Abstract
In the mouse genome, 20 connexin genes have been detected that code for proteins of high sequence identity in the two extracellular loops, especially six conserved cysteine residues. The mouse connexin23 (Cx23) gene (Gje1) differs from all other connexin genes in vertebrates, since it codes for a protein that contains only 4 instead of 6 cysteine residues in the extracellular loops. Recently, two zebrafish connexin genes (Cx23a and Cx23b) have been identified, and a mouse mutant in the Gje1 gene has been described that exhibits a developmental defect in the lens. Here, we have compared the Cx23 gene in different mammalian species and found no transcripts in cDNA libraries of primates. Furthermore, all primate genomes analyzed contain stop codons in the Cx23 sequence, indicating inactivation of the orthologous primate GJE1 gene. No Cx23 mRNA was found in human eye. In order to analyze the properties of mouse Cx23 channels, we isolated HeLa cell clones stably expressing wild-type mCx23 or mCx23 fused to eGFP. Cells expressing Cx23-eGFP demonstrated its insertion in the plasma membrane but no punctate staining in contacting membranes characteristic for junctional plaques. In addition, we tested whether Cx23 forms functional gap junction channels electrophysiologically in cell pairs as well as by microinjection of neurobiotin and found that mouse Cx23 did not form gap junction channels in HeLa cells. However, there was a significant release of ATP from different Cx23 HeLa cell clones, even in the presence of normal culture medium with high calcium ion concentration, suggesting a hemichannel-based function of Cx23. Therefore, Cx23 seems to share functional properties with pannexin (hemi) channels rather than gap junction channels of other connexins.
Keywords: Cx23, Hemichannels, Pannexins, Pseudogene
Introduction
Gap junction channels not only allow intercellular electrical signal transfer but also cell-to-cell diffusion of ions, metabolites, nutrients, or second messengers (Willecke et al., 2002). Six connexin subunits thereby form a connexon or hemichannel which can dock to another hemichannel on apposed plasma membranes of neighboring cells to form a functional gap junction channel. Up to now, 20 connexin genes have been identified in the mouse and 21 in the human genome most of which can be considered to be orthologs (Söhl and Willecke, 2003). Connexins appear to share a common membrane topology of four transmembrane domains, intracellular C- and N-terminal regions, one intracellular and two extracellular loops, each of which contains three canonical cysteine residues forming intramolecular disulfide bounds (Foote et al., 1998).
In addition to connexins, vertebrates express another, closely related family of three proteins, called pannexins. They exhibit the same predicted protein structure but differ in both extracellular loops (Baranova et al., 2004), which are much larger, with pannexins containing only two instead of three conserved cysteine residues per loop as known from connexins (Panchin, 2005). However, the ability of pannexins to form functional gap junction channels is still under debate (Shestopalov and Panchin, 2008). Recently, it was demonstrated that the extracellular loops of pannexin-1 and pannexin-3 are glycosylated, which possibly inhibits docking of pannexin hemichannels (Boassa et al., 2007; Penuela et al., 2007) that were suggested to be involved in paracrine signaling via release of ATP and generation of calcium waves (Locovei et al., 2006). In this regard, connexins were also proposed to function as hemichannels in paracrine signaling in addition to their role as intercellular gap junction channels. Connexin hemi-channels were found to be regulated by membrane potential, phosphorylation and redox status. Furthermore, they were shown, at least in vitro, to open under conditions of low extracellular calcium concentrations (Evans et al., 2006; Spray et al., 2006).
The Cx23 gene was originally identified as cDNA (618 nucleotides open reading frame (ORF)) in a mouse transcriptome analysis (Gustincich et al., 2003), and was later recognized to code for a connexin protein with unusual four instead of six cysteine residues in its two presumptive extracellular loops (Söhl and Willecke, 2004). More recently, two orthologs of mouse Cx23 were detected in zebrafish (Cx23a and Cx23b), which show high sequence similarity to other connexins, but contain only two cysteine residues in each extracellular loop, as previously described for pannexins (Iovine et al., 2008).
In mouse lens, already three connexins were found to be expressed, Cx46 and Cx50 in fiber cells and Cx50 as well as Cx43 in lens epithelium (Gong et al., 2007). Deficiency as well as mutations in Cx50 and Cx46 result in impaired lens transparency and cataract formation in mice and humans (Gong et al., 1997; White et al., 1998). Apparently, zebrafish Cx23a, but not zebrafish Cx23b, is likely to be the fourth connexin expressed in lens during development (Iovine et al., 2008). Most recently, a mutation in mouse Cx23 was associated with complete disruption of lens formation, indicating that Cx23 plays an important role in lens development of the mouse (Puk et al., 2008). Previously, it has been reported that the formation of gap junctional plaques in lens fibers is abolished in Cx46/Cx50 double knockout mice, indicating that at least one of these two connexins is necessary for gap junctional communication in the lens fiber cells (Xia et al., 2006).
Here, we have studied Cx23 orthologs in the genome of other mammalian species. In humans, the orthologous gene (GJE1) contains a stop mutation at the beginning of the third exon and is not expressed in the lens of human eye. A closer examination of the genomes of other primates revealed stop mutations also in the GJE1 sequences of chimpanzee and rhesus monkey, suggesting functional impairment of Cx23 expression in primates. Mouse Cx23 does not form functional gap junction channels in transfected HeLa cells, although it is transported to the plasma membrane. On the other hand, we demonstrate hemichannel activity via ATP release in these cells under normal extracellular calcium concentrations. This suggests a gap junction-independent function of mouse Cx23, possibly similar as proposed for pannexins.
Materials and methods
Data acquisition
The Ensemble Gene Report (AK018698) predicted a mouse cDNA clone isolated from the RIKEN consortium (D230044M03Rik) to be homologous to a genomic mouse clone, which contained a connexin-like sequence coding for a protein of approximately 23 kDa (Gustincich et al., 2003; Söhl and Willecke, 2004). We have used this novel cDNA for a detailed BlastN search in the HUSAR/EMBL/Heidelberg derived human, rat and chimpanzee databases as well as a BlastN search on NCBI and found different additional genomic clones highly homologous to the first cDNA sequence (see Table 1). Additional transcripts were identified in rabbit, cow and zebrafish.
Table 1.
Cx23 cDNA and genomic sequences of different species
| Species | cDNA | Tissue | Chromosome (Chr) | Nucleotides (nt) | Protein (aa) | Accession no. |
|---|---|---|---|---|---|---|
| Mouse | Yes | Eye ED12 | Chr 10 | 618 | 205 | NW_001030408 |
| Rat | No | Chr 1p12 | 618 | 205 | NW_047544 | |
| Rabbit | Yes | Lens | Not sequenced | – | 205 | – |
| Opossum | No | Chr 2 | 618 | 205 | NW_00158187 | |
| Cat | No | Shotgun sequence | 618 | 205 | AANG01374118 | |
| Dog | No | Chr 1 | 618 | 205 | NW_876269 | |
| Cow | Yes (partly) | Lens | Chr 9 | 618 | 205 | NW_001495598 |
| Horse | No | Chr31 | 615 | 204 | NW_001799710 | |
| Human | No | Chr 6q24 | 606 | 201 | NW_001838990 | |
| Chimpanzee | No | Chr 5 | 606 | 201 | NW_001236564 | |
| Rhesus monkey | No | Chr 4 | 2 stop | – | NW_001116522 | |
| Zebrafish (Cx23b) | Yes | Embryo | Chr 20 | 624 | 207 | NW_001512129 |
| Zebrafish (Cx23a) | Chr 17 | 618 | 205 | NW_001511510 | ||
| Zebrafish (Cx23a) | Chr 2 | 618a | (205) | NW_001512972 |
Cx23 cDNA is found in EST databases of mouse, rabbit, cow and zebrafish. Nearly all genomes analyzed contained orthologous sequences to mouse Cx23. Predicted Cx23 sequences of human and chimpanzee are four amino acid residues shorter than mouse Cx23. In zebrafish, Cx23a is duplicated on Chr2 and Chr17.
End of exon 2 not completely sequenced (10–15 nucleotides n).
Tissue preparation
Adult 3-month-old male mice (C57BL/6) were deeply anesthetized with 2 ml Forene® airway application for 5 min according to the manufacturer’s instructions. Mice were decapitated and tissues were quickly removed and frozen in liquid nitrogen. All animal experiments were performed according to the instructions of federal and local authorities for animal welfare.
Tissues from a human eye were prepared from a postmortem biopsy cooled at 4 °C for 16 h. The female patient was 93 years old and died from colon cancer. Retina, lens, iris diaphragm and optical nerve stump were prepared and quickly frozen in liquid nitrogen.
Northern blot analysis
Total RNA from the different mouse and human tissues was prepared with TRIzol® reagent (GibcoBRL, Eggenstein, Germany) according to the manufacturer’s instructions. RNA (20 μg) was electrophoresed and transferred to Hybond™N nylon membranes (Amersham International, Amersham, Bucks., UK) by capillary diffusion in 20 × SSC. Membranes were probed by using the corresponding hybridization fragments of mouse Cx23 cDNA (618 nucleotides) (see Results). Subsequently, the amounts of total RNA on the blots were normalized by probing for glyceraldehyde-phosphate dehydrogenase (GAPDH) (Hanauer and Mandel, 1984). Probes were 32P labeled, using random-primed labeling (Multiprime Labelling Kit, Amersham) to a specific activity of 0.5–1.0 × 109 cpm/μg DNA and added to fresh QuikHyb® hybridization solution (Stratagene, La Jolla, CA, USA) at 1.25 × 106 cpm/ml. Hybridization at high stringency was carried out for 1 h at 68 °C. The filters were finally washed for 30 min in 0.1 × SSC/0.1% SDS at 60 °C and exposed to XAR X-ray film (Eastman Kodak, Rochester, NY, USA) with intensifying screen at −70 °C for three weeks. Equal loading of total RNA was additionally verified by ethidium bromide staining (18S and 28S rRNA; not shown).
RT-PCR analysis
Reverse transcription of total RNA from mouse and human tissues was performed according to (Söhl et al., 1998). Aliquots of the transcribed cDNA (approximately 0.1 ng) from tissue and cells were amplified using corresponding combinations of primers for mCx23 and hCx23: Upstream primer specific for mouse and human: Cx23KXhoIUSP 5′-ccgctcgagcggccaccatgtctctaaattacatcaagaac, downstream primer specific for mouse: mCx23EcoSTOP 5′-ggaattcctcatcgtttaaagagaaaacatgatc and downstream primer specific for human: hCx23Eco-DSP 5′-ggaattccctcattgtctgaatggaaagtataatc. Reaction mixtures (50 μl) contained 20 mM Tris-HCl (pH 8.4), 250 μM dNTPs, 1.25 mM MgCl2, 50 mM KCl, 2 μM of each primer, and 1 unit Taq DNA polymerase (Promega, Madison, Wisconsin, USA). PCR was carried out for 40 cycles using a PTC-200 thermal cycler (MJ Research, Watertown, MA, USA) with the following program: first denaturing step at 94 °C for 3 min, denaturing at 94 °C for 1 min, annealing at 55 °C for 1 min, elongation at 72 °C for 2 min, and final elongation for 7 min. After gel electrophoresis in a 1% agarose gel, the ethidium bromide stained fragments were documented. Fragments of interest were excised from the gel, purified by using the QiaQuick® purification procedure for PCR fragments (Qiagen, Hilden, Germany) and finally subcloned into the vector system suited for cloning PCR fragments (Promega). Fragments were commercially sequenced by AGOWA, Berlin, Germany.
Human DNA sequencing
The genomic region of hCx23 around the splice acceptor of exon 3 was PCR-amplified from human genomic DNA (gift of Dr. S. Dhein, Klinik für Herzchirurgie, Leipzig, Germany). Fragments were excised after gel electrophoresis, purified by using the Perfectprep® Gel Cleanup procedure for PCR fragments (Eppendorf, Hamburg, Germany), subcloned into the pGEM-Teasy vector system suitable for cloning PCR fragments (Promega) and commercially sequenced by AGOWA. The following upstream and downstream primers were used: hCx23 USP1 5′-tcatcattatcagtcattgagagg and hCx23 DSP1 5′-gccagaatgctattgccgctag.
In situ hybridization
Probes for Cx23 were generated by digesting the complete cDNA of mCx23 in the pGEM-Teasy vector (see RT-PCR analysis) with XhoI for the antisense and SpeI for the sense probe, followed by in vitro transcription (T7 or Sp6 RNA polymerase, respectively), using NTPs and digoxigenin-labeled UTP (Roche, Mannheim, Germany). C57BL/6 mice were sacrificed by cervical dislocation, eyes were removed, frozen on powdered dry ice and stored at −80 °C until sectioned. Twenty-μm cryosections were obtained and subjected to in situ hybridization using the IsHyb in situ Hybridization Kit (BioChain, Hayward, CA, USA; #K2191050) according to the manufacturer’s instructions.
Cloning of Cx23 expression vectors
Mouse Cx23 was amplified via PCR using mCx23EcoGo (5′-ggaattcctcttcgtttaaagagaaaacatg-3′) and Cx23-KXhoIUSP (see RT-PCR analysis). The fragment was cloned into the pMJ-Green vector (Maxeiner et al., 2003) using EcoRI and XhoI restriction enzymes in frame with eGFP resulting in expression of an eGFP fusion protein (i.e., Cx23eGFP). Additionally, a construct for expression of Cx23 and an IRES driven eGFP cDNA was cloned using pMJ-Green, pIRES-eGFP (Dobrowolski et al., 2007) and a PCR fragment of the complete Cx23 cDNA amplified with mCx23EcoSTOP and Cx23KXhoIUSP (see RT-PCR analysis). pMJ-Green was cleaved by XhoI and NotI removing its eGFP sequence, the Cx23 cDNA was cut by EcoRI and XhoI and the IRES-eGFP sequence was cut out of p-IRES-eGFP by NotI and EcoRI. All three fragments were ligated resulting in the final pMJ-Cx23-IRES-eGFP vector.
HeLa cell culture
Experiments were performed using HeLa cells (Human cervix carcinoma cells, ATCC CCL2) grown in Dulbeco’s modified Eagles medium supplemented with 10% fetal calf serum (Gibco BRL), 100 μg/ml streptomycin and 100 units/ml penicillin. The medium for the HeLa transfectants contained in addition 1 μg/ml puromycin (Sigma). The cells were passaged for two generations, diluted 1:10 and maintained in the 10% CO2 incubator in a moist atmosphere at 37 °C. HeLa cells were transfected with 24 μg pMJ-Cx23eGFP or pMJ-Cx23-IRES-eGFP using Lipofectamine2000 (Invitrogen, Carlsbad, CA), following the protocol of the manufacturer. Clones were picked after two weeks and grown under selective conditions.
Microinjection
The ability of Cx23 to form gap junctional channels was tested via microinjection of neurobiotin using iontophoresis. HeLa cells were grown to confluence and injected with 6% neurobiotin in 1 M KCl. Cells were fixed in 2% glutaraldehyde for 5 min, washed three times with PBS and permeabilized with 0.2% Triton-X-100 overnight at 4 °C. Neurobiotin was detected using horseradish peroxidase-conjugated avidin D (Vector Laboratories, Burlingame, CA) for 90 min at room temperature. After washing with PBS, diaminobenzidine (DAB) was used for 1 min to detect the horseradish peroxidase. Neurobiotin-positive cells were manually counted. At least 50 injections were analyzed per Cx23-expressing cell line and statistically evaluated (i.e., standard error of the mean, unpaired Student’s t-test) using Microsoft (Redmound, WA) Excel.
Electrophysiological measurements
Experiments were performed in modified Krebs-Ringer (MKR) solution containing (in mM): NaCl, 140; KCl, 4; CaCl2, 2; MgCl2, 1; glucose, 5; pyruvate, 2; and HEPES, 5 (pH 7.4). Electrodes were filled with pipette solution containing (in mM): KCl, 140; NaAsp, 10; MgATP, 3; MgCl2, 1; CaCl2, 0.2; EGTA, 2; and HEPES, 5 (pH 7.3). For simultaneous electrophysiological and fluorescence recordings, cells were grown on glass coverslips and transferred to an experimental chamber mounted on the stage of an inverted microscope (Olympus IX70) equipped with a fluorescence imaging system (UltraVIEW, Perkin Elmer, Life Sciences, Boston, MA). Cells were perfused with MKR solution at room temperature. Patch pipettes were made from patch-clamp capillary glass tubes with filaments (A-M Systems Inc., Carlsborg, WA, USA) using a horizontal puller Model P-97 (Sutter Instruments Co., USA). Junctional conductance (gj) was measured in selected cell pairs using the dual whole-cell patch-clamp system (for more details see (Rackauskas et al., 2007)). Briefly, each cell of a pair was voltage clamped independently with a separate patch-clamp amplifier. Transjunctional voltage (Vj) was induced by changing the voltage in cell-1 (ΔV1) and keeping the other constant, Vj = ΔV1. Junctional current (Ij) was measured as the change in current in the unstepped cell-2, Ij = ΔI2. Thus, gj was obtained from the ratio, −Ij/Vj, where negative sign indicates that junctional current measured in cell-2 is oppositely oriented to the one measured in cell-1.
ATP release
In order to measure the hemichannel activity in Cx23 transfectants, cells were cultured to a confluence of 50–70% on six-well plates (Falcon, Becton Dickinson, Lincoln Park, NJ) and incubated in 500 μl modified Hank’s balanced salt solution (HBSS; Sigma Aldrich) with or without Ca2+, Mg2+ and additional 1 mM ethyleneglycol-tetraacetic acid (EGTA) for 20 min at 37 °C. After incubation, 100 μl of the supernatant were collected and incubated with 100 μl nucleotide-releasing reagent (ViaLight HS kit, Cambrex, Rockland, ME, USA) for 20 min. ATP concentration was determined after addition of 20 μl ATP monitoring reagent using a Berthold Microplate LB96V luminometer to measure luciferase activity for 10 s. The luminometric results were normalized to whole protein amount. The expression level of Cx23 in HeLa cell transfectants was determined by immunoblot analyses and was further used in these studies to normalize the ATP release.
Results
Sequence analyses of Cx23 (Gje1) in different species
In a previous study on sequence similarities in gene families of channel-forming proteins (i.e., gap junctions), a novel putative member of the connexin family (GenBank PC17005) was discovered to be expressed in a cDNA contig library derived from mouse eyeball at postnatal day 12 which was assembled by the RIKEN GSC Genome Exploration Research Group (Gustincich et al., 2003). This expressed sequence tag (EST) (D230044M03Rik) was linked to a distinct genomic locus on mouse chromosome 10 in the Ensemble ID Gene Report that is schematically shown in Fig. 1A after a corresponding NCBI-mediated database search. Its predicted ORF is distributed on 3 exons and comprises 618 nucleotides (nt). The calculated theoretical molecular mass of the corresponding 205 amino acid residues (aa) is 23,830 Dalton (23 kDa), which was used to tentatively designate this connexin as mouse Cx23, in accordance with the previous common convention (Söhl and Willecke, 2003). Recently, the nomenclature of mouse (http://www.informatics.jax.org) and human (http://genenames.org/genefamily/gjphp) connexin genes was extended. The mouse Cx23 gene is now designated as Gje1, the human Cx23 gene as GJE1.
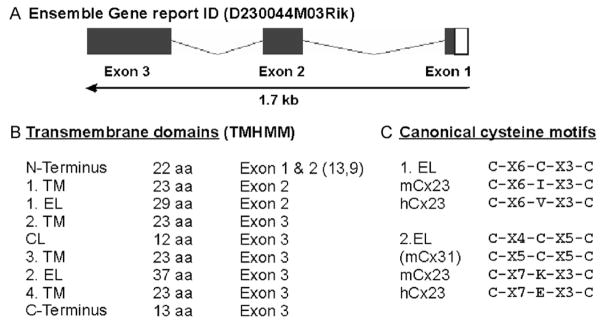
Fig. 1.
Sequence analyses of mouse Cx23. (A) Schematic drawing of the mouse Cx23 gene structure (1.7 kb) as deduced from the Ensemble Gene Report. The coding region of 618 nucleotides is distributed over three exons. (B) Size and exon-specific distribution of nine different putative domains of mouse Cx23 corresponding to transmembrane topology predicted by the HUSAR-derived subprogram TMHMM. (C) Comparison of the canonical cysteine motifs known from connexin genes (1.EL and 2.EL) with the cysteine motifs of mouse and human Cx23. The central cysteine in both presumably extracellular loops of Cx23 is missing. The distribution of the other conserved amino acid residues between the two flanking cysteines in the second extracellular loop is changed. C, cysteine, K, lysine, E, glutamate X, miscellaneous amino acid. EL, extracellular loop; CL, cytoplasmic loop; TM, transmembrane region.
More detailed sequence analyses (Fig. 1B) indicated that (i) the N-terminus is located on exon 1 and 2, (ii) the first transmembrane region and the first extracellular loop are encoded by exon 2 and (iii) that all subsequent domains are distributed on exon 3. Both the cytoplasmic loop (12 aa) and C-terminus (13 aa) are remarkably short compared to other connexins (Söhl and Willecke, 2003), whereas the second extracellular loop (37 aa) is unusually long. The predicted splice pattern and the location of the transmembrane domains were verified using the appropriate HUSAR-derived subprograms (Gene Prediction and TMHMM). However, the cysteine motifs found in both extracellular loops diverge from the canonical cysteine motifs known to be unique for connexin proteins (Fig. 1C). In each motif, the central cysteine is missing which is similar to the motifs of innexin (Phelan and Starich, 2001) or pannexin proteins (Panchin et al., 2000). Moreover, the distance between both residual cysteines of the second extracellular loop is enlarged. A sequence comparison of mCx23 using the HUSAR-based BLAST algorithm with all other mouse gene sequences indicated that mouse Cx23 was predominantly aligned to connexins. Alignment of the mCx23 protein sequence with those of the other 19 known mouse connexin proteins (Söhl and Willecke, 2003) identified 4 cysteines (i.e., the two residual ones in each of the cysteine motifs) and 4 other amino acid residues as 100% conserved.
Additional database searches in genomic libraries of rat, rabbit, cat, dog, cow, horse, human, chimpanzee, rhesus monkey and zebrafish revealed the presence of putative Cx23 orthologs in most species (Table 1), however, ESTs orthologuous to mCx23 have only been found in databases of rabbit (EB384662), cow (EG705869) and zebrafish (Iovine et al., 2008).
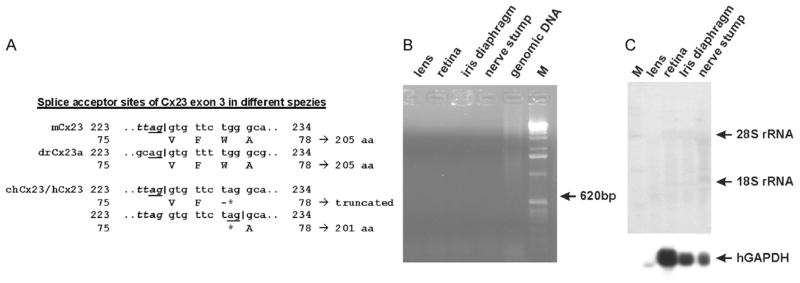
The deduced ORF of the human and chimpanzee Cx23 genes is both 12 nucleotides or four amino acid residues shorter than the mouse or zebrafish ortholog (Table 1), although two Cx23 protein sequences (A6NN92 and ENSP0000356620) suggest human Cx23 to comprise 205 amino acid residues. However, both sequences have been annotated by the Ensembl automatic analysis pipeline using either a GeneWise/Exonerate model from a database protein or a set of aligned cDNAs even from other species followed by an ORF prediction (Curwen et al., 2004). A direct comparison of mouse vs. human Cx23 resulted in 83% nucleotide and peptide identity as well as 89% peptide similarity. The shortening of the predicted sequence of primate Cx23 is apparently caused by the use of a putative different splice acceptor site of exon 3. If spliced similar to mouse Cx23, a nonsense mutation within codon 77 (tgg to tag) would otherwise terminate translation three amino acid residues after the mouse inherent splice acceptor site of exon 3. This might abrogate the functional expression of human and chimpanzee Cx23. The surrounding of the newly emerged amber stop codon 77 is predicted to be used instead as the novel splice acceptor of exon 3, which would result in a protein shortened by three amino acid residues (see Fig. 2A). We have confirmed this DNA difference by sequencing of the corresponding human genomic DNA. Interestingly, whereas the Cx23 sequences in chimpanzee and human are the same, in the Cx23 ORF of the rhesus monkey two point mutations will cause a stop of translation after codon 29 and codon 63, which were not found in Cx23 sequences of other species.
Fig. 2.
Cx23 in human lens. (A) Major difference between mouse, human, chimpanzee and zebrafish Cx23a splice acceptor site of exon 3. The underlined “ag” nt are conserved to 100% in the canonical splice acceptor motif. The asterisk indicates the g-to-a transition. This can either lead to a severely truncated version of the protein caused by a stop codon at that position, or the newly emerged “ag” can be used as a new splice acceptor site resulting in a protein that is four amino acid residues shorter than the mouse ortholog. (B) RT-PCR analysis of the hCx23 expression in four different human tissues. Amplification of a fragment indicating hCx23 expression failed, although the corresponding β-actin spliced amplicon (243 bp) was amplified from human retina cDNA and the other tissues (not shown). M: RNA ladder (Gibco-BRL). (C) Northern blot hybridization of total RNA from four different tissues of the human eye. No band indicating human Cx23 expression was detected even after three weeks of exposure, whereas rehybridization to a probe coding for the human glyceraldehyde-phosphate dehydrogenase (GAPDH) (Hanauer and Mandel, 1984) led to a specific hybridization signal of about 1.4 kb. Due to lower metabolic activity, the amounts of hGAPDH and 28S rRNA are lower in RNA from lens.
Cloning and transcription analysis of connexin23
In order to clarify the Cx23 expression profile in human and mouse, we have designed linker primers covering both the start and stop codon of either mouse or human Cx23. These primers were applied in an intron-spanning RT-PCR, to detect Cx23 expression in embryonic or adult mouse as well as human tissues. We only found a weak RT-PCR amplicon of about 600 bp in RNA of the total embryo (ED11.5), and in adult retina but the most abundant amplification occurred in adult mouse lens (Fig. 3), although this RT-PCR cannot be strictly used for quantification. However, all other tissues tested were found to be negative for Cx23 expression (Fig. 3). Both amplicons, obtained from lens and retina, were sequenced and the suggested splice pattern was confirmed. To test if the stop codon 77 in human Cx23 leads either to a truncated protein or a shift in the splicing pattern by using a newly generated splice acceptor site, we analyzed the human lens and retina for Cx23 expression. Neither Northern blot hybridization (Fig. 2B) nor RT-PCR analyses (Fig. 2C) showed any evidence for a transcript of the human Cx23 gene in any of these tissues.
Fig. 3.
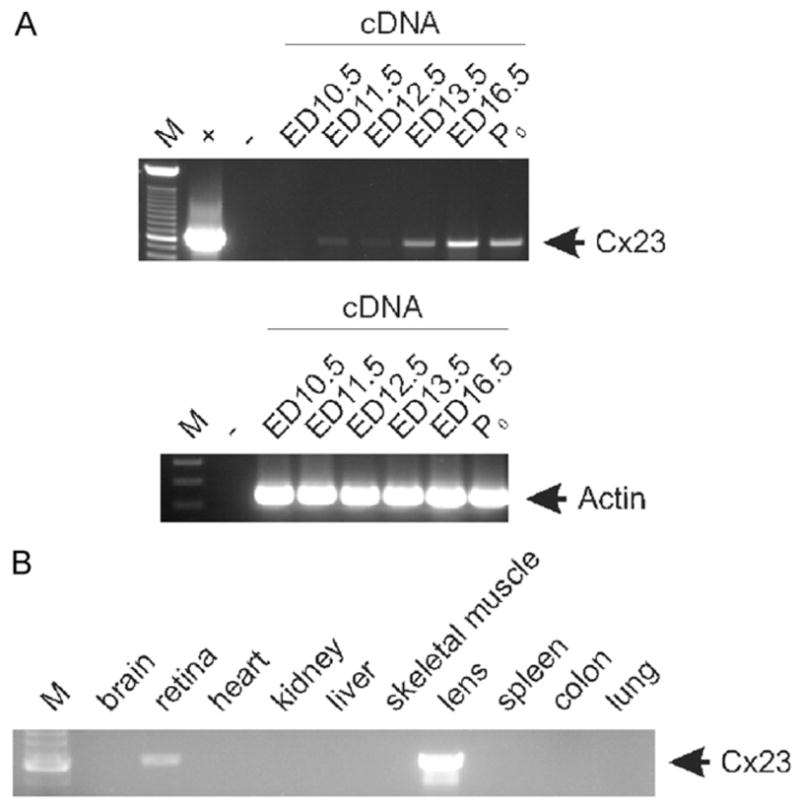
RT-PCR analysis of Cx23 expression in different mouse tissues. An amplicon of about 600 bp specific for mouse Cx23 was only weakly detected in adult retina and embryo starting at ED 11.5 (A), but highly abundant in the adult lens (B). All other tissues tested were found negative for Cx23 RNA. Amplification of the corresponding β-actin spliced amplicon (243 bp) from embryonic cDNA demonstrates integrity of cDNA synthesis without traces of genomic DNA. M: 100 bp ladder (Invitrogen); +: plasmid cDNA of Cx23; −: water control.
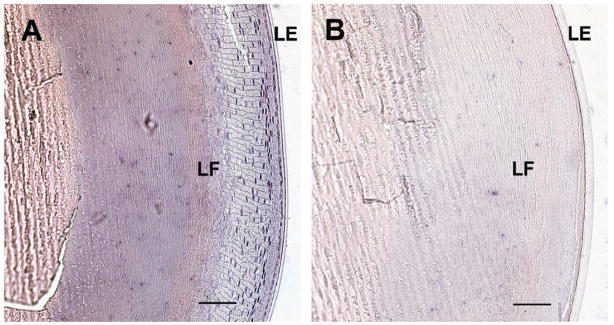
In order to determine cell-type specific transcription of mCx23 in the mouse eye, we performed non-radioactive in situ hybridizations with corresponding sense and anti-sense probes. Fig. 4A illustrates that a Cx23-specific hybridization signal was detected after incubation with the anti-sense probe in lens epithelial cells that start to get integrated at the lens equator as fibers and to a minor degree in lens epithelium. In control experiments using the corresponding sense probe no background staining was seen in total lens (Fig. 4B). We never found any specific signals in the retina, regardless of which primers (sense or anti-sense) were used. Together, these results imply Cx23 expression only in newly formed lens fiber cells of the mouse, but not in the human lens.
Fig. 4.
Non-radioactive in situ hybridization of cryosections from adult mouse eye. (A) Lens and attached lens epithelium hybridized to a Cx23 anti-sense probe of complete Cx23 cDNA. Strong staining can be seen in the nuclei of lens fiber and lens epithelial cells that start to be integrated as lens fibers (LF), whereas the lens epithelium (LE) shows less staining. (B) Lens and lens epithelium hybridized to sense control probe for comparison. Bars: 100 μm.
Function of mCx23 in transfected HeLa cells
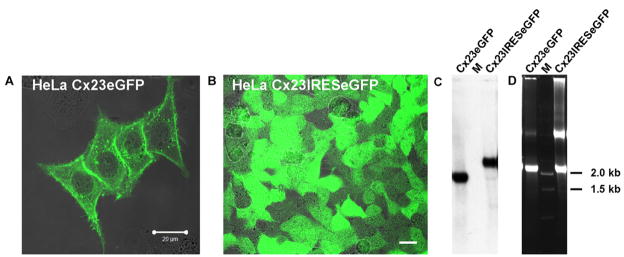
Cx23 contains only four instead of the six conserved extracellular cysteine residues of all other connexins. We wanted to determine whether Cx23 has channel properties different from the already characterized connexins and, therefore, generated HeLa cell lines stably over-expressing Cx23-eGFP or Cx23-IRES-eGFP. Expression of mCx23 and eGFP was confirmed by Northern blot analyses and fluorescence microscopy (Fig. 5). The Cx23eGFP fusion protein is ubiquitously located in the plasma membrane (Fig. 5A), however, it does not exhibit the typical punctate staining in the junctional membrane resembling junctional plaques typical of most other connexin isoforms.
Fig. 5.
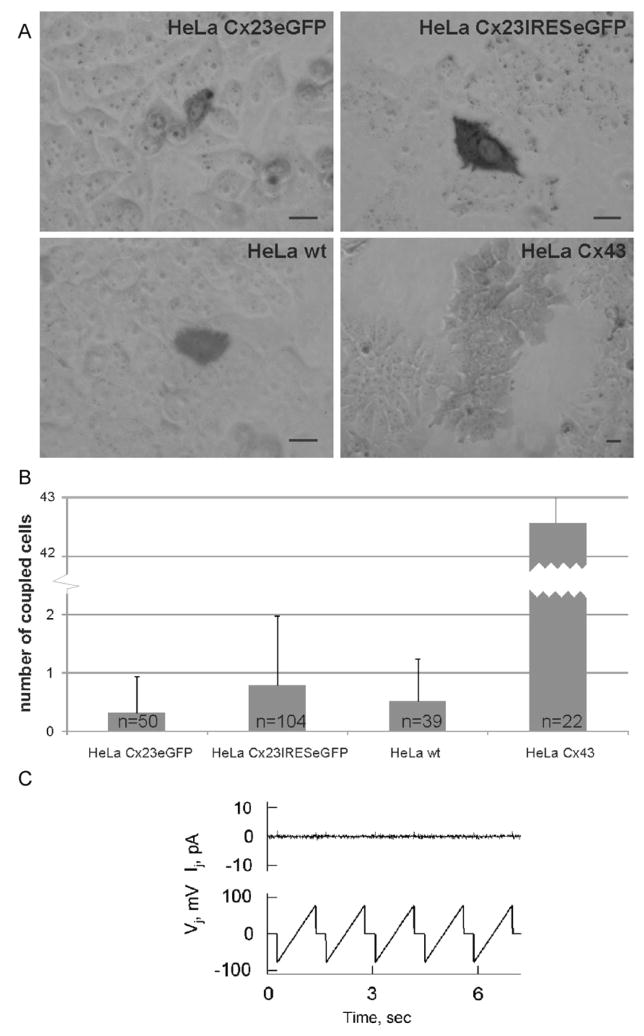
Expression analyses of Cx23 in stably transfected HeLa cells. Fluorescence microscopy of Cx23eGFP and Cx23-IRES-eGFP HeLa cells shows expression of the Cx23eGFP fusion protein in the plasma membranes of HeLa cells (A) and cytoplasmic eGFP in Cx23-IRES-eGFP HeLa cells (B). Bars: 20 μm. Northern blot analysis (C) of total RNA (D) from Cx23eGFP and Cx23-IRES-eGFP HeLa cells confirms expression of the corresponding mRNAs (1.8 kb for Cx23eGFP, 2.3 kb for Cx23-IRES-eGFP) in stably transfected HeLa cell lines. M: RNA ladder (Gibco-BRL).
To determine if Cx23 forms functional gap juntion channels, we injected neurobiotin into HeLa-Cx23eGFP, HeLa-Cx23-IRES-eGFP, HeLa-Cx43, and compared the results with injections into HeLa wild-type cells. As shown in Fig. 6, neither HeLa-Cx23eGFP nor HeLa-Cx23 showed tracer transfer significantly different from HeLa parental cells (p23eGFP = 0.179; p23IRES = 0.198), whereas Cx43-expressing HeLa cells exhibit effective transfer of neurobiotin into many neighboring cells. This suggests that Cx23 does not form functional gap junction channels in HeLa cells.
Fig. 6.
Analyses of cell–cell coupling in Cx23-expressing HeLa cells. (A) Tracer transfer was tested after microinjection of neurobiotin in a single cell of monolayers formed by HeLa-Cx23eGFP, HeLa-Cx23IRES-eGFP, HeLa-Cx43, or HeLa parental cells. Bars: 20 μm. (B) Neurobiotin-positive cells per injection were counted and statistically evaluated. Neither Cx23eGFP- nor Cx23-IRES-eGFP-expressing HeLa cells showed coupling that differed significantly from HeLa parental cells (p23eGFP = 0.179, p23IRES = 0.198). HeLa-Cx43 cells showed robust transfer of neurobiotin. (C) Absence of electrical coupling is demonstrated by dual whole-cell voltage clamp analysis of a representative cell pair expressing Cx23eGFP. Repeated voltage ramps of ±75 mV applied to one cell (Vj trace) produced no changes in junctional current (Ij) recorded in the neighboring cell.
To confirm these results, eight Cx23eGFP- and four Cx23-IRES-eGFP-expressing cell pairs were examined by using the dual whole-cell voltage clamp system. Seven of eight examined HeLa-Cx23eGFP cell pairs demonstrated full absence of electrical cell–cell coupling, as demonstrated in Fig. 6C. One cell pair was electrically coupled with gj = 0.3 nS. More close examination of this case revealed that the voltage-sensitive gating and unitary gating transitions exhibited could be attributed to Cx45, which is expressed intrinsically in HeLa wild-type cells. None of the examined cell pairs showed punctate fluorescent signals in the plasma membrane. All four HeLa-Cx23-IRES-eGFP cell pairs examined did not exhibit electrical cell–cell coupling. Therefore, we conclude that Cx23 protein either fused or not fused with eGFP is not able to form functional gap junction channels in HeLa cell transfectants.
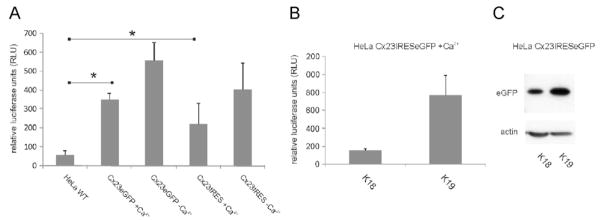
In order to identify another possible function of Cx23 apart from gap junction formation, we analyzed the activity of Cx23 hemichannels via tracer uptake and ATP release. There was no difference in uptake of propidium iodide or 4′,6-diamidino-2-phenylindole (DAPI) between Cx23-expressing HeLa cells and HeLa parental cells in the presence or absence of extracellular calcium (not shown). The extracellular ATP concentration of different Cx23-expressing HeLa clones was measured using a luciferase-based detection assay. ATP concentration in the extracellular media of Cx23-expressing cells was significantly higher (p23eGFP = 0.0187; p23IRES = 0.0043) than for HeLa wild-type cells even in the presence of normal extracellular calcium concentrations (Fig. 7). ATP release of Cx23-expressing HeLa cells was increased in the absence of calcium compared to normal calcium conditions, but this increase was not significant (p23eGFP = 0.1; p23IRES = 0.36). Thus, mouse Cx23 can form functional, calcium-independent hemi-channels in transfected HeLa cells.
Fig. 7.
ATP release from Cx23 expressing HeLa cells. (A) Two independent clones for Cx23eGFP and Cx23-IRES-eGFP were tested for release of ATP and adjusted to their expression level via Western blot analyses for eGFP. The combined data from Cx23eGFP- and Cx23-IRES-eGFP-expressing HeLa cells showed release of significantly higher amounts of ATP into the extracellular medium relative to HeLa parental cells (p23eGFP = 0.018; p23IRES = 0.004). Zero extracellular calcium concentration leads to a weak, statistically non-significant increase in ATP release in Cx23-expressing HeLa cells (p23eGFP = 0.1, p23IRES = 0.36). (B) Increased expression of Cx23 in different Cx23-IRES-eGFP HeLa cell clones led to an elevated release of ATP (clone K19 versus K18). (C) As an approximation for the expression level of Cx23 in different Cx23-IRES-eGFP-expressing cell lines, the level of eGFP was determined via Western blot analyses with β-actin serving as loading control.
Discussion
Sequence analyses of mCx23
Twenty different connexin genes were identified in the mouse genome and subsequently characterized with regard to their expression pattern and function (Willecke et al., 2002). Among them, Cx23 or Gje1 exhibits the shortest protein sequence (Söhl and Willecke, 2003). Some other sequence peculiarities are obvious: first, cytoplasmic loop and C-terminus are only half as long as the N-terminus or the other domains. This feature is quite unusual when compared to other connexins, since their cytoplasmic loops are usually about 30 to 55 amino acid residues long, and their C-termini range from 18 (mCx26) to 262 (mCx57) amino acid residues (Hombach et al., 2004; Willecke et al., 2002). The very short cytoplasmic domains of Cx23 are unlikely to function according to a ball and chain model as proposed for the closure of a Cx43 channel or hemichannels (Homma et al., 1998; Liu et al., 2006), suggesting that at least some connexin channels might be regulated by a different mechanism. The short cytoplasmic domains will also compromise the availability of potential antigenic epitopes for Cx23-specific antibodies.
Second, both extracellular loops contain cysteine motifs, with the central cysteine residue missing when compared to the canonical motifs of other connexins (Söhl and Willecke, 2003). Furthermore, the cysteine motif in the putative second extracellular loop of Cx23 is enlarged due to the insertion of additional amino acid residues resembling the corresponding motifs of innexins (Phelan and Starich, 2001) or pannexins (Panchin et al., 2000). Previous phylogenetic analyses revealed that based on sequence similarities, Cx23 is a member of the connexin gene family rather than the pannexin family (Iovine et al., 2008). Thus, Cx23 was placed at the trunk of the connexin gene family tree and could be a remnant of the early development of the connexin gene family.
Conservation of Cx23 in vertebrates
The gene structure of Cx23 with three exons of 39, 183 and 396 bp is highly conserved between species in which Cx23 transcripts are expressed and can also be found in the genome of many others. Is the Cx23 gene in primates an exception to this? If spliced according to the otherwise conserved splicing pattern, a stop mutation is located at the beginning of the third exon in human and chimpanzee. On the other hand, this could possibly generate a new splice acceptor site resulting in a four amino acid residues shorter protein. Nevertheless, RT-PCR and Northern blot hybridization analyses failed to detect such transcript in human, and no expressed sequence of Cx23 was reported in human or chimpanzee (NCBI). Thus, the mutation in human and chimpanzee is most likely a stop mutation and does not generate a new splice acceptor site. The genomic sequence of Cx23 in rhesus monkey contains two stop mutations in exon 2. We conclude that Cx23 is a pseudogene in primates. Its function could redundantly be taken over by one of the other lens connexins or pannexins. The reason why Cx23, which is highly conserved even in distantly related species like mouse, cow and zebrafish, has obviously been inactivated by mutations in primates, remains to be elucidated.
Expression pattern
The gene coding for Cx23 in zebrafish and mouse was previously shown to be expressed in the developing lens (Iovine et al., 2008; Puk et al., 2008). Transcript analyses in this study confirmed the expression of Cx23 during lens development starting with ED11.5, consistent with the beginning of lens fiber cell differentiation (Puk et al., 2008) and in the adult lens. A weak signal was also detected by RT-PCR analyses of the adult retina which, however, was not supported by in situ hybridization and is most likely due to contaminating lens cells in the retina preparation. Therefore, in addition to Cx46 (Gja3) and Cx50 (Gja8) (Gong et al., 1997; White et al., 1998), Cx23 is the third connexin expressed in lens fiber cells. Mutations or disruption of both former genes lead to cataracts and impaired lens development in mouse (Gong et al., 2007), whereas a homozygous point mutation in mCx23 leads to a complete disruption of lens development (Puk et al., 2008). This suggests an important role for Cx23 in lens development, at least in the mouse.
Channel function
The function of gap junctional channels and hemi-channels formed by mCx23 was tested in stably transfected HeLa cells. A fusion protein of Cx23 and eGFP was transported to the plasma membrane, but did not evoke the typical punctate fluorescence pattern of gap junction channels (Elfgang et al., 1995). The detectable signals were instead equally distributed on the plasma membrane similar to pannexins in NRK cells (Penuela et al., 2007). Although zebrafish Cx23a was recently shown to allow weak neurobiotin transfer between transfected HeLa cells in about 50% of the injected cell pairs (Iovine et al., 2008), we could not detect mCx23 gap junction-mediated coupling by using tracer transfer and dual whole-cell voltage clamp analyses. This is reminiscent to NRK cells in which glycosylation of both extracellular loops impedes the formation of pannexin-mediated gap junction channels (Penuela et al., 2007). Further studies are required to test whether mouse Cx23 is also unable to form functional gap junction channels in lens fibers.
Several connexins and pannexins were shown to form functional hemichannels in Xenopus oocytes or mammalian cell lines, i.e., Cx30, Cx45, Cx46, Cx50 and Px1 (Dahl and Locovei, 2006; Valiunas and Weingart, 2000; Valiunas, 2002) which are thought to play a pivotal role in paracrine signaling. Most connexin hemichannels, except Cx32 (De Vuyst et al., 2006), mouse Cx30.2 and its putative human ortholog, Cx31.9 (Bukauskas et al., 2006), are dependent on low extracellular calcium concentration for opening, whereas hemichannels formed of pannexins are not sensitive to extracellular calcium (Dahl and Locovei, 2006). HeLa cells expressing Cx23 exhibited release of ATP at a significantly higher level than HeLa wild-type cells. In addition, ATP release was not sensitive to extracellular calcium concentration. Dye uptake could not be observed. This difference between ATP release and dye uptake through Cx23 hemichannels, which was not reported for Cx43 (Kang et al., 2008), might have two reasons: The detection of ATP is much more sensitive compared to the simple observation of dye uptake. On the other hand, the Cx23 hemichannel might be selective. Negatively charged ATP (MW 507; charge z = −4) could be favored over positively charged molecules used for dye uptake like DAPI (MW 279; z = +2) or propidium iodide (MW 415; z = +2). The selectivity of Cx23 hemichannels should be investigated in further studies. However, the calcium-independent release of ATP, as demonstrated here, suggests that Cx23 may play its vital role in differentiation and elongation of lens fiber cells during lens development in the mouse rather by a hemichannel-mediated mechanism than as gap junctional channel. The functional role of Cx23 in this process and the impact of different mutations on its function need to be determined in the future.
Acknowledgments
We would like to thank Prof. Dr. Antonia Joussen and Dr. Norbert Kociok from the Center of Ophthalmology, University of Cologne, for providing tissue biopsies from a human eye. We further thank Joana Fischer and Petra Kußmann for excellent technical assistance. We also thank Alexandra Gellhaus, PhD, Institute of Molecular Biology, and Melanie Friederichs, PhD, ZMB-Developmental Biology, University of Duisburg-Essen, for help with in situ hybridizations. This work was supported by a grant of the German Research Association (Wi 270/22-5,6) and SFB 645, Projekt B2, to K. Willecke and by NIH Grants R01NS036706 and RO1HL084464 to F.F. Bukauskas.
References
- Baranova A, Ivanov D, Petrash N, Pestova A, Skoblov M, Kelmanson I, Shagin D, Nazarenko S, Geraymovych E, Litvin O, Tiunova A, Born TL, Usman N, Staroverov D, Lukyanov S, Panchin Y. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics. 2004;83:706–716. doi: 10.1016/j.ygeno.2003.09.025. [DOI] [PubMed] [Google Scholar]
- Boassa D, Ambrosi C, Qiu F, Dahl G, Gaietta G, Sosinsky G. Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J Biol Chem. 2007;282:31733–31743. doi: 10.1074/jbc.M702422200. [DOI] [PubMed] [Google Scholar]
- Bukauskas FF, Kreuzberg MM, Rackauskas M, Bukauskiene A, Bennett MV, Verselis VK, Willecke K. Properties of mouse connexin 30.2 and human connexin 31.9 hemichannels: implications for atrioventricular conduction in the heart. Proc Natl Acad Sci USA. 2006;103:9726–9731. doi: 10.1073/pnas.0603372103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SM, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl G, Locovei S. Pannexin: to gap or not to gap, is that a question? IUBMB Life. 2006;58:409–419. doi: 10.1080/15216540600794526. [DOI] [PubMed] [Google Scholar]
- De Vuyst E, Decrock E, Cabooter L, Dubyak GR, Naus CC, Evans WH, Leybaert L. Intracellular calcium changes trigger connexin 32 hemichannel opening. EMBO J. 2006;25:34–44. doi: 10.1038/sj.emboj.7600908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolski R, Sommershof A, Willecke K. Some oculodentodigital dysplasia-associated Cx43 mutations cause increased hemichannel activity in addition to deficient gap junction channels. J Membr Biol. 2007;219:9–17. doi: 10.1007/s00232-007-9055-7. [DOI] [PubMed] [Google Scholar]
- Elfgang C, Eckert R, Lichtenberg-Frate H, Butterweck A, Traub O, Klein RA, Hulser DF, Willecke K. Specific permeability and selective formation of gap junction channels in connexin-transfected HeLa cells. J Cell Biol. 1995;129:805–817. doi: 10.1083/jcb.129.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WH, De Vuyst E, Leybaert L. The gap junction cellular internet: connexin hemichannels enter the signalling limelight. Biochem J. 2006;397:1–14. doi: 10.1042/BJ20060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote CI, Zhou L, Zhu X, Nicholson BJ. The pattern of disulfide linkages in the extracellular loop regions of connexin 32 suggests a model for the docking interface of gap junctions. J Cell Biol. 1998;140:1187–1197. doi: 10.1083/jcb.140.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Li E, Klier G, Huang Q, Wu Y, Lei H, Kumar NM, Horwitz J, Gilula NB. Disruption of alpha3 connexin gene leads to proteolysis and cataractogenesis in mice. Cell. 1997;91:833–843. doi: 10.1016/s0092-8674(00)80471-7. [DOI] [PubMed] [Google Scholar]
- Gong X, Cheng C, Xia CH. Connexins in lens development and cataractogenesis. J Membr Biol. 2007;218:9–12. doi: 10.1007/s00232-007-9033-0. [DOI] [PubMed] [Google Scholar]
- Gustincich S, Batalov S, Beisel KW, Bono H, Carninci P, Fletcher CF, Grimmond S, Hirokawa N, Jarvis ED, Jegla T, Kawasawa Y, LeMieux J, Miki H, Raviola E, Teasdale RD, Tominaga N, Yagi K, Zimmer A, Hayashizaki Y, Okazaki Y. Analysis of the mouse transcriptome for genes involved in the function of the nervous system. Genome Res. 2003;13:1395–1401. doi: 10.1101/gr.1135303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauer A, Mandel JL. The glyceraldehyde 3 phosphate dehydrogenase gene family: structure of a human cDNA and of an X chromosome linked pseudogene; amazing complexity of the gene family in mouse. EMBO J. 1984;3:2627–2633. doi: 10.1002/j.1460-2075.1984.tb02185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach S, Janssen-Bienhold U, Söhl G, Schubert T, Bussow H, Ott T, Weiler R, Willecke K. Functional expression of connexin57 in horizontal cells of the mouse retina. Eur J Neurosci. 2004;19:2633–2640. doi: 10.1111/j.0953-816X.2004.03360.x. [DOI] [PubMed] [Google Scholar]
- Homma N, Alvarado JL, Coombs W, Stergiopoulos K, Taffet SM, Lau AF, Delmar M. A particle-receptor model for the insulin-induced closure of connexin43 channels. Circ Res. 1998;83:27–32. doi: 10.1161/01.res.83.1.27. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Gumpert AM, Falk MM, Mendelson TC. Cx23, a connexin with only four extracellular-loop cysteines, forms functional gap junction channels and hemichannels. FEBS Lett. 2008;582:165–170. doi: 10.1016/j.febslet.2007.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Kang N, Lovatt D, Torres A, Zhao Z, Lin J, Nedergaard M. Connexin43 hemichannels are permeable to ATP. J Neurosci. 2008;28:4702–4711. doi: 10.1523/JNEUROSCI.5048-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Arce FT, Ramachandran S, Lal R. Nanomechanics of hemichannel conformations: connexin flexibility underlying channel opening and closing. J Biol Chem. 2006;281:23207–23217. doi: 10.1074/jbc.M605048200. [DOI] [PubMed] [Google Scholar]
- Locovei S, Wang J, Dahl G. Activation of pannexin 1 channels by ATP through P2Y receptors and by cytoplasmic calcium. FEBS Lett. 2006;580:239–244. doi: 10.1016/j.febslet.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Maxeiner S, Kruger O, Schilling K, Traub O, Urschel S, Willecke K. Spatiotemporal transcription of connexin45 during brain development results in neuronal expression in adult mice. Neuroscience. 2003;119:689–700. doi: 10.1016/s0306-4522(03)00077-0. [DOI] [PubMed] [Google Scholar]
- Panchin YV. Evolution of gap junction proteins – the pannexin alternative. J Exp Biol. 2005;208:1415–1419. doi: 10.1242/jeb.01547. [DOI] [PubMed] [Google Scholar]
- Panchin Y, Kelmanson I, Matz M, Lukyanov K, Usman N, Lukyanov S. A ubiquitous family of putative gap junction molecules. Curr Biol. 2000;10:R473–R474. doi: 10.1016/s0960-9822(00)00576-5. [DOI] [PubMed] [Google Scholar]
- Penuela S, Bhalla R, Gong XQ, Cowan KN, Celetti SJ, Cowan BJ, Bai D, Shao Q, Laird DW. Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J Cell Sci. 2007;120:3772–3783. doi: 10.1242/jcs.009514. [DOI] [PubMed] [Google Scholar]
- Phelan P, Starich TA. Innexins get into the gap. BioEssays. 2001;23:388–396. doi: 10.1002/bies.1057. [DOI] [PubMed] [Google Scholar]
- Puk O, Loster J, Dalke C, Soewarto D, Fuchs H, Budde B, Nurnberg P, Wolf E, de Angelis MH, Graw J. Mutation in a novel connexin-like gene (Gjf1) in the mouse affects early lens development and causes a variable small-eye phenotype. Invest Ophthalmol Vis Sci. 2008;49:1525–1532. doi: 10.1167/iovs.07-1033. [DOI] [PubMed] [Google Scholar]
- Rackauskas M, Kreuzberg MM, Pranevicius M, Willecke K, Verselis VK, Buckauskas FF. Gating properties of heterotypic gap junction channels formed of connexins 40, 43, and 45. Biophys J. 2007;92:1952–1965. doi: 10.1529/biophysj.106.099358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestopalov VI, Panchin Y. Pannexins and gap junction protein diversity. Cell Mol Life Sci. 2008;65:376–394. doi: 10.1007/s00018-007-7200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söhl G, Willecke K. An update on connexin genes and their nomenclature in mouse and man. Cell Commun Adhes. 2003;10:173–180. doi: 10.1080/cac.10.4-6.173.180. [DOI] [PubMed] [Google Scholar]
- Söhl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res. 2004;62:228–232. doi: 10.1016/j.cardiores.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Söhl G, Degen J, Teubner B, Willecke K. The murine gap junction gene connexin36 is highly expressed in mouse retina and regulated during brain development. FEBS Lett. 1998;428:27–31. doi: 10.1016/s0014-5793(98)00479-7. [DOI] [PubMed] [Google Scholar]
- Spray DC, Ye ZC, Ransom BR. Functional connexin “hemichannels”: a critical appraisal. Glia. 2006;54:758–773. doi: 10.1002/glia.20429. [DOI] [PubMed] [Google Scholar]
- Valiunas V. Biophysical properties of connexin-45 gap junction hemichannels studied in vertebrate cells. J Gen Physiol. 2002;119:147–164. doi: 10.1085/jgp.119.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiunas V, Weingart R. Electrical properties of gap junction hemichannels identified in transfected HeLa cells. Pflügers Arch. 2000;440:366–379. doi: 10.1007/s004240000294. [DOI] [PubMed] [Google Scholar]
- White TW, Goodenough DA, Paul DL. Targeted ablation of connexin50 in mice results in microphthalmia and zonular pulverulent cataracts. J Cell Biol. 1998;143:815–825. doi: 10.1083/jcb.143.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke K, Eiberger J, Degen J, Eckardt D, Romualdi A, Guldenagel M, Deutsch U, Söhl G. Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem. 2002;383:725–737. doi: 10.1515/BC.2002.076. [DOI] [PubMed] [Google Scholar]
- Xia CH, Cheng C, Huang Q, Cheung D, Li L, Dunia I, Benedetti LE, Horwitz J, Gong X. Absence of alpha3 (Cx46) and alpha8 (Cx50) connexins leads to cataracts by affecting lens inner fiber cells. Exp Eye Res. 2006;83:688–696. doi: 10.1016/j.exer.2006.03.013. [DOI] [PubMed] [Google Scholar]