Abstract
We synthesized reversible terminators with tethered inhibitors for next generation sequencing. These were efficiently incorporated with high fidelity while preventing incorporation of additional nucleotides and were used to sequence canine bacterial artificial chromosomes in a single-molecule system that provided even coverage for over 99% of the region sequenced. This single-molecule approach generated high quality sequence data without the need for target amplification and thus avoided concomitant biases.
Highly parallel sequencing technologies have revolutionized biology by providing orders of magnitude more DNA sequence data than previously possible1. Most of these technologies require synthesizing DNA from an existing template using either a polymerase or ligase2. The first commercialized technologies required amplification of the template DNA prior to sequencing, but this can introduce a host of biases caused by differential behavior from many factors3, making even representation or accurate quantitation of samples difficult. Single-molecule sequencing4,5 can eliminate biases introduced by amplification.
Any method employing polymerase-based sequencing-by-synthesis encounters the problem of how to count bases within homopolymers (sequences that repeat the same base). One strategy relies on nucleotide analogs that are capable of being incorporated once but block subsequent additions. If inhibition is efficient and reversible, the nucleotides could be used to step through homopolymer regions one base at a time. Modifications of the base with a fluorescent label and the 3′ hydroxyl with a blocker that prevents extension have been described6,7. These molecules allow stepwise addition through a homopolymer repeat but require removal of both modifications. Here, we report a different strategy, creating four analogs modified at only a single position. Each contain three features: i) free 3′-OH maintaining natural interactions at the polymerase active site, ii) base modified with a propargylamine connected to a cleavable linker, and iii) fluorescent dye tethered to an inhibitor, attached via the cleavable linker. We call these “Virtual Terminator” nucleotides since they are efficiently incorporated yet block incorporation of a second nucleotide on a homopolymer template, despite possessing a free 3′ hydroxyl. A similar approach but with just a single nucleotide has been described8.
In a previous study demonstrating the feasibility of directly sequencing many single DNA molecules bound to a surface5, M13 bacteriophage was sequenced except for homopolymers longer than three. This first generation of nucleotides (Supplementary Figure 1) was modified such that the Cy5 dye was attached to the base with a linker containing a disulfide bond (Cy5-12ss-dNTP analogs,). Cy5 served as the fluorescent marker during the imaging phase of sequencing and could then be removed by cleavage of the disulfide bond prior to the next incorporation event. Despite the large size of the linker and fluorescent tags, the DNA polymerase was able to efficiently incorporate them, albeit at a slower rate than natural nucleotides. These nucleotides maintained a low misincorporation rate but were not homopolymer-competent. The Cy5-containing nucleotides described herein were tested for their ability to incorporate efficiently while preventing a second round of synthesis. These were all modified via the same linker attachment site present in the commercially-available nucleotides All of our nucleotides were synthesized as described in the Supplementary Note according to reaction schemes shown in Supplementary Figures 2 and 3. To determine the biochemical properties of our nucleotide analogs (Supplementary Table 1), we analyzed incorporation into primer-template DNA.
One measure of the efficiency with which a nucleotide can be incorporated is the polymerization rate divided by the nucleotide dissociation rate (kPol/KD)9. This provided a measure of the likelihood that a given nucleotide will dissociate from the active site versus move onto the next step and be incorporated. KDs and kPols are provided for selected analogs (Table 1). The novel analogs did not incorporate as fast as Cy5-12ss-dNTPs but this rate was still sufficiently fast for sequencing.
Table 1. Incorporation rates for selected analogs.
For selected analogs listed in Supplementary Table 1, the KD, kpol, and ratio of kpol/kD are provided. Measurements were carried out as described in Online Methods. Compound structures are shown in Supplementary Table 1 and Supplementary Figure 1. The “type” column is a shorthand nomenclature which indicates the incorporated nucleotide connected via the tether (*) to the inhibitory component.
| Analog | Type | KD(μM) | kpol (s-1) | kpol/KD |
|---|---|---|---|---|
| 17 | U*pU | 4.93 (+/-0.9) | 0.86 (+/-0.07) | 0.17 |
| 18 | U*U | 1.9 | 0.87 | 0.46 |
| 19 | G*pCp | 3.98 (+/-1.0) | 0.7 (+/-0.07) | 0.18 |
| 20 | A*pCp | 13.5 (+/-0.9) | 0.99 (+/-0.12) | 0.07 |
| 21 | U*pCp | 12.1 (+/-0.7) | 1.04 (+/-0.03) | 0.09 |
| 23 | C*pC | 4.14 (+/-0.7) | 0.77 (+/-0.06) | 0.19 |
| 25 | 12ss-dUTP | 4.9 (+/-0.6) | 2.4 (+/-0.11) | 0.49 |
| 29 | A*pU | 2.7 (+/-1.0) | 0.57 (+/-0.06) | 0.21 |
| 30 | U**pU | 4.4 (+/-1.3) | 0.92 (+/-0.12) | 0.21 |
| 31 | G***pU | 2.0 (+/-0.4) | 0.98 (+/-0.27) | 0.49 |
| 32 | C****pC | 3.6 (+/-0.9) | 0.67 (+/-0.06) | 0.19 |
Nucleotides must be incorporated in the correct position and also have very little misincorporation. Fidelity was assessed by determining the pre-steady-state rate of incorporation into primer-templates that coded for incorrect additions. Select analogs were tested against three misincorporation templates, one for each possible mispair. Despite being incorporated at lower efficiency, these analogs were added with fidelity similar to Cy5-12ss-dNTPs (Supplementary Table 2) and natural nucleotides10.
To determine whether the novel analogs can function as reversible terminators, we performed experiments similar to those described above except that the template encoded two base homopolymers. The rates of both first and second base addition were measured and the first then divided by the second (k1/k2, Supplementary Table 3). This described the effectiveness of the analog as a homopolymer run-through inhibitor normalized to incorporation efficiency at the first base. We observed a striking correlation between the number of phosphates on the inhibitory base and its effectiveness as a reversible terminator. Analogs lacking phosphates on the inhibitory base gave low k1/k2 values, indicating limited usefulness as a homopolymer inhibitor. In contrast, monophosphates on the inhibitory moiety had higher k1/k2 while the bisphosphate analogs showed even greater effectiveness as terminators. These biphosphate analogs provide the right combination of incorporation at correct positions and not at incorrect and homopolymer sites.
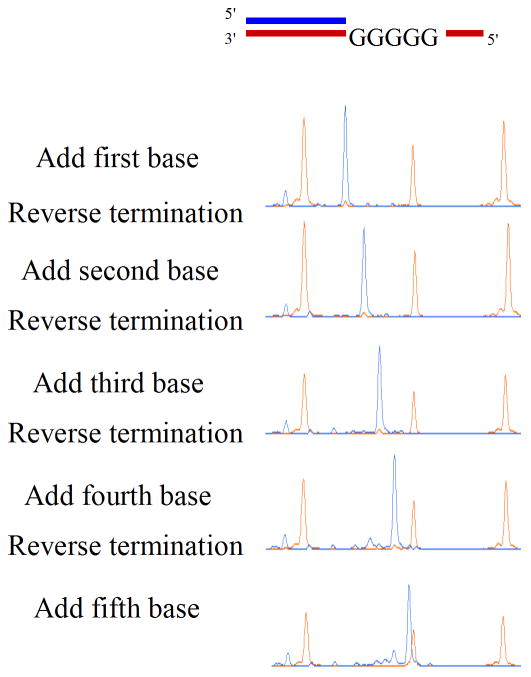
Critical to the utility of reversible terminators is the ability to reverse the inhibition prior to subsequent base additions. We used a template with five consecutive Cs and performed base addition cycles followed by removal of the inhibitor-dye. Five cycles of addition-cleavage on such a template resulted in an almost perfectly synchronous walk through the homopolymer (Figure 1). Thus, these analogs were highly effective reversible terminators.
Figure 1. Virtual Terminator nucleotide base-by-base incorporation in a G5 homopolymer.
The substrate used for testing homopolymer sequencing is shown along with successive cycles of addition of compound 22 in a solution phase reaction. Removal of the inhibitor-dye was accomplished by cleavage of the disulfide using TCEP, a reducing agent, followed by treatment with iodoacetamide to cap the free thiol. After each cycle, an aliquot of the reaction is run on an ABI3730 sequencing machine to achieve single base resolution of DNA. Length markers are shown in orange and the DNA being synthesized is shown in blue.
To test the Virtual Terminator nucleotides with mammalian DNA, canine BAC AC187329 was resequenced. This previously-sequenced BAC contains 194 kb of complex mammalian sequence. Two pass, single-molecule sequencing in which each molecule is sequenced twice, as described previously5 and in Online Methods, was performed with a low-capacity prototype sequencing instrument. Image analysis software allowed matching of reads from the same DNA molecule in the two passes. Comparison of two sequences from each molecule yielded a high confidence consensus sequence for that molecule which can then be combined with other reads to generate a final sequence. It was possible to use just single pass sequence data to generate a high quality consensus sequence with the higher error rate of the individual read offset by the higher coverage obtainable via single pass.
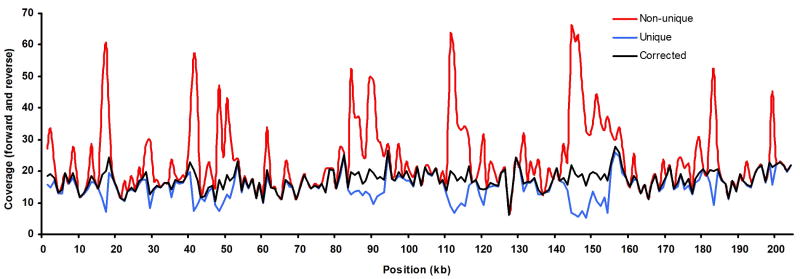
Prior to alignment of sequence reads to the BAC reference, filtering of raw reads was carried out to eliminate artifacts and non-informative strands, as described in Online Methods. After filtering, high quality reads from 123,418 DNA strands were obtained that met criteria for both passes. An even larger number of DNA strands had high quality reads that met criteria for just one pass. Alignment of the two pass sequences to the reference yielded a median coverage of 15 (Figure 2). If only uniquely aligned sequences were included, even coverage of nearly the entire sequence was generated. The only low coverage positions corresponded to repetitive regions which are not capable of yielding unique alignments. If all uncovered regions were combined, they amounted to only 0.2% of the entire sequence.
Figure 2. Sequence coverage of BAC.
Depth of coverage for the BAC is shown across its length. When all reads (non-unique) were mapped to the sequence (red), repeat regions received higher coverage due to multiply aligning reads. When only uniquely aligning sequences were included (blue), the repeat regions were under-represented. When a fractional correction was made to multiply aligning sequences (black), even coverage was obtained across the length of the BAC. When only unique alignments were used, the longest uncovered stretch of DNA was 279 bp, corresponding to a repeat region. If all alignments were used, the longest uncovered region was a highly AT rich 103 bp segment that included 28 consecutive AT nucleotides and many other shorter AT runs.
The average per nucleotide error rate for all strands ≥ 15 in length was 0.58% with little variation as a function of read length (Supplementary Figure 4). The length-independence of error rate was a natural property of single-molecule sequencing in which it is impossible to dephase the sequence since each molecule is read individually. If incorporation was missed during one cycle, it can occur in the next with no loss of information. Most errors were deletions, likely caused by incorporation without detection. Such deletions could result from either chemical or optical imperfections.
Because these analogs were designed to overcome issues with homopolymer sequencing, it was of special interest to determine how well those regions sequenced. Over 98% of all 38,353 homopolymers in this BAC had coverage of ≥ 10 reads and can be called for length. Of those called, over 99.99% are called correctly. Only when the homopolymer length reached ten were fewer than half of homopolymers not covered with a sufficient number of reads for a call. Increased sequencing depth, as is generated by the commercial instrument, would alleviate this problem.
Single-molecule sequencing provides tremendous advantages in experimental design relative to the more classical sequencing of ensembles of molecules generated by amplification. Turning this potential into a practical method for sequencing DNA has required several advances as described here. Previous work demonstrated the feasibility of this approach but homopolymers were difficult to call accurately. These novel nucleotides have been modified to include a tethered inhibitor to take advantage of interactions in the active site so that additional nucleotides may be prevented from entering and being incorporated. Highly efficient, fast cleavage of the dye and tethered inhibitor at the proper time (and not before) using mild conditions was also achieved. The analogs presented here include all these properties and thus enabled single-molecule sequencing with complex mammalian DNA at a scale not previously possible. The orders of magnitude higher throughput in single molecule sequencers, compared to Next generation sequencers that require target amplification promise a scalable method for achieving the $1000 genome. This work indicates that only technical optimization and not new technology is required to achieve that end.
Supplementary Material
Figure 1: Cy5-12ss-dNTP analogs
Figure 2: Scheme 1 for nucleotide analog synthesis
Figure 3: Scheme 2 for nucleotide analog synthesi
Figure 4: Error rate
Table 1: Nucleotide analogs
Table 2: Misincorporation rates for two analogs
Table 3: Termination ability of analogs to prevent second base incorporation
Acknowledgments
We thank all of the past and present colleagues at Helicos who have contributed to this work. This publication was made possible by grant number R01 HG004144 from the National Human Genetics Research Institute (NHGRI) at the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NHGRI.
Footnotes
Author contributions: JM, JWE, EK-O, SM, AR, and SMS contributed to the synthesis and/or design of novel compounds. JB, EB, and PM contributed to the design and execution of biochemical assays. MC, MJ, LK, GML, AP, and KS contributed to the sequencing of DNA samples. MC, MJ, LK, DL, GML and AP contributed to the analysis of sequencing data. JB, JM, EK-O, SMS, and JFT contributed to the writing of the manuscript. JB, PRB, MC, JWE, MJ, and SMS contributed to the supervision and management of the project.
References
- 1.Kahvejian A, Quackenbush J, Thompson JF. Nat Biotechnol. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braslavsky I, Hebert B, Kartalov E, Quake SR. Proc Natl Acad Sci U S A. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris TD, et al. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 6.Bentley DR, et al. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ju J, et al. Proc Natl Acad Sci U S A. 2006;103:19635–19640. doi: 10.1073/pnas.0609513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu W, et al. Nucl Acids Res. 2007;35:6339–6349. doi: 10.1093/nar/gkm689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuchta RD, et al. Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 10.Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. J Biol Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1: Cy5-12ss-dNTP analogs
Figure 2: Scheme 1 for nucleotide analog synthesis
Figure 3: Scheme 2 for nucleotide analog synthesi
Figure 4: Error rate
Table 1: Nucleotide analogs
Table 2: Misincorporation rates for two analogs
Table 3: Termination ability of analogs to prevent second base incorporation