Abstract
Background
Single-nucleotide polymorphisms (SNPs) in 2 distinct regions of the gene for the sortilin-related receptor (SORL1) (bounded by consecutively numbered SNPs 8−10 and 22−25) were shown to be associated with Alzheimer disease (AD) in multiple ethnically diverse samples.
Objective
To test the hypothesis that SORL1 is associated with brain magnetic resonance imaging (MRI) measurements of atrophy and/or vascular disease.
Design, Setting, and Patients
We evaluated the association of 30 SNPs spanning SORL1 with MRI measures of general cerebral atrophy, hippocampal atrophy, white matter hyperintensities, and overall cerebrovascular disease in 44 African American and 182 white sibships from the MIRAGE Study. We performed single-and 3-SNP haplotype association analyses using family-based tests. Haplotypes found to be significantly associated with at least 1 MRI trait were tested for association with 6 pathological traits in a separate sample of 69 white patients with autopsy-confirmed AD.
Results
In white patients, white matter hyperintensities were associated with multiple markers in the region encompassing SNPs 6 to 10, whereas cerebral and hippocampal atrophy were associated with markers from the region including SNPs 21 to 26. Examination of specific 3-SNP haplotypes from these 2 regions in the autopsy-confirmed cases of AD revealed association of white matter disease with SNPs 8 to 10 and association of hippocampal atrophy with SNPs 22 to 26. The haplotype CGC at SNPs 8 to 10 was associated with fewer white matter changes in the clinical (P<.001) and autopsy (P=.02) samples.
Conclusions
Variants of SORL1 previously associated with AD are also associated with MRI and neuropathological measures of neurodegenerative and cerebrovascular disease. These findings not only support the hypothesis that multiple areas in SORL1 are of functional importance but also raise the possibility that multiple SORL1 variants influence amyloid precursor protein or endothelial lipoprotein processing or both in different regions of the brain.
CONVERGING LINES OF EVIdence have implicated 1 member of the vacuolar proteinsorting 10 family—the sortilin-related receptor, low-density lipoprotein receptor class A repeat-containing protein (SORL1) (GenBank 6653)—in the pathogenesis of Alzheimerdisease (AD).1-3 Pathological studies4,5 have documented reduced SORL1 expression in the brains of some patients with AD. Association between the risk of AD and multiple SORL1 single-nucleotide polymorphisms (SNPs) has been demonstrated in several studies3,6-10 that included populations of diverse ethnic background, used various study designs(ie, clinic case-control, family-based, or population-based), and relied on clinical or pathological criteria for AD, whereas the evidence of the association was equivocal in another study.11
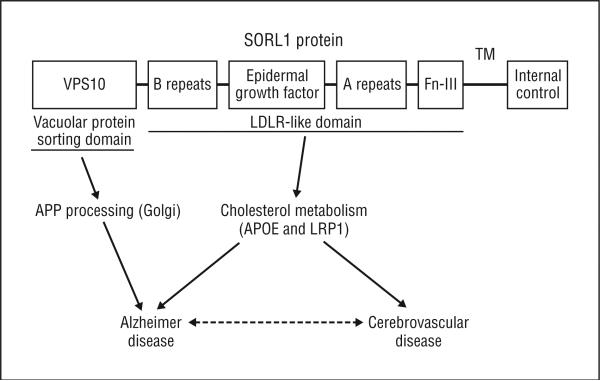
One hypothesis for the role of SORL1 in the pathogenesis of AD is that SORL1 modulates subcellular trafficking of the amyloid precursor protein (APP). Reduced expression of SORL1 may lead to routing of APP into compartments where it is cleaved by the presenilin 1 complex to generate the amyloid β peptide.3 The role of SORL1 in APP processing is strongly supported by the fact that SORL1 interacts with endogenous APP holoproteins3,12 but does not bind to APP C-terminal fragments or to other type I membrane proteins.3 The SORL1 protein also belongs to a superfamily of low-density lipoprotein receptors (SorLA/LR11) that bind apolipoprotein E (APOE) and are implicated in cholesterol metabolism13 and atherogenesis.14 Human macrophages exposed to SORL1 have elevated lipid levels, and mice genetically deficient for SORL1 show signs of thickening arteries and macrophage infiltration relative to SORL1 knockout controls. These observations suggest a role for SORL1 in the development of atherosclerosis14 and raise the intriguing possibility that, like APOE and other cholesterol metabolism genes (eg, paraoxonase15), SORL1 may also increase dementia risk through effects on cerebrovascular abnormalities. Indeed, cerebrovascular disease increases the odds of dementia16 and potentially interacts with AD pathologic mechanisms to alter memory.17 Neurodegenerative and cerebrovascular processes can be linked to the known protein structure of SORL1 (Figure).
Figure.
Inferred biochemical basis for the role of the sortilin-related receptor SORL1 in Alzheimer disease and cerebrovascular disease. The SORL1 protein contains several functional domains. The N-terminus region of the protein contains the vacuolar protein sorting 10 (VPS10) domain, which may be the region of the gene directly involved in amyloid precursor protein (APP) processing, although the APP binding domain is not yet known. SORL1 also contains a low-density lipoprotein receptor (LDLR)–like domain that is important in cholesterol metabolism. APOE indicates apolipoprotein E; Fn-III, fibronectin type III region; LRP1, low-density lipoprotein–related protein 1; and TM, transmembrane region.
The possibility of multiple mechanisms of action associated with distinct SORL1 polymorphisms is supported by previous association studies. The SNPs associated with AD are located in 2 regions in SORL1 separated by a mean (SD) of 100 (15) kilobases that contain areas of tight linkage disequilibrium in white and African American subjects.3,18 Associations with SNPs in the region near the 5′ end have been reported in white European subjects, Hispanic subjects from the Dominican Republic, and Israeli-Arab subjects,3,6 whereas SNPs in the region closer to the 3′ end show association in African American, Han Chinese, and some white European and Hispanic subjects.3,6,8,10 Furthermore, associations with a specific 3-SNP haplotype in each of these locations have been replicated in multiple data sets representing several ethnic groups.3,6 These results suggest the existence of AD predisposing variants in different functional domains of SORL1 and thus raise the possibility for variable effects of intragenic polymorphisms on the biochemical properties of SORL1, on its cell-type–specific patterns of expression, or on both.
To further elucidate how SORL1 gene variants influence processes leading to AD, we evaluated the association of SORL1 SNPs with brain magnetic resonance imaging (MRI) findings in a multiethnic group of families containing at least 1 sibpair discordant for AD. In addition, we compared SORL1 haplotypes with analogous measures of severity of AD and vascular abnormalities in a separate group of deceased individuals with pathologically confirmed AD.
METHODS
SUBJECT RECRUITMENT AND CLASSIFICATION
Subjects included for the genetic comparisons with brain MRI traits are participants of the Multi-Institutional Research in Alzheimer's Genetic Epidemiology (MIRAGE) Study, an ongoing, family-based genetic epidemiological study of AD described in detail elsewhere.19 Briefly, patients with AD and their siblings were recruited from 14 clinical centers in the United States, Canada, Germany, and Greece. Sibships were ascertained through a single affected proband with probable AD according to the criteria of the National Institute of Neurological and Communicative Disorders and Stroke and Alzheimer's Disease and Related Disorders Association.20 The cognitive status of individuals identified as nondemented was confirmed by a score of 86 or higher on the modified Telephone Interview of Cognitive Status.21 Age at onset of AD was defined as the age at which the earliest symptoms were reported by proxy. Only subjects who self-reported their ethnicity as white of European descent or African American and gave written informed consent are included in this study. Study protocols were approved by institutional review boards at each recruitment site. Families with at least 1 affected and 1 unaffected sibling with clinical, MRI, and genotype data available for analysis were included in this study. An independent sample of 69 patients with autopsy-confirmed AD (mean [SD] age at death, 75.0 [7.5] years) was identified through the Alzheimer Disease Center of Boston University and was obtained from the Edith Norse Rogers Veterans Affairs Medical Center, Bedford, Massachusetts. This group is a subsample of patients included in a previous neuropathological study of AD22 with complete neuropathological data and DNA specimens available for analysis.
MRI TRAITS
In the living subjects (those with AD and the unaffected siblings), MRI scans of the brain were obtained with 1.5-T magnetic field strength scanners using a standard protocol of 3-dimensional T1-weighted high-resolution sequence, a double spin-echo sequence, and a fluid-attenuated inversion recovery sequence. Semiquantitative measures of neurodegeneration and cerebrovascular disease were derived from digitized brain MRIs using methods previously described.23 These measures were designed to be simple to use and have been shown to correlate linearly with image quantification.24 The MRIs were evaluated by a single rater (C.D.) blinded to age, sex, and affection status to reduce the interrater variability common to semiquantitative methods. Previous work has found that MRI ratings of white matter hyperintensities (WMHs) and MRI infarcts are correlated with cerebrovascular abnormalities but not with AD pathologic features.25 We used WMHs and a severity rating of cerebrovascular disease (CVR) (a combined measure of WMH and infarction) as indicators of brain cerebrovascular disease. Bilateral medial temporal atrophy (MTA) is also significantly correlated with AD pathologic features among individuals clinically diagnosed as having AD26 and was used as an indicator of the AD process. A measure of generalized cerebral atrophy (CA) was included in this analysis. Cerebral atrophy and WMH were rated on a scale of 0 to 100. When an infarct was observed, a CVR of 0 to 100 was assigned. When no infarct was observed, the CVR was set equal to the WMH score. We rated MTA on an ordinal scale of 0 to 4 according to previously described methods.27 We demonstrated previously that CA, MTA, WMH, and CVR have moderate to high heritability.28
NEUROPATHOLOGICAL TRAITS
Brains from the subjects with autopsy-confirmed AD were assessed by a single neuropathologist (A.C.M.). A more detailed description of the brain samples, including staining techniques and scoring, is described elsewhere.22 Tissue was scored by region for detailed abnormalities, including evidence of atherosclerosis, infarct, and atrophy using an ordinal (0 [none] to 3 [extensive]) or a binary (present or absent) scale. Traits reflecting vascular, amyloid, white matter, or hippocampal damage or atrophy were derived from existing tissue measurements. A summary variable for atherosclerosis was defined as the sum of ordinal measures of atherosclerosis recorded for the internal carotid, anterior, middle, posterior cerebral, basilar, and vertebral arteries. A summary measure of nonacute infarcts was defined as the sum of binary measurements recorded for the cerebrum, cerebellum, and basal ganglia. White matter disease was defined (1) as the sum of ordinal measures of white matter atrophy in the frontal, parietal, temporal, and occipital regions (nWM1) and (2) as the sum of ordinal and binary measures of general cerebral disease, astrocytosis, demyelination, arteriolar sclerosis, and microinfarcts observed in the white matter (nWM2). Measures of amyloid angiopathy and hippocampal atrophy (HA) were also included in this analysis.
GENOTYPING
Genotyping of genomic DNA extracted from peripheral blood lymphocytes or frozen brain tissue was performed using SNP assays on a real-time platform (7900; Applied Biosystems, Inc, Foster City, California) using the manufacturer's protocols. Twenty-nine SNPs tested in the original report3 and 1 additional SNP (designated 22b) were selected for genotyping for this study (Table 1). Duplicate wells were scattered on DNA template plates. The duplicate discordance rate did not exceed 5% and was persistently localized to 2 samples, which were subsequently excluded from all analyses. The overall genotype call rate was found to be more than 95% for all SNPs typed. The SNPs were assessed for Hardy-Weinberg equilibrium in unrelated, unaffected siblings within each ethnic group and excluded from further analysis in that ethnic group if the test result was significant at the .05 level. The APOE genotyping was performed as described previously.19
Table 1.
SORL1 SNP Characteristics
| Minor Allele (Frequency) |
|||||
|---|---|---|---|---|---|
| Marker No. | SNP Database No. | Alleles | Physical Map Location, bp | White Subjects | African American Subjects |
| 1 | rs4935774 | A/C | 120826964 | C (0.23) | C (0.46) |
| 2 | rs578506 | C/G | 120828687 | C (0.46) | C (0.13) |
| 3 | rs582446 | A/G | 120833069 | G (0.12) | G (0.43) |
| 4 | rs661057 | C/T | 120834164 | C (0.42) | C (0.29) |
| 5 | rs11218304 | C/T | 120854321 | C (0.39) | C (0.09) |
| 6 | rs560573 | A/T | 120866094 | A (0.36) | A (0.34) |
| 7 | rs12364988 | A/G | 120872836 | A (0.47) | G (0.41) |
| 8 | rs668387 | T/C | 120873131 | T (0.41) | T (0.36) |
| 9 | rs689021 | A/G | 120876330 | A (0.42) | A (0.35) |
| 10 | rs641120 | T/C | 120886175 | T (0.41) | T (0.33) |
| 11 | rs4935775 | C/A | 120894712 | C (0.42) | C (0.10) |
| 12 | rs12285364 | T/C | 120898436 | T (0.05) | T (0.09) |
| 13 | rs2298813 | A/G | 120898894 | A (0.05) | A (0.09) |
| 14 | rs11600231 | C/T | 120911918 | C (0.09) | C (0.11) |
| 15 | rs2276346 | T/G | 120919686 | T (0.36) | T (0.09) |
| 16 | SORL1-T833T | T/A | 120931165 | T (0.07) | T (0.01) |
| 17 | rs556349 | G/T | 120931417 | T (0.33) | G (0.32) |
| 18 | rs11218340 | T/A | 120936564 | T (0.08) | T (0.06) |
| 19 | rs2070045 | G/T | 120953300 | G (0.23) | G (0.08) |
| 20 | rs3824966 | G/C | 120953393 | G (0.23) | G (0.08 |
| 21 | SORL1-ex26 | G/C | 120959359 | C (0.09) | C (0.27) |
| 22 | rs1699102 | C/T | 120962172 | C (0.33) | T (0.47) |
| 22b | rs2276412 | C/T | 120966056 | T (0.02) | T (0.16) |
| 23 | rs3824968 | T/A | 120981132 | A (0.31) | A (0.13) |
| 24 | rs2282649 | C/T | 120984168 | T (0.28) | T (0.11) |
| 25 | rs1010159 | C/T | 120988611 | C (0.37) | T (0.46) |
| 26 | rs1784933 | G/A | 120994626 | G (0.06) | G (0.39) |
| 27 | rs1614735 | C/A | 120998211 | A (0.50) | C (0.16) |
| 28 | rs1133174 | A/G | 121006965 | A (0.45) | G (0.25) |
| 29 | rs1131497 | G/C | 121007955 | G (0.43) | G (0.19) |
Abbreviations: bp, base pair; SNP, single-nucleotide polymorphism; SORL1, sortilin-related receptor gene.
STATISTICAL ANALYSIS
MRI Traits
Each MRI trait was evaluated for statistical normality within subgroups of AD patients and unaffected siblings using Kolmogorov-Smirnov and Anderson-Darling statistics and normal probability plots implemented in commercially available software (SAS, version 9.1.3).29 Variables with skewed distributions were natural log–transformed and reassessed for normality. Association analyses of MRI traits and SNPs, adjusting for potential confounders, were conducted using transmission disequilibrium tests,30,31 assuming an additive genetic model under the null hypothesis of no linkage and no association between SNP markers and traits. This procedure inherently accounts for population admixture by analyzing the family as a unit rather than as independent individuals. Covariate adjustment was incorporated into association testing through generalized estimating equations. The residuals of traits after adjustment for age at MRI and disease status were used as continuous outcomes. All association analyses were performed separately for the African American and white families. Disease duration, APOE genotype, and sex were also considered covariates but did not change results and therefore were not included in analyses. Haplotype association analyses were performed using sliding windows of 3 contiguous SNPs for each trait. A haplotype was considered to be associated with the MRI trait if the global P value was significant at the .05 level. Individual haplotypes with less than 5% frequency were excluded from global testing procedures. Haplotype-specific P values were also calculated for tests, with global P values significant at less than .05.
Replication Using Neuropathological Traits
Six summary measures from autopsy-derived neuropathological traits were analyzed as ordinal traits using a proportional odds model. To constrain the number of statistical comparisons, each trait was evaluated for association with sliding windows of 3 contiguous SNPs from each SORL1 region that was significantly associated with MRI traits and with AD in previous studies. We used haplo.score32 to assess overall haplotype distribution differences for each trait under an additive genetic model. Association of individual haplotypes with each neuropathological trait was evaluated for each haplotype with at least 5% frequency.
RESULTS
A total of 44 African American and 182 white families, each of which included at least 1 affected and 1 unaffected sibling with clinical, MRI, and genotype data available for analysis, were included in this study. Characteristics of these subjects are displayed in Table 2. The African American and white families had approximately the same proportion of unaffected siblings. Within each ethnic group, the mean age at disease onset in individuals with AD was approximately the same as the mean age at the MRI examination in the unaffected siblings. Mean ages at onset and MRI examination were similar across ethnic groups. A higher proportion of white participants were male relative to the African American participants.
Table 2.
Characteristics of MIRAGE Families With at Least 1 Member With AD and 1 Unaffected Sibling
| African American Subjects | White Subjects | |
|---|---|---|
| Total No. of individuals | 101 | 414 |
| No. of families with ≥1 unaffected-affected pair | 42 | 182 |
| No. of individuals with AD | 44 | 189 |
| No. of unaffected siblings | 57 | 225 |
| No. of families with >1 individual with AD | 2 | 7 |
| No. of families with >1 unaffected sibling | 9 | 34 |
| Mean (SD) age at disease onset in individuals with AD, y | 70.6 (9.0) | 67.2 (9.1) |
| No. male/total No. (%) of individuals with AD | 15/44 (34) | 86/189 (46) |
| Mean (SD) age at MRI in unaffected siblings, y | 71.3 (9.5) | 70.1 (9.0) |
| No. male/total No. (%) of unaffected siblings | 18/57 (32) | 88/225 (39) |
Abbreviations: AD, Alzheimer disease; MIRAGE, Multi-Institutional Research in Alzheimer's Genetic Epidemiology; MRI, magnetic resonance imaging.
ASSOCIATIONS WITH MRI TRAITS
All SNPs were in Hardy-Weinberg equilibrium in the unrelated white siblings unaffected by AD. In the unrelated African American siblings without AD, SNPs 4, 6, 10, 12, 21, and 23 were not in Hardy-Weinberg equilibrium and were thus excluded from association analyses in this ethnic group. Initial evaluation of potential confounders, namely sex and APOE genotype, showed little effect on association estimates. The reported models were adjusted for age at MRI and disease status according to previously described methods.23
Table 3 indicates that significant associations at the α = .05 significance level were observed in the white families for WMH with SNPs 6, 8, 9, 10, and 15 and for CVR with SNPs 1, 8, 9, 10, and 18. There was a marginally significant association of MTA with SNP 11. Cerebral atrophy was associated with 2 more distally located SNPs (16 and 21). None of the traits were associated with SORL1 SNPs in the African American families (results not shown); however, there were relatively few families informative (ie, with sibling genotypes that allow for inference of differential transmission of marker alleles from a parent) for these analyses (9−33 families).
Table 3.
SORL1 SNPs Showing Association With at Least 1 MRI Trait in the MIRAGE White Families
|
P Value (No. of Informative Families)a |
||||
|---|---|---|---|---|
| SNP | WMH | CVR | CA | MTA |
| 1 | .053 (73) | .046 (73) | .43 (73) | .21 (Tb/73) |
| 6 | .03 (66) | .16 (66) | .65 (66) | .18 (Tb/66) |
| 8 | .001 (81) | .006 (81) | .35 (81) | .34 (Cb/81) |
| 9 | <.001 (76) | .002 (76) | .44 (76) | .29 (Gb/76) |
| 10 | .006 (78) | .02 (78) | .94 (78) | .16 (Cb/78) |
| 11 | .08 (76) | .42 (76) | .57 (Tb/76) | .050 (Tb/76) |
| 15 | .04 (Gb/80) | .47 (80) | .42 (Gb/80) | .12 (80) |
| 16 | .33 (Ab/31) | .21 (Ab/31) | .004 (31) | .36 (Ab/31) |
| 18 | .15 (29) | .03 (29) | .45 (29) | .98 (29) |
| 21 | .38 (Gb/38) | .35 (Gb/38) | .02 (38) | .24 (38) |
Abbreviations: CA, cerebral atrophy; CVR, severity rating of cerebrovascular disease; MIRAGE, Multi-Institutional Research in Alzheimer's Genetic Epidemiology; MRI, magnetic resonance imaging; MTA, medial temporal atrophy; SNP, single-nucleotide polymorphism; SORL1, sortilin-related receptor gene; WMH, white matter hyperintensity.
Significant results are given in boldface type.
Indicates major allele associated with increasing MRI trait abnormality.
In white families, 3-SNP haplotypes in regions constituting SNPs 1 to 3, 6 to 10, 21 to 25, and 27 to 29 were associated with at least 1 MRI trait (Table 4). The most frequent haplotypes for SNPs 6 to 8 (TGC) and 7 to 9 (GCG) were associated with increased CVR and WMH. Another haplotype for SNPs 7 to 9 (ATA) was associated with decreased CA. Haplotype CGC in SNPs 8 to 10 was associated with decreased WMH and CVR. Cerebral atrophy and MTA were associated with haplo-types in the region spanning SNPs 21 to 29.
Table 4.
SORL1 Haplotypes Showing Association With at Least 1 MRI Trait in the MIRAGE White Families
| SNP, Haplotype | Frequency, % (No. of Families) | Trait | P/R (Haplotype-Specific P Value) | Global P Value |
|---|---|---|---|---|
| 1−3 | WMH | .049 | ||
| TGA | 33 (101) | P (.03) | ||
| CGA | 16 (103) | R (.09) | ||
| TCA | 35 (127) | R (.40) | ||
| 6−8 | WMH/CVR | <.001/.04 | ||
| TGC | 31 (143) | R (.003)/R (.02) | ||
| TAC | 16 (92) | P (.73)/P (.35) | ||
| AAT | 15 (133) | R (.38)/R (.40) | ||
| AGC | 6 (67) | R (.68)/R (.94) | ||
| AAC | 6 (76) | P (.52)/R (.15) | ||
| TGT | 8 (71) | P (.06)/R (.13) | ||
| AGT | 9 (73) | R (.20)/R (.39) | ||
| TAT | 10 (88) | P (.16)/P (.3) | ||
| 7−9 | WMH/CVR/CA | .02/.047/.02 | ||
| GCG | 30 (122) | R (<.001)/R (.001)/P (.56) | ||
| ACG | 15 (102) | P (.80)/P (.5)/P (.57) | ||
| GTA | 10 (82) | R (.09)/P (.21)/P (.44) | ||
| ATA | 18 (134) | R (.16)/R (.22)/P (.026) | ||
| ACA | 7 (88) | P (.99)/P (.95)/P (.20) | ||
| GCA | 6 (72) | R (.44)/R (.19)/P (.32) | ||
| ATG | 6 (85) | P (.72)/P (.98)/R (.14) | ||
| GTG | 6 (69) | R (.50)/R (.51)/R (.78) | ||
| 8−10 | WMH/CVR | .009/.003 | ||
| CGC | 38 (166) | P (<.001)/P (.002) | ||
| TAT | 22 (165) | R (.02)/P (.046) | ||
| TGC | 7 (76) | R (.51)/R (.60) | ||
| CAC | 7 (79) | P (.82)/R (.23) | ||
| TAC | 7 (80) | R (.16)/R (.27) | ||
| CGT | 7 (75) | R (.69)/R (.66) | ||
| TGT | 6 (75) | P (.66)/R (.74) | ||
| CAT | 7 (77) | P (.60)/R (.82) | ||
| 21−22b | CA | .02 | ||
| GTC | 58 (92) | P (.97) | ||
| CTC | 8 (39) | R (.003) | ||
| GCC | 31 (76) | R (.12) | ||
| 23−25 | CA | .006 | ||
| TCT | 48 (114) | P (.49) | ||
| ACT | 6 (123) | R (.41) | ||
| TCC | 11 (137) | R (.09) | ||
| ACC | 8 (129) | R (.11) | ||
| TTC | 6 (122) | R (.04) | ||
| ATC | 13 (104) | R (.11) | ||
| 27−29 | MTA | .05 | ||
| GGC | 30 (98) | P (.005) | ||
| TGC | 18 (93) | R (.42) | ||
| GAC | 5 (80) | R (.23) | ||
| TAC | 6 (76) | R (.21) | ||
| GAG | 12 (86) | R (.92) | ||
| TAG | 22 (90) | R (.52) |
Abbreviations: CA, cerebral atrophy; CVR, severity rating of cerebrovascular disease; MIRAGE, Multi-Institutional Research in Alzheimer's Genetic Epidemiology: MRI, magnetic resonance imaging; MTA, medial temporal atrophy; P, protective; R, risk; SNP, single-nucleotide polymorphism; SORL1, sortilin-related receptor gene; WMH, white matter hyperintensity.
ASSOCIATIONS WITH NEUROPATHOLOGICAL TRAITS
Based on the association results for MRI traits in the MIRAGE sample and for AD in previous studies, SORL1 SNPs 1 to 3, 8 to 10, and 22 to 26 (ie, six 3-SNP combinations) were selected for haplotype analysis with the 6 neuropathology traits in the autopsy sample. The overall haplotype distribution for SNPs 8 to 10 was associated with nWM1 (Table 5). The CGC haplotype was associated with decreased nWM1 and the TAT haplotype was associated with increased nWM1. The nHA measure was associated with each of the sliding window 3-SNP haplotypes in the region of SNPs 22 to 26 (global P=.01, P=.009, P=.008, and P=.02). We present the results for individual haplo-types for those sets of SNPs with global P<.01 (SNPs 22b-24 and 23−25). Inspection of these results revealed that the SNPs 23−24 TC core haplotype is consistently associated with increased nHA and the AT core haplotype is associated with decreased nHA (Table 5). There was no association of any of the overall haplotype distributions with summary measures of nonacute infarcts, amyloid angiopathy, or atherosclerosis or for nWM2.
Table 5.
SORL1 Haplotypes Showing Association With at Least 1 Neuropathological Trait in the Autopsy Sample
| SNP, Haplotype | Frequency, % | Trait | P/R (Haplotype-Specific P Value) | Global P Value |
|---|---|---|---|---|
| 8−10 | nWM1 | .03 | ||
| CGC | 60 | P (.02) | ||
| TAT | 36 | R (.009) | ||
| 22b-24 | nHA | .009 | ||
| CTC | 70 | R (.01) | ||
| CAT | 21 | P (<.001) | ||
| CAC | 5 | R (.63) | ||
| 23−25 | nHA | .008 | ||
| TCT | 66 | R (.01) | ||
| ATC | 21 | P (<.001) | ||
| TCC | 8 | R (.62) |
Abbreviations: nHA, sum of all measures of hippocampal atrophy; nWM1, the sum of ordinal measures of white matter atrophy in the frontal, parietal, temporal, and occipital regions; P, protective; R, risk; SNP, single-nucleotide polymorphism; SORL1, sortilin-related receptor gene.
REVISITING THE ASSOCIATION OF SORL1 WITH AD IN THE MIRAGE STUDY
Among the 9 data sets included in the original genetic association study of SORL1 and AD, the MIRAGE white family data set is 1 of 2 that did not show significant association with any SNPs or haplotypes.3 To assess whether this is a result of heterogeneity (eg, only some families associated with SORL1,orsome families associated with a variant in one region of SORL1 and other families associated with a variant in another region) ascribable to the degree of cerebrovascular disease, we evaluated the association of SORL1 3-SNP haplotypes with AD among subgroups of the MIRAGE white families with high and low WMH findings. Families for which the WMH score of a sibling of the individual with AD was greater than the median WMH score of 7 among all individuals with AD were classified as having high WMH, and the remaining families were classified as having low WMH. Table 6 indicates that, among families with low WMH, the combination of SNPs 16 to 18 was associated with AD (global P = .03). The AGA haplotype was associated with increased risk of AD (P=.007), and the ATT haplotype is protective (P=.03). Further analysis of SNPs in this haplotype revealed that the association signal is attributable primarily to SNP 17 (P = .008). None of the SORL1 haplotypes were significant in the families with high WMH.
Table 6.
Association of SORL1 Haplotypes With AD in MIRAGE White Families Stratified by Amount of WMHs
|
Low-WMH Familiesa(n=94) |
High-WMH Familiesa(n=90) |
|||||||
|---|---|---|---|---|---|---|---|---|
| SNP, Haplotype | Global P Value | Frequency, % | P Value | R/P | Global P Value | Frequency, % | P Value | R/P |
| 16−18 | .03 | .84 | ||||||
| AGA | 64.0 | .007 | R | 64.0 | >.99 | R | ||
| ATA | 23.9 | .13 | P | 24.2 | .89 | P | ||
| ATT | 6.1 | .03 | P | 5.2 | .70 | R | ||
| TGA | 5.8 | .89 | P | 6.2 | .34 | P | ||
Abbreviations: AD, Alzheimer disease; MIRAGE, Multi-Institutional Research in Alzheimer's Genetic Epidemiology; P, protective; R, risk; SNP, single-nucleotide polymorphism; SORL1, sortilin-related receptor gene; WMH, white matter hyperintensity.
Defined in the “Revisiting the Association of SORL1 With AD in the MIRAGE Study” subsection of the “Results” section.
COMMENT
We observed that SORL1 variants previously associated with risk of the AD clinical phenotype were also associated with MRI measures of brain atrophy and cerebrovascular disease in white families containing at least 1 AD-affected and 1 unaffected sibling. In particular, measures of white matter disease with and without inclusion of cerebral infarcts were associated with multiple SORL1 markers in the region encompassing SNPs 6 to 10. By comparison, measures of CA and HA were associated primarily with markers in a distinctly separate region of SORL1 that includes SNPs 21 to 26. The MRI association findings were investigated further by comparing the distributions of 3-SNP haplotypes from these 2 SORL1 regions with several analogous neuropathological traits in a series of autopsy-confirmed AD brains. Specifically, a measure of white matter disease was associated with SNPs 8 to 10 and HA was associated with SNPs 22 to 26. The SNP 23-to-25 haplo-type ATC, which was significantly associated with less HA in the sample of AD brains, was previously shown to be associated with decreased risk of AD in several white data sets.3 Moreover, the SNP 23-to-25 haplotype TTC was significantly associated with increased risk of AD in the study by Rogaeva et al3 and with greater HA in the MIRAGE families. Finally, we found that adjusting for the extent of presumed cerebrovascular disease (as measured by the amount of WMH) resulted in the detection of significant association between SORL1 and AD that was previously not identified in this data set using the conventional clinical phenotype.3 We believe these results extend current knowledge regarding the role of SORL1 as a genetic risk factor for clinical AD through the use of unique endophenotypes associated with separate domains of the SORL1 gene that are presumed to have a different biological function (Figure).
Although unique, these findings are consistent with evolving evidence that the clinical phenotype of AD may result from multiple diseases,16 particularly the interaction of AD with cerebrovascular disease leading to changes in memory function.17 One study17 suggests that AD-related neurodegenerative processes and cerebrovascular infarcts contribute independently to the odds of dementia. Magnetic resonance imaging studies support the pathological data. Among older individuals, WMH burden is significantly increased in association with advancing age, risk factors for stroke, and the presence of infarction.33,34 White matter hyperintensities are also increased in volume and distribution among patients with AD compared with cognitively normal control subjects23,35 and are associated with pathological evidence of cerebrovascular disease.25 Consistent with the pathological data, a population-based study showed that HA and WMH, independently and synergistically, substantially increase the odds of dementia.36 In addition, infarction on MRI is associated with 2-fold increased odds of dementia.37 These converging lines of evidence suggest that AD-related dementia may be the consequence of multiple pathological processes.
Our genetic data support a multifactorial hypothesis for AD. When the MIRAGE families in this study were stratified according to the extent of WMH burden in individuals with AD, a significant association between SORL1 and AD was identified, whereas previous association studies of this cohort found no association.3 Among the families in which a sibling of the individual with AD was classified as having low WMH, AD was associated with a haplotype from a region (bounded by SNPs 16−18) that also showed association with AD in other data sets.3,8
We acknowledge several limitations in our study. First, our clinical sample, which was ascertained through sibling pairs discordant for AD, is probably enriched for brain atrophy compared with a general population of elderly adults. Subjects with severe cerebrovascular disease are probably poorly represented in our sample because this presentation is an exclusionary criterion for the diagnosis of probable AD,20 and siblings would be less likely to volunteer if they were ill with conditions such as heart disease or diabetes that might be more associated with cerebrovascular abnormalities. To address the possibility that the genetic basis of the MRI traits may differ in those who do and do not have AD, we adjusted our association models for the confounding effects of AD status and age. Second, few of the results in our family sample would be considered significant after adjustment using a Bonferroni correction for testing 30 individual SNPs (threshold P=.05/30=.0017) and 28 haplotypes (threshold global P=.05/28=.0018). Because the transmission disequilibrium test is one of the most conservative but robust methods for genetic association analysis,38 other approaches can be considered for assessing the impact of multiple comparisons on false-positive results. The expected number of results with P<.05 for each trait in our study would be 1.5 for single-SNP analyses (ie, 30×.05) and 1.4 for haplotype analyses (ie, 28×.05), assuming a 1-tailed test. Two or more significant single SNP (Table 4) and global haplotype (Table 5) comparisons were observed for 3 of the traits (WMH, CVR, and CA), suggesting that our findings are not caused by chance alone. The results in the autopsy sample are less sensitive to multiple testing because we tested only 6 SNP combinations. Third, the pattern of haplotype associations for SNPs 23 to 25 with HA in the autopsy sample is not consistent with the pattern in the MRI sample and, thus, these results may not represent a true replication. This inconsistency may be explained by differences in haplotypic frequencies between the 2 samples and by differences in the composition of the study samples, one of which includes controls and mildly to moderately affected individuals with AD and the other of which contains only individuals with end-stage AD. Nonetheless, the patterns of association of the specific haplotypes with HA in each data set are consistent with haplotypic association patterns for AD observed in several other white populations.3
We also acknowledge that results in the white families may not be generalizable to other ethnic groups. None of the single-SNP association tests in the African American sample yielded significant results, perhaps owing to the small number of these families compared with the much larger samples of white families. Haplotype analysis was attempted in the African American families, but sample sizes were too small to be conclusive (data not shown). The clinical and autopsy samples do not overlap, which precludes the possibility of directly correlating MRI and neuropatho-logical measures within the same subjects. Because none of the SNPs tested in this study are known to affect SORL1 protein structure or expression, further experiments demonstrating function of these and perhaps additional SORL1 variants will be necessary. A much larger number of SNPs will likely be needed to cover adequately the haplotype structure of this large gene.3,18 Another concern is that semi-quantitative MRI measures are simplified constructs of cerebrovascular and neurodegenerative changes and may not truly represent these domains in the manner we have suggested. However, we have shown previously in this sample that these measures are highly heritable after controlling for APOE genotype.28 Finally, the MRI and neuropatho-logical traits in this study are snapshots of progressive disease. Alzheimer disease may depend on both static factors (eg, inherited predispositions) and dynamic processes (eg, cumulative injury over time). Controlling for APOE geno-type and age in our analyses lessens but only partially addresses this concern.
Our findings also do not provide a simple explanation of how polymorphisms in the SORL1 gene might lead to expressed dementia. In the SNP 8-to-10 haplotype, GCG has been associated with increased risk of AD in previously published data sets,3,6 but in the present study this same haplotype was associated with decreased WMH in the MIRAGE white families and was not associated with CA or HA as expected. These apparently incongruent association patterns are consistent with the idea that the SNPs assayed in this study are not coding specifically for the functional variants, but that there is a variant in this region of SORL1 that is directly related to AD pathogenesis (and perhaps to a process leading to increased white matter disease). This explanation is also supported by the findings of AD association in some data sets with other SNP 8 to 10 haplotypes6 and with SNPs adjacent to, but not including, 8 to 10.9 An alternative explanation for the association of haplotype CGC with the increased risk of AD and decreased risk of WMD is that different biological variants in SORL1 may influence AD pathogenesis through multiple and possibly synergistic pathways. Regions of the low-density lipoprotein domain bind APOE,39 which may be involved in the pathogenesis of AD by mechanisms not yet fully understood,40,41 and γ-secretase cleavage of APP may regulate cholesterol metabolism in the central nervous system via the lipoprotein receptor LRP1.42 These associations in multiple regions of the gene could reflect the mosaic structure of the SORL1 protein (Figure).43 Thus, the mechanism of action of SORL1 on AD-related neurodegenerative processes may reflect the increased formation of amyloid β peptide1-3 and impaired cholesterol metabolism, which may also influence the formation of amyloid β peptide, cerebrovascular disease, or both. More detailed genetic and functional studies will be necessary to sort out these complex multidimensional interactions among SORL1, AD, and cerebrovascular disease. These future studies may capitalize on data emerging from genome-wide association studies of the MIRAGE and other cohorts44 to evaluate interaction of SORL1 with other genes on brain changes associated with AD.
In summary, our study shows that SORL1 variants previously associated with AD are also associated with MRI and neuropathological measures of neurodegenerative and cerebrovascular disease. These findings support not only the hypothesis of multiple areas in SORL1 of functional importance, but also the possibility that SORL1 variants result in differential influence on APP, endothelial lipo-protein processing, or both.
Funding/Support
This study was supported in part by grants R01-AG09029, R01-HG/AG02213, K24-AG027841, and P30-AG13846 from the National Institutes of Health, and by the Wellcome Trust (Dr St. George-Hyslop).
Footnotes
Financial Disclosure: None reported.
REFERENCES
- 1.Andersen OM, Reiche J, Schmidt V, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102(38):13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Offe K, Dodson SE, Shoemaker JT, et al. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26(5):1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogaeva E, Meng Y, Lee JH, et al. The sortilin-related receptor SORL1 is functionally and genetically associated with Alzheimer's disease. Nat Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scherzer CR, Offe K, Gearing M, et al. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61(8):1200–1205. doi: 10.1001/archneur.61.8.1200. [published correction appears in Arch Neurol. 2007;64(4):557] [DOI] [PubMed] [Google Scholar]
- 5.Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65(9):866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JH, Cheng R, Schupf N, et al. The association between genetic variants in SORL1 and Alzheimer's disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64(4):501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JH, Cheng R, Honig LS, Vonsattel J-PG, Clark L, Mayeux R. The association between genetic variants in SORL1 and autopsy-confirmed Alzheimer's disease. Neurology. 2008;70(11):887–889. doi: 10.1212/01.wnl.0000280581.39755.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng Y, Lee JH, Cheng R, St George-Hyslop P, Mayeux R, Farrer LA. Association between SORL1 and Alzheimer's disease in a genome-wide study. Neuroreport. 2007;18(17):1761–1764. doi: 10.1097/WNR.0b013e3282f13e7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webster JA, Myers AJ, Pearson JV, et al. Sorl1 as an Alzheimer's disease predisposition gene? Neurodegener Dis. 2008;5(2):60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- 10.Tan E, Lee J, Chen CP, Teo Y, Zhao Y, Lee WL. SORL1 haplotypes modulate risk of Alzheimer's disease in Chinese. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2007.10.013. [published online ahead of print December 4, 2007] doi:10.1016/j.neurobiolaging.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Rowland C, Catanese J, et al. SORL1 variants and risk of late-onset Alzheimer's disease. Neurobiol Dis. 2008;29(2):293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt V, Sporbert A, Rohe M, et al. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282(45):32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- 13.Sugiyama T, Kumagai H, Morikawa Y, et al. A novel low-density lipoprotein receptor–related protein mediating cellular uptake of apolipoprotein E-enriched β-VLDL in vitro. Biochemistry. 2000;39(51):15817–15825. doi: 10.1021/bi001583s. [DOI] [PubMed] [Google Scholar]
- 14.Ohwaki K, Bujo H, Jiang M, Yamazaki H, Schneider WJ, Saito Y. A secreted soluble form of LR11, specifically expressed in intimal smooth muscle cells, accelerates formation of lipid-laden macrophages. Arterioscler Thromb Vasc Biol. 2007;27(5):1050–1056. doi: 10.1161/ATVBAHA.106.137091. [DOI] [PubMed] [Google Scholar]
- 15.Erlich PM, Lunetta KL, Cupples LA, et al. MIRAGE Study Group Polymorphisms in the PON gene cluster are associated with Alzheimer disease. Hum Mol Genet. 2006;15(1):77–85. doi: 10.1093/hmg/ddi428. [DOI] [PubMed] [Google Scholar]
- 16.Schneider JA, Boyle PA, Arvanitakis Z, Bienias JL, Bennett DA. Subcortical infarcts, Alzheimer's disease pathology, and memory function in older persons. Ann Neurol. 2007;62(1):59–66. doi: 10.1002/ana.21142. [DOI] [PubMed] [Google Scholar]
- 17.Schneider JA, Wilson RS, Bienias JL, Evans DA, Bennett DA. Cerebral infarctions and the likelihood of dementia from Alzheimer disease pathology. Neurology. 2004;62(7):1148–1155. doi: 10.1212/01.wnl.0000118211.78503.f5. [DOI] [PubMed] [Google Scholar]
- 18.Thorisson GA, Smith AV, Krishnan L, Stein LD. The international HapMap Project Web site. Genome Res. 2005;15(11):1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green RC, Cupples LA, Go RCPG, et al. MIRAGE Study Group Risk of dementia among white and African American relatives of patients with Alzheimer's disease. JAMA. 2002;287(3):329–336. doi: 10.1001/jama.287.3.329. [DOI] [PubMed] [Google Scholar]
- 20.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of the Department of Health and Human Services Task Force on Alzheimer's disease. Neurology. 1984;34(7):939–945. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 21.Roccaforte WH, Burke WJ, Bayer BL, Wengel SP. Validation of a telephone version of the Mini-Mental State Examination. J Am Geriatr Soc. 1992;40(7):697–702. doi: 10.1111/j.1532-5415.1992.tb01962.x. [DOI] [PubMed] [Google Scholar]
- 22.Yip AG, McKee AC, Green RC, et al. APOE, vascular pathology and the AD brain. Neurology. 2005;65(2):259–265. doi: 10.1212/01.wnl.0000168863.49053.4d. [DOI] [PubMed] [Google Scholar]
- 23.T Cuenco K, Green RC, Zhang J, et al. Magnetic resonance imaging traits in siblings discordant for Alzheimer disease. J Neuroimaging. 2008;18(3):268–275. doi: 10.1111/j.1552-6569.2007.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeCarli C, Maisog J, Murphy DG, Teichberg D, Rapoport SI, Horwitz N. Method for quantification of brain, ventricular, and subarachnoid CSF volumes from MR images. J Comput Assist Tomogr. 1992;16(2):274–284. doi: 10.1097/00004728-199203000-00018. [DOI] [PubMed] [Google Scholar]
- 25.Jagust WJ, Zheng L, Harvey DJ, et al. Neuropathological basis of magnetic resonance images in aging and dementia. Ann Neurol. 2008;63(1):72–80. doi: 10.1002/ana.21296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jack CR, Jr, Dickson DW, Parisi JE, et al. Antemortem MRI findings correlate with hippocampal neuropathology in typical aging and dementia. Neurology. 2002;58(5):750–757. doi: 10.1212/wnl.58.5.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheltens P, Leys D, Barkhof F, et al. Atrophy of medial temporal lobes on MRI in “probable” Alzheimer's disease and normal aging: diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry. 1992;55(10):967–972. doi: 10.1136/jnnp.55.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lunetta KL, Erlich PM, T Cuenco K, et al. MIRAGE Study Group Heritability of magnetic resonance imaging (MRI) traits in Alzheimer disease cases and their siblings in the MIRAGE Study. Alzheimer Dis Assoc Disord. 2007;21(2):85–91. doi: 10.1097/WAD.0b013e3180653bf7. [DOI] [PubMed] [Google Scholar]
- 29.SAS Institute Inc. Statistical Analysis System. SAS Institute Inc; Cary, NC: 2003. [Google Scholar]
- 30.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26(1):61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 31.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74(2):367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeCarli C, Massaro J, Harvey D, et al. Measures of brain morphology and infarction in the Framingham Heart Study: establishing what is normal. Neurobiol Aging. 2005;26(4):491–510. doi: 10.1016/j.neurobiolaging.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Jeerakathil T, Wolf PA, Beiser A, et al. Stroke risk profile predicts white matter hyperintensity volume: the Framingham Study. Stroke. 2004;35(8):1857–1861. doi: 10.1161/01.STR.0000135226.53499.85. [DOI] [PubMed] [Google Scholar]
- 35.Yoshita M, Fletcher E, Harvey D, et al. Extent and distribution of white matter hyperintensities in normal aging, MCI, and AD. Neurology. 2006;67(12):2192–2198. doi: 10.1212/01.wnl.0000249119.95747.1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu CC, Mungas D, Petkov CI, et al. Brain structure and cognition in a community sample of elderly Latinos. Neurology. 2002;59(3):383–391. doi: 10.1212/wnl.59.3.383. [DOI] [PubMed] [Google Scholar]
- 37.Vermeer SE, Prins ND, den Heijer T, Hofman A, Koudstaal PJ, Breteler MM. Silent brain infarcts and the risk of dementia and cognitive decline. N Engl J Med. 2003;348(13):1215–1222. doi: 10.1056/NEJMoa022066. [DOI] [PubMed] [Google Scholar]
- 38.Van Steen K, McQueen MB, Herbert A, et al. Genomic screening and replication using the same data set in family-based association testing. Nat Genet. 2005;37(7):683–691. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- 39.Yamazaki H, Bujo H, Kusunoki J, et al. Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J Biol Chem. 1996;271(40):24761–24768. doi: 10.1074/jbc.271.40.24761. [DOI] [PubMed] [Google Scholar]
- 40.Holtzman DM. In vivo effects of ApoE and clusterin on amyloid-β metabolism and neuropathology. J Mol Neurosci. 2004;23(3):247–254. doi: 10.1385/JMN:23:3:247. [DOI] [PubMed] [Google Scholar]
- 41.Xu PT, Li YJ, Qin XJ, et al. A SAGE study of apolipoprotein E3/3, E3/4 and E4/4 allele-specific gene expression in hippocampus in Alzheimer disease. Mol Cell Neurosci. 2007;36(3):313–331. doi: 10.1016/j.mcn.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Q, Zerbinatti CV, Zhang J, et al. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56(1):66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobsen L, Madsen P, Jacobsen C, Nielsen MS, Gliemann J, Petersen CM. Activation and functional characterization of the mosaic receptor SorLA/LR11. J Biol Chem. 2001;276(25):22788–22796. doi: 10.1074/jbc.M100857200. [DOI] [PubMed] [Google Scholar]
- 44.Seshadri S, DeStefano AL, Au R, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham study. BMC Genet. 2007;8(suppl 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]