Abstract
Reliable identification and assignment of cis-regulatory elements in promoter regions is a challenging problem in biology. The sophistication of transcriptional regulation in higher eukaryotes, particularly in metazoans, could be an important factor contributing to their organismal complexity. Here we present an integrated approach where networks of co-expressed genes are combined with gene ontology–derived functional networks to discover clusters of genes that share both similar expression patterns and functions. Regulatory elements are identified in the promoter regions of these gene clusters using a Gibbs sampling algorithm implemented in the A-GLAM software package. Using this approach, we analyze the cell-cycle co-expression network of the yeast Saccharomyces cerevisiae, showing that this approach correctly identifies cis-regulatory elements present in clusters of co-expressed genes.
Keywords: Promoter sequences, transcription factor–binding sites, co-expression, networks, gene ontology, Gibbs sampling
1. Introduction
The identification and classification of the entire collection of transcription factor–binding sites (TFBSs) are among the greatest challenges in systems biology. Recently, large-scale efforts involving genome mapping and identification of TFBS in lower eukaryotes, such as the yeast Saccharomyces cerevisiae, have been successful (1). On the other hand, similar efforts in vertebrates have proven difficult due to the presence of repetitive elements and an increased regulatory complexity (2–4). The accurate prediction and identification of regulatory elements in higher eukaryotes remains a challenge for computational biology, despite recent progress in the development of algorithms for this purpose (5). Typically, computational methods for identifying cis-regulatory elements in promoter sequences fall into two classes, enumerative and alignment techniques (6). We have developed algorithms that use enumerative approaches to identify cis-regulatory elements statistically significantly over-represented in promoter regions (7). Subsequently, we developed an algorithm that combines both enumeration and alignment techniques to identify statistically significant cis-regulatory elements positionally clustered relative to a specific genomic landmark (8).
Here, we will present a systems biology framework to study cis-regulatory elements in networks of co-expressed genes. This approach includes a network comparison operation, namely the intersection between co-expression and functional networks to reduce complexity and false positives due to co-expression linkage but absence of functional linkage. First, co-expression (9, 10) and functional networks (11, 12) are created using user-selected thresholds. Second, the construction of a single network is obtained from the intersection between co-expression and functional networks (13). Third, the highly interconnected regions in the intersection network are identified (14). Fourth, upstream regions of the gene clusters that are linked by both co-expression and function are extracted. Fifth, candidate cis-regulatory elements using A-GLAM (8) present in dense cluster regions of the intersection network are identified. In principle, the calculation of intersections for other types of networks with co-expression and/or functional networks could also be used to identify groups of co-regulated genes of interest (15) that may share cis-regulatory elements.
2. Materials
2.1. Hardware Requirements
Personal computer with at least 512 MB of random access memory (RAM) connected to the Internet.
Access to a Linux or UNIX workstation.
2.2. Software Requirements
The latest version of the Java Runtime Environment (JRE) freely available at http://www.java.com/.
The latest version of Cytoscape – a bioinformatics software platform for visualizing molecular interaction networks (13) freely available at http://www.cytoscape.org/.
The latest version of the MCODE plug-in for Cytoscape – finds clusters or highly interconnected regions in any network loaded into Cytoscape (14) freely available at http://cbio.mskcc.org/~bader/software/mcode/.
A modern version of the Perl programming language installed on the Linux or UNIX workstation freely available at http://www.perl.com/.
The A-GLAM package (8) freely available at ftp://ftp.ncbi.nih.gov/pub/spouge/papers/archive/AGLAM/.
3. Methods
The size of co-expression networks depends on the number of nodes in the network and the threshold used to define an edge between two nodes. There are a number of distance measures that are often used to compare gene expression profiles (16).
Here we use the Pearson correlation coefficient (PCC) as a metric to measure the similarity between expression profiles and to construct gene co-expression networks (17, 18). We establish a link by an edge between two genes, represented by nodes, if the PCC value is higher or equal to 0.7; this is an arbitrary cut-off that can be adjusted depending on the dataset used. The microarray dataset used here is the yeast cell-cycle progression experiment from Cho et al. (9) and Spellman et al. (10). The semantic similarity method (11) was used to quantitatively assess the functional relationships between S. cerevisiae genes.
The A-GLAM software package uses a Gibbs sampling algorithm to identify functional motifs (such as TFBSs, mRNA splicing control elements, or signals for mRNA 3’-cleavage and polyadenylation) in a set of sequences. Gibbs sampling (or more descriptively, successive substitution sampling) is a respected Markov-chain Monte Carlo procedure for discovering sequence motifs (19). Briefly, A-GLAM takes a set of sequences as input. The Gibbs sampling step in A-GLAM uses simulated annealing to maximize an ‘overall score’, a figure of merit corresponding to a Bayesian marginal log-odds score. The overall score is given by
| [1] |
.
In Eq. [1], m! = m(m – 1)…1 denotes a factorial; aj, the pseudocounts for nucleic acid j in each position; a = a1 + a2 + a3 + a4, the total pseudo-counts in each position; cij, the count of nucleic acid j in position i; and c = ci1 + ci2 + ci3 + ci4, the total number of aligned windows, which is independent of the position i. The rationale behind the overall score s in A-GLAM is explained in detail elsewhere (8).
To initialize its annealing maximization, A-GLAM places a single window of arbitrary size and position at every sequence, generating a gapless multiple alignment of the windowed subsequences. It then proceeds through a series of iterations; on each iteration step, A-GLAM proposes a set of adjustments to the alignment. The proposal step is either a repositioning step or a resizing step. In a repositioning step, a single sequence is chosen uniformly at random from the alignment; and the set of adjustments include all possible positions in the sequence where the alignment window would fit without overhanging the ends of the sequence. In a resizing step, either the right or the left end of the alignment window is selected; and the set of proposed adjustments includes expanding or contracting the corresponding end of all alignment windows by one position at a time. Each adjustment leads to a different value of the overall score s. Then, A-GLAM accepts one of the adjustments randomly, with probability proportional to exp(s/T). A-GLAM may even exclude a sequence if doing so would improve alignment quality. The temperature T is gradually lowered to T = 0, with the intent of finding the gapless multiple alignment of the windows maximizing s. The maximization implicitly determines the final window size. The randomness in the algorithm helps it avoid local maxima and find the global maximum of s. Due to the stochastic nature of the procedure, finding the optimum alignment is not guaranteed. Therefore, A-GLAM repeats this procedure ten times from different starting points (ten runs). The idea is that if several of the runs converge to the same best alignment, the user has increased confidence that it is indeed the optimum alignment. The steps (below) corresponding to E-values and post-processing were then carried out with the PSSM corresponding to the best of the ten scores s.
The individual score and its E-value in A-GLAM
The Gibbs sampling step produces an alignment whose overall score s is given by Eq. [1]. Consider a window of length w that is about to be added to A-GLAM’s alignment. Let δi(j) equal 1 if the window has nucleic acid j in positioni, and 0 otherwise. The addition of the new window changes the overall score by
| [2] |
.
The score change corresponds to scoring the new window according to a position-specific scoring matrix (PSSM) that assigns the ‘individual score’
| [3] |
to nucleic acid j in position i. Equation [3] represents a log-odds score for nucleic acid j in position i under an alternative hypothesis with probability (cij + aj)/(c + a) and a null hypothesis with probability pij. PSI-BLAST (20) uses Eq. [3] to calculate E-values. The derivation through Eq. [2] confirms the PSSM in Eq. [3] as the natural choice for evaluating individual sequences.
The assignment of an E-value to a subsequence with a particular individual score is done as follows: consider the alignment sequence containing the subsequence. Let n be the sequence length, and recall that w is the window size. If ΔSi denotes the quantity in Eq. [2] if the final letter in the window falls at position i of the alignment sequence, then ΔS* = max{ΔSi : i = w,…,n} is the maximum individual score over all sequence positions i. We assigned an E-value to the actual value ΔS* = Δs*, as follows. Staden’s method (21) yields ℙ{ΔSi Δs*} (independent of i) under the null hypothesis of bases chosen independently and randomly from the frequency distribution {pj}. The E-value E = (n – w + 1)ℙ{ΔSi Δs*} is therefore the expected number of sequence positions with an individual score exceeding Δs*. The factor n – w + 1 in E is essentially a multiple test correction.
More recently, the A-GLAM package has been improved to allow the identification of multiple instances of an element within a target sequence (22). The optional ‘scanning step’ after Gibbs sampling produces a PSSM given by Eq. [3]. The new scanning step resembles an iterative PSI-BLAST search based on the PSSM. First, it assigns an ‘individual score’ to each subsequence of appropriate length within the input sequences using the initial PSSM. Second, it computes an E-value from each individual score to assess the agreement between the corresponding subsequence and the PSSM. Third, it permits subsequences with E-values falling below a threshold to contribute to the underlying PSSM, which is then updated using the Bayesian calculus. A-GLAM iterates its scanning step to convergence, at which point no new subsequences contribute to the PSSM. After convergence, A-GLAM reports predicted regulatory elements within each sequence in the order of increasing E-values; users then have a statistical evaluation of the predicted elements in a convenient presentation. Thus, although the Gibbs sampling step in A-GLAM finds at most one regulatory element per input sequence, the scanning step can now rapidly locate further instances of the element in each sequence.
3.1. Co-expression Network Construction
The yeast cell-cycle-regulated expression data are obtained from http://cellcycle-www.stanford.edu/ (see Note 1).
Pairwise Pearson correlation coefficient (PCC) values are calculated using a subroutine implemented in the Perl programming language (23) (see Note 2).
The co-expression network is constructed with all gene pairs with a PCC greater or equal to 0.7 and is formatted according to the simple interaction file (SIF) described in the Cytoscape manual available at http://www.cytoscape.org/ (see Note 3).
The co-expression network can be loaded in Cytoscape, which is an open-source software for integrating biomolecular interaction networks. Cytoscape is available for a variety of operating systems, including Windows, Linux, Unix, and Mac OS X.
3.2. Functional Similarity Network Construction
Gene ontology (GO) annotations for yeast gene products come from the Saccharomyces Genome Database (SGD) and were downloaded from http://www.geneontology.org/cgi-bin/downloadGOGA.pl/gene_association.sgd.gz. The evidence supporting such annotations is captured by evidence codes, including TAS (Traceable Author Statement) and IEA (Inferred from Electronic Annotation). While TAS refers to peer-reviewed papers and indicates strong evidence, IEA denotes automated predictions, not curated by experts, i.e., generally less reliable annotations. For this reason, IEA annotations were excluded from this study.
- Functional relationships between S. cerevisiae genes were assessed quantitatively using a semantic similarity method (11) based on the gene ontology (GO). We first computed semantic similarity among GO terms from the Biological Process hierarchy using the Lin metric. This metric is based on information content and defines term–term similarity, i.e., the semantic similarity sim(ci, cj) between two terms ci and cj as
, where S(ci,cj) represents the set of ancestor terms shared by both ci and cj, ‘max’ represents the maximum operator, and p(c) is the probability of finding c or any of its descendants in the SGD database. It generates normalized values between 0 and 1. Gene–gene similarity results from the aggregation of term–term similarity values between the annotation terms of these two genes. In practice, given a pair of gene products, gk and gp, with sets of annotations Ak and Ap comprising m and n terms, respectively, the gene–gene similarity, SIM(gk, gp), is defined as the highest average (inter-set) similarity between terms from Ai and Aj:[4]
, where sim(ci,cj) may be calculated using Eq. [1]. This aggregation method (12) can be understood as a variant of the Dice similarity.[5] The functional similarity network is constructed using semantic similarity greater or equal to 0.7 and is formatted according to the simple interaction file (SIF).
Functional relationships in a group of genes can be further explored in Cytoscape using the BiNGO plug-in (24). Here we have used the hypergeometric test to assess the statistical significance (p < 0.05) and the Benjamini & Hochberg False Discovery Rate (FDR) correction (25).
3.3. Intersection Network Construction
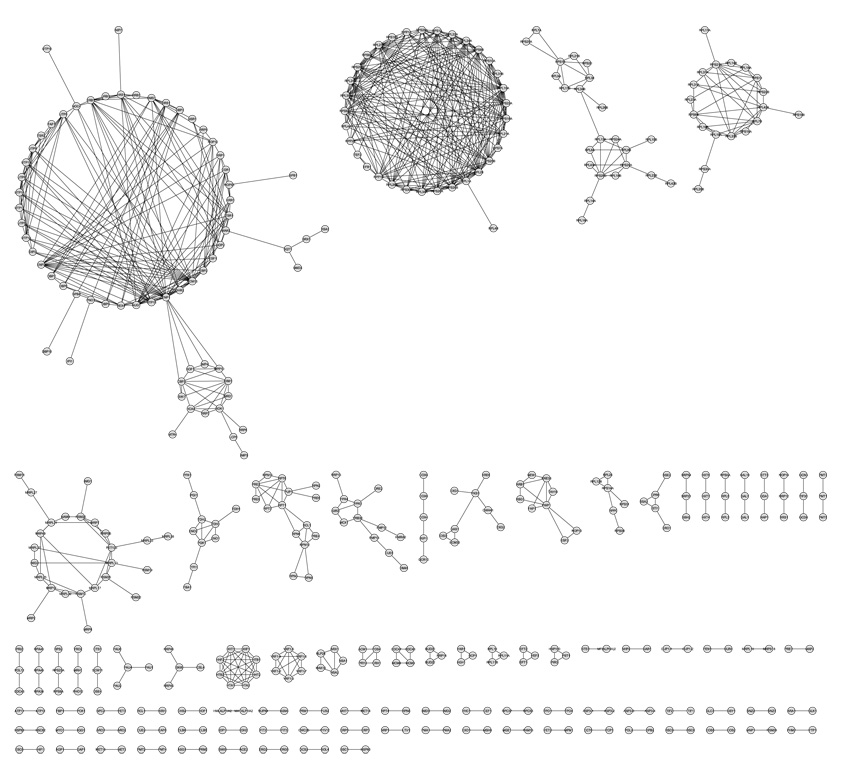
The yeast co-expression and functional similarity networks are loaded in Cytoscape and the intersection network can be obtained by using the Graph Merge plug-in, freely available at the Cytoscape Web site. The nodes that are connected by having similar expression profiles and GO annotations are present in the intersection network (Fig. 1.1) (see Note 4).
The intersection network can be visualized using a variety of layouts in Cytoscape. A circular layout of the intersection network using the yFiles Layouts plug-in is depicted in Fig. 1.1.
Fig. 1.1. Yeast cell-cycle gene co-expression and GO intersection network.
The intersection network topology is shown for yeast genes, represented by nodes linked by one or more edges as described in the text. An edge represents both co-expression and functional linkage between the nodes connected.
3.4. Identification of Highly Interconnected Regions
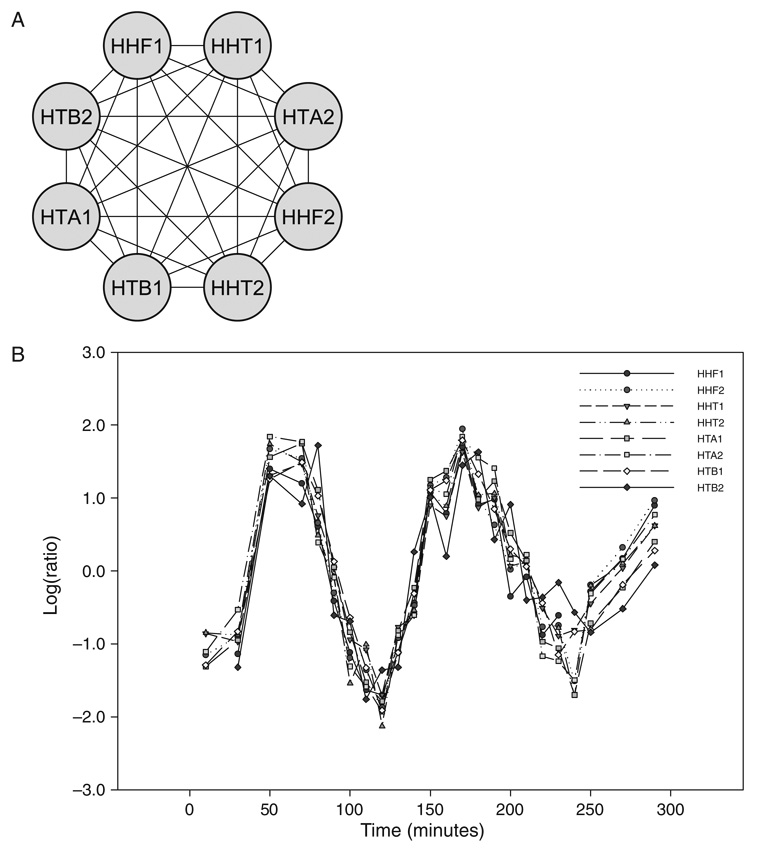
The identification of dense gene clusters in the intersection network is done using the MCODE Cytoscape plug-in (14) (see Note 5). The clusters identified share similar expression patterns and functions as described by GO (Fig. 1.2).
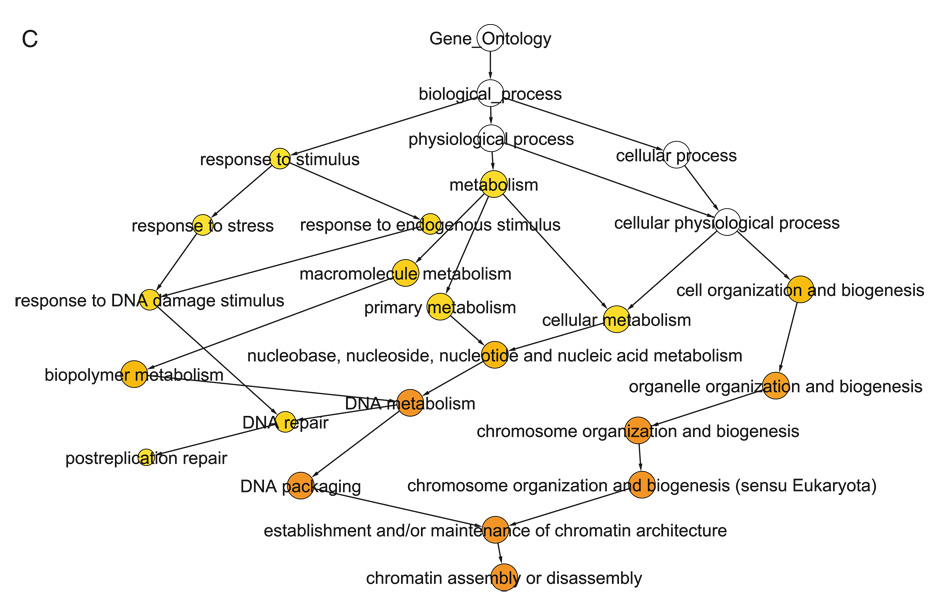
Fig. 1.2. Core histone gene cluster in the intersection network.
A. Highly connected cluster identified by MCODE corresponds to eight core histone genes present in the yeast genome. The eight nodes are connected by 28 co-expression and functional edges. B. Expression profiles of the core histone genes over the cell cycle. C. Over-represented GO terms in the Biological Process category for the core histone genes. The statistical significance of each GO term is related to the intensity of the colored circles (see Note 5).
3.5. Identification of Proximal Promoter Regions
The Saccharomyces Genome Database (SGD) maintains the most current annotations of the yeast genome (see http://www.yeastgenome.org/). The SGD FTP site contains the DNA sequences annotated as intergenic regions in FASTA format (available at ftp://genome-ftp.stanford.edu/pub/yeast/sequence/genomic_sequence/intergenic/), indicating the 5’ and 3’ flanking features. Additionally, a tab-delimited file with the annotated features of the genome is necessary to determine the orientation of the intergenic regions relative to the genes (available at ftp://genome-ftp.stanford.edu/pub/yeast/chromosomal_feature/). The two files can be used to extract upstream intergenic regions (26) for the genes present in the intersection network clusters (see Note 6).
3.6. Identification of cis-Regulatory Elements in Promoter Regions
Construct FASTA files for each of the gene clusters identified by MCODE.
Install the A-GLAM package (see Note 7).
-
The A-GLAM package has a number of options that can be used to adjust search parameters (see Note 8).
$ aglam
Usage summary: aglam [options] myseqs.fa
Options:
- h help: print documentation
- n end each run after this many iterations without improvement (10,000)
- r number of alignment runs (10)
- a minimum alignment width (3)
- b maximum alignment width (10,000)
- j examine only one strand
- i word seed query ()
- f input file containing positions of the motifs ()
- z turn off ZOOPS (force every sequence to participate in the alignment)
- v print all alignments in full
- e turn off sorting individual sequences in an alignment on p-value
- q pretend residue abundances = 1/4
- d frequency of width-adjusting moves (1)
- p pseudocount weight (1.5)
- u use uniform pseudocounts: each pseudocount = p/4
- t initial temperature (0.9)
- c cooling factor (1)
- m use modified Lam schedule (default = geometric schedule)
- s seed for random number generator (1)
- w print progress information after each iteration
- l find multiple instances of motifs in each sequence
- k add instances of motifs that satisfy the cutoff e-value (0)
- g number of iterations to be carried out in the post-processing step (1,000)
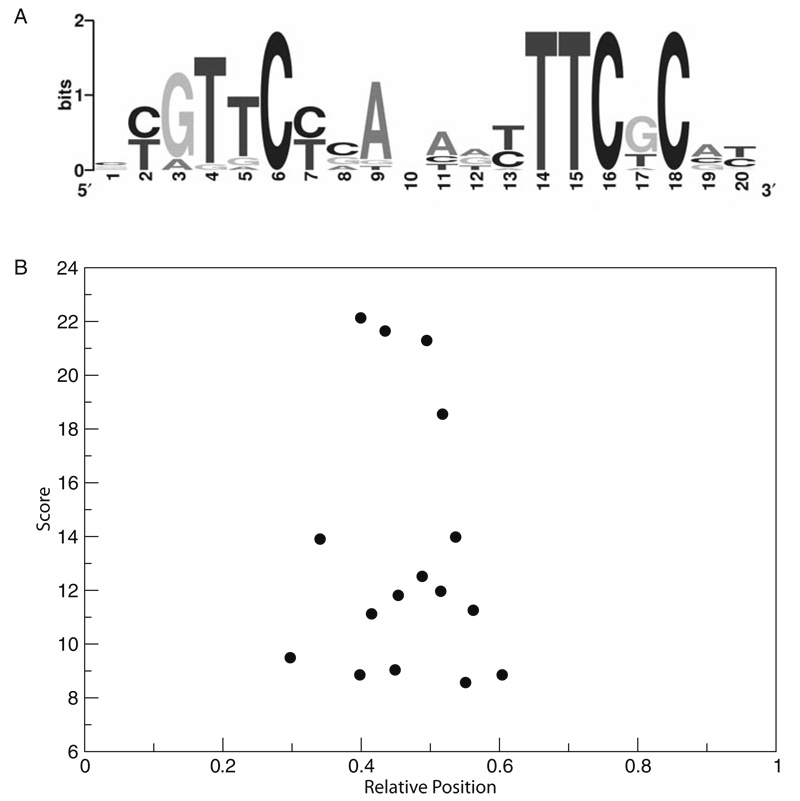
Run A-GLAM to identify the regulatory elements present in the gene clusters with similar expression patterns and GO annotations (see Note 9). A-GLAM correctly identifies an experimentally characterized element known to regulate core histone genes in yeast (27). The alignments produced by A-GLAM can be represented by sequence logos (28, 29) and the positional preferences of the elements can be evaluated by plotting the score against relative positions, normalized by sequence length, in the promoter sequences (Fig. 1.3).
Fig. 1.3. Core histone regulatory element identified with A-GLAM.
A. Sequence logo representation of the motif obtained from the ungapped multiple sequence alignment identified by A-GLAM (see Note 9). B. Positional preference plot for the elements identified by A-GLAM where the score in bits is plotted against the relative position of the element in the upstream regions of the core histone genes.
4. Notes
The yeast cell cycle data from the Web site include the experiments from Cho et al. (9) and Spellman et al. (10).
- The following Perl code can be used to calculate the PCC:
my$r = correlation(\@{$values{$probe1}}, \@{$values {$probe2}}); sub covariance { my ($array1ref,$array2ref) = @_; my ($i,$result); for ($i = 0;$i < @$array1ref;$i++) {$result +=$array1 ref->[$i] *$array2ref->[$i]; } $result /= @$array1ref; $result -= mean($array1ref) * mean($array2ref); } sub correlation { my ($array1ref,$array2ref) = @_; my ($sum1,$sum2); my ($sum1_squared, $sum2_squared); foreach (@$array1ref) {$sum1 +=$_;$sum1_squared +=$_ ** 2 } foreach (@$array2ref) {$sum2 +=$_;$sum2_squared +=$_ ** 2 } return (@$array1ref ** 2) * covariance($array1ref, $array2ref) /sqrt(((@$array1ref *$sum1_squared) - ($sum1 ** 2)) *((@$array1ref *$sum2_squared) - ($sum2 ** 2))); } sub mean { my ($arrayref) = @_; my$result; foreach (@$arrayref) {$result +=$_ } return$result / @$arrayref; } - The simple interaction file (SIF or .sif format) consists of lines where each node, representing a protein, is connected by an edge to a different protein in the network. Lines from the simple interaction file from the co-expression network:
- RPL12A pp THR1
- RPL12A pp TIF2
- RPL12A pp TIF1
- RPL12A pp GUK1
- RPL12A pp URA5
- RPL12A pp RPL1B
- RPL12A pp SSH1
- RPL12A pp SNU13
- RPL12A pp RPL23B
- SHU1 pp DON1
Two or more networks can be used to calculate their intersection as needed to select for connections that meet certain criteria. The researcher can overlay protein–protein interactions, co-expression and functional networks to identify the protein complexes created under specific experimental conditions.
-
The MCODE plug-in ranks the clusters according to the average number of connections per protein in the complex (Score). The top five clusters identified by MCODE in the intersection network are shown below:
Cluster Score Proteins Interactions 1 6.6 15 99 2 3.5 8 28 3 2.267 15 34 4 2 5 10 5 2 5 10 The BiNGO plug-in can be used to determine the GO terms statistically over-represented in a group of genes. Here we show the results for cluster 2:- Selected statistical test : Hypergeometric test
- Selected correction : Benjamini & Hochberg False Discovery Rate (FDR) correction
- Selected significance level : 0.05
- Testing option : Test cluster versus complete annotation
-
The selected cluster :HHT1 HHF1 HTA1 HHT2 HHF2 HTA2 HTB1 HTB2
- Number of genes selected : 8
- Total number of genes in annotation : 5932
There are a number of Web sites that facilitate the extraction of promoter sequences. A service for the extraction of human, mouse, and rat promoters is freely available at http://bio wulf.bu.edu/zlab/promoser/
-
The A-GLAM package is currently available in source code and binary forms for the Linux operating system (see ftp://ftp.ncbi.nih.gov/pub/spouge/papers/archive/AGLAM/).
GO ID P-value Corrected P-value Description 6333 4.9168E-15 1.2292E-13 Chromatin assembly or disassembly 6325 2.2510E-12 1.8758E-11 Establishment and/or maintenance of chromatin architecture 6323 2.2510E-12 1.8758E-11 DNA packaging 7001 2.0415E-10 1.2759E-9 Chromosome organization and biogenesis (sensu Eukaryota) 51276 2.5897E-10 1.2949E-9 Chromosome organization and biogenesis 6259 5.9413E-9 2.4756E-8 DNA metabolism 6996 6.9565E-7 2.4845E-6 Organelle organization and biogenesis Installation of the Linux binary: Get the executable from the FTP site and set execute permissions.- $chmod +x aglam
Installation from source: Unpack the glam archive and compile A-GLAM.-
$tar –zxvf aglam.tar.gz$cd aglam$make aglam
-
Possible scenarios and options to modify A-GLAM’s behavior.
- $aglam <myseqs.fa>
This command simply uses the standard Gibbs sampling procedure to find sequence motifs in “myseqs.fa”.- $aglam <myseqs.fa> -n 20000 -a 5 -b 15 -j
This tells the program to search only the given strand of the sequences to find motifs of length between 5 and 15 bp. The flag n specifies the number of iterations performed in each of the ten runs. Low values of n are adequate when the problem size is small, i.e., when the sequences are short and more importantly there are few of them, but high values of n are needed for large problems. In addition, smaller values of n are sufficient when there is a strong alignment to be found, but larger values are necessary when there is not, e.g., for finding the optimal alignment of random sequences. You will have to choose n on a case-by-case basis. This parameter also controls the tradeoff between speed and accuracy.- $aglam <myseqs.fa> -i TATA
This important option sets the program to run in a “seed”-oriented mode. The above command restricts the search to sequences that are TATA-like. Unlike the procedure followed in the standard Gibbs sampling algorithm, however, A-GLAM continues to align one exact copy of the “seed” in all “seed sequences”. Therefore, A-GLAM uses the seed sequences to direct its search in the remaining non-seed “target sequences”. Using this option leads to the global optimum quickly.- $aglam <myseqs.fa> -f <positions.dat>
The above command uses an extra option that allows A-GLAM to take a set of positions from an input file “positions.dat”. Like with the “-” flag, this option provides “seeds” for the A-GLAM alignment. Using this command restricts the Gibbs sampling step to aligning the original list of windows specified by the positions in the file. The seed sequences then direct the search in the remaining non-seed sequences.- $aglam <myseqs.fa> -l –k 0.05 –g 2000
Usable only with version 1.1. This tells the program to find multiple motif instances in each input sequence, via the scanning step (described above). Those instances that receive an E-value less than 0.05 are included in the PSSM. The search for multiple motifs is carried on until either (a) no new motifs are present or (b) the user-specified number of iterations (in this case, it is 2,000) is attained, whichever comes first.
- A-GLAM uses sequences in FASTA format as input. Cluster number 2, identified by MCODE, is composed of eight genes linked by 28 co-expression and GO connections. Interestingly, the intergenic regions of the same cluster are shared between the genes in the cluster:
>B:235796–236494, Chr 2 from 235796–236494, between YBL003C and YBL002W TATATATTAAATTTGCTCTTGTTCTGTACTTTCCTAATTCTTATGTA AAAAGACAAGAAT TTATGATACTATTTAATAACAAAAAACTACCTAAGAAAAGCATCATGCAG TCGAAATTGA AATCGAAAAGTAAAACTTTAACGGAACATGTTTGAAATTCTAAGAAAGC ATACATCTTCA TCCCTTATATATAGAGTTATGTTTGATATTAGTAGTCATGTTGTAATCT CTGGCCTAAGT ATACGTAACGAAAATGGTAGCACGTCGCGTTTATGGCCCCCAGGTTAAT GTGTTCTCTGA AATTCGCATCACTTTGAGAAATAATGGGAACACCTTACGCGTGAGCTGT GCCCACCGCTT CGCCTAATAAAGCGGTGTTCTCAAAATTTCTCCCCGTTTTCAGGATCAC GAGCGCCATCT AGTTCTGGTAAAATCGCGCTTACAAGAACAAAGAAAAGAAACATCGCGT AATGCAACAGT GAGACACTTGCCGTCATATATAAGGTTTTGGATCAGTAACCGTTATTTG AGCATAACACA GGTTTTTAAATATATTATTATATATCATGGTATATGTGTAAAATTTTTT TGCTGACTGGT TTTGTTTATTTATTTAGCTTTTTAAAAATTTTACTTTCTTCTTGTTAAT TTTTTCTGATT GCTCTATACTCAAACCAACAACAACTTACTCTACAACTA >D:914709–915525, Chr 4 from 914709–915525, between YDR224C and YDR225W TGTATGTGTGTATGGTTTATTTGTGGTTTGACTTGTCTATATAGGATAA ATTTAATATAA CAATAATCGAAAATGCGGAAAGAGAAACGTCTTTAATAAATCTGACCAT CTGAGATGATC AAATCATGTTGTTTATATACATCAAGAAAACAGAGATGCCCCTTTCTTA CCAATCGTTAC AAGATAACCAACCAAGGTAGTATTTGCCACTACTAAGGCCAATTCTCTT GATTTTAAATC CATCGTTCTCATTTTTTCGCGGAAGAAAGGGTGCAACGCGCGAAAAAGT GAGAACAGCCT TCCCTTTCGGGCGACATTGAGCGTCTAACCATAGTTAACGACCCAACCG CGTTTTCTTCA AATTTGAACTCGCCGAGCTCACAAATAATTCATTAGCGCTGTTCCAAAA TTTTCGCCTCA CTGTGCGAAGCTATTGGAATGGAGTG TATTTGGTGGCTCAAAAAAAGAGCACAATAGTTA ACTCGTCGTTGTTGAAGAAACGCCCGTAGAGATATGTGGTTTCTCATGC TGTTATTTGTT ATTGCCCACTTTGTTGATTTCAAAATCTTTTCTCACCCCCTTCCCCGTT CACGAAGCCAG CCAGTGGATCGTAAATACTAGCAATAAGTCTTGACCTAAAAAATATATA AATAAGACTCC TAATCAGCTTGTAGATTTTCTGGTCTTGTTGAACCATCATCTATTTACT TCCAATCTGTA CTTCTCTTCTTGATACTACATCATCATACGGATTTGGTTATTTCTCAGT GAATAAACAAC TTCAAAACAAACAAATTTCATACATATAAAATATAAA >N:576052–576727, Chr 14 from 576052–576727, between YNL031C and YNL030W TGTGGAGTGTTTGCTTGGATCCTTTAGTAAAAGGGGAAGAACAGTTGGAA GGGCCAAAGT GGAAGTCACAAAACAGTGGTCCTATATAAAAGAACAAGAAAAAGATTATT TATATACAAC TGCGGTCACAAGAAGCAACGCGAGAGAGCACAACACGCTGTTATCACGCA AACTATGTTT TGACACCGAGCCATAGCCGTGATTGTGCGTCACATTGGGCGATAATGAAC GCTAAATGAC CAACTCCCATCCGTAGGAGCCCCTTAGGGCGTGCCAATAGTTTCACGCGC TTAATGCGAA GTGCTCGGAACGGACAACTGTGGTCGTTTGGCACCGGGAAAGTGGTACTA GACCGAGAGT TTCGCATTTGTATGGCAGGACGTTCTGGGAGCTTCGCGTCTCAAGCTTTT TCGGGCGCGA AATGCAGACCAGACCAGAACAAAACAACTGACAAGAAGGCGTTTAATTTA ATATGTTGTT CACTCGCGCCTGGGCTGTTGTTATTCGGCTAGATACATACGTGTTTGTGC GTATGTAGTT ATATCATATATAAGTATATTAGGATGAGGCGGTGAAAGAGATTTTTTTT TTTTCGCTTAA TTTATTCTTTTCTCTATCTTTTTTCCTACATCTTGTTCAAAAGAGTAGC AAAAACAACAA TCAATACAATAAAATA >B:255683–256328, Chr 2 from 255683–256328, between YBR009C and YBR010W ATTTTACTATATTATATTTGTTGCTTGTTTTTGTTTGTTGCTTTAGTAC TATAGAGTACA ATAATGCGACGGAAACCATCATATAGAAAAAATATCTCGGTATTTATAG GAAAAAGAATT AGACCTTTTCCACAACCAATTTATCCATCAAATTGGTCTTTACCCAATG AATGGGGAAGG GGGGGTGGCAATTTACCACCGTATTCGCGGGCATTTGCTAAAGTAAACA ACTTCGGTTTT TACCACTAACCATTATGGGGAGAAGCGCTCGGAACAGTTTTACTATGTG AAGATGCGAAG TTTTCAGAACGCGGTTTCCAAATTCGGCGGGGAGATACAAAAAAGATTT TTGCTCTCGTT CTCACATTTTCGCATTGTCCCATACATTATCGTTCTCACAATTTCTCAC ATTTCCTTGCT CTGCACCTTTGCGATCCTGGCCGTAATATCTCTCCTTGACTTTTAGCGT GGAAGATAACG AAATGCCCGGGCGATTTTTCTTTTTGGTACCCTCCACGGCTCCTTGTTG AAATACATATA TAAAAGACTGTGTATTCTTCGGGATACATCTCTTTCCTCAACCTTTTAT ATTCTTTCTTT CTAGTTAATAAGAAAAACATCTAACATAAATATATAAACGCAAACAA-GLAM has a number of useful command line options that can be adjusted to improve ab initio motif finding; in this example we have restricted the search to motifs no larger than 20 bp.- $aglam -b 20 -1 02.fa
A-GLAM: Anchored Gapless Local Alignment of Multiple Sequences Compiled on Jun 2 2006 Run 1... 11724 iterations Run 2... 10879 iterations Run 3... 10878 iterations Run 4... 10336 iterations Run 5... 10181 iterations Run 6... 10637 iterations Run 7... 10116 iterations Run 8... 11534 iterations Run 9... 10097 iterations Run 10... 10239 iterations ! The sequence file was [ 02 .fa] ! Reading the file took [ 0] secs ! Sequences in file [4] ! Maximum possible alignment width [ 1292] ! Score [243.4] bits ! Motif Width [ 20] ! Runs [ 10] ! Best possible alignment: >B:235796–236494, Chr 2 from 235796–236494, between YBL003C and YBL002W 365 AGGCGAAGCGGTGGGCACAG 346 − (21.29360) (2.820982e-08) 394 GGGAGAAATTTTGAGAACAC 375 − (13.97930) (5.205043e-04) 309 ATGCGAATTTCAGAGAACAC 2 90 − (11.12770) (5.771870e-03) 314 TTGAGAAATAATGGGAACAC 333 + (9.034960) (2.714569e-02) >D:914709–915525, Chr4 from 914709–915525, between YDR224C and YDR225W 423 GTGCGAAGCTATTGGAATGG 442 + (18.55810) (2.256236e-06) 278 GCGCGAAAAAGTGAGAACAG 297 + (13.90430) (6.4 9552 6e-04) 418 AGGCGAAAATTTTGGAACAG 399 − (12.51460) (2.007017e-03) 262 CCGCGAAAAAATGAGAACGA 243 − (9.499530) (2.2 99132e-02) >N:576052–576727, Chr 14 from 576052–576727, between YNL031C and YNL030W 294 ATGCGAAGTGCTCGGAACGG 313 + (21.65330) (1.526033e-08) 367 ATGCGAAACTCTCGGTCTAG 348 − (11.95760) (2.781407e-03) 399 ACGCGAAGCTCCCAGAACGT 380 − (11.25120) (5.253971e-03) 288 GCGTGAAACTATTGGCACGC 269 − (8.853600) (3.961768e-02) >B:255683–256328, Chr 2 from 255683–256328, between YBR009C and YBR010W 258 GGGAGAAGCGCTCGGAACAG 277 + (22.13350) (6.281785e-09) 293 ATGCGAAGTTTTCAGAACGC 312 + (11.81510) (3.041439e-03) 409 GTGAGAAATTGTGAGAACGA 390 − (8.852760) (3.780865e-02) 375 ATGCGAAAATGTGAGAACGA 356 − (8.564750) (4.774790e-02) ! 16 sequences in alignment ! Residue abundances:Pseudocounts ! A = 0.312544:0.468816 C = 0.187456:0.281184 ! G = 0.187456:0.281184 T = 0.312544:0.468816 ! Total Time to find best alignment [15.87] secs
Acknowledgments
The authors would like to thank King Jordan for important suggestions and helpful discussions and Alex Brick for his assistance in obtaining intergenic regions during his internship at NCBI. This research was supported by the Intramural Research Program of the NIH, NLM, NCBI.
References
- 1.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431(7004):99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16(5):595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116(4):499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 4.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8(7):764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 5.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23(1):137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 6.Ohler U, Niemann H. Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet. 2001;17(2):56–60. doi: 10.1016/s0168-9525(00)02174-0. [DOI] [PubMed] [Google Scholar]
- 7.Marino-Ramirez L, Spouge JL, Kanga GC, Landsman D. Statistical analysis of over-represented words in human promoter sequences. Nucleic Acids Res. 2004;32(3):949–958. doi: 10.1093/nar/gkh246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tharakaraman K, Marino-Ramirez L, Sheetlin S, Landsman D, Spouge JL. Alignments anchored on genomic landmarks can aid in the identification of regulatory elements. Bioinformatics. 2005;21 Suppl 1:i440–i448. doi: 10.1093/bioinformatics/bti1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2(1):65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 10.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9(12):3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lord PW, Stevens RD, Brass A, Goble CA. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19(10):1275–1283. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- 12.Azuaje F, Wang H, Bodenreider O. Ontology-driven similarity approaches to supporting gene functional assessment. Proceedings of the ISMB’2005 SIG Meeting on Bio-Ontologies; Detroit, MI. 2005. pp. 9–10. [Google Scholar]
- 13.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsaparas P, Marino-Ramirez L, Bodenreider O, Koonin EV, Jordan IK. Global similarity and local divergence in human and mouse gene co-expression networks. BMC Evol Biol. 2006;6:70. doi: 10.1186/1471-2148-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babu MM. Grant RP. Computational Genomics: Theory and Application. Cambridge, UK: Horizon Bioscience; 2004. An introduction to microarray data analysis; pp. 225–249. [Google Scholar]
- 17.Jordan IK, Marino-Ramirez L, Koonin EV. Evolutionary significance of gene expression divergence. Gene. 2005;345(1):119–126. doi: 10.1016/j.gene.2004.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan IK, Marino-Ramirez L, Wolf YI, Koonin EV. Conservation and coevolution in the scale-free human gene coexpression network. Mol Biol Evol. 2004;21(11):2058–2070. doi: 10.1093/molbev/msh222. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootton JC. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262(5131):208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staden R. Methods for calculating the probabilities of finding patterns in sequences. Comput Appl Biosci. 1989;5(2):89–96. doi: 10.1093/bioinformatics/5.2.89. [DOI] [PubMed] [Google Scholar]
- 22.Tharakaraman K, Marino-Ramirez L, Sheetlin S, Landsman D, Spouge JL. Scanning sequences after Gibbs sampling to find multiple occurrences of functional elements. BMC Bioinformatics. 2006;7:408. doi: 10.1186/1471-2105-7-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orwant J, Hietaniemi J, Macdonald J. Mastering Algorithms with Perl. Sebastopol, CA: O’Reilly; 1999. [Google Scholar]
- 24.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 26.Marino-Ramirez L, Jordan IK, Landsman D. Multiple independent evolutionary solutions to core histone gene regulation. Genome biology. 2006;7(12):R122. doi: 10.1186/gb-2006-7-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eriksson PR, Mendiratta G, McLaughlin NB, Wolfsberg TG, Marino-Ramirez L, Pompa TA, Jainerin M, Landsman D, Shen CH, Clark DJ. Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements. Mol Cell Biol. 2005;25(20):9127–9137. doi: 10.1128/MCB.25.20.9127-9137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18(20):6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]