
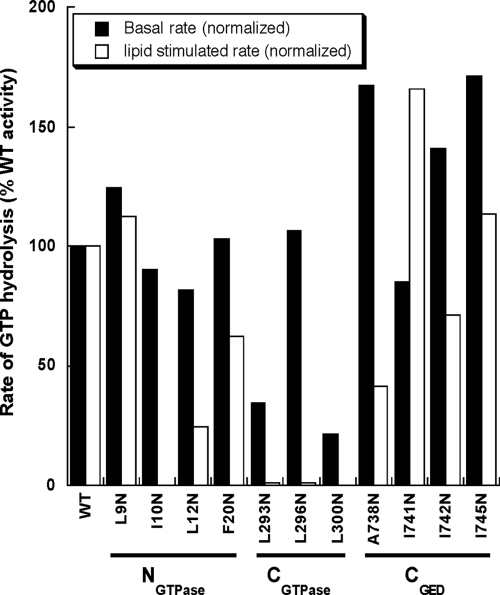
Preliminary screening experiments identified mutations in each interface helix that differentially affect dynamin GTPase activity (Figure 4), thereby providing strong evidence that our putative three-helix bundle at the GTPase-GED interface is functionally relevant. Comparison of the normalized rates to wild-type dynamin reveals two classes of interface mutants. The first class (I10N, L293N, L296N, and L300N) is severely defective in stimulated GTPase activity, with some mutants such as L293N and L300N also exhibiting a basal defect. The second class (L12N, F20N, and A738N) is partially defective in stimulated turnover, with phenotypes ranging from 40 to 60% of the normal activation. Interestingly, A738 corresponds to the residue mutated in a Sushi allele (A738T) that was shown to rescue function of a dynamin GTPase domain mutant in vivo (Narayanan et al., 2005). The small increase in basal GTPase activities observed for some mutants in this preliminary screen were not reproducible and may reflect small amounts of aggregated species present in individual preps.
Figure 4.
Basal and assembly-stimulated GTPase activities of interface mutants in vitro. Normalized activity for each mutant is expressed as a percentage of wild-type activity. All mutations were assayed in the context of full-length dynamin at a concentration of 0.5 μM in buffer containing 20 mM HEPES, pH 7.5, 150 mM KCl, 1 mM MgCl2, and 500 μM GTP. Activity was measured on the soluble fraction of each sample. The data presented here derive from an initial screen aimed at identifying potentially interesting interface mutant phenotypes.
Class I Interface Mutations Destabilize Dynamin Structure
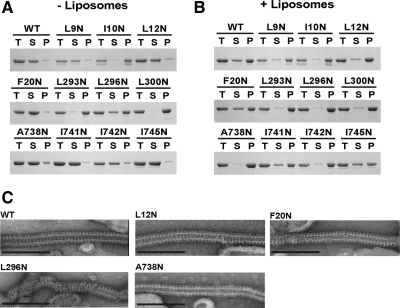
Because the GTPase-GED interface is primarily hydrophobic in nature, it is possible that mutations in this region could have significant effects on the structural integrity and proper folding of dynamin. We analyzed the interface mutants by sedimentation and EM to determine if the observed phenotypes were related to structural irregularities (Figure 5). In the absence of liposomes, wild-type dynamin exists as tetramers in solution that remain in the soluble fraction after sedimentation (Figure 5A). Addition of PIP2-containing liposomes shifts the protein to the pellet due to self-assembly on the charged membrane template (Figure 5B). Under these conditions, wild-type dynamin forms large, decorated tubes that are visible by negative-stain EM (Figure 5C). Analysis of the interface mutants shows that the class I mutations pellet even in the absence of liposomes (Figure 5A). When visualized by EM, these proteins form large, amorphous aggregates on the grid (data not shown). Only L296N retains the capacity for self-assembly and tubulation, which likely reflects the small amount of the protein that is still soluble and unassembled in the absence of liposomes (Figure 5, A and B). The length and morphology of these L296N decorated tubes differ significantly from wild-type dynamin, as evidenced by the incomplete pattern of decoration. On the basis of these observations, we conclude that the class I interface mutants (I10N, L293N, L296N, and L300N) destabilize dynamin structure and that the observed defects in stimulated GTPase activity result from aggregation of the protein and the inability to self-assemble properly. These structural defects could also explain the basal hydrolysis defects observed in L293N and L300N.
Figure 5.
Self-assembly properties of interface mutants. (A) Sedimentation of interface mutants (1 μM) in the absence of liposomes. After sedimentation, supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE. T, total amount of protein in the initial mixture. (B) Sedimentation of interface mutants (1 μM) in the presence of 0.1-μm PIP2-containing liposomes (300 μM). (C) Representative electron micrographs of interface mutants assembled on 0.4-μm PIP2-containing liposomes. Scale bar, 200 nm.
Class II Interface Mutations Uncouple Stimulated GTPase Activity from Dynamin Assembly
In contrast to the class I mutants, the class II mutants (L12N, F20N, and A738N) remain in the soluble fraction when subjected to centrifugation in the absence of liposomes (Figure 5A). When incubated with PIP2-containing liposomes, these proteins sediment and assemble normally, forming decorated tubes that are indistinguishable from wild-type dynamin (Figure 5, B and C). At this low resolution, the packing along these tubes is apparently unaltered by changes in the GTPase-GED interface, indicating that these proteins are competent for self-assembly. The specificity of these biochemical phenotypes implies that the structural integrity of these proteins is preserved and that these mutations uncouple stimulated GTPase activity from dynamin assembly.
To investigate this further, we determined the kinetic parameters (kcat and Km) of the basal and assembly stimulated GTPase activities of the class II interface mutants. These studies were performed on several independently purified batches of WT and mutant dynamin to confirm the reproducibility of our findings. Although all three mutants exhibit robust basal rates of GTP hydrolysis (Table 1), the L12N and A738N mutants are reduced relative to WT. Importantly, the Km for basal hydrolysis, which is a close approximation of GTP-binding affinity (Table 1), is unaffected, further confirming that the GTPase domains of these mutants are properly folded. The stimulated GTPase activity was diminished in each mutant (Table 1), although we observe significant batch-to-batch variation under these conditions. Though the measured Km values for stimulated activity are a less accurate representation of binding affinity because of variable and more rapid rates of GTP hydrolysis, the mutants still do not appear to disrupt GTP binding relative to WT. Together these data suggest that the GG interface plays a more selective role in modulating assembly-stimulated GTPase activity in addition to its critical function in stabilizing dynamin structure.
Table 1.
Michaelis-Menten kinetic parameters for basal and assembly- stimulated GTPase activities of wildtype dynamin and GTPase-GED interface mutants
| Dynamin | Basal |
Lipid stimulated |
||
|---|---|---|---|---|
| kcat (min−1) | Km (μM) | kcat (min−1) | Km (μM) | |
| WT | 1.54 ± 0.13 | 65.43 ± 1.64 | 179.8 ± 46.7 | 153.58 ± 55.7 |
| L12N | 0.93 ± 0.21 | 37.72 ± 7.99 | 92.24 ± 34.6 | 75.97 ± 28.4 |
| F20N | 1.55 ± 0.15 | 51.18 ± 3.91 | 121.58 ± 30.5 | 97.12 ± 18.6 |
| A738N | 0.72 ± 0. 4 | 19.48 ± 1.20 | 107.74 ± 18.6 | 78.84 ± 24.6 |
These data represent the average of at least three independent experiments with multiple independently purified batches of protein.
Class II Interface Mutants Reduce Dynamin-Catalyzed Membrane Fission In Vitro
Dynamin's role in CME depends on its ability to catalyze membrane fission, which has recently been shown to require localized curvature imposed by the assembly of GTPase-limited, short dynamin collars (Pucadyil and Schmid, 2008; Bashkirov et al., 2008). Longer dynamin assemblies that can only assemble in the absence of GTP were shown to stabilize underlying tubular membranes and were unable to mediate membrane fission. Thus, dynamin-catalyzed fission critically depends on GTPase-regulated cycles of dynamin assembly and disassembly (Ramachandran and Schmid, 2008). In this context, we anticipated that the assembly-stimulated defects exhibited by class II mutants would translate into a reduction in membrane fission by dynamin. To test this prediction, we used a recently developed assay that measures vesicle formation from supported bilayers with excess membrane reservoir (SUPER templates; Pucadyil and Schmid, 2008) and examined the ability of the L12N, F20N, and A738N mutants to catalyze membrane fission. SUPER templates containing fluorescent lipid were mixed with dynamin in the presence or absence of GTP and then centrifuged at low speed. Fission was detected as an increase in the fluorescence intensity of the supernatant resulting from the liberation of lipid vesicles (Figure 6). As can be seen, L12N is significantly impaired in its ability to catalyze membrane fission and vesicle release, whereas A738N only shows a partial defect and F20N appears to function as effectively as WT. Unexpectedly, the degree to which each mutant can facilitate fission in vitro does not fully correlate with the severity of its assembly-stimulated defect, suggesting that other aspects of dynamin function not reflected in its in vitro GTPase activities but differentially dependent on the GTPase-GED interface are important for dynamin-catalyzed fission.
Figure 6.
Membrane fission by class II interface mutants in vitro. Fluorescent SUPER templates were prepared as described in Materials and Methods. ■, fission by 0.5 μM dynamin in the absence of nucleotide; □, fission in the presence of 1 mM GTP.
Class II Interface Mutants Affect the Late Stages of Endocytosis In Vivo
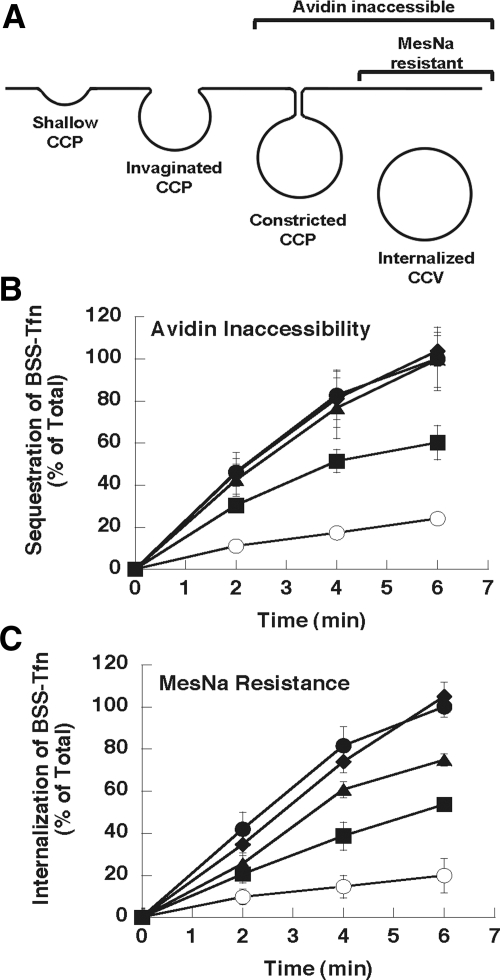
Because L12N reduced stimulated activity and fission in vitro, we wanted to know if this change at the GTPase-GED interface also adversely affects CME in vivo. To examine this possibility, we performed experiments in Dyn2 KO cells reconstituted with GFP-tagged WT, L12N, F20N, or A738N Dyn2 proteins. Stable cell lines expressing these mutants at levels equivalent to endogenous Dyn2 were generated, and then endogenous Dyn2 was excised by introduction of Cre recombinase (Liu et al., 2008). We chose this system to avoid the potential of stronger and/or nonspecific defects caused by overexpression of these mutants in the presence of endogenous Dyn2. The uptake of biotinylated-Tfn (BSS-Tfn) was determined either by inaccessibility to avidin, a large bulky probe, or to MesNa, a small, membrane-impermeant reducing agent. Sequestration of BSS-Tfn from avidin assesses the formation of both constricted coated pits and coated vesicles, whereas MesNa resistance is only acquired after BSS-Tfn has been internalized into sealed coated vesicles (Figure 7A). We were unable to detect an in vivo defect in Dyn2 KO cells reconstituted with the A738N mutant. Although not correlating with impaired GTPase and fission activities of this mutant, this finding was consistent with the fact that mutation of this residue can restore function of the GTP-binding defective, shits2 dynamin mutant. In contrast, the in vivo phenotype of the L12N mutation correlated well with its in vitro properties. L12N partially blocked uptake into both the avidin- and MesNa-inaccessible compartments; however, there was a greater degree of inhibition when assessed by MesNa resistance than avidin inaccessibility (Figure 7, B and C). These data indicate that L12N partially impairs the later stages of endocytosis, leading to an accumulation of BSS-Tfn in constricted coated pits that are inaccessible to avidin but remain accessible to MesNa as would be predicted from its in vitro defects. We also detect a slight inhibition of uptake for F20N in the MesNa assay (Figure 7C), although not in the avidin assay (Figure 7B). These phenotypes are consistent with a late effect on CME, resulting from reduction of either the rate or efficiency of membrane fission.
Figure 7.
Class II interface mutants disrupt clathrin-mediated endocytosis in vivo. (A) Diagram of endocytosis intermediates and their accessibility to the avidin and MesNa probes. (B and C) Dynamin-2 knockout (Dyn2 KO) cells expressing either GFP (○) or the following GFP-tagged Dyn2 constructs: WT (•), L12N (■), F20N (▴), or A738N (♦) were generated as described in Materials and Methods. Endocytosis in these cells was quantified using biotinylated transferrin (BSS-Tfn) as the ligand and measuring its uptake into an avidin- (B) or MesNa- (C) inaccessible compartment. The amount of uptake is expressed as a percentage of the total surface-bound BSS-Tfn.
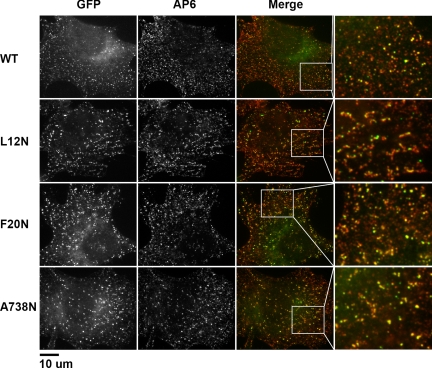
To gain further insight into the stage at which these mutations impaired CME, we inspected the clathrin-coated pit (CCP) intermediates in our reconstituted Dyn2 KO cells using indirect immunofluorescence (Figure 8). Permeabilized cells expressing GFP-tagged Dyn2 constructs (WT, L12N, F20N, and A738N) were fixed and stained with the anti-ΑP2 antibody AP6. We found there to be no detectable difference in the number of coated pits between the class II interface mutants and WT Dyn2; however, short, tubule-like formations with associated dynamin puncta and decorated with AP2 were present in cells reconstituted with L12N (Figure 8, enlarged boxes). Although the resolution of these experiments precludes a detailed description of these structures, the pattern of staining indicates that AP2 can bind along the length of these tubules. The accumulation of tubular structures in L12N is consistent with a defect in fission and correlates with its stronger inhibition of Tfn uptake in the MesNa assay.
Figure 8.
L12N alters the morphology of clathrin-coated pits in reconstituted Dyn2 KO cells. Clathrin-coated pits were visualized by indirect immunofluorescence using the AP6 mAb directed against the α-adaptins of AP2. Dynamin-2 constructs (WT, L12N, F20N, or A738N) were localized using GFP fluorescence. White boxes in each merged image indicate sections magnified in the panels on the right.
To further explore the possibility that these structures represent delayed fission events we examined the endocytic intermediates in these cells by high-resolution EM. Plastic-embedded thin sections of reconstituted Dyn2 KO cells were imaged and the CCP intermediates for each mutant were counted and scored according to their morphology as either shallow, invaginated, or constricted (Figure 9). Invaginated pits are visibly connected to the membrane in a single thin section and thus are assumed to correspond to avidin-accessible structures (Figure 9A). Given that uncoating is rapid and that endocytic vesicles rapidly translocate away from the cell surface, coated vesicle structures closely apposed to the plasma membrane are assumed to represent constricted intermediates, whose narrow necks are not captured in these individual thin sections (Figure 9A). This classification has been used and validated extensively in past studies, under conditions in which CME is known to be inhibited (Schmid and Carter, 1990; Sandvig et al., 1987).
Figure 9.
L12N increases the number of late endocytic intermediates in reconstituted Dyn2 KO cells. (A) Thin sections of Dyn2 KO cells reconstituted with GFP-tagged Dyn2 proteins (WT, L12N, and F20N and A738N, as indicated) were prepared and imaged as described in Materials and Methods. Scale bar, 200 nm. Endocytic intermediates were counted and scored as shallow (S), invaginated (I), or constricted (C) CCPs as indicated. (B) Quantitation of CCP intermediates expressed as % of total CCPs scored (n = 79, 90, 87, and 61 for WT, L12N, F20N, and A738N, respectively). **p < 0.01 in t test.
Based on this quantification, we observe a statistically significant increase in the number of constricted CCPs in L12N versus WT (Figure 9B). This increase in late intermediates is seen at the expense of the invaginated and shallow CCPs, suggesting that L12N specifically hinders the late stages of clathrin-mediated endocytosis by delaying dynamin-dependent fission. Such a delay would impede the formation of coated vesicles, consistent with the stronger inhibition of Tfn uptake by L12N as detected by MesNa resistance. F20N also showed a slight inhibition of endocytosis in the MesNa assay, but we did not detect any significant change in the distribution of CME intermediates for this mutant by our EM analysis (Figure 9B). Similarly, we detect no increase in late CME intermediates in cells expressing A738N, which mirrors the WT behavior of this mutant in the Tfn uptake assays. Coated vesicle-like structures accumulated adjacent to the plasma membrane only in cells expressing the L12N mutant. That this mutant shows the greatest defect in our in vitro membrane fission assay and the strongest defect in vivo supports our assumption that these structures are indeed constricted CCPs and not detached CCVs. Together these data suggest that the GG interface is critical for coordinating dynamin activity at late stages of CCV formation.
DISCUSSION
We have used a combination of functional, structural, and mutant analyses to define the GTPase-GED interface in dynamin. We show that this region is comprised of the N- and C-termini of the GTPase domain and the C-terminus of GED, which we speculate form a three-helix bundle on the back of the GTPase domain to bury a large number of conserved hydrophobic residues. Although previous yeast two-hybrid analysis established that GED interacts with the CGTPase (Smirnova et al., 1999), our chemical cross-linking results represent the first direct evidence of a physical interaction between GED and the NGTPase.
Our findings demonstrate that this GED docking is essential for the structural integrity of dynamin. Major perturbations to the GG interface, as demonstrated by the class I mutants, destabilize the protein and cause aggregation. This is further underscored by the folding of our minimal GG fusion. When only the GTPase domain is expressed, the hydrophobic interactions at the GTPase-GED interface are not satisfied and the construct precipitates. By tethering the C-terminus of GED to the GTPase domain, we overcome this obstacle and salvage solubility. This requirement for CGED-GTPase interactions is likely conserved throughout the dynamin family given the high degree of sequence similarity at this interface. Moreover, a six-amino acid C-terminal deletion of the GED in Mx strongly inhibits its GTPase activity (Schwemmle et al., 1995). Whereas the GTPase-GED interactions are clearly intrapolypeptide in our monomeric GG construct, we cannot rule our interpolypeptide interactions in the context of the full-length tetrameric dynamin.
GED docking also plays an important role in regulating dynamin's functional properties. The L12N mutation at the GTPase-GED interface selectively impairs dynamin's assembly-stimulated turnover and fission activity in vitro without disrupting dynamin structure. This distinguishes it from mutations such as I690K, K694A, and R725A, whose primary defect is in self-assembly, leading secondarily to a defect in assembly-stimulated GTPase activity (Sever et al., 2000; Marks et al., 2001; Song et al., 2004). The reduced activities associated with L12N shift the kinetics of endocytosis in vivo such that fission is now rate limiting, as evidenced by the accumulation of late intermediates and a more severe defect in internalization than in constriction. This behavior is unique among inhibitory dynamin mutants, as all previously identified dominant-negative dynamin mutants inhibit Tfn uptake into avidin-inaccessible and MesNa-resistant coated pits equally, suggesting that they block the earlier stages of CCV formation. Such substitutions lead to the accumulation of deeply invaginated CCPs that nonetheless remain biochemically accessible to both avidin and MesNa (van der Bliek et al., 1993; Damke et al., 1994; Damke et al., 2001, Song et al., 2004). L12N thus represents a new class of hypomorphic dynamin mutants.
It is important to note that the in vitro phenotypes of the class II interface mutants do not strictly correlate with their in vivo effects. In particular, the biochemical properties of the A738N mutant are similar to those of L12N, but only the L12N mutant impairs CME in vivo. The ability of A738N to support CME is consistent with the ability of the second site A738T Sushi mutation to restore function to a dynamin mutant defective in GTP binding (Narayanan et al., 2005). Moreover, this illustrates the underlying complexity of dynamin activation. The GTPase-GED interface is on the back side of the GTPase domain relative to its active site, yet it can directly modulate assembly-stimulated GTP hydrolysis. Importantly, the class II interface mutants only moderately affect this activity, suggesting that other mechanisms besides GED docking must also be involved in precipitating the 100-fold stimulation of GTP hydrolysis upon self-assembly.
Taken together, our results suggest that the NGTPase, CGTPase, and CGED form an intramolecular signaling module, which we term the bundle signaling element (BSE), that can sense and transmit the conformational changes associated with dynamin assembly to the GTPase domain. This activity indirectly promotes stimulated GTP hydrolysis, possibly by triggering further conformational changes in the GTPase domain. Although dynamin activation through the BSE occurs in a back-to-front manner, it is feasible that this module can also function in the opposite direction to facilitate communication between the GTPase domain and the membrane surface. Real-time fluorescence experiments have shown that conformational changes in the nucleotide-binding pocket are coupled to membrane binding through dynamin's pleckstrin homology (PH) domain despite a large separation of these two regions (Ramachandran and Schmid, 2008). A conformational coupling between the GTPase and PH domains has also been detected by tryptophan fluorescence measurements (Solomaha and Palfrey, 2005). The BSE could relay this information from the GTPase domain, thus providing a general mechanism for coordinating membrane binding, dynamin assembly, stimulated GTP hydrolysis, and the subsequent disassembly of the polymer. High-resolution structural studies will be necessary to elucidate the details of this intramolecular communication.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vasyl Lukiyanchuk for assistance in cloning and purification, Malcolm Wood for preparation of thin sections for EM analysis, Sunia Trauger for the mass spectrometry analysis, and Petra Verdino for insightful discussions. This work was supported by National Institutes of Health Grants GM042455 and MH061345 to S.L.S. J.S.C. was supported by a Ruth Kirschstein individual predoctoral fellowship from the National Institute of Mental Health Grant MH081419 and by the American Heart Association predoctoral fellowship (AHA-0515009Y. Y.-W.L. was supported by a fellowship from the National Science Council of Taiwan and from the Muscular Dystrophy Association (MDA-114824). A portion of the electron microscopy imaging was carried out at the National Resource for Automated Microscopy, which is supported by the NIH through the National Center for Research Resources' P41 program (RR17573).
Abbreviations used:
- Dyn 2
dynamin 2
- BSS-Tfn
biotinylated-transferrin
- GED
GTPase effector domain
- GG
minimal GTPase-GED fusion
- MTS
methanthiosulfonate
- PIP2
l-α-phosphatidylinositol-4,5-bisphosphate.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0318) on June 10, 2009.
REFERENCES
- Barylko B., Binns D., Lin K. M., Atkinson M. A., Jameson D. M., Yin H. L., Albanesi J. P. Synergistic activation of dynamin GTPase by Grb2 and phosphoinositides. J. Biol. Chem. 1998;273:3791–3797. doi: 10.1074/jbc.273.6.3791. [DOI] [PubMed] [Google Scholar]
- Bashkirov P. V., Akimov S. A., Evseev A. I., Schmid S. L., Zimmerberg J., Frolov V. A. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. J., Zhang P., Egelman E. H., Hinshaw J. E. The stalk region of dynamin drives the constriction of dynamin tubes. Nat. Struct. Mol. Biol. 2004;11:574–575. doi: 10.1038/nsmb762. [DOI] [PubMed] [Google Scholar]
- Chugh J., Chatterjee A., Kumar A., Mishra R. K., Mittal R., Hosur R. V. Structural characterization of the large soluble oligomers of the GTPase effector domain of dynamin. FEBS J. 2006;273:388–397. doi: 10.1111/j.1742-4658.2005.05072.x. [DOI] [PubMed] [Google Scholar]
- Conner S. D., Schmid S. L. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Dalmas O., Orelle C., Foucher A. E., Geourjon C., Crouzy S., Di Pietro A., Jault J. M. The Q-loop disengages from the first intracellular loop during the catalytic cycle of the multidrug ABC transporter BmrA. J. Biol. Chem. 2005;280:36857–36864. doi: 10.1074/jbc.M503266200. [DOI] [PubMed] [Google Scholar]
- Damke H., Baba T., Warnock D. E., Schmid S. L. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H., Binns D. D., Ueda H., Schmid S. L., Baba T. Dynamin GTPase domain mutants block endocytic vesicle formation at morphologically distinct stages. Mol. Biol. Cell. 2001;12:2578–2589. doi: 10.1091/mbc.12.9.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J. E. Dynamin and its role in membrane fission. Annu. Rev. Cell Dev. Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon G. L., Bruice T. W. Novel sulfhydryl reagents. Methods Enzymol. 1977;47:407–430. doi: 10.1016/0076-6879(77)47042-3. [DOI] [PubMed] [Google Scholar]
- Leonard M., Song B. D., Ramachandran R., Schimd S. L. Robust colorimetric assays for dynamin's basal and stimulated GTPase activities. Methods Enzymol. 2005;404:490–503. doi: 10.1016/S0076-6879(05)04043-7. [DOI] [PubMed] [Google Scholar]
- Liu Y. W., Surka M. C., Schroeter T., Lukiyanchuk V., Schmid S. L. Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells. Mol. Biol. Cell. 2008;19:5347–5359. doi: 10.1091/mbc.E08-08-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo T. W., Clarke D. M. Determining the dimensions of the drug-binding domain of human P-glycoprotein using thiol cross-linking compounds as molecular rulers. J. Biol. Chem. 2001;276:36877–36880. doi: 10.1074/jbc.C100467200. [DOI] [PubMed] [Google Scholar]
- Marks B., Stowell M.H.B., Vallis Y., Mills I. G., Gibson A., Hopkins C. R., McMahon H. T. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- Mears J. A., Ray P., Hinshaw J. E. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–1202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan R., Leonard M., Song B. D., Schmid S. L., Ramaswami M. An internal GAP domain negatively regulates presynaptic dynamin in vivo: a two-step model for dynamin function. J. Cell Biol. 2005;169:117–126. doi: 10.1083/jcb.200502042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann H. H., Knetsch M.L.W., Scherer A., Manstein D. J., Kull F. J. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 2001;20:5813–5821. doi: 10.1093/emboj/20.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C. S., et al. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77:153–161. doi: 10.1016/s0304-3991(99)00043-1. [DOI] [PubMed] [Google Scholar]
- Pucadyil T., Schmid S. L. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke G. J., McMahon H. T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Ramachandran R., Surka M., Chappie J. S., Fowler D. M., Foss T. R., Song B. D., Schmid S. L. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R., Schmid S. L. Real-time detection reveals that effectors couple dynamin's GTP-dependent conformational changes to the membrane. EMBO J. 2008;27:27–37. doi: 10.1038/sj.emboj.7601961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold T. F., Eschenburg S., Becker A., Leonard M., Schmid S. L., Vallee R. B., Kull F. J., Manstein D. J. Crystal structure of the GTPase domain of rat dynamin1. Proc. Natl. Acad. Sci. USA. 2005;102:13093–13098. doi: 10.1073/pnas.0506491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittinger K., Walker P. A., Eccleston J. F., Nurmahomed K., Owen D., Laue E., Gamblin S. J., Smerdon S. J. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997a;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- Rittinger K., Walker P. A., Eccleston J. F., Smerdon S. J., Gamblin S. J. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997b;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- Sandvig K., Olsnes S., Petersen O. W., van Deurs B. Acidification of the cytosol inhibits endocytosis from coated pits. J. Cell Biol. 1987;105:679–689. doi: 10.1083/jcb.105.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein. A., Schmitz F., Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Schmid S. L., Carter L. L. ATP is required for receptor-mediated endocytosis in intact cells. J. Cell Biol. 1990;111:2307–2318. doi: 10.1083/jcb.111.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmle M., Richter M. F., Herrmann C., Nassar N., Staeheli P. Unexpected structural requirements for GTPase activity of the interferon-induced MxA protein. J. Biol. Chem. 1995;270:13518–13523. [PubMed] [Google Scholar]
- Seewald M. J., Körner C., Wittinghofer A., Vetter I. R. RanGAP mediates GTP hydrolysis without an arginine finger. Nature. 2002;415:662–666. doi: 10.1038/415662a. [DOI] [PubMed] [Google Scholar]
- Sever S., Muhlberg A. B., Schmid S. L. Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- Sever S., Damke H., Schmid S. L. Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J. Cell Biol. 2000;150:1137–1148. doi: 10.1083/jcb.150.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Shurland D. L., Newman-Smith E. D., Pishvaee B., van der Bliek A. M. A model for dynamin self-assembly based on binding between three different protein domains. J. Biol. Chem. 1999;274:14942–14947. doi: 10.1074/jbc.274.21.14942. [DOI] [PubMed] [Google Scholar]
- Solomaha E., Palfrey H. C. Conformational changes in dynamin on GTP binding and oligomerization reported by intrinsic and extrinsic fluorescence. Biochem. J. 2005;391:601–611. doi: 10.1042/BJ20050707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B. D., Schmid S. L. A molecular motor or a regulator? Dynamin's in a class of its own. Biochemistry. 2003;42:1369–1376. doi: 10.1021/bi027062h. [DOI] [PubMed] [Google Scholar]
- Song B. D., Yarar D., Schmid S. L. An assembly-incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo. Mol. Biol. Cell. 2004;15:2243–2252. doi: 10.1091/mbc.E04-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell M. H., Marks B., Wigge P., McMahon H. T. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat. Cell Biol. 1999;1:27–32. doi: 10.1038/8997. [DOI] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C. S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tesmer J.J.G., Berman D. M., Gilman A. G., Sprang S. R. Structure of RGS4 bound to AlF4-activated Giα1: stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- van der Bliek A. M., Redelmeier T. E., Damke H., Tisdale E. J., Meyerowitz E. M., Schmid S. L. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J. Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D., Waterman-Storer C. M., Schmid S. L. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol. Biol. Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Hinshaw J. E. Three-dimensional reconstruction of dynamin in the constricted state. Nat. Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.