Abstract
While many advanced mixed-effects models have been proposed and are used in fMRI, the simplest, ordinary least squares (OLS), is still the one that is most widely used. A survey of 90 papers found that 92% of group fMRI analyses used OLS. Despite the widespread use, this simple approach has never been thoroughly justified and evaluated; for example, the typical reference for the method is a conference abstract, (Holmes and Friston, 1998), which has been referenced over 400 times.
In this work we fully derive the simplified method in a general setting and carefully identify the homogeneity assumptions it is based on. We examine the specificity (Type I error rate) of the OLS method under heterogeneity in the one-sample case and find that the OLS method is valid, with only slight conservativeness. Surprisingly, a Satterthwaite approximation for effective degrees of freedom only makes the method more conservative, instead of more accurate. While other authors have highlighted the inferior power of the OLS method relative to optimal mixed effects methods under heterogeneity, we revisit these results and find the power differences very modest.
While statistical methods that make the best use of the data are always to be preferred, software or other practical concerns may require the use of the simple OLS group modeling. In such cases, we find that group mean inferences will be valid under the null hypothesis and will have nearly optimal sensitivity under the alternative.
Keywords: Functional Magnetic Resonance Imaging, ordinary least squares, general linear model, specificity, hypothesis testing, Two-Stage Summary Statistics, Study Design
1 Introduction
The analysis of multisubject fMRI data presents a number of challenges, in particular the need to account for two sources of variance: “measurement error” variability in the estimated response in each subject, and the “individual differences” variability in the true response between subjects. Appropriately modeling these within- and between-subject variances have motivated a number of papers on the best way to perform group modeling of fMRI data (Friston et al., 2002, 2005; Woolrich et al., 2004; Worsley et al., 2002; Beckmann et al., 2003; Mumford and Nichols, 2006; Penny and Holmes, 2006). Most of these methods individually weight data from each subject, down-weighting subjects with relatively high intrasubject variability, possibly even shrinking intrasubject estimates towards a population estimate. We refer to such weighted methods generically as generalized least squares (GLS) (Searle, 1971). Although the GLS methods make fewer assumptions about the distribution of the data, they are computationally more complicated to employ.
While several software packages have implemented such voxel-wise GLS methods1, the simpler method, of simply modeling 1st level contrast data with an un-weighted, ordinary least squares (OLS) analysis is still in widespread use. For example, a small survey of 90 papers matching the keyword “group fmri” anywhere in the text in NeuroImage, Human Brain Mapping and Cerebral Cortex 2 found that 92% of group fMRI analyses used OLS. Further, OLS also forms the core of other methods. For example permutations are generally based on OLS (though see Mriaux et al. (2006)) and region of interest analyses typically are based on OLS.
The original reference for OLS analysis for group fMRI is a conference abstract, (Holmes and Friston, 1998), which has been referenced 448 times according to Google Scholar. While there are publications on GLS methods that compare to OLS, these comparisons have focused on the sensitivity but not the specificity (the ability to control false positives accurately) of the OLS model. Friston et al. (2005) compared thresholded test statistics from both the GLS and OLS models to determine how robust the OLS model was to violations of the assumption of homoscedasticity; their study focused on a single real dataset and did not consider the specificity of OLS when the assumptions were violated. Likewise Beckmann et al. (2003) compared the power of OLS and GLS under heteroscedasticity and they showed a moderate increase in power when using GLS. Instead of formally estimating power, their work looked at the percent change in the Z statistic, which is an approximation to the t-statistic that is used in standard analyses.
In this paper we provide a detailed description of OLS as it is typically used, focusing on the assumptions of this model and how well they hold for fMRI data. In particular, we highlight that the OLS approach always provides unbiased estimates of effect magnitude and, for the frequently-used one-sample model, unbiased variance estimates. The other possible problem caused by heterogeneity is disturbance of the distributional accuracy of t- or F-statistics, which can affect p-value accuracy. The traditional solution is to alter the degrees-of-freedom (DF) as part of a Satterthwaite adjustment (Satterthwaite, 1946). Satterthwaite has been found to be useful with the OLS model to protect against false positives in single subject fMRI analysis when data are temporally autocorrelated (Worsley and Friston, 1995; Kiebel et al., 2003), and we consider the performance of the Satterthwaite approximation in group fMRI analysis.
2 Methods
2.1 Model for Group fMRI data
Group fMRI data are typically analyzed in a two-stage process. In the 1st level intrasubject models are fit independently to each subject, and in the 2nd level summary measures from each subject are modeled.
2.1.1 First Level: Within-subject Model
For a given voxel, there is a first stage model for each subject k:
| (1) |
where Yk is the T × 1 vector containing the blood oxygen level dependent (BOLD) time series, Xk is the T × p design matrix containing p regressors of interest, βk is a p × 1 vector of parameters and εk is the T × 1 vector error term. Since fMRI data are temporally autocorrelated, we allow non-independent errors and write , where is the within-subject error variance and Vk is the temporal correlation of the error. For a review of temporal autocorrelation models used for Vk, see Mumford and Nichols (2006). We assume normally distributed errors and, at this first level, regard the βk as fixed effects; hence the model can be concisely written as , where “~” denotes the distribution of a random variable. Note that model (Xk) and noise ( & Vk) are subject-specific.
While many regressors are needed to fit complex experimental designs and nuisance effects, an individual research question is usually addressed with 1-dimensional contrast3 c, forming a linear combination of parameter estimates cβk. The effect magnitude cβk and its variance are estimated with generalized least squares (GLS) as cβ̂k,
| (2) |
and its sample variance, for fixed true βk, is
| (3) |
| (4) |
Though not recommended, ordinary least squares (OLS) can be used instead of GLS at the first level by (incorrectly) assuming Vk = IT. This will produce biased standard errors ( ) and effect magnitude estimates that have sub-optimal precision (Var(cβ̂k) higher than with GLS). As will be seen below, biased standard errors only affect 2nd level GLS not OLS.
2.1.2 Second Level: Between-subject Model
At the “2nd level” we would ideally regress the true subject responses, γ = {cβk}k, on a group model
| (5) |
where XG is N × pG group-level design matrix, βG is the group level parameter vector and εG is the group error vector with , where is the between-subject variance and I is the identity matrix. Crucially, the true subject responses are now regarded as random, with . While XG is often just a column of ones (for a one-sample t-test) it can take any form in general.
2.1.3 Second Level: Estimation with OLS
Unfortunately, we only have the estimated contrasts, YG = {cβ̂k}k, and so the OLS 2nd level model in practice has the form
| (6) |
where is the mixed-effects error, , containing variation from both imperfect intrasubject fit (YG − γ) and the distribution of true responses in the population (εG). Specifically we have , where , where diag is the diagonal matrix operator, and the β subscript denotes that this is an intrasubject variance.
Write the OLS estimate of βG, , where − denotes pseudo inverse, and is the estimate of the mixed-effects error variance.
For the OLS approach, it is assumed that the first level variance is homogeneous, for all i, j and therefore the second level error variance can be expressed as , where is the common within-subject variance. Therefore the variance can be simplified to , where is the combined within- and between-subject variance term. Since there is just a single variance term, this model is much easier to estimate and does not require iterative maximization techniques.
Without the homogeneity assumption the OLS estimates may not have optimal precision, though they are unbiased (E(β̂OLS) = βG). While the standard errors are not unbiased in general, in the widely-used one-sample t-test the standard errors are unbiased (See Appendix A for full details).
2.1.4 Second Level: Estimation with GLS
In the GLS approach to the multistage mixed model, the assumption at the second stage is that . For fMRI software packages that use a GLS approach, such as FSL or fmristat, the estimates of the variance and correlation Vk from the first level analysis are used for , where the diagonal elements correspond to the individual estimated variances from equation 3, and is estimated as part of an iterative model estimation algorithm, such as restricted maximum likelihood (Harville, 1974).4 GLS estimates of group-level effects are unbiased and have minimum variance among all linear unbiased estimates (Searle, 1971).
2.1.5 Model for Evaluating Heteroscedasticity in the Second Level Model
This note focuses on the one-sample t-test under the OLS approach, so from this point on we can assume that x1,…, xN are the N first level contrast estimates from N subjects, previously referred to as YG. The OLS test statistic is given by
| (7) |
where orresponds to and β̂OLS and
| (8) |
corresponds to . Inference is carried out by comparing T to a t-distribution with N − 1 degrees of freedom (TN−1).
2.1.6 Satterthwaite Correction to Address Heteroscedasticity
Although the OLS variance estimate is unbiased in the case of the one-sample t-test (See Appendix A), the distributional assumptions change under heteroscedasticity. Under homoscedasticity the sample variance S2 (equation 8) is proportional to a random variable, where the degrees of freedom also define the t-distribution used to test the null hypothesis of the t-statistic (equation 7). Under heteroscedasticity the sample variance is only approximately proportional to a χ2 random variable. The motivation of the Satterthwaite approach is to estimate the effective degrees of freedom (eDF) such that the sample variance is proportional to .
The Satterthwaite degree of freedom approximation is based on matching the first and second moments of S2 and a scaled χ2 distribution, solving for the χ2 degrees-of-freedom νSAT (Satterthwaite, 1946),
Using the values in Table 1 and after some algebra, one finds that
Table 1.
First two moments of a random variable and S2
| Moment | S2 | ||
|---|---|---|---|
| First | ν |
|
|
| Second | ν(ν + 2) |
|
| (9) |
In the real data analyses below, we use the FSL estimates for to compute νSAT. (FSL’s FEAT analysis software uses GLS to find these quantities and saves these estimates as var_filtered_func_data and stats/mean_random_effects_var1.)
2.2 Real data analysis
2.2.1 Data and variance estimation
Our data are from a finger tapping experiment of the right hand involving 12 normal subjects (Johansen-Berg et al., 2002). This was a block design study consisting of blocks of rest and 3 pseudorandomly cued tasks: tapping of the index finger, sequentially tapping fingers, randomly tapping fingers.
The data were analyzed using the fMRIB software library (FSL), using a two-level “FLAME” model. This model produced 12 contrast estimates, and 12 subject specific mixed-effects variance estimates for each of the 226,000 voxels. The contrast of interest for our study tested the difference between the response when randomly tapping the fingers and sequentially tapping the fingers. For each voxel the p-value for the OLS statistic was calculated using the Satterthwaite correction and two other methods that are described in the following sections.
2.2.2 Permutation test
The permutation p-values were obtained by comparing the test statistic, T, to an empirical t-distribution based on permutations. Since we are making inference on contrasts of parameter estimates involving differences, the order of differencing does not matter under the null hypothesis of no activation; for example β1 − β2 is equivalent to β2 − β1. Therefore, by permuting the signs on the contrasts we can construct the null distribution of the contrast (Nichols and Holmes, 2002). Specifically, all possible 2N permutations are created by the different +/− combinations of the N contrasts and a test statistic for each permutation is calculated and Pperm = % of the 2N permuted test statistics as large or larger than the test statistic T.
2.2.3 Monte Carlo simulation details
Since the true null distribution is unknown under the heteroscedastic case, we used Monte Carlo to calculate “correct” p-values at each voxel, assuming the variances of the contrast for each subject, , to be known. For each realization, we generated 10,000 sets of N(0, ) data for each of the i subjects and used it to calculate 10,000 test statistics. PMC = % of 10,000 test statistics as large or larger than the test statistic, T, for that voxel.
2.3 Simulated data analysis methods
Simulations were used to study different sample sizes with differing numbers of outliers and varying degrees of outlying variances. To simulate outlying within-subject variances we used a mixture of χ2 random variables where, with probability 0.9, the variance was chosen from a distribution and, with a probability of 0.1 it was chosen from a distribution, where and are the within-subject variances for the non-outlying and outlying subjects, respectively. Between-subject variances were chosen such that the overall variance for each simulation was kept constant. Therefore across different variance settings a given effect size would correspond to equivalent statistical power.
Variances that have been found in real data are shown in Table 2 and the range of values for , and were chosen to include these values. The details are described in Appendix B. While the overall standard deviation was fixed, varying sample sizes required different effect magnitudes Δ to maintain 80% power across simulations. Specifically, Δ was set to 28.14, 19.04, and 15.34, for 10, 20, and 30 subjects, respectively.
Table 2.
Average estimated variances from real data analyses with outlying variances where variance estimates resulted from a mixed effects analysis using FSL. Averages shown are for voxels within the interquartile range of the nonzero between-subject variances. For comparison purposes, numbers were scaled such that was 400 across studies. The last column corresponds to the values on the x-axis of Figure 3. Data used in the rows of the table came from Stover et al. (2006), Harley et al. (2009), Foerde et al. (2006), Cazalis et al. (2004), and Harley et al. (2009), respectively.
| Study type | # subjects | # outliers | mean | mean | mean | ||
|---|---|---|---|---|---|---|---|
| Blocked | 16 | 1 | 1138.1 | 400 | 3558.5 | 67.3 | |
| Blocked | 20 | 2 | 179.6 | 400 | 1134.4 | 55.9 | |
| Blocked | 15 | 1 | 465.3 | 400 | 1499.5 | 56.0 | |
| Event Related | 14 | 2 | 971.3 | 400 | 910.4 | 27.1 | |
| Event Related | 20 | 2 | 488.1 | 400 | 1992.4 | 64.2 |
3 Results
3.1 Real Data
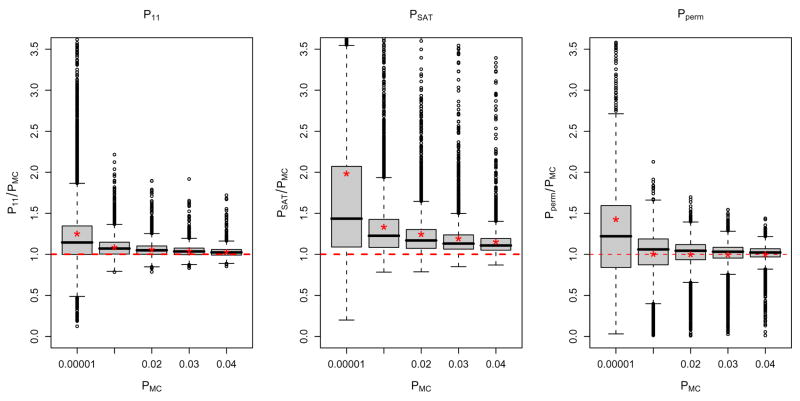
Using the subject-specific first level contrast estimates we calculated TOLS (eq 7) and obtained four different P-values: P11 found with the usual t-distribution with N − 1 = 11 DF, PSAT found with a t-distribution with Satterthwaite DF (νSAT ), PMC created with the Monte Carlo simulated t-distribution, and Pperm computed with the permutation driven null distribution. To evaluate the accuracy of the p-values at potential signal voxels, we compared both P11 and PSAT to PMC in the voxels that were found to be significant with PMC < 0.05. The left two panels of Figure 1 show the boxplots of the ratios of P11/PMC and PSAT/PMC over different ranges of PMC. The mean corresponding to each boxplot is indicated by a red star. Surprisingly, P11 is slightly conservative and PSAT is even more conservative, having values larger than PMC on average. Since νSAT ≤ 11 this indicates that the true degrees of freedom of the null distribution tends to be larger than N − 1. P-values that are too large indicate that the null distribution used has tails that are too heavy, in particular, that both DF values of νSAT and 11 are too small and the effective degrees of freedom that best match the true distribution are yet larger than the nominal DF.
Figure 1.
Comparisons of P11, PSAT, and Pperm with PMC over values of PMC < 0.05. The x-axis show the lower bound of the interval for PMC and the red stars indicate the means of the distributions.
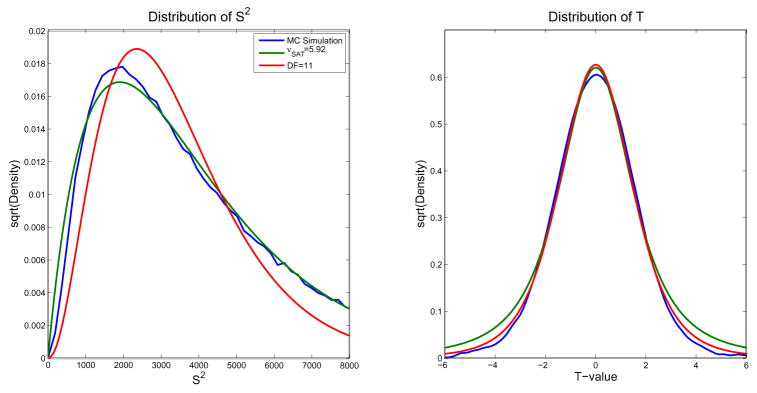
In order to understand the poor performance of PSAT, we examined the implied distribution of S2 and T for each method. Figure 2 shows these two distributions, where the blue line is the probability density estimate based on the Monte Carlo simulation values. The distribution of S2 under 11 and νSAT degrees of freedom (or in general νmethod) are χ2 distributions scaled by E(S2)/νmethod, which is a gamma distribution with shape parameter νmethod/2 and scale parameter 2E(S2)/νmethod. The distribution of S2 based on the Monte Carlo simulation is quite similar to the distribution of S2 under νSAT, with the mean and variance of the two distributions matching. Even so, the corresponding t-distributions do not agree in the tails of the distribution, which is important since it affects the p-values of interest. This is due to the distributions of S2 not matching well for values near zero. There is a much higher probability of obtaining near-zero values of S2 under νSAT, which leads to a higher probability of getting larger values in the corresponding t-distribution, explaining the fatter tails.
Figure 2.
The distribution of S2 (left) and T (right). Although the mean and variance of the distributions of S2 for the MC simulation and νSAT are similar, the lower tails do not match as well as ν11. The larger lower tail of the distribution of S2 causes the tails of the distribution of T to be too large.
The distribution of S2 based on 11 DF does not look as similar to that based on the Monte Carlo simulation overall, but for the values of S2 that we are interested in, the lower values, the distributions are much closer and hence p-values are more similar. While nonparametric inferences are known to be exact, we confirmed this by comparing Pperm to PMC. In the right panel of Figure 1, the mean of the ratio PMC/Pperm is nearly 1 (means marked with asterisks), except for the smallest P-values which likely are exhibiting discreteness-induced conservativeness. One possible limitation of our Monte Carlo simulations is that they assume that the FLAME-derived variance estimates are the true values of the variances in the real data. To assess this assumption, we repeated the Monte Carlo simulation using t-statistic values based on samples from a Normal distribution with known variances and obtained similar results (see Supplementary Material), suggesting the FLAME variance estimates are accurate.
3.2 Simulations
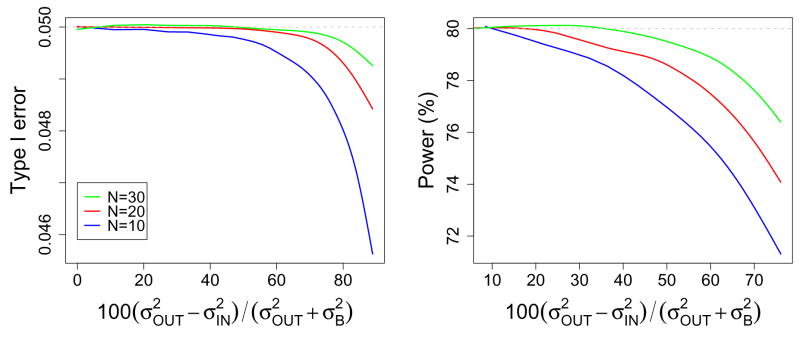
Simulations were used to study type I error rate and power over a range of degrees of outlying variance when using TOLS versus TGLS and a null distribution tN−1. Figure 3 shows the type I error rate and power as a function of the percent difference between the mixed effects variance for the outliers and non-outliers, . The left panel of Figure 3 supports the finding of our real data analysis, in that when using OLS the type I error rate is close to the desired level and may be slightly conservative (though note the very tight range of the y-axis). The type I error is most conservative for smaller sample sizes and larger outlying variances. Note in each sample size there were 10% outliers.
Figure 3.
Type I error rate (left) and power (right) as a function of the % difference in the mixed effects variances of outlying and non-outlying variances, , for sample sizes of 10, 20 and 30. Overall variance was held constant for each sample size over the range of the x axis to ensure power for each sample size reflected the same effect size.
The right panel of Figure 3 shows the power under the OLS model, where the true power (under GLS) was 80% in each case. As found in Beckmann et al. (2003) and Friston et al. (2005) power can be lost when using OLS under heteroscedasticity. The decrease was as high as 9% and was worst for the smaller sample size and larger outlying variances.
The x-axis range of Figure 3 can be compared to the values found from real data shown in the last column of Table 2. We searched over 12 data analyses that were analyzed using the Feat analysis tool of FSL, including event related and blocked designs and found that 5 of these studies had subjects with outlying variances. This was determined by subsetting voxels with nonzero between-subject variances and plotting boxplots of the voxel-averaged within-subject variances. In cases where outlying variances were found, the within-subject variance distributions were similar for voxels where the between-subject variance was 0. Table 2 lists the average within-subject variances for outlying and non-outlying subjects within the interquartile range of the nonzero between-subject variances, scaled such that for all studies.
4 Discussion
We hypothesized that when using a one-sample t-test for a group contrast mean of fMRI data, simply using N − 1 degrees of freedom would lead to invalid p-values due to the heterogeneity of variances. Surprisingly, in our data analysis, we found N − 1 degrees of freedom to be slightly conservative and hence valid when compared to p-values calculated with a Monte Carlo simulation or permutation test. The 2 moment matching Satterthwaite approximation was even more conservative than using N − 1 degrees of freedom. Although the Satterthwaite approximation has been shown to give valid hypothesis tests in single subject fMRI analysis (Kiebel et al., 2003), it did not perform well in the given situation where observations are uncorrelated, but have heterogeneous variance. This is even more surprising since the Satterthwaite approximation was originally developed to handle cases of heteroscedasticity, but suggests that the approach may not perform well with low degrees-of-freedom. As shown in our comparison of distributions of S2 and T in Figure 2, the Satterthwaite approximation only matches the first two moments of the distribution, which does not ensure the left tails of the distributions of S2 for Satterthwaite and the true distribution match; hence the tails of the T distributions will not match. Therefore the Satterthwaite approximation tends to be too conservative in this application.
A three moment matching eDF approach of Scariano and Davenport (1986) was also considered, but similar to the Satterthwaite approximation the effective degrees of freedom are always less than N − 1 and so this method was not considered. The permutation test is the only test that does not assume the T test statistics follow a specific distribution, and its only limitation is discrete P-values for very small sample sizes (e.g. for 6 subjects all P-values are multiples of 1/26 = 0.015625).
Our simulation study allowed us to study type I error and power under a range of sample sizes and outlying variances that are representative of real data findings. Our findings for type I error supported our real data analysis finding that the OLS-based hypothesis test on the sample mean was slightly conservative under heteroscedasticity. Although the true type I error rate was most conservative for small sample sizes and/or the presence of very large outliers, the smallest we found in our simulations was 0.0456 when the goal type I error rate was 0.05.
Although previous studies by Beckmann et al. (2003) and Friston et al. (2005) implied there was a loss in power under the OLS model, they did not formally quantify the loss. Our results show that although there is a loss in power, it was not found to be larger than a 9% in our simulations and this was for a small sample size of 10 subjects with a very large outlying variance.
Although we have shown the OLS model is robust to violations of the heteroscedasticity assumption for the 1-sample t-test, it is probably not the case that this result would also hold in the case of simple linear regression as the slope of a line is easily influenced by outliers. It is likely that in the case of simple linear regression the GLS model should be used to ensure outliers are properly down-weighted.
Finally, we note that these results for fMRI also inform group analyses in other modalities. In particular, in PET or EEG, where it may be impractical or undesirable to estimate intrasubject variance, it is useful to know that the OLS model is performing well in the face of any potential heteroscedasticity.
In conclusion, while a weighted, GLS mixed effects model is the more optimal modeling approach, we find “plain old” OLS surprisingly robust for the widely-used one-sample model. We have provided evidence that an OLS model used with varying designs or outlier-induced heteroscedasticity actually controls false positive risk and has near-optimal power.
Supplementary Material
Appendices
A Bias of OLS variance estimators
In this appendix we find the bias of the OLS variance estimators under (unmodeled) heterogeneous variance. Writing the second level model originally shown in equation 6 using a simplified notation we have,
| (10) |
where Y is the N-vector of contrast data fed up from the first level, X is N × p second-level design matrix, and β are the group-level parameters, and ε are the second-level errors. We assume that Var(ε) = V σ2, where σ2 is the average mixed effects variance, V = diag(vi), Σi vi = 1 are the scaling factors for each subject, allowing for heteroscedasticity. Under this model of the variance we are interested in the properties of the OLS estimators:
| (11) |
| (12) |
| (13) |
First essential result is that E(β̂OLS) = β, that is, the estimates of the regression coefficients are unbiased. However, the estimates of error and parameter variance are not necessarily unbiased. First
| (14) |
| (15) |
where H = X(X′X)−1X′ is the so-called hat matrix, and hi is the ith diagonal element of H, the leverage of observation i. A basic result of linear models gives Σi(1 − hi) = N − p (Harville, 2008). Thus if the variance scaling factors are not all equal (to unity), then the requirement for unbiased is that all leverages hi are all equal. A one-sample model has equal leverage values, as does any balanced ANOVA design. Except for this balanced ANOVA case, a regression model will not have all equal leverages.
The estimator variance is
| (16) |
while
| (17) |
If there is no bias in , is unbiased when
| (18) |
This is true for a one-sample problem, but not even for a balanced ANOVA model. However, for balanced ANOVA models and typical contrasts of interest, there may be no bias. For example, for a balanced two sample t-test, with design matrix
| (19) |
and contrast c = [−1 1] can be shown to have unbiased for cVar(β̂OLS)c′.
B Simulation details
Using GLS, the group mean and estimated variance have the following form,
| (20) |
| (21) |
where is the sum of the within- and between-subject variance for subject i. When the variance is known, this yields the z statistic, . Assuming 90% and 10% of the within-subject variances are and , respectively, that is the between-subject variance and that xi = Δ for all subjects,
| (22) |
which corresponds to a test statistic for a group mean whose value is Δ with N subjects and a standard deviation of
| (23) |
For our simulations we chose a constant across all simulations and varied the outlying variance across the range between 400–3600 and the between subject variance was chosen so that the overall standard deviation in equation 23 was held constant at 33. This range included variance combinations that were found in real data analyses with outlying variances shown in Table 2.
Footnotes
FSL (http://www.fmrib.ox.ac.uk/fsl/), fmristat (www.math.mcgill.ca/keith/fmristat/); SPM (www.fil.ion.ucl.ac.uk/spm/), but only through the hidden spm mfx function.
The 30 most recent papers from each journal were used including early views of in press articles with dates ranging between December 2007- March 2009.
In fullest generality, the contrast C and even the number of parameters P may vary between subjects. The only requirement is that the contrast of parameter estimates has the same units and interpretation across all subjects.
In standard mixed model estimation both the within- and between-subject variances are estimated iteratively, but this approach is computationally too intensive for fMRI data and so the within-subject variance is simply set to the value of the first stage variance estimate.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beckmann CF, Jenkinson M, Smith SM. General multilevel linear modeling for group analysis in FMRI. NeuroImage. 2003;20:1052–1063. doi: 10.1016/S1053-8119(03)00435-X. [DOI] [PubMed] [Google Scholar]
- Cazalis F, Tom S, Reger M, Stover E, Turner K, Poldrack R. Event-related fmri study of mirror reading skill acquisition. Oral presentation at the October, 2004 meeting of the Society for Neuroscience.2004. [Google Scholar]
- Foerde K, Knowlton BJ, Poldrack RA. Modulation of competing memory systems by distraction. Proc Natl Acad Sci USA. 2006;103:11778–11783. doi: 10.1073/pnas.0602659103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K, Stephan K, Lund T, Morcom A, Kiebel S. Mixed-effects and fmri studies. NeuroImage. 2005;24:244–252. doi: 10.1016/j.neuroimage.2004.08.055. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Penny W, Phillips C, Kiebel S, Hinton G, Ashburner J. Classical and Bayesian inference in neuroimaging: theory. NeuroImage. 2002;16:465–483. doi: 10.1006/nimg.2002.1090. [DOI] [PubMed] [Google Scholar]
- Harley E, Pope W, Villablanca J, Mumford J, Suh R, Mazziotta JDE, Engel S. Engagement of fusiform cortex and disengagement of lateral occipital cortex in the acquistion of radiological expertise. Cerebral Cortex. 2009 doi: 10.1093/cercor/bhp051. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harville D. Bayesian inference for variance components using only error contrasts. Biometrika. 1974;61:383–385. [Google Scholar]
- Harville D. Matrix Algebra From a Satistician’s Perspective. Springer; 2008. [Google Scholar]
- Holmes A, Friston K. Generalisability, random effects & population inference. NeuroImage 7 (4 (2/3)), S754, proceedings of Fourth International Conference on Functional Mapping of the Human Brain; June 7–12, 1998; Montreal, Canada. 1998. [Google Scholar]
- Johansen-Berg H, Rushworth MFS, Bogdanovic MD, Kischka U, Wimalaratna S, Matthews PM. The role of ipsilateral premotor cortex in hand movement after stroke. Proc Natl Acad Sci USA. 2002;99:14518–23. doi: 10.1073/pnas.222536799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebel SJ, Glaser DE, Friston KJ. A heuristic for the degrees of freedom of statistics based on multiple variance parameters. Neuroimage. 2003;20:591–600. doi: 10.1016/s1053-8119(03)00308-2. [DOI] [PubMed] [Google Scholar]
- Mumford JA, Nichols T. Modeling and inference of multisubject fMRI data. IEEE Eng Med Biol Mag. 2006;25:42–51. doi: 10.1109/memb.2006.1607668. [DOI] [PubMed] [Google Scholar]
- Mriaux S, Roche A, Dehaene-Lambertz G, Thirion B, Poline JB. Combined permutation test and mixed-effect model for group average analysis in fMRI. Hum Brain Mapp. 2006;27:402–410. doi: 10.1002/hbm.20251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny W, Holmes A. Random effects analysis. In: Friston K, Ashburner J, Kiebel S, Nichols T, Penny W, editors. Statistical Parametric Mapping: The analysis of functional brain images. Elsevier; London: 2006. [Google Scholar]
- Satterthwaite F. An approximate distribution of estimates of variance components. Biometrics Bulletin. 1946;2:110–114. [PubMed] [Google Scholar]
- Scariano S, Davenport J. A four moment approach and other practical solutions to the Behrens-Fisher problem. Communications in Statistics-Theory and Methods. 1986;15:1467–1505. [Google Scholar]
- Searle SR. Linear Models. John Wiley & Sons; 1971. [Google Scholar]
- Stover E, Trepel C, Fox C, Poldrack R. The neural correlates of decision making under risk: an fmri study. NeuroImage 31 (Supp 1), S157, the 12th Annual Meeting of the Organization of Human Brain Mapping; June 11–15, 2006; Florence, Italy. 2006. [Google Scholar]
- Woolrich MW, Behrens TECF, Jenkinson M, Smith SM. Multilevel linear modelling for FMRI group analysis using Bayesian inference. NeuroImage. 2004;21:1732–1747. doi: 10.1016/j.neuroimage.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Friston KJ. Analysis of fMRI time-series revisited–again. NeuroImage. 1995;2:173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Liao CH, Aston J, Petre V, Duncan GH, Morales F, Evans AC. A general statistical analysis for fMRI data. NeuroImage. 2002;15:1–15. doi: 10.1006/nimg.2001.0933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.