Abstract
B7-H3, a surface immunomodulatory glycoprotein, inhibits natural killer cells and T cells. The monoclonal antibody 8H9 is specific for 4Ig-B7-H3, the long and principal form of B7-H3. Early results from radioimmunotherapy using 8H9 have shown promise in patients with metastatic solid tumors to the central nervous system. While B7-H3 transcript was ubiquitously expressed in a wide spectrum of human solid tumors as well as human normal tissues, B7-H3 protein was preferentially expressed only in tumor tissues. By quantitative RT-PCR, all 3 isoforms of microRNA miR-29 (a, b, c) were highly expressed in normal tissues. However, they were down-regulated in a broad spectrum of solid tumors, including neuroblastoma, sarcomas, brain tumors, and tumor cell lines. B7-H3 protein expression was correlated inversely with miR-29 levels in both cell lines and tumor tissues tested. Using luciferase reporter assay, miR-29a was shown to directly target B7-H3 3′UTR, and knock-in and knock-down of miR-29a led to down-regulation and up-regulation, respectively, of B7-H3 protein expression. The ability of miR-29 to control B7-H3 protein expression has implications in immune escape by solid tumors. Differential modulation of this key immunoinhibitory molecule in tumor versus normal tissues may advance both cell mediated immunotherapy, as well as antibody-based targeted strategies using the B7-H3 specific monoclonal antibody 8H9.
Keywords: miR-29, B7-H3, 8H9, immunotherapy, solid tumors
Introduction
Human B7-H3 (also named as CD276) is a member of the B7/CD28 immunoglobulin superfamily which provides crucial costimulatory signals that regulate T cell functions in tumor surveillance, infections, and autoimmune diseases (1). B7-H3 was initially identified as a type I transmembrane protein with its extracellular region containing only one V-like and C-like Ig domain (2Ig-B7-H3) (2), similar to all other B7 family members. Subsequently, a second dominantly expressed form of human B7-H3 that contains tandemly duplicated V-like and C-like Ig domain (4Ig-B7-H3) was found (3, 4). The inhibitory role of B7-H3 was supported by the reports that both 2Ig and 4Ig forms of human B7-H3 inhibited T cell proliferation and cytokine production (3). B7-H3 preferentially down-regulated TH1-mediated immune response in B7-H3-deficient mice (5), and 4Ig-B7-H3 inhibited NK-mediated lysis of neuroblastoma cells by interacting with a putative inhibitory receptor on the surface of NK cells (6). In more recent studies of patients with prostate cancer, tumor B7-H3 expression was strongly correlated with disease spread at time of surgery, increased risk of clinical cancer recurrence and cancer-specific death (7, 8). Moreover, tumor B7-H3 expression was correlated with patient poor survival in both clear cell renal cell carcinoma and urothelial cell carcinoma (9, 10). Earlier reports have also implicated a positive immunologic function of B7-H3, e.g. human B7-H3 (2Ig form) promoted T cell activation and IFN-γ production by binding to a putative receptor on activated T cells (2). Furthermore, antitumor response was enhanced by B7-H3 expression in murine tumor models (11). B7-H3 positivity in gastric carcinoma was also correlated with increased survival (12). It is very possible that B7-H3 has both coinhibitory as well as costimulatory properties depending on the receptors (13).
MicroRNAs (miRNAs) represent a class of naturally occurring small noncoding RNAs which function as gene regulators. Mature miRNAs are ∼ 22 nucleotide (nt) molecules cleaved from ∼ 70-100 nt hairpin pre-miRNA precursors (14). Single-stranded miRNAs bind to the 3′UTR of target mRNAs primarily through base pairing of the “seed region” of the miRNA (nt 2-7) to the cognate target (15), thereby regulating gene expression post-transcriptionally by decreasing protein translation, increasing degradation of the target message, or both (16). There is increasing evidence that miRNAs play critical roles in tumorigenesis by functioning as oncogenes or tumor suppressors (17).
Our laboratory has developed monoclonal antibody (MoAb) 8H9 which targets a 58 kDa glycoprotein broadly expressed in human solid tumors, including embryonal tumors and carcinomas (18). It has shown favorable tumor uptake in xenograft models. In early phase human clinical trials, it appears to prolong survival among high risk patients with solid tumors suffering from central nervous system (CNS) metastasis (19, 20). In this report, we described the identification of 4Ig-B7-H3 as the target for MoAb 8H9. We further investigated B7-H3 expression at the mRNA and protein level for both human tumors and normal tissues. miRNA miR-29 was found to regulate B7-H3 protein expression with potential implications for immune-based therapy of human solid tumors.
Materials and Methods
Cell Culture and Human Tissues
Human neuroblastoma cell line LAN-1 was provided by Dr. Robert Seeger (Children's Hospital of Los Angeles, Los Angeles, CA), and NB1691 by Dr. Peter Houghton (St. Jude Children's Research Hospital, Memphis, TN). Human Burkitt's lymphoma cell line Daudi and adenocarcinoma cell line HeLa were purchased from American Type Culture Collection (Bethesda, MD). All cell lines were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2mM glutamine, 100 U/ml penicillin, and 100 ug/ml streptomycin at 37°C in a 5% CO2 incubator. Normal tissues as well as solid tumor samples of different histological types obtained at Memorial Sloan-Kettering Cancer Center (MSKCC) were snap frozen in liquid nitrogen. Written informed consent was obtained from the patients and/or their guardians in accordance to the guidelines of the institutional review board of MSKCC.
Monoclonal Antibodies
MoAbs 8H9 (murine IgG1) and 3E7 (murine IgG2b specific for L1-CAM) were produced against human neuroblastoma in our laboratory. They were purified by protein A (GE Healthcare, Piscataway, NJ) affinity chromatography before use. MAB1027 (anti-B7-H3 MoAb) was purchased from R&D System, Minneapolis, MN.
Whole Cell Lysates and Western blot
8H9-positive cell line LAN-1 and 8H9-negative cell line Daudi were grown to ∼ 80% confluence. Cells were harvested using 2 mM EDTA and washed with ice-cold PBS. Cells were lysed on ice (20 min) in Triton Lysis Buffer (50 mM Tris-HCl, pH 7.2, 50 mM NaCl, 10% glycerol, 1% Triton X-100, and protease inhibitor cocktail tablets). The lysates were clarified by centrifugation at 14,000 rpm for 20 min at 4°C. ∼25-50 ug whole cell lysates were analyzed by SDS-PAGE under nonreducing condition using Tris-Glycine Ready Gel System (Bio-Rad, Hercules, CA). After electrophoresis, samples were transferred onto Immun-Blot PVDF Membrane (Bio-Rad), blocked for one hour at room temperature (RT) with 10% dry milk in Tris-Buffered Saline Tween-20 (TBST), and incubated with primary antibodies (8H9 at 5-20 ug/ml, and MAB1027 at 5 ug/ml) for 3 hours at RT. The membrane was then washed with TBST, and incubated with secondary peroxidase-conjugated AffiniPure Goat Anti-Mouse IgG (H+L) (Jackson ImmunoResearch, West Grove, PA). Bands were detected with SuperSignal West Pico Chemiluminescent Substrate (PIERCE, Rockford, IL).
Subcellular Fractionation
For crude membrane preparation, LAN-1 cells were pipetted off the tissue culture dish, washed with ice-cold PBS, and lysed on ice in sucrose buffer (0.25 M sucrose, 5 mM Tris-HCl, pH 7.2, and protease inhibitor cocktail tablets) with a Dounce homogenizer (Kontes, Vineland, NJ). Upon centrifugation for 10 min at 1,000 g to pellet all nuclei, the supernatant was then ultracentrifuged at 100,000 g for 30 min in a Beckman L-70K (25,000 rpm, SW41Ti rotor) to separate the membrane particulate from the cytosolic fraction. The cytosolic fraction was adjusted to 1% Triton, while crude nuclear and membrane fractions were resuspended in Triton Lysis Buffer and clarified before use.
8H9 Antigen Affinity Purification
8H9 antigen was purified from LAN-1 cell extracts by immuno-affinity chromatography using MoAb 8H9. The 8H9 affinity column was prepared by covalently conjugating Fc portion of 8H9 to protein G on the gel matrix using Protein G IgG Plus Orientation Kit (PIERCE) according to the manufacturer's instructions. 4 mg of LAN-1 whole cell lysates or equivalent membrane fraction prepared as described above were incubated overnight at 4°C with 20 ul 8H9-protein G Sepharose (3 mg bound 8H9/ml beads). After extensive washing with Triton Lysis Buffer, the column was eluted sequentially with 50 mM Tris-HCl, pH 7.2 containing 1M NaCl (E-NaCl), 0.1 M Glycine-HCl, pH 2.8 (E-2.8) and pH 2.0 (E-2.0), SDS Sample Buffer (E-SDS: 62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.005% Bromophenol blue), and SDS Sample Buffer plus boiling in water for 5 min (E-SDS-B). A small aliquot of eluates was monitored for the presence of 8H9 antigen by Western blot using 8H9 antibody. 25% of the eluate was also analyzed by silver staining (SilverQuest Silver Staining Kit, Invitrogen). Finally, 50% of the 8H9 antigen-positive eluate (E-2.0 fraction) was analyzed by colloidal Coomassie blue staining (GelCode Blue Stain Reagent, PIERCE), and the 8H9 antigen-positive band was collected and used for mass spectrometric identification by the Microchemistry and Proteomics Core Facility at MSKCC.
Detection of B7-H3 mRNA by quantitative (q)RT-PCR
Total RNA from normal tissues, as well as solid tumors detailed in Fig. 3 were isolated using Trizol Reagent (Invitrogen) according to the manufacturer's instructions. One ul of cDNA synthesized from 1 ug of total RNA was used for real-time qPCR using Applied Biosystems (ABI) Sequence Detection System 7300 (Foster City, CA). B7-H3 (CD276) gene expression assay reagent (Hs00228846_m1) as well as 2 endogenous controls: hypoxanthine phosphoribosyltransferase 1 (HPRT1, 4326321E) and succinate dehydrogenase complex, subunit A, flavoprotein (Fp) (SDHA, Hs00188166_ml) were purchased from ABI. Each sample was quantified using the comparative CT method (ABI) as a relative fold-difference compared to peripheral blood mononuclear cells (PBMC).
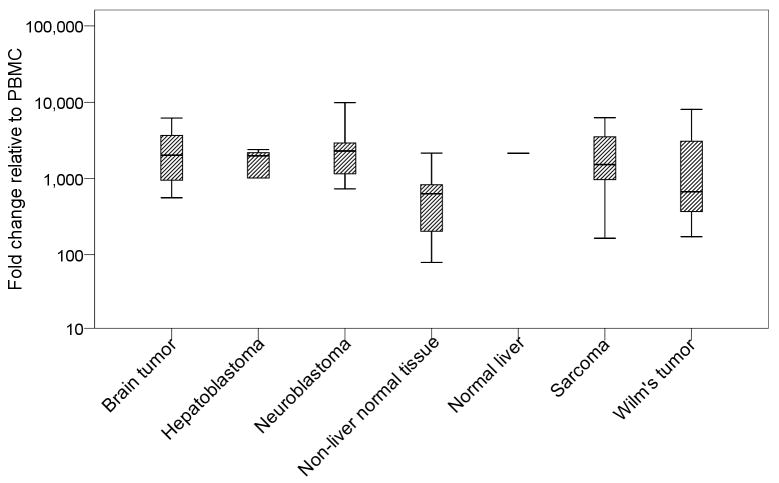
Fig. 3. B7-H3 transcript was ubiquitously expressed in solid tumors and normal human tissues.
mRNA levels were measured by qRT-PCR. Geometrical mean of HPRT1 and SDHA transcript levels served as the endogenous control, and expression levels in log scale were fold change relative to peripheral blood mononuclear cells (PBMC); number of samples for each histological type detailed in Results, and normal liver tissue (8H9 positive) was singled out for direct comparison.
Quantitation of miR-29
miR-29a, miR-29b, miR-29c, as well as the endogenous control RNU48 were purchased from ABI. A two-step RT-PCR with reverse transcription using a miRNA-specific primer, followed by qPCR with TaqMan probes was carried out according to the manufacturer's instructions.
Protein expression by immunofluorescence and flow cytometry
These procedures were previously described (18). The blocking experiment was carried out by incubating 0 ∼ 10 ug of recombinant human 4Ig-B7-H3 (R&D System) with 1 ug 8H9 or 0.1 ug 3E7 (as negative control) for 30 min at room temperature, before mixing with 106 M14 melanoma cells for immunofluorescence staining and flow cytometry analyses.
Luciferase Reporter Assay
Oligonucleotides corresponding to the miR-29a binding site in the B7-H3 3′UTR or a single-base mutant (illustrated in Fig 6A) were synthesized (Integrated DNA Technologies, Coralville, IA) and inserted into the XbaI site immediately downstream from the stop codon of firefly luciferase of the pGL3-control vector (Promega, Madison, WI). HeLa cells were cotransfected in 24-well plates using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol, with 50 ng of the firefly luciferase reporter, 1 ng of the renilla luciferase reporter (pRL-CMV vector, Promega) as transfection control, and 100 nM (final) miRNA mimics (ThermoFisher Scientific, Lafayette, CO). Firefly and renilla luciferase activities were measured sequentially using dual-luciferase assays (Promega) 24 hrs after the transfection. The experiments were performed in triplicate.
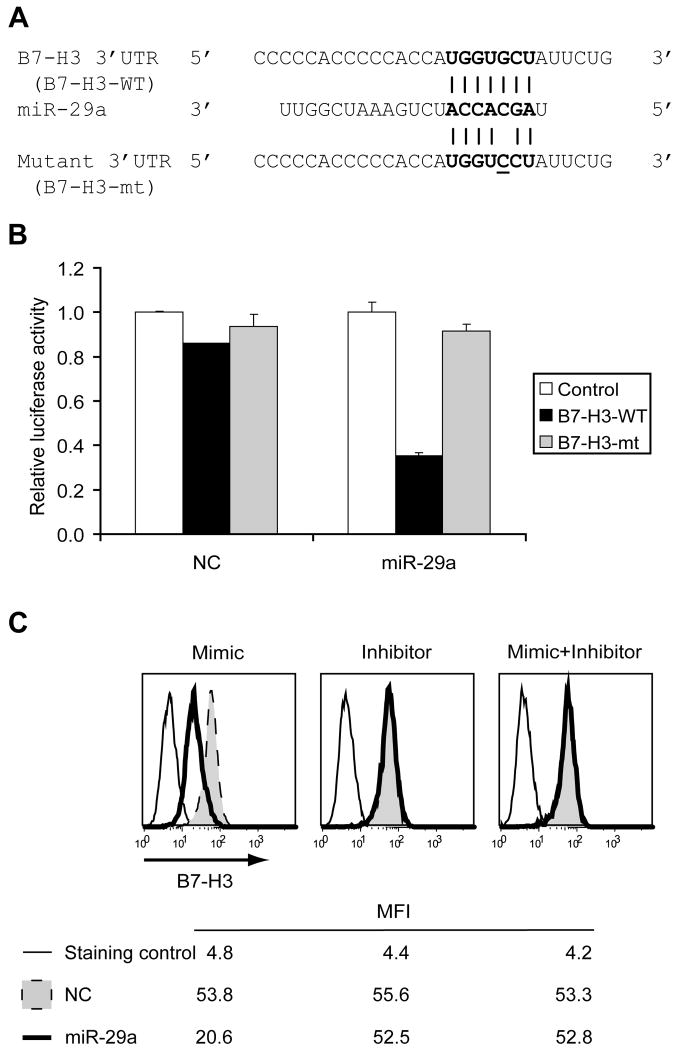
Fig. 6. miR-29a directly targets B7-H3 3′UTR and downregulates B7-H3 protein expression.
(A) Alignment of miR-29a with the target site derived from B7-H3 3′UTR. Note the seed complementarity at the 5′ end of miR-29a (bolded). A single-base mutant (underlined) was also synthesized. (B) Luciferase activity in HeLa cells transiently cotransfected with firefly luciferase constructs and miRNA mimics. Luciferase vectors were parental (Control), with the B7-H3 3′UTR insert (B7-H3-WT), or with the mutant insert (B7-H3-mt). miRNA mimics were miR-29a or NC (negative control). Luciferase values were normalized to Control independently for NC and miR-29a because each of these miRNAs differentially influenced the values of the renilla luciferase transfection control. Mean + SD. (C) B7-H3 expression in NB1691 cells transiently transfected with miRNA mimics, inhibitors, or mimics plus inhibitors. miRNAs were miR-29a or NC (negative control). Protein expression was measured by 8H9 immunofluorescence staining, and analyzed by FACS. Representative FACS histograms with mean fluorescence intensity (MFI) from three independent experiments were shown, and SD of the mean of the triplicates was < 5%. A non-specific MoAb TIB114 served as the staining control.
NB1691 Transient Transfection
NB1691 cells were transfected in 6-well plates using DharmaFECT reagent (ThermoFisher Scientific) according to the manufacturer's instructions, with 100 nM (final) miRNA mimics, inhibitors, or mimics plus inhibitors (ThermoFisher Scientific). 24 hrs after transfection, cells were treated with 1 mg/ml Pronase E (E. Merck, Darmstadt, Germany) for 30 min at 37°C to strip off B7-H3 protein already on the cell surface, and another 48 hrs later newly expressed B7-H3 protein level were measured by 8H9 immunofluorescence staining followed by flow cytometry analyses as previously described (18).
Results
Affinity purification of the antigen targeted by MoAb 8H9
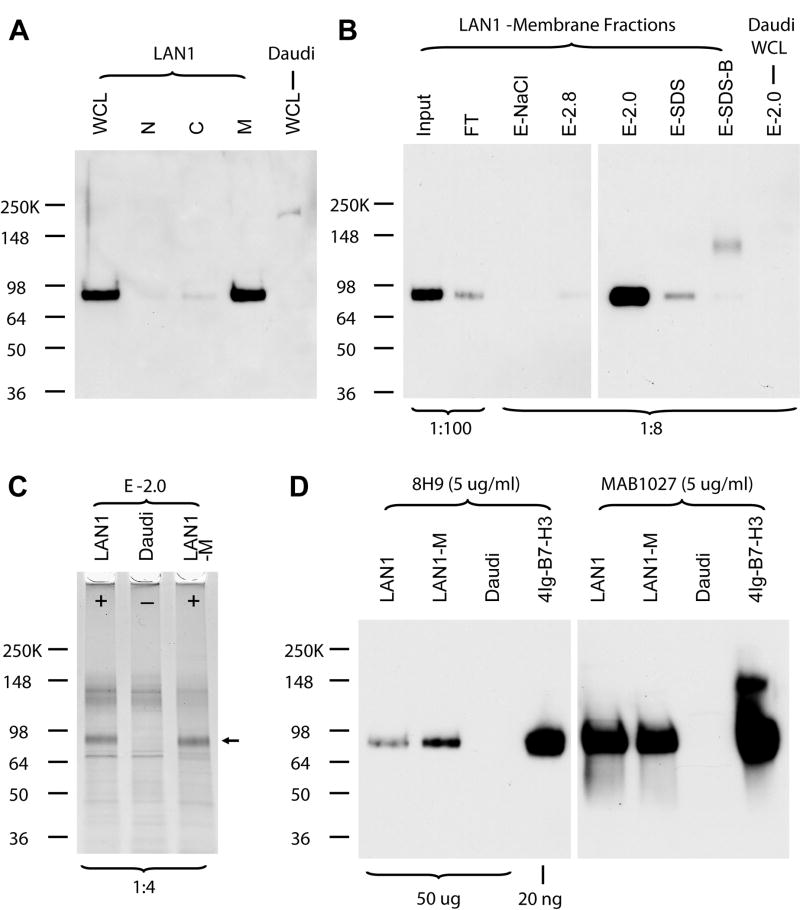
To monitor its affinity purification, the antigen was detected by Western blotting under nonreducing conditions using Tris-Glycine SDS-PAGE. Fig 1A showed a single band (∼ 90 KD) was detected in 8H9-positive cell line LAN-1, but not 8H9-negative cell line Daudi. Data was not shown for 2 other 8H9-positive cell lines HTB82 and U2OS, as well as control mIgG1 MoAb 5F9, which did not detect a band at the same size. Binding to 8H9 was lost under reducing conditions (data not shown), suggesting 8H9 recognizes a conformation-sensitive epitope. After subcellular fractionation, this antigen was found to be predominantly in the membrane fraction (Fig 1A).
Fig. 1. Identification of 8H9 antigen as 4Ig-B7-H3.
(A) Western blot detection of 8H9 antigen using 8H9. The highly glycosylated 8H9 antigen migrated at ∼ 90 KD on 4-15% Tris-Glycine SDS PAGE under nonreducing conditions. LAN1 - 8H9 positive cell line; Daudi – 8H9 negative cell line; whole cell lysates (WCL), nuclei fraction (N), cytosolic fraction (C), and membrane fraction (M). The purification procedure was monitored by (B) 8H9 Western blot using Invitrogen SeeBlue Plus2 Pre-Stained Standard as the protein molecular weight marker and (C) silver staining. Fractions designation detailed in Methods. Amount of aliquots loaded (ratio to total amount) were marked at the bottom of the panels. (D) Confirmation of 8H9 antigen as 4Ig-B7-H3 by Western blot using both 8H9 and MAB1027.
To identify this antigen, protein affinity purification in combination of mass spectrometric analysis was utilized. After incubating either LAN-1 (and Daudi as negative control) whole cell lysates or LAN-1 membrane fraction with 8H9-protein G Sepharose overnight, a significant portion (> 50%) of 8H9 antigen was bound to the Sepharose (Fig 1B, Input vs. flowthrough (FT)). 8H9 antigen was eluted specifically and predominantly in 0.1 M Glycine-HCl, pH 2.0 (E-2.0) as monitored by Western blot analysis (Fig 1B), suggesting a very strong interaction between 8H9 antibody and its antigen. After silver staining the same eluate, a clear band (arrow position) was detected accordingly only in LAN-1 cell extracts but not in Daudi cell extracts (Fig 1C). ∼ 10 ng of the band (visible with colloidal Coomassie staining, data not shown) was collected and used for mass spectrometric identification.
8H9 Antigen identification
In-gel tryptic digested eluate was analyzed and mass spectrometric sequencing (MALDI-TOF-MS/MS) confirmed two peptides [P1: NPVLQQDAHSSVTITPQR and P2: SPTGAVEVQVPEDPVVALVGTDATLR]. These peptides unequivocally matched the 8H9-reactive molecule to 4Ig-B7-H3 (CD276, NCBI# 74757248), the long and principal form of B7-H3 in human tissues.
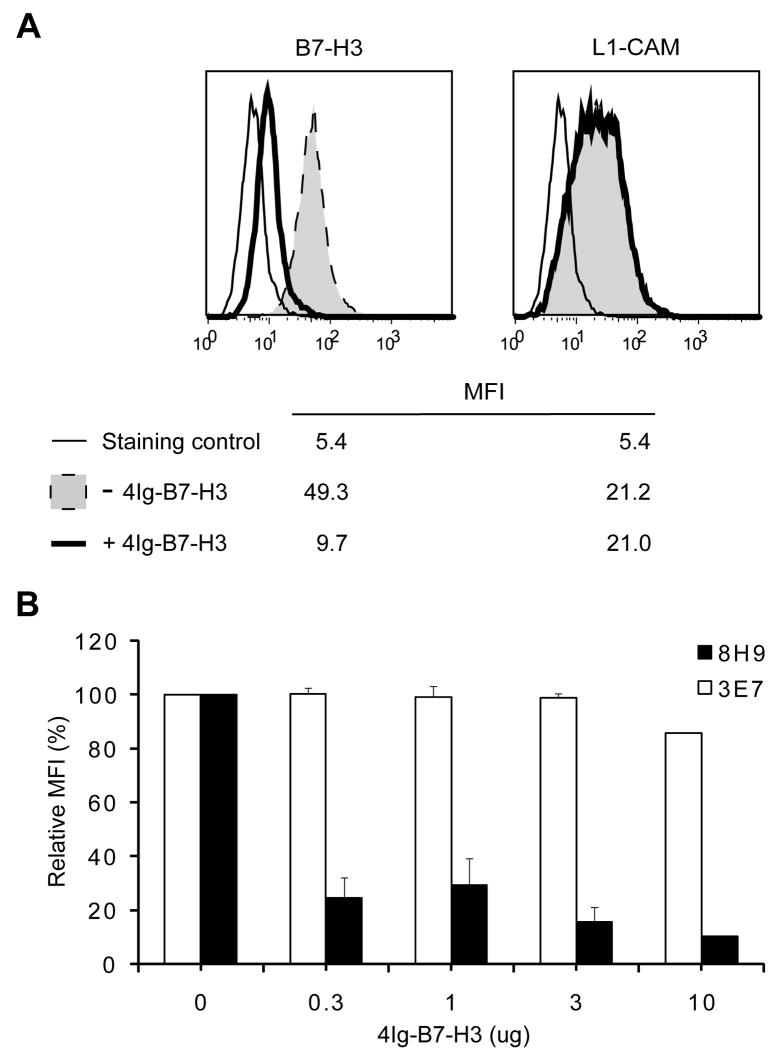
As further confirmation, both 8H9 and MAB1027 recognized a single band at ∼ 90 KD on Tris-Glycine SDS-PAGE under nonreducing conditions, when either 8H9-positive cell extracts or recombinant human 4Ig-B7-H3 protein was used (Fig 1D). This band migrated to ∼58 KD, the calculated molecular mass for 4Ig-B7-H3, after N-Glycanase treatment (data not shown). Our affinity purification strategy only identified 4Ig form from LAN-1 cell extracts, although 8H9 was also able to recognize the 2Ig form of recombinant B7-H3 protein by Western blot analysis (data not shown). Moreover, 8H9 binding measured by FACS analyses to the natively expressed B7-H3 on M14 melanoma cells was blocked by recombinant human 4Ig-B7-H3 (Fig 2). In contrast, control MoAb 3E7 was not affected by the presence of recombinant human 4Ig-B7-H3 (Fig 2).
Fig. 2. Blocking of 8H9 binding to natively expressed B7-H3 on M14 cells by recombinant human 4Ig-B7-H3.
(A) Either 0 ug (− 4Ig-B7-H3) or 3 ug (+ 4Ig-B7-H3) recombinant human 4Ig-B7-H3 was used for blocking. Natively expressed B7-H3 was detected by 8H9, while L1-CAM by 3E7. Representative FACS histograms with mean fluorescence intensity (MFI) from three independent experiments were shown. A non-specific MoAb TIB114 served as the staining control. (B) Various amounts of recombinant human 4Ig-B7-H3 (as indicated) were used for blocking. Bar graphs with MFI relative to 0 ug 4Ig-B7-H3 were shown. Mean + SD.
B7-H3 mRNA and protein expression
To systematically study the expression pattern of B7-H3, 18 normal tissues, including adrenal, cerebellum, frontal lobe, heart, ileum, kidney, liver, lung, lymph node, pancreas, pons, sigmoid colon, skeletal muscle, spinal cord, spleen, stomach, testes, and thyroid, were tested by qRT-PCR for their level of B7-H3 mRNA expression. Solid tumors, including 15 brain tumors, 5 hepatoblastomas, 41 neuroblastomas, 11 sarcomas, and 5 Wilm's tumors were also analyzed. Expression level as fold change relative to PBMC was tabulated in Fig 3. B7-H3 transcript was found to be ubiquitously expressed in solid tumors of different histological types, as well as normal human tissues.
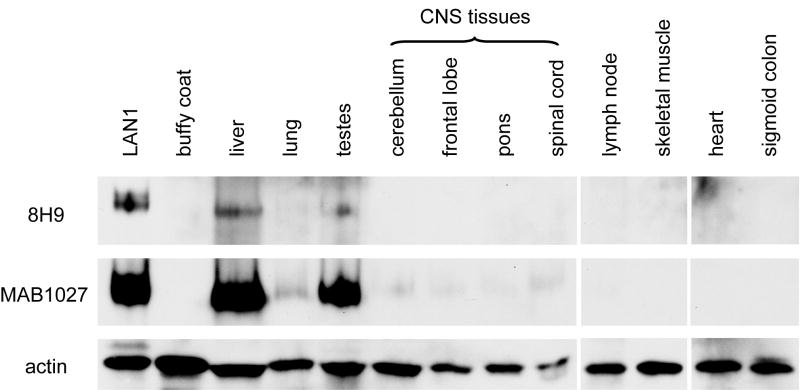
In contrast to the ubiquitous abundance of mRNA expression, there was a marked differential protein expression in solid tumors versus normal tissues. While solid tumors were strongly 8H9-positive by immunohistochemical staining, reflecting abundant B7-H3 protein expression, B7-H3 was undetectable in most normal tissues, including normal CNS tissues (18). Western blot analysis using 8H9 also confirmed this finding (Fig 4), even after longer film exposure (data not shown). By contrast, the commercial antibody MAB1027 showed much higher sensitivity to B7-H3 by both immunohistochemistry (data not shown) and Western blot analysis (Fig 1 and 4).
Fig. 4. B7-H3 protein was not detected by 8H9 in most normal tissues, including normal CNS tissues.
Protein levels were detected by Western blot using 8H9 or MAB1027. Actin levels served as loading control.
Relationship between B7-H3 protein and miR-29 expression among solid tumors and normal tissues
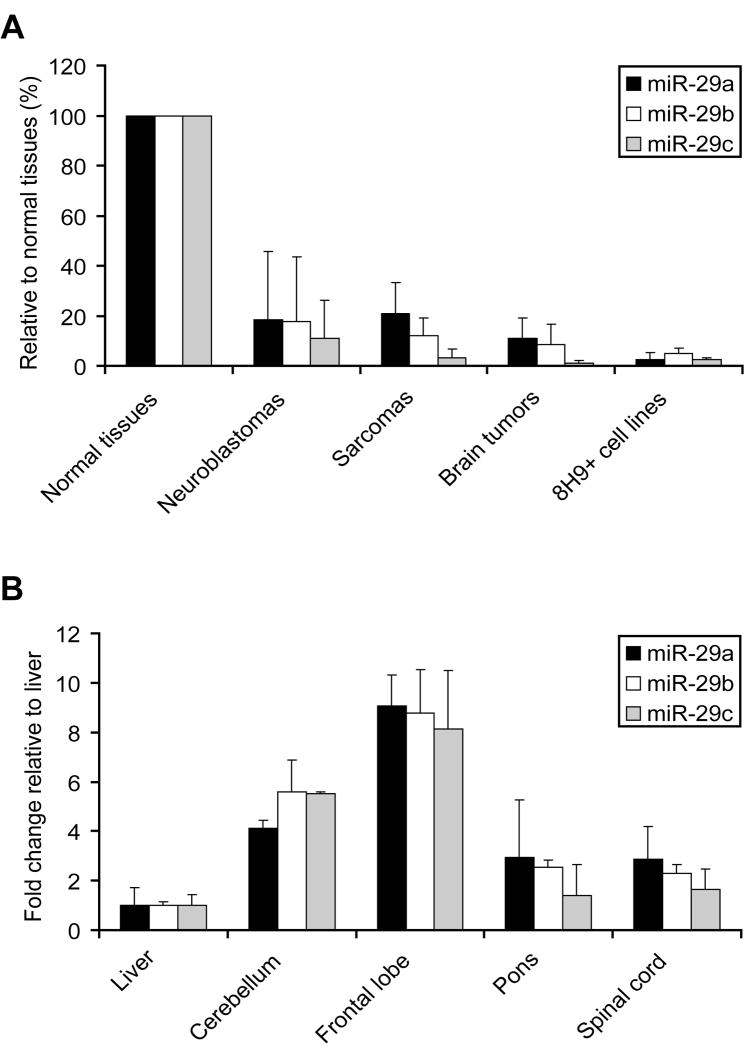
To test if miRNA can modulate differential B7-H3 overexpression in solid tumors, we used TargetScan (21) to identify miR-29 as the only broadly conserved miRNA family among vertebrates with a conserved target site in the B7-H3 3′UTR. All three isoforms of miR-29 (a, b, c) share the same seed complementarity (nt 2-7) between miR-29 and B7-H3 3′UTR (position 1339-1346), suggesting all isoforms potentially target B7-H3. If miR-29 is a potent regulator of B7-H3 protein expression, then miR-29 level should be downregulated in B7-H3 overexpressed tumor tissues. Indeed, when compared to the average of 18 normal tissues (normalized as 100%) by qRT-PCR, a broad spectrum of solid tumors, as well as 8H9-positive tumor cell lines had much lower miR-29 expression in all 3 isoforms (Fig 5A). Among normal tissues, miR-29 levels in 8H9-positive liver were 2 to 9 fold (the sum of all three isoforms) lower than the levels in 8H9-negative CNS tissues (cerebellum, frontal lobe, pons and spinal cord) (Fig 5B). All these data suggest an inverse correlation between B7-H3 protein and miR-29 expression level.
Fig. 5. Inverse correlation between B7-H3 protein and miR-29s expression among solid tumors and normal tissues.
miR-29 (a, b, c) levels were measured by qRT-PCR. RNU48 level served as the endogenous control, and miR-29 levels were normalized to the mean of 18 normal tissues (A): 18 normal tissues (detailed in the Results), 40 neuroblastomas, 8 sarcomas, 8 brain tumors, and 8 8H9+ (positive) tumor cell lines. (B) normal CNS tissues were normalized to liver.
miR-29a directly targets B7-H3 3′UTR
To test if miR-29 acts directly on B7-H3 expression, we performed a luciferase reporter assay. We chose miR-29a for these studies, since all three isoforms of miR-29 share the same seed complementarity to B7-H3 3′UTR, and have the similar inverse relationship between B7-H3 protein and miR-29 levels. The alignment of miR-29a with the B7-H3 3′UTR target site is illustrated in Fig 6A. The target site was cloned into the 3′UTR of the firefly luciferase gene and cotransfected with miR-29a into HeLa cells. As shown in Fig 6B, cotransfection of WT luciferase construct (B7-H3-WT) with NC (negative control) miRNA only moderately reduced luciferase activity with respect to parental luciferase construct (Control), presumably due to the existence of low levels of endogenous miR-29 in HeLa cells. However, cotransfection of B7-H3-WT with miR-29a significantly reduced luciferase activity, reducing by > 60% when compared to control level. This repression was reversed by a single base mutation in the binding site (B7-H3-mt). These results suggest that complementary site in the B7-H3 3′UTR is a direct target of miR-29a mediated post-transcriptional gene silencing.
Neuroblastoma cell line NB1691 showed strong B7-H3 protein expression by 8H9 staining with low endogenous miR-29a. Overexpression of miR-29a in NB1691 substantially reduced B7-H3 protein expression with a reduction of ∼ 60% when compared with negative control NC (Fig 6C). B7-H3 mRNA level was not affected (data not shown), suggesting that modulation of B7-H3 protein levels by miR-29a was primarily due to repressed translation, and not mRNA degradation. And this repression was also reversed when miR-29a inhibitor was being cotransfected together (miR-29a mimic+inhibitor). Similar findings were obtained when another neuroblastoma cell line LAN1 was tested (data not shown). As an additional control for this experiment, the level of another cell surface antigen L1-CAM was found to remain unchanged (data not shown), suggesting that results shown in Fig 6C were not due to cellular non-specific effects. These data provide direct evidence that miR-29a targeted B7-H3 mRNA and was able to modulate B7-H3 protein expression.
Discussion
In this study, we described the identification of 4Ig-B7-H3 as the target antigen for 8H9, a MoAb in clinical trial for solid tumors metastatic to the CNS (NCT00089245, http://clinicaltrials.gov/ct2/show/NCT00089245?term=8H9&rank=2). Furthermore, we provided evidence that B7-H3 protein overexpression in tumor tissue was highly correlated with decreased expression of miR-29 as compared to normal tissues, and B7-H3 protein level could be modulated through manipulating miR-29 level in cultured cell lines. The ability of miR-29 to control 4Ig-B7-H3 protein expression has implications in immune escape by solid tumors, and may allow differential modulation of this key immunoinhibitory molecule in tumor versus normal tissues, for both cell mediated immunotherapy as well as for antibody-based targeted strategies using 8H9.
Both negative and positive immunologic functions for B7-H3 have been reported. These contradictory findings could be explained by the existence of antagonistic B7-H3 receptors. Indeed, a costimulatory receptor for mouse B7-H3 that preferentially enhances mouse CD8+ T cell activation [TREM-like transcript 2 (TLT-2)], has been identified recently (22). The association of B7-H3 expression and worse clinical outcome in prostate cancer, clear cell renal cell carcinoma and urothelial cell carcinoma strongly implicates its potential role in inhibiting immune surveillance (7-10). Yet, coinhibitory receptor(s) for B7-H3 still remains to be found.
By western and immunohistochemistry both in this report and in previous publication (18), there was a clear differential expression of B7-H3 in solid tumors versus normal tissues. Since (a) 4Ig-B7-H3 inhibits NK-mediated lysis of neuroblastoma cells (6), and (b) most neuroblastomas do not express surface HLA-class I molecules, thereby escaping the attack by the cytotoxic T lymphocytes, it is reasonable to hypothesize that overexpression of B7-H3 on the surface of neuroblastoma and possibly other solid tumors will protect tumor cells from NK/T cell-mediated lysis, thus escaping the immune surveillance. Blockade of the inhibitory effects using MoAbs like 8H9 or suppression of B7-H3 protein expression using miRNAs like miR-29 (discussed below) could potentially enhance immune response to tumors.
The discrepancy between the ubiquitous expression at the mRNA level (Fig 3) versus the differential expression (tumor compared to normal tissues) at the protein level (Fig 4) suggests a post-transcriptional control. The regulation of protein expression is complex. Protein turnover through ubiquitination pathway is a classic mechanism for protein modulation. However, after binding to 8H9, B7-H3 was not modulated significantly even after 48 hrs of incubation at 37 °C (18). Aberrant glycosylation of cell surface antigens cannot be the reason, since the peptide-specific (not conformation dependent) antibody MAB1027 still detected the large differential between tumor and normal tissues, which did not change after N-Glycanase treatment (data not shown).
An emerging paradigm for protein regulation is through miRNA (14). miR-29, although not been studied as extensively as some other miRNAs, is known to be downregulated in chronic lymphocytic leukemia, cholangiocarcinoma, lung cancer and rhabdomyosarcoma (23-26). Our study extended these findings of the downregulation of miR-29 family to a broad spectrum of solid tumors, including neuroblastoma. Moreover, this downregulation was inversely correlated with high protein expression of B7-H3, an immune modulator which appears to be overexpressed in many human solid tumors including neuroblastoma, sarcomas, brain tumors, and many carcinomas including breast cancer, non-small-cell lung cancer, prostate cancer, clear cell renal cell carcinoma and urothelial cell carcinoma (6-10, 18, 27). The interaction between miR-29 and B7-H3 is direct, although only about half of the protein expression was repressed by miR-29 overexpression in our in vitro assays (Fig 6B and 6C). This is probably due to the fact that only a single conserved miR-29 binding site is in B7-H3 3′UTR (predicted by the software), and multiple miRNA binding sites are probably required for efficient translational repression in most cases (28). TargetScan software also predicted dozens of poorly conserved sites for the conserved miRNA families, plus sites for poorly conserved miRNA families in the human B7-H3 3′UTR. At this point, we cannot rule out the possibility of involvement of other miRNA families in the regulation of B7-H3 expression, alone or in cooperation with miR-29 family. The fact that there was near absence of B7-H3 protein (e.g. in spinal Cord, Fig. 4) when only a small increase in miR-29 expression was found (Fig. 5B) implicates the involvement of other miRNAs and/or regulatory factors in the control of B7-H3 expression, possibly in a tissue-specific manner. One can speculate that these other miRNAs are also aberrant in tumor cells when compared to normal tissues. In fact, other miRNA families (miR-9, 34a, 125a/b and 184) have been shown to be involved in the control of proliferation, differentiation and apoptosis of human neuroblastoma (29-31). But it does appear that since miR-29 binding site on B7-H3 is conserved in evolution, it probably plays a critical role in determining the differential protein expression of B7-H3 in tumor tissues versus normal tissues. Recent studies of NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma (26) suggest that miR-29 may act as a tumor suppressor through its promyogenic function. An additional role of miR-29 as “tumor suppressor” in vivo may be through modulating B7-H3 protein expression, and engaging tumor surveillance by NK cells and T cells.
In summary, it is worth noting that this report is the first demonstration of miR-29 modulation of an immunomodulatory molecule. Whether upregulation of miR-29 can sensitize B7-H3 overexpressed tumor cells to NK/T cell killing will need to be investigated. Nevertheless, modulating B7-H3 protein expression by miR-29 should improve therapeutic potential of MoAbs like 8H9. Although most normal tissues were negative for 8H9 staining, we did see positive staining in the liver. In patient imaging studies, moderate uptake of 8H9 in the liver was observed (NCT00582608, http://clinicaltrials.gov/ct2/show/NCT00582608?term=8H9&rank=1). Our in vitro studies suggest that suppressing B7-H3 protein expression in the liver maybe possible by increasing endogenous miR-29 or by administering miR-29 mimics, thereby improving the therapeutic index of MoAb targeting to B7-H3.
Acknowledgments
We thank Dr. Peter Houghton of St. Jude Children's Research Hospital for providing us the cell line NB1691; NCI P30-CA08748 which provides partial support for the Microchemistry and Proteomics Core Facility at MSKCC; and Dr. Hediye Erdjument-Bromage for the mass spectrometry analysis. We also thank Yi Feng for her excellent technical support.
Sources of support: Supported in part by grants from the National Institutes of Health CA106450, Robert Steel Foundation, and Hope Street Kids.
Footnotes
Potential conflicts of interest: 8H9 was licensed by Memorial Sloan-Kettering Cancer Center to United Therapeutics, Inc., Silver Spring, MD.
References
- 1.Flies DB, Chen L. The new B7s: playing a pivotal role in tumor immunity. J Immunother. 2007;30:251–60. doi: 10.1097/CJI.0b013e31802e085a. [DOI] [PubMed] [Google Scholar]
- 2.Chapoval AI, Ni J, Lau JS, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol. 2001;2:269–74. doi: 10.1038/85339. [DOI] [PubMed] [Google Scholar]
- 3.Steinberger P, Majdic O, Derdak SV, et al. Molecular characterization of human 4Ig-B7-H3, a member of the B7 family with four Ig-like domains. J Immunol. 2004;172:2352–9. doi: 10.4049/jimmunol.172.4.2352. [DOI] [PubMed] [Google Scholar]
- 4.Sun M, Richards S, Prasad DV, Mai XM, Rudensky A, Dong C. Characterization of mouse and human B7-H3 genes. J Immunol. 2002;168:6294–7. doi: 10.4049/jimmunol.168.12.6294. [DOI] [PubMed] [Google Scholar]
- 5.Suh WK, Gajewska BU, Okada H, et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat Immunol. 2003;4:899–906. doi: 10.1038/ni967. [DOI] [PubMed] [Google Scholar]
- 6.Castriconi R, Dondero A, Augugliaro R, et al. Identification of 4Ig-B7-H3 as a neuroblastoma-associated molecule that exerts a protective role from an NK cell-mediated lysis. Proc Natl Acad Sci U S A. 2004;101:12640–5. doi: 10.1073/pnas.0405025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth TJ, Sheinin Y, Lohse CM, et al. B7-H3 ligand expression by prostate cancer: a novel marker of prognosis and potential target for therapy. Cancer Res. 2007;67:7893–900. doi: 10.1158/0008-5472.CAN-07-1068. [DOI] [PubMed] [Google Scholar]
- 8.Zang X, Thompson RH, Al-Ahmadie HA, et al. B7-H3 and B7× are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc Natl Acad Sci U S A. 2007;104:19458–63. doi: 10.1073/pnas.0709802104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crispen PL, Sheinin Y, Roth TJ, et al. Tumor cell and tumor vasculature expression of B7-H3 predict survival in clear cell renal cell carcinoma. Clin Cancer Res. 2008;14:5150–7. doi: 10.1158/1078-0432.CCR-08-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boorjian SA, Sheinin Y, Crispen PL, et al. T-cell coregulatory molecule expression in urothelial cell carcinoma: clinicopathologic correlations and association with survival. Clin Cancer Res. 2008;14:4800–8. doi: 10.1158/1078-0432.CCR-08-0731. [DOI] [PubMed] [Google Scholar]
- 11.Sun X, Vale M, Leung E, Kanwar JR, Gupta R, Krissansen GW. Mouse B7-H3 induces antitumor immunity. Gene Ther. 2003;10:1728–34. doi: 10.1038/sj.gt.3302070. [DOI] [PubMed] [Google Scholar]
- 12.Wu CP, Jiang JT, Tan M, et al. Relationship between co-stimulatory molecule B7-H3 expression and gastric carcinoma histology and prognosis. World J Gastroenterol. 2006;12:457–9. doi: 10.3748/wjg.v12.i3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmeyer KA, Ray A, Zang X. The contrasting role of B7-H3. Proc Natl Acad Sci U S A. 2008;105:10277–8. doi: 10.1073/pnas.0805458105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 17.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18:103–10. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Modak S, Kramer K, Gultekin SH, Guo HF, Cheung NK. Monoclonal antibody 8H9 targets a novel cell surface antigen expressed by a wide spectrum of human solid tumors. Cancer Res. 2001;61:4048–54. [PubMed] [Google Scholar]
- 19.Kramer K, Modak S, Kushner BH, et al. Radioimmunotherapy of metastatic cancer to the central nervous system: Phase I study of intrathecal 131I-8H9. American Association for Cancer Research. 2007 LB-4 (Presentation) [Google Scholar]
- 20.Kramer K, Kushner BH, Modak S, et al. Effective Intrathecal Radioimmunotherapy-Based Salvage Regimen for Metastatic Central Nervous System (CNS) Neuroblastoma (NB). ISPNO 2008; 2008. [Google Scholar]
- 21.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 22.Hashiguchi M, Kobori H, Ritprajak P, Kamimura Y, Kozono H, Azuma M. Triggering receptor expressed on myeloid cell-like transcript 2 (TLT-2) is a counter-receptor for B7-H3 and enhances T cell responses. Proc Natl Acad Sci U S A. 2008;105:10495–500. doi: 10.1073/pnas.0802423105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pekarsky Y, Santanam U, Cimmino A, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–3. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 24.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Garzon R, Sun H, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–81. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Wang Y, Zhao J, et al. B7-H3 and B7-H4 expression in non-small-cell lung cancer. Lung Cancer. 2006;53:143–51. doi: 10.1016/j.lungcan.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 29.Laneve P, Di Marcotullio L, Gioia U, et al. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc Natl Acad Sci U S A. 2007;104:7957–62. doi: 10.1073/pnas.0700071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–22. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–83. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]