Abstract
P53 regulates an array of target genes, which mediates p53 tumor suppression by inducing cell cycle arrest, apoptosis, and cell survival. G protein-coupled receptors (GPCRs) belong to a superfamily of cell-surface molecules and are known to regulate cell proliferation, migration and survival. Here, we found that G protein-coupled receptor 87 (GPR87) was up-regulated by p53 and by DNA damage in a p53-dependent manner. We also found that p53 directly regulated GPR87 potentially via a p53-responsive element in the GPR87 gene. To investigate the role of GPR87 in the p53 pathway, we generated multiple RKO and MCF7 cell lines in that GPR87 can be inducibly overexpressed or knocked down by a tetracycline-inducible system. We found that overexpression of GPR87 had little effect on cell growth. However, GPR87 knockdown sensitized cancer cells to DNA damage-induced growth suppression via enhanced p53 stabilization and activation. Importantly, the pro-survival activity of GPR87 can be reversed by knockdown of p53. Together, our results suggested that GPR87 is essential for p53-dependent cell survival in response to DNA damage. Thus, due to its expression on the cell surface and its role in cell survival, GPR87 may be explored as a novel therapeutic target for cancer treatment and prevention.
Keywords: GPR87, p53, DNA damage, growth suppression
Introduction
G protein-coupled receptors (GPCRs) constitute the largest and most diverse family of cell surface receptors. GPCRs contain a seven membrane-spanning helix connected by three intracellular loops with an extracellular amino terminus and an intracellular carboxy terminus (1). All GPCRs can trigger the exchange of guanosine diphosphate (GDP) to guanosine triphosphate (GTP), which in turn leads to activation and subsequent dissociation of α subunit and βγ dimer of the G protein complex (2). GPCR signaling is subject to extensive regulations through receptor desensitization and sequestration, G protein activation, and enzymatic degradation of second messengers. Additionally, protein-protein interactions modulate GPCR signaling, including the formation of GPCR homo- and hetero-dimers, the interaction with receptor activity-modifying proteins, and the binding of various scaffolding proteins to intracellular receptor domains (3).
Many GPCRs are overexpressed in tumor tissues (4). In addition, a number of GPCR ligands, such as bioactive peptides, biogenic amines, and chemokines, are highly expressed in the tumor microenvironment (5). Thus, GPCR signaling promotes cell growth and survival, angiogenesis, metastasis, and drug resistance (6–11). Due to the large number of GPCRs, their expression on the cell surface, and their diverse biological functions, GPCRs become the prime target for therapeutic intervention, accounting for 50% of all drug targets (12).
Under the normal condition, p53 is maintained at low levels via proteasomal degradation (13, 14). In response to DNA damage and other stresses, p53 is stabilized and activated (15), which then induces a plethora of downstream target genes (16), including p21 (WAF1/CIP1), Mdm2, ASC, and HB-EGF. These p53 target genes mediate p53 tumor suppression by inducing multiple cellular responses, including cell cycle arrest, apoptosis, DNA repair, and metabolism.
Here, we found that G protein-coupled receptor (GPR87) is regulated by p53 and DNA damage in a p53-dependent manner. We also found that knockdown of GPR87 sensitizes tumor cells to DNA damage-induced growth suppression in a p53-dependent manner. Our study revealed that GPR87 mediates p53 pro-survival function. Since GPR87 is a cell surface receptor, it might be explored as a target for cancer therapy.
Materials and Methods
Cell culture
MCF7, RKO, HCT116, MCF7-p53-KD, P53-null HCT116, RKO-E6 and RKO-p53-KD were previously described (17–18). The MCF7 cell line which inducibly expresses p53 was as previously described (19–21). RKO and MCF7 cell lines, which inducibly express GPR87, were generated as previously described (22). To generate inducible GPR87-knockdown cell lines, pBabe-H1-siGPR87 (below for detail) was transfected into MCF7 (MCF7-TR-7) or RKO (RKO-TR-13) cells in which a tetracycline repressor is expressed by pcDNA6 (23). GPR87-knockdown cell lines were selected with puromycin and confirmed by RT-PCR. To generate inducible GPR87-knockdown cell lines with stable p53 knockdown, pBabe-H1-siGPR87 was cotransfected with pBabe-U6-sip53 into RKO cells in which a tetracycline repressor is expressed by pcDNA6. The dual GPR87/p53-knockdown cell lines were selected with puromycin.
Plasmids
GPR87 was amplified by PCR using an EST clone (GenBank #BC112941) as a template with forward primer, 5’-ACAAGCTTAGAATGGGGTTCAACTTGACGC-3’, and reverse primer, 5’-ACCTCGAGAGGCCTACACATCAGTGTAATC-3’. The siRNA-resistant GPR87 was generated by two-step PCR with two sets of primers. The siRNA targeting sequence (GCATCTTGCTGAATGGTTT) in GPR87 was replaced with a sequence with five substitutions (GCATCTTGtTaAAcGGccT; lower case letter represents substitution) without changing the sequence of amino acids. The first set of primers are: forward primer 5’- AGCATCTTGtTaAAc GGccTAGCAGTGTGGATCTTCTTC C-3’, and reverse primer, 5’-ACCTCGAGAGGCCTACACATCAGTGTAATC-3’. The other set of primers are: forward primer, 5’-ACAAGCTTAGAATGGGGTTCAACTTGACGC-3’, and reverse primer, 5’-ACACTGCTAggCCgTTtAaCAAGATGCTTGCCACAAATAT-3’.
To generate inducible siRNA against GPR87 under the control of the tetracycline-regulated H1 promoter, two 64-base oligos were annealed and then cloned into pBabe-H1 siRNA expression vector, and the resulting plasmid designed pBabe-H1-siGPR87. The sense oligo is 5’-GATCCCCGCATCTTGCTGAATGGTTTTTCAAGAGAAAACCATTCAGCAAGATGCTTTTTGGAAA, and the antisense oligo is 5’-AGCTTTTCCAAAAAGCATCTTGCTGAATGGTTTTCTCTTGAAAAACCATTCAGCAAGATGCGGG-3’, with the siRNA targeting region underlined. We would like to mention that the siRNA-targeting sequence is unique to GPR87, but not to other members of the GPR family. The pBabe-U6-sip53 was previously described (24).
The luciferase reporter under the control of the p21 promoter, pGL2-p21A, was as previously described (25). A 433-bp DNA fragment containing a potential p53-RE in the GPR87 gene (nt +476 to +909) was amplified from the genomic DNA purified from MCF7 cells with forward primer, 5’-AAGGATCCATTCTGATCCCTGGAGAAGTCATG-3’, and reverse primer, 5’-AAAAGCTTCAACAGGGTGTCTGGCTTTTTTCC-3’. The DNA fragment was confirmed by sequencing and then cloned upstream of the minimum c-fos promoter in the luciferase reporter O-Fluc (26). The resulting plasmid is designated as GPR87-W. GPR87-M was similarly generated with four nucleotide substitutions in the p53 responsive element (C and G at nt +751 and +754 were replaced with T, whereas C and G at nt +761 and +764 were replaced with T and A, respectively).
PCR primers
The primers to detect GPR87 are: forward primer 5’-GCCAGGAAAGAACACCACCC-3’ and reverse primer 5’-CGATCAGAATCAACCAGCACG-3’. The primers to detect actin are: forward primer 5’-CTGAAGTACCCCATCGAGCACGGCA-3’ and reverse primer 5’-GGATAGCACAGCCTGGATAGCAACG-3’.
Luciferase assay
The dual luciferase assay was done in triplicate according to the manufacturer’s instructions (Promega). The luciferase activity was measured 24 h following the transfection as previously described (23). The fold increase in relative luciferase activity is a product of the luciferase activity induced by wild-type or mutant p53 divided by that induced by empty pcDNA3 vector.
Microarray assay, Northern blot analysis, and RT-PCR
Total RNA was isolated from cells using Trizol reagent (Invitrogen). The U133-plus GeneChip was purchased from Affymetrix. GeneChip analysis was performed at UC Davis Gene Expression Facility according to the manufacturer’s instruction. Northern blot analysis and preparation of p21 and GAPDH probes were as previously described (27). The GPR87 probe was prepared from EST clone #BC112941. cDNA was generated using iScriptTM cDNA Synthesis Kit (Bio-Rad). Real-time PCR was performed using RealMaster mix with SYBR Green and realplex2 Mastercycler (Eppendorf). The data were analyzed with the Fit Points Method of the Light Cycler software.
Immunocytochemical staining
Cells seeded on a slide chamber with or without tetracycline for 24 h were fixed with 3% of paraformaldehyde and permeabilized with 0.1% Triton X-100. Cells were incubated with 1% BSA for 30 min and then incubated with primary antibody in 1% BSA overnight, followed with secondary antibody for 30 min at room temperature. The slides were mounted in DAPI-mounting medium for 15 min and sealed with nail polish.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was done as described (24, 28–29). To amplify the potential p53-RE in the GPR87 gene, PCR was done with forward primer, 5’- CCACAAAGAACAGGGCAATC −3’, and reverse primer, 5’-ACTGAGATCTCCATGCTTGC-3’. The primers to amplify p53-RE in the p21 gene and the region in the GAPDH gene were previously described (17).
Colony formation assay
MCF7 (400 cells/well) and RKO (200 cells/well) cells in a six-well plate were cultured with or without tetracycline (1.0 µg/mL) along with or without doxorubicin (10 ng/mL) or camptothecin (10 nM) for ~14 d and then fixed with methanol/glacial acetic acid (7:1), followed by staining with 0.1% crystal violet.
DNA histogram analysis
Cells were induced or uninduced to knock down GPR87 for 72 hrs, followed by mock-treatment or treatment with doxorubicin (250 ng/mL) for 24 h. The preparation of cells and FACS analysis were carried out as previously reported (29).
Western blot analysis
Western blotting was performed as described (24, 28). Antibodies used were anti-GPR87 (Abcam), anti-p53 (DO-1, PAb1801, PAb240, and PAb421), anti-actin (Sigma), anti-GAPDH (Santa Cruz), anti-p21 (C-19) (Santa Cruz), anti-Mdm2 (Santa Cruz), anti-PARP (BD Pharmingen), and anti-Myc (9E10) (Cell Signaling).
Statistical analysis
Data were presented as means +/− standard deviation (SD). Statistical significance was determined by Student’s t test. Values of p<0.05 were considered significant.
Results
GPR87 is a novel target gene of p53
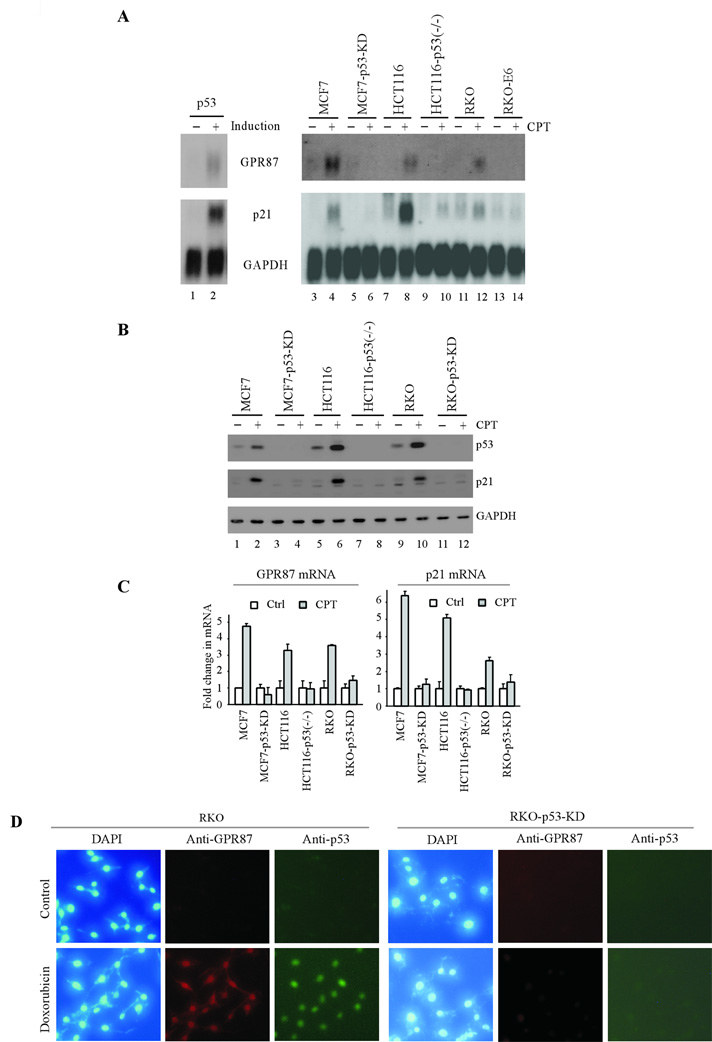
To identify novel target genes regulated by p53, microarray assay was performed with RNAs purified from MCF7 cells uninduced or induced to express p53. Many well-defined p53 target genes were found to be upregulated, including p21, Mdm2, and FDXR. Among these is GPR87. To confirm this, Northern blot analysis was performed and showed that the level of GPR87 transcript was increased by wild-type p53 in MCF7 cells (Fig. 1A, lanes 1–2). The levels of the p21 and GAPDH transcripts were examined as positive and loading controls, respectively. It is well-known that DNA damage stabilizes and activates p53, leading to induction of p53 target genes. Thus, we tested whether GPR87 can be induced by DNA damage in cells harboring wild-type p53. We found that like p21, GPR87 was up-regulated in MCF7, HCT116, and RKO cells treated with camptothecin, an inhibitor of DNA topoisomerase I (Fig. 1A, compare lanes 3, 7, and 11 with 4, 8, and 12, respectively). However, little if any up-regulation of GPR87 and p21 was observed in p53-knockdown MCF7, p53-null HCT116, and p53-null-like RKO (RKO-E6) cells (Fig. 1A, compare lanes 5, 9, and 13 with 6, 10, and 14, respectively).
Figure 1.
GPR87 is induced by p53. (A) Northern blots were prepared with 10 µg of total RNAs purified from MCF7 cells, which were uninduced (–) or induced (+) to express p53 for 24 h, or from MCF7, HCT116, and RKO cells, which were mock-treated (−) or treated (+) with 250 nM camptothecin for 24 h. The blots were probed with cDNAs derived from the GRP87, p21, and GAPDH genes, respectively. (B) Western blots were prepared using extracts from MCF7, HCT116, and RKO cells, which were mock-treated (−) or treated (+) with 250 nM camptothecin for 12 h. p53, p21, and GAPDH were detected by their respective antibodies. (C) Real-time RT-PCR was performed with RNAs purified from MCF7, HCT116, and RKO cells, which were treated as in (B). The levels of GPR87 and p21 transcripts were normalized with the levels of actin transcript. (D) Immunocytochemistry was performed using RKO and RKO-p53-KD cells untreated (Control) or treated with 250 ng/mL of doxorubicin for 12 h. The nuclei, GPR87, and p53 were measured by DAPI, anti-GPR87, and anti-p53, respectively.
To quantify induction of GPR87, real-time PCR was performed. To make sure that p53 was properly activated, Western blot analysis was performed and showed that the level of p53 was increased along with induction of p21 upon treatment with camptothecin in p53-proficient MCF7, HCT116, and RKO cells (Fig. 1B, compare lanes 2, 6, and 10 with 1, 5, and 9, respectively). In contrast, p53 and p21 were undetectable in p53-knockdown MCF7 and RKO cells and p53-null HCT116 cells regardless of treatment with camptothecin (Fig. 1B, lanes 3–4, 7–8, and 11–12). Next, Real-time PCR was performed with total RNAs purified from same groups of cells as in Fig. 1B. We found that the level of GPR87 transcript was increased in p53-proficient but not p53-deficient cells upon treatment with camptothecin (~4.7 fold in MCF7 cells; ~3.5 fold in RKO and HCT116 cells) (Fig. 1C). Similarly, the level of p21 was induced in p53-proficient but not p53-deficient cells (Fig. 1C).
To determine whether the increased levels of GPR87 transcript correlate with an increase in the levels of GPR87 protein, RKO and p53-KD RKO cells were mock-treated or treated with 250 ng/mL of doxorubicin for 12 h. Immunostaining was performed with anti-GPR87 to measure the expression of GPR87 since the antibody was not suitable for Western blot analysis. We found that p53 and GPR87 were detected in RKO but not p53-KD RKO cells upon treatment with doxorubicin (Fig. 1D). Interestingly, GPR87 was found to be highly expressed in nucleus.
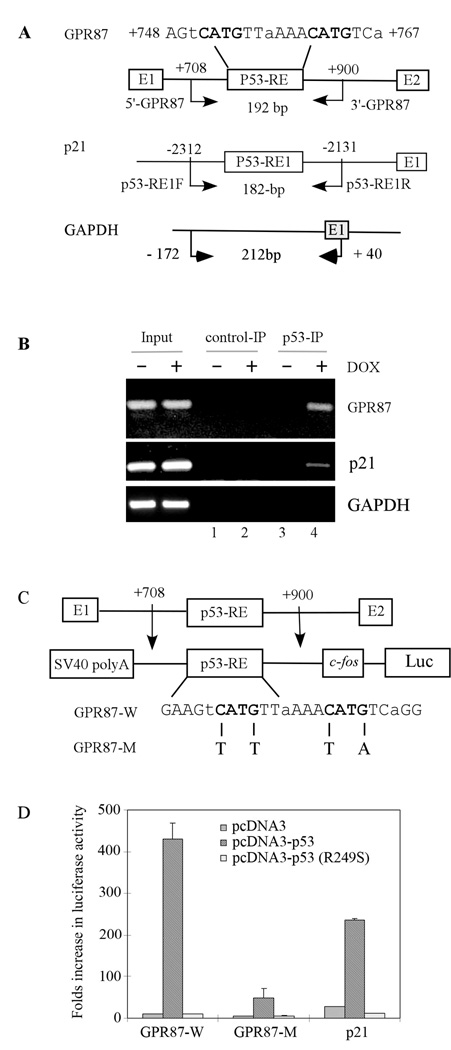
P53 regulates its targets through binding to p53-REs in the promoter or intron. Thus, we searched for p53-REs in the GPR87 genomic locus and found one potential binding site located at nt +748 to +767 in intron 1 with the sequence of AGtCATGTTaAAACATGTCa (Fig. 2A, top panel, lowercase letters represent mismatch). Thus, ChIP assay was performed to examine whether p53 binds to the potential p53-RE in the GPR87 gene (Fig. 2A). The binding of p53 to the p53-RE in the p21 gene was used as a control (Fig. 2A). To test this, RKO cells were untreated (−) or treated (+) with doxorubicin to activate p53 followed by cross-linking with formaldehyde and immnoprecipitation with anti-p53 or rabbit IgG as a negative control. We found that when activated, p53 bound to p53-REs in the GPR87 and p21 genes but not to the GAPDH promoter (Fig. 2B, compare lanes 3–4). In contrast, no DNA fragments were detected in control IgG immunoprecipitates (Fig. 2B, control-IP, lanes 1–2).
Figure 2.
Identification of potential p53-RE in the GPR87 gene. (A) Schematic presentation of the GPR87, p21, and GAPDH promoters along with the location of the p53-RE and primers used for ChIP assay. (B) p53 binds to the p53-RE in the GPR87 and p21 genes. ChIP assay was performed with anti-p53 to detect p53-DNA complexes along with a control IgG for non-specific binding. (C) Schematic presentation of the GPR87 locus containing a potential p53-RE located in intron 1 along with the location of nucleotide substitutions in GPR87-M. (D) Wild-type, but not mutated, p53-RE in the GPR87 gene was responsive to wild-type but not mutant p53. The luciferase assay was performed as described in “Materials and Methods”.
To test whether the potential p53-RE in the GPR87 gene is responsive to p53, a 433-bp DNA fragment (from nt +476 to +909) was cloned upstream of the minimal c-fos promoter in O-Fluc luciferase reporter (26). In addition, a mutant p53-RE was generated and similarly cloned into O-Fluc (Fig. 2C). The luciferase reporter under the control of the p21 promoter (29) was used as a control. We showed that like the p53-RE in the p21 promoter, the potential p53-RE in the GPR87 gene (GPR87-W) was responsive to wild-type but not mutant p53 (Fig. 2D). In contrast, the luciferase activity for GPR87-M was not significantly increased by wild-type p53 (Fig. 2D).
Ectopic expression of GPR87 has no effect on cell proliferation
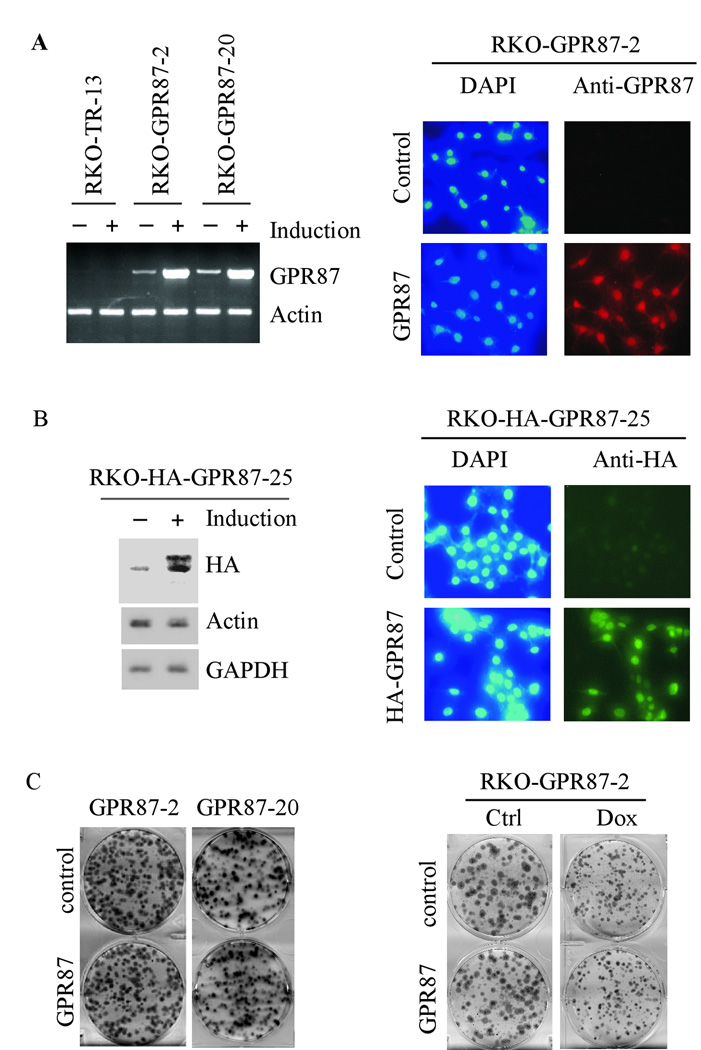
The GPR87 gene is found to be over-expressed in lung and bladder carcinomas (30), suggesting that GPR87 is likely to promote cell growth. To test this, we generated RKO cell lines that inducibly express GPR87 as measured by RT-PCR (Fig. 3A, right panel) and immunostaining (Fig. 3A, left panel). RKO-TR-13 is the control parental cell line (Fig. 3A, left panel). We also generated one RKO cell line that inducibly expresses HA-tagged GPR87 as measured by Western blotting (Fig. 3B, left panel) and immunostaining (Fig. 3B, right panel) with anti-HA. Next, colony formation assay was performed and showed that under both normal and stress conditions, GPR87 had little if any effect on the number and size of the colonies in RKO cells (Fig. 3C). Similarly, we found that GPR87 had no effect on cell proliferation in MCF7 cells (data not shown).
Figure 3.
Over-expression of GPR87 has no effect on cell proliferation in RKO cells. (A) Left panel: generation of RKO cell lines that inducibly express GPR87. RKO-TR-13 is a control cell line. The levels of GPR87 transcript were quantified by RT-PCR in RKO cells uninduced (−) or induced (+) to express GPR87 for 24 h. Right panel: RKO cells uninduced or induced to express GPR87 for 24 h were stained with anti-GPR87 along with DAPI as described in “Materials and Methods”. (B) Left panel: generation of RKO cell lines that inducibly express HA-tagged GPR87. The level of HA-tagged GPR87 in RKO cells was measured by Western blotting with anti-HA. Right panel: the expression of HA-tagged GPR87 in RKO cells was stained by anti-HA. (C) Left panel: colony formation assays were performed with RKO cells uninduced (control) or induced (GPR87) to express GPR87 over a 14-day period. Right panel: colony formation assays were performed with RKO cells uninduced (control) or induced (GPR87) to express GPR87 along with mock-treatment (Ctrl) or treatment with 10 ng/mL of doxorubicin over a 14-day period.
Lack of GPR87 sensitizes tumor cells to DNA damage-induced growth suppression
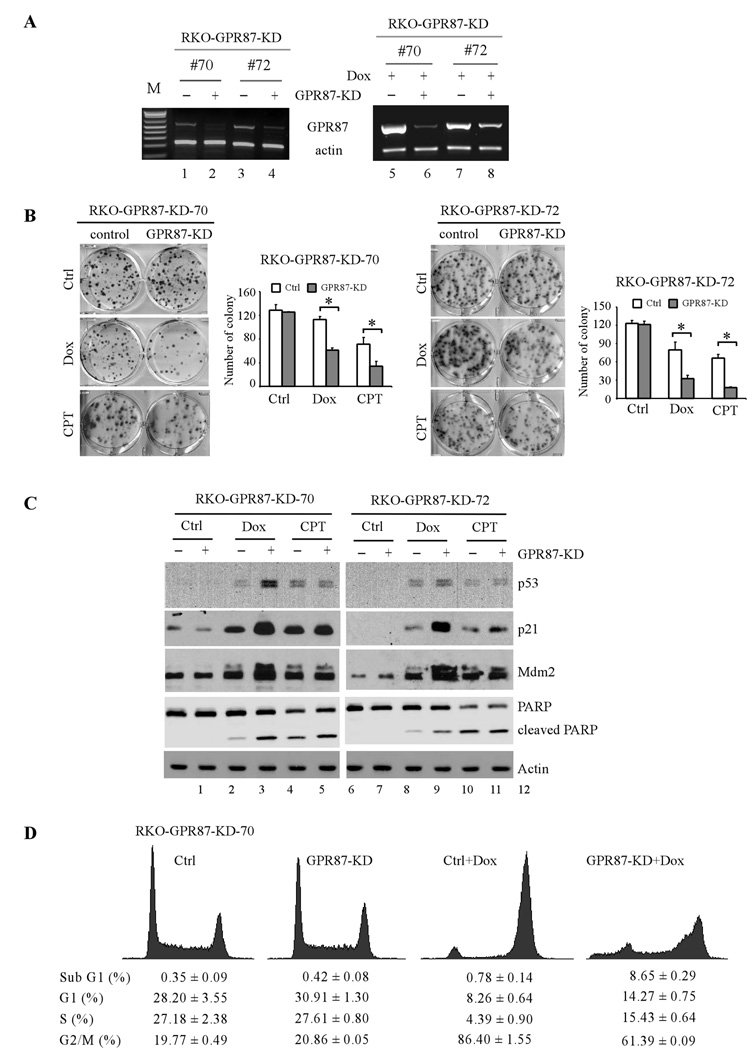
To examine the physiological role of GPR87, we generated RKO cell lines in which endogenous GPR87 can be inducibly knocked down by siRNA under the control of a tetracycline-regulated promoter. Two representative cell lines (#70 and #72) were used for further characterization. Upon induction of siRNA against GPR87 for 3 d, the levels of GPR87 transcript were markedly reduced (Fig. 4A, left panel). In addition, upon treatment with doxorubicin, which leads to p53 induction of GPR87 (Fig. 1), GPR87 was still efficiently knocked down by siRNA (Fig. 4A, right panel). Next, colony formation assay was performed and showed that GPR87 knockdown alone had little effect on cell growth (Fig. 4B, Ctrl panels). However, GPR87 knockdown remarkably decreased cell survival upon treatment with doxorubicin or camptothecin (Fig. 4B, Dox and CPT panels).
Figure 4.
GPR87 is required for cell proliferation and survival upon DNA damage. (A) Left panel: RT-PCR was performed to measure the levels of GPR87 transcript in GPR87-KD RKO cells (clone #70 and #72) upon induction of siRNA for 3 days. Right panel: The experiment was performed as in the left panel with RKO cells uninduced (−) or induced (+) to knock down GPR87 for 3 days along with or without treatment of 250 ng/mL of doxorubicin for 24 h. (B) GPR87-knockdown enhances DNA damage-induced growth inhibition. Colony formation assay was performed with RKO clone # 70 and #72, which were uninduced (control) or induced (GPR87-KD) to knock down GPR87 along with mock-treatment (Ctrl) or treatment with 10 ng/mL of doxorubicin (Dox) or 10 nM camptothecin (CPT), and then cultured over a 14-day period. The average number of colonies for each treatment condition was calculated from three separate experiments. (C) Western blots were prepared using extracts from RKO cells that were uninduced (−) or induced (+) to knock down GPR87 for 3 d, followed by mock-treatment (Ctrl) or treatment with 250 ng/mL of doxorubicin (Dox) or 250 nM camptothecin (CPT) for 24 h. The blots were probed with antibodies against p53, p21, Mdm2, PARP, and actin, respectively. (D) DNA damage-induced apoptosis is enhanced by GPR87-KD. DNA histogram analysis was performed with RKO cells, which were uninduced (−) or induced (+) to knock down GPR87 for 3 d, followed by mock-treatment (Ctrl) or treatment with 250 ng/mL of doxorubicin (Dox) for 24 h.
Since p53 is a major mediator of the DNA damage response, we examined whether p53 is involved in the GPR87 signaling pathway. Upon treatment with doxorubicin or camptothecin, p53 was stabilized and activated to induce p21 and Mdm2 (Fig. 4C). Interestingly, GPR87 knockdown markedly enhanced p53 stabilization, induction of p21 and Mdm2, and DNA damage-induced PARP cleavage (Fig. 4C, compare lanes 3 and 5 with 4 and 6, respectively; also compare lanes 9 and 11 with 10 and 12, respectively). Consistent with this, DNA histogram analysis showed that upon GPR87 knockdown, the extent of sub-G1 cells induced by treatment with doxorubicin was increased from 0.78 to 8.65% (Fig. 4D).
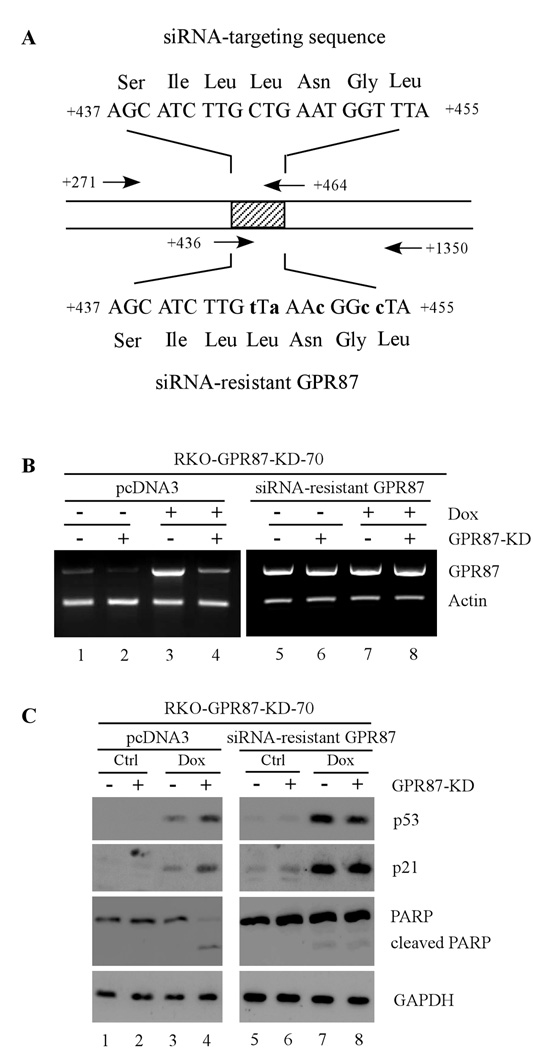
To further test this, siRNA-resistant GPR87 was generated (Fig. 5A) and transfected into RKO cells. We showed that upon inducible expression of siRNA against GPR87, endogenous GPR87 was knocked down (Fig. 5B, compare lanes 1 and 3 with 2 and 4, respectively) whereas exogenous siRNA-resistant GPR87 was still expressed (Fig. 5B, compare lanes 5 and 7 with 6 and 8, respectively). Interestingly, we showed that such a replacement abrogated the enhancement of p53 activation, induction of p21, and enhanced PARP cleavage by GPR87 knockdown upon DNA damage (Fig. 5C, compare lanes 3 and 7 with 4 and 8, respectively).
Figure 5.
Expression of siRNA-resistant GPR87 abrogates GPR87 knockdown to enhance DNA damage-induced apoptosis. (A) Schematic presentation of the siRNA-targeting sequence and the siRNA-resistant GPR87 sequence. (B) RKO cells were uninduced (−) or induced (+) to knock down GPR87 for 3 d, and then transfected with a control vector or a vector expressing siRNA-resistant GPR87 for 24 h, followed by treatment with 250 ng/mL of doxorubicin for 24 h. The level of GPR87 transcript was measured by RT-PCR. (C) Western blots were prepared with extracts from RKO cells that were treated as in (B). The blots were probed with antibodies against p53, p21, PARP and GAPDH, respectively.
To rule out potential non-specific effect by siRNA, we generated RKO and MCF7 cell lines that inducibly express siRNA against bacterial LacZ. The inducible expression of siRNA against LacZ was confirmed by reduced expression of HA-tagged LacZ in cells transfected with pcDNA3-HA-LacZ (supplemental Fig. 1A, left panel). We showed that expression of siRNA against LacZ had no effect on GPR87 expression (supplemental Fig. 1A, right panel), cell proliferation (supplemental Fig. 1B), and p53 activation (supplemental Fig. 1C).
To rule out potential cell type-specific effect, we generated MCF7 cell lines in that GPR87 can be inducibly knocked down. We showed that GPR87 was efficiently knocked down in mock- and doxorubicin-treated MCF7 cells (supplemental Fig. 2A). Similarly, we found that knockdown of GPR87 alone had no significant effect on colony formation, but enhanced growth suppression induced by treatment with doxorubicin or camptothecin (supplemental Fig. 2B). Furthermore, p53 stabilization, induction of p21 and Mdm2, and PARP cleavage were enhanced by GPR87 knockdown (supplemental Fig. 2C).
To examine whether GPR87 has similar effect in immortalized but not transformed cells, GPR87 was transiently knocked by siRNA in MCF10A cells (supplemental Fig. 3A). We showed that knockdown of GPR87 sensitized MCF10A cells to DNA damage-induced p53 stabilization and subsequent induction of p21 (supplemental Fig. 3B, compare lanes 1–3 with 4–6, respectively).
P53 is required for GPR87 pro-survival activity
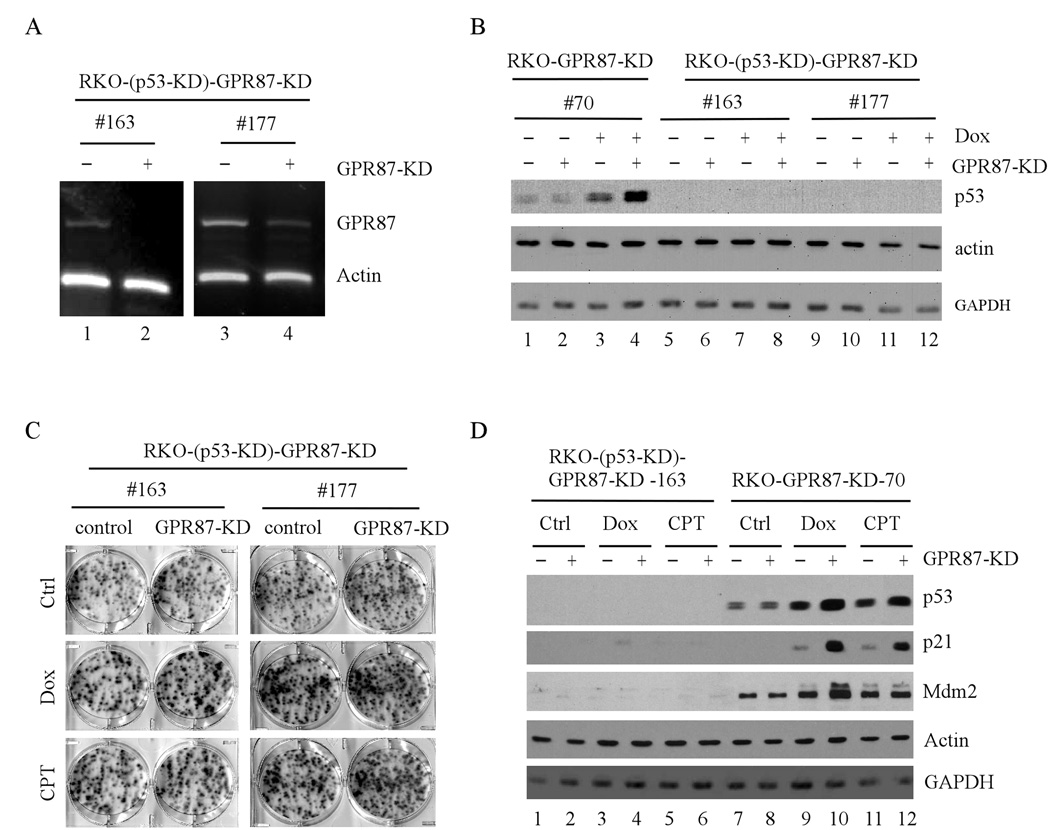
To examine whether p53 is required for GPR87 function, we generated RKO cell lines in which GPR87 can be inducibly knocked down and endogenous wild-type p53 was stably knocked down by siRNA. Two representative cell lines (clone #163 and #177) were identified in that the levels of GPR87 transcript can be inducibly knocked down (Fig. 6A, compare lanes 1 and 3 with 2 and 4, respectively) whereas the levels of p53 protein were undetectable at both mock-treated and doxorubicin-treated conditions (Fig. 6B, lanes 5–8 and 9–12). As a control, the levels of p53 protein were stabilized upon DNA damage, which was further enhanced upon GPR87 knockdown in RKO-GPR87-KD #70 cells (Fig. 6B, compare lanes 1 and 3 with 2 and 4, respectively). Next, colony formation assay was performed and showed that stable p53 knockdown had no significant effect on cell survival regardless of GPR87 knockdown (Fig. 6C, Ctrl panel). However, the enhanced sensitivity to DNA damage by GPR87 knockdown was abrogated when p53 was constitutively knocked down in both clones (#163 and #177) (Fig. 6C, Dox and CPT panels). We also showed that little if any induction of p21 and Mdm2 were detected in cells with stable p53 knockdown upon DNA damage (Fig. 6D, lanes 1–6). As a control, DNA damage induction of p21 and mdm2 was enhanced by GPR87 knockdown in RKO cells (clone #70) (Fig. 6D, lanes 7–12). Finally, we examined the activation of Akt, a signaling pathway shared by many G protein-coupled receptors. We found that GPR87 knockdown decreased Akt activation upon DNA damage (supplemental Fig. 4, compare lanes 1, 3, 5, 7 and 9 with 2, 4, 6, 8, and 10, respectively). Since Akt is known to antagonize p53 activation (31), the decreased activation of Akt may play a role in the enhanced activation of p53 by GPR87 knockdown upon DNA damage.
Figure 6.
p53 is required for GPR87 pro-survival activity. (A) Generation of p53-KD RKO cell lines in which GPR87 can be inducibly knocked down. The level of GPR87 transcript was measured in RKO cells (clone #163 and #177) uninduced (−) or induced (+) to knock down GPR87 for 3 d. (B) p53 was stably knocked down in inducible GPR87 knockdown RKO cell lines. Western blots were prepared with extracts from RKO cells that were uninduced (−) or induced (+) to knock down GPR87 for 3 d, followed by treatment with 250 ng/mL of doxorubicin for 24 h. The blots were probed with antibodies against p53, actin, and GAPDH, respectively. RKO-GPR87-KD #70 cells were similarly treated and used as a control for p53 stabilization upon DNA damage. (C) Colony formation assay was performed with RKO cells (clone #163 and #177), which were uninduced (control) or induced (GPR87-KD) to knock down GPR87 along with mock-treatment (Ctrl) or treatment with 10 ng /mL of doxorubicin (DOX ) or 10 nM camptothecin (CPT). (D) Western blots were prepared with extracts from p53-proficient (lanes 7–12) and p53-knockdown (lanes 1–6) RKO cells, which were uninduced (−) or induced (+) to knock down GPR87 for 3 d, followed by mock-treatment (Ctrl) or treatment with 250 ng/mL of doxorubicin (Dox) or 250 nM camptothecin (CPT) for 24 h. The blots were probed with antibodies against p53, p21, Mdm2, actin, and GAPDH, respectively.
Discussion
Here, we found that GPR87 is regulated by DNA damage in a p53-dependent manner. We also found that GPR87 is required for maintaining cell viability following DNA damage. Most importantly, the requirement of GPR87 for cell survival is p53-dependent and p53 is hyperactivated by DNA damage when GPR87 is knocked down. Consistent with this, GPR87 was shown to be over-expressed in squamous cell carcinoma in the lung, bladder, and head and neck (30, 32). Interestingly, transient knockdown of over-expressed GPR87 in head and neck cancer cells was found to inhibit tumor cell survival, suggesting that GPR87 is potentially responsible for these tumor cell transformations. Together, these results indicate that GPR87 is a pro-survival factor and serves as a mediator of the p53 pro-survival pathway. However, we would like to mention that over-expression of GPR87 alone has no significant effect on cell proliferation (Fig. 3). This is not surprising since the signals elicited by GPR87 are dependent on activation of GPR87 via its ligand(s). Conversely, we found that in MCF7 and RKO cells with limited basal expression levels of GPR87, knockdown of GPR87 alone has limited effect on cell survival, suggesting that GPR87 signal is not responsible for MCF7 and RKO cell transformation.
Upon activation by their ligands, GPCRs elicit many growth signals via the coupled G proteins, including activation of the MAPK pathway and the PI3K pathway (4, 33). In this study, we found that GPR87 is induced by DNA damage in a p53-dependent manner, suggesting that many survival pathways can be activated by DNA damage via GPR87. Importantly but not surprisingly, we found that knockdown of GPR87 sensitizes tumor cells to DNA damage via p53, suggesting that p53 is also a downstream effector of the GPR87 signaling pathway. Thus, how p53 is activated by lack of GPR87? Since p53 is primarily stabilized and activated post-translationally, two major pathways are likely to be involved in p53 activation (34). First, GPR87 is considered to be a member of lysophosphatidic acid receptors, which would signal to AKT for pro-survival activity (4, 31, 33). PI3K-KT is known to be a negative regulator of p53 (31). Thus, upon knockdown of GPR87, PI3K-KT would not be activated and as a result, p53 is stabilized and activated. Indeed, we found that GPR87 knockdown inhibits Akt activation upon DNA damage (supplemental Fig. 4). Alternatively, anti-survival signals that are normally repressed by GPR87 would be activated upon GPR87 knockdown, such as checkpoint kinases (ATM, ChK2) (16), leading to p53 stabilization and activation. Together, p53 and GPR87 appear to be involved in a feedback loop reminiscent of p53-MDM2 relationship except that unlike MDM2, GPR87 does not lead p53 to degradation; instead GPR87 knockdown up-regulates p53 activation. Thus, future studies are needed to further address p53 activation upon knockdown of GPR87, especially given that an antagonist to GPR87 might be explored as an agent for cancer therapy.
GPCRs are the largest family of cell surface receptors and transduce a diverse array of signals for cell survival and death. As a result, GPCRs have always been explored for drug targets. Indeed, ~50% of all current therapeutic agents directly or indirectly target GPCRs (1). As a survival factor and target of p53, GPR87 might be explored as a novel target for cancer therapeutic agent. Since lack of GPR87 can sensitize DNA damage-induced growth suppression, an antagonist to GPR87 would be a good adjuvant agent for current cancer therapeutic agents, most of which are DNA damage agents.
Supplementary Material
Acknowledgments
We thank members of the Chen laboratory for suggestions.
This work is supported in part by NIH grants (CA076069 and CA102188).
Footnotes
Disclosure of potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Spiegel AM, Weinstein LS. Inherited diseases involving g proteins and g protein-coupled receptors. Annu Rev Med. 2004;55:27–39. doi: 10.1146/annurev.med.55.091902.103843. [DOI] [PubMed] [Google Scholar]
- 3.Luttrell LM. Transmembrane signaling by G protein-coupled receptors. Methods Mol Biol. 2006;332:3–49. doi: 10.1385/1-59745-048-0:1. [DOI] [PubMed] [Google Scholar]
- 4.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 5.Muller JM. Potential inhibition of the neuro-neoplastic interactions: the clue of a GPCR-targeted therapy. Prog Exp Tumor Res. 2007;39:130–153. doi: 10.1159/000100074. [DOI] [PubMed] [Google Scholar]
- 6.Dahmane N, Sanchez P, Gitton Y. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128:5201–5212. doi: 10.1242/dev.128.24.5201. [DOI] [PubMed] [Google Scholar]
- 7.Addison CL, Daniel TO, Burdick MD. The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J Immunol. 2000;165:5269–5277. doi: 10.4049/jimmunol.165.9.5269. [DOI] [PubMed] [Google Scholar]
- 8.Lango MN, Dyer KF, Lui VW. Gastrin-releasing peptide receptor-mediated autocrine growth in squamous cell carcinoma of the head and neck. J Natl Cancer Inst. 2002;94:375–383. doi: 10.1093/jnci/94.5.375. [DOI] [PubMed] [Google Scholar]
- 9.Darmoul D, Gratio V, Devaud H, Lehy T, Laburthe M. Aberrant expression and activation of the thrombin receptor protease-activated receptor-1 induces cell proliferation and motility in human colon cancer cells. Am J Pathol. 2003;162:1503–1513. doi: 10.1016/S0002-9440(10)64283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boire A, Covic L, Agarwal A, Jacques S, Sherifi S, Kuliopulos A. PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell. 2005;120:303–313. doi: 10.1016/j.cell.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Carmeci C, Thompson DA, Ring HZ, Francke U, Weigel RJ. Identification of a gene (GPR30) with homology to the G-protein-coupled receptor superfamily associated with estrogen receptor expression in breast cancer. Genomics. 1997;45:607–617. doi: 10.1006/geno.1997.4972. [DOI] [PubMed] [Google Scholar]
- 12.Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci. 2001;22:368–376. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 13.Vousden KH. Activation of the p53 tumor suppressor protein. Biochim Biophys Acta. 2002;1602:47–59. doi: 10.1016/s0304-419x(02)00035-5. [DOI] [PubMed] [Google Scholar]
- 14.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 16.Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci. 2004;61:822–842. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol. 2006;26:1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunz F, Dutriaux A, Lengauer C. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 19.Liu G, Nozell S, Xiao H, Chen X. DeltaNp73beta is active in transactivation and growth suppression. Mol Cell Biol. 2004;24:487–501. doi: 10.1128/MCB.24.2.487-501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nozell S, Wu Y, McNaughton K. Characterization of p73 functional domains necessary for transactivation and growth suppression. Oncogene. 2003;22:4333–4347. doi: 10.1038/sj.onc.1206470. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J, Zhang S, Jiang J, Chen X. Definition of the p53 functional domains necessary for inducing apoptosis. J Biol Chem. 2000;275:39927–39934. doi: 10.1074/jbc.M005676200. [DOI] [PubMed] [Google Scholar]
- 22.Dohn M, Nozell S, Willis A, Chen X. Tumor suppressor gene-inducible cell lines. Methods Mol Biol. 2003;223:221–235. doi: 10.1385/1-59259-329-1:221. [DOI] [PubMed] [Google Scholar]
- 23.Yan W, Chen X. GPX2, a direct target of p63, inhibits oxidative stress-induced apoptosis in a p53-dependent manner. J Biol Chem. 2006;281:7856–7862. doi: 10.1074/jbc.M512655200. [DOI] [PubMed] [Google Scholar]
- 24.Liu G, Xia T, Chen X. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J Biol Chem. 2003;278:17557–17565. doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]
- 25.Nozell S, Chen X. p21B, a variant of p21(Waf1/Cip1), is induced by the p53 family. Oncogene. 2002;21:1285–1294. doi: 10.1038/sj.onc.1205191. [DOI] [PubMed] [Google Scholar]
- 26.Johansen FE, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Bargonetti J, Prives C. p53, through p21 (WAF1/CIP1), induces cyclin D1 synthesis. Cancer Res. 1995;55:4257–4263. [PubMed] [Google Scholar]
- 28.Harms KL, Chen X. The C terminus of p53 family proteins is a cell fate determinant. Mol Cell Biol. 2005;25:2014–2030. doi: 10.1128/MCB.25.5.2014-2030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Shu L, Chen X. Syntaxin 6, a Regulator of the Protein Trafficking Machinery and a Target of the p53 Family, Is Required for Cell Adhesion and Survival. J Biol Chem. 2008;283:30689–30698. doi: 10.1074/jbc.M801711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glatt S, Halbauer D, Heindl S. hGPR87 contributes to viability of human tumor cells. Int J Cancer. 2008;122:2008–2016. doi: 10.1002/ijc.23349. [DOI] [PubMed] [Google Scholar]
- 31.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 32.Gugger M, White R, Song S. GPR87 is an overexpressed G-protein coupled receptor in squamous cell carcinoma of the lung. Dis Markers. 2008;24:41–50. doi: 10.1155/2008/857474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spiegelberg BD, Hamm HE. Roles of G-protein-coupled receptor signaling in cancer biology and gene transcription. Curr Opin Genet Dev. 2007;17:40–44. doi: 10.1016/j.gde.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.