Summary
Chaperonin action is controlled by cycles of nucleotide binding and hydrolysis. Here we examine the effects of nucleotide binding on an archaeal group 2 chaperonin. In contrast to the ordered apo state of the group 1 chaperonin GroEL, the unliganded form of the homo-16mer Methanococcus maripaludis group 2 chaperonin is very open and flexible, with intersubunit contacts only in the central double belt of equatorial domains. The intermediate and apical domains are free of contacts and deviate significantly from the overall 8-fold symmetry. Nucleotide binding results in three distinct, ordered 8-fold symmetric conformations, open, partially closed and fully closed. The partially closed ring encloses a 40% larger volume than the GroEL-GroES folding chamber, enabling it to encapsulate proteins up to 80 kDa, in contrast to the fully closed form whose cavities are 20% smaller than the GroEL-GroES chamber.
Keywords: molecular chaperones, allostery, cryo-electron microscopy, image processing, single particle analysis, atomic structure fitting
Introduction
The chaperonins are double ring molecules that are found in all forms of life and are essential for cell viability, as they provide an isolated environment for ATP dependent protein folding (Bukau and Horwich, 1998; Frydman, 2001; Horwich et al., 2007). The chaperonins are found in two sub-groups: group 1 in eubacteria, mitochondria and chloroplasts and group 2 in archaea and in the eukaryotic cytosol (Bukau and Horwich, 1998; Gutsche et al., 1999; Spiess et al., 2004). The sequence and structural similarity between the two groups suggest they share a common mechanism of action. The most detailed structural and mechanistic information on the group 1 chaperonins comes from Escherichia coli GroEL, for which multiple conformations have been identified by both X-ray crystallography and cryo-electron microscopy (cryo-EM) (Braig et al., 1994; Xu et al., 1997; Ranson et al., 2001 & 2006). Limited structural information has been determined for the group 2 chaperonins from X-ray and cryo-EM structures of the thermosome and cryo-EM structures of CCT and TF55 (Ditzel et al., 1998; Llorca et al., 1998 & 1999a; Schoehn et al., 2000a & 2000b). Their open states are defined only at very low resolution, so that the domain positions and orientations are unclear.
Chaperonins of both groups contain two back-to-back rings of identical or closely related subunits, with 7-fold rings for the group 1 and 8- or 9-fold rings for group 2. The subunits contain equatorial, intermediate and apical domains connected by flexible hinge regions. Negative allosteric communication takes place between the two rings through the back-to-back equatorial domain contacts (Yifrach and Horovitz, 1996; Kafri et al., 2001; Kusmierczyk and Martin, 2003a; Bigotti and Clarke, 2005). These are mediated via cross-ring subunit contacts, 1:2 in group 1 chaperonins, in which the two rings are offset, and 1:1 organisation for the group 2 chaperonins, where the rings are in register (Braig et al., 1994; Ditzel et al., 1998).
The effects of nucleotide binding on the global conformation of group 2 chaperonins are not understood in detail. In the case of group 1, from cryo-EM studies it is known that nucleotide binding to apo GroEL releases an inter-subunit salt bridge that allows mobility of the substrate-binding apical domains and also causes small rotations of the equatorial domains that distort the inter-ring interface (Ranson et al., 2001). An asymmetric conformation is also observed in group 2, most likely caused by nucleotide binding (Llorca et al., 1999a; Schoehn et al., 2000a; Gutsche et al., 2001). Protease protection studies show that transition state analogue ADP•AlF3 is required to close the rings of CCT, suggesting that ATP hydrolysis may be necessary for this step (Meyer et al., 2003), unlike the formation of the closed GroEL-GroES chamber which only requires nucleotide binding.
In group 1 the apical domains contain the hydrophobic binding sites that bind both substrate protein and the co-chaperonin (Fenton et al., 1994; Xu et al., 1997). In group 2 the apical domains also contain substrate binding sites, but the nature of these sites is still unclear (Llorca et al., 1999b; Spiess et al., 2006). When the co-chaperonin GroES (hsp10/cpn10) binds to the apical domains of the ATP-bound group 1 chaperonins, it causes them to elevate and twist clockwise more than 100°, creating a hydrophilic chamber for protein folding (Sigler et al., 1998). The group 2 chaperonins do not require a co-chaperonin to form the folding chamber since they have an insertion in their apical domains that forms a built-in lid structure (Klumpp et al., 1997; Ditzel et al., 1998). However, the subunit conformations and movements between the apo, substrate-binding conformation and the ATP-driven substrate-folding conformation in the group 2 chaperonins are not well characterised. The only state of group 2 observed by crystallography is a fully closed form analogous to a compressed GroES-GroEL-GroES “football” (Schmidt et al., 1994). In this crystal structure the folding chamber is 20% smaller in volume than that of GroEL-GroES, suggesting that the group 2 chaperonins might not accommodate large substrate proteins (Ditzel et al., 1998). Other reports have suggested that the group 2 chaperonins can assist in the folding of proteins up to 100 kDa, which would be too large for encapsulation in the folding chamber of the closed complex observed by X-ray crystallography (Spiess et al., 2004).
In this study we examine the effects of nucleotide binding to a homo-oligomeric group 2 chaperonin, from the mesophilic archaeon Methanococcus maripaludis (Mm). The open, partially closed, and fully closed conformations of Mm were modelled by rigid body fitting of the domains of the thermosome crystal structure into cryo-EM densities. The models reveal a novel ATP-induced ordering of the positions of the mobile apical domains in the group 2 chaperonin cycle.
Results and Discussion
The unliganded form of the Mm chaperonin is open and flexible
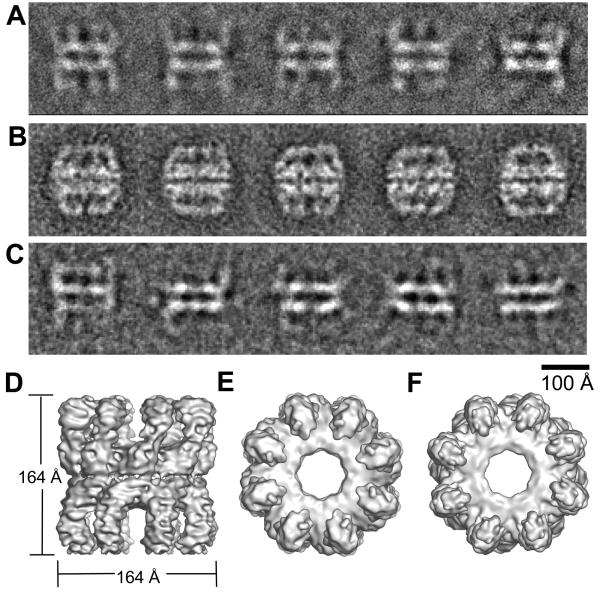
Cryo-EM images of unliganded Mm chaperonin revealed a disordered, open conformation (Figure 1A). At low pH, Mm loses activity (Kusmierczyk and Martin, 2003b), and a closed conformation is observed in uranyl acetate at pH 4 (Figure 1B), but the open form is also observed in negative stain at pH 8 (Figure 1C). In many of the cryo images and class averages of the open form, only the equatorial domains are clearly visible (Figure 1A). The other domains are in variable orientations and do not give consistent projections. A 3D map of apo-Mm chaperonin was reconstructed from cryo images of the 4000 most ordered particles (showing the most density for intermediate and apical domains, and therefore expected to have approximate 8-fold symmetry) by angular reconstitution and projection matching (Figure 1D-F). The complex has a very open structure with the ring of subunits held together only by interactions between the equatorial domains. The apical and intermediate domain densities are incomplete, confirming that these domains are moving, through rigid body domain rotations about flexible hinge points, leading to significant deviations from 8-fold symmetry in this region. In contrast, apo-GroEL has intersubunit contacts between intermediate and apical domains (Braig et al., 1994). The lack of intersubunit contacts at the intermediate and apical domain level in apo-Mm underlies the inherent flexibility of the complex.
Figure 1. The unliganded state of the Mm complex in different stains and in cryo-EM.
Averaged views of the Mm complex, without nucleotide, unstained in ice (A), stained with 2% uranyl acetate (B), and stained with 2% sodium vanadate (C). Cryo-EM 3D reconstruction of the apo-Mm complex viewed as a surface from the side (D), top (E) and bottom (F).
Nucleotide binding yields three distinct conformations of the group 2 chaperonin
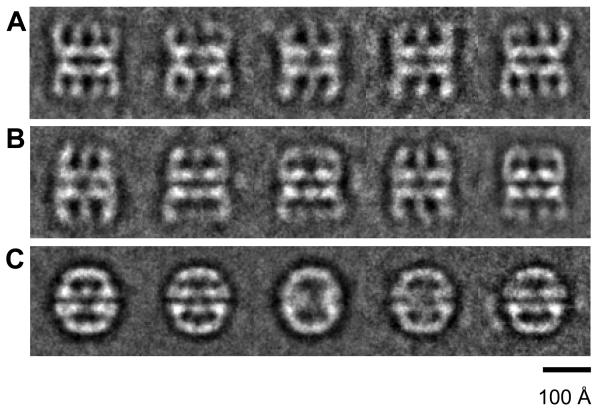
At 37°C and 150 mM KCl, pH 7.4, the Mm ATPase is fully active and Mm is able to refold rhodanese (not shown; see Kusmierczyk and Martin, 2003a). Therefore, all grids were prepared under those conditions. To examine the conformational changes induced by ATP binding, we imaged the Mm complex in the presence of the ATP analogue, ADP•AlF3 (another analogue, AMPPNP, supports protein folding by Mm (Kusmierczyk and Martin, 2003b)). The Mm-ADP•AlF3 side view images showed that the addition of nucleotide caused the complex to become more ordered, since the class averages showed clearly defined projections for the intermediate and apical domains (Figure 2A-C). Notably the open form of Mm in nucleotide has much less mobility in its intermediate and apical domains than in the open form of the apo complex (compare figures 1A and 2A), similar to an earlier conclusion deduced from negatively stained views of CCT (Rivenzon-Segal et al., 2005). Within the side view population of these double-ring complexes there were three discernable conformations: open-open, partially closed-open (bullet) and closed-closed forms. The distribution of side views between the three conformations was approximately 35% open, 50% bullets and 15% closed. A very similar distribution of these three conformations was seen in side views of the complex in the presence of ATP (Supplementary Figure 1A). In contrast, in ADP only open and partially closed states were observed, and the closed conformation was not seen (Supplementary Figure 1B). The observation of multiple conformations in the presence of nucleotides is consistent with transient kinetic studies reporting multiple states (Horovitz and Willison, 2005; Cliff et al., 2006).
Figure 2. Nucleotide-bound states of Mm.
Cryo-EM class averages of the open (A), bullet (B) and closed (C) conformations of the Mm-ADP•AlF3 complex. Each class average contains approximately 30 aligned images.
3D reconstruction and atomic structure fitting of the three Mm-ADP•AlF3 conformations
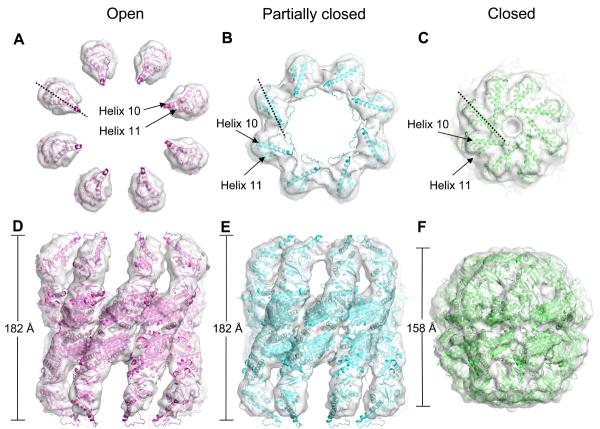
Using classification of side views it was possible to separate the Mm-ADP•AlF3 cryo-EM data set into distinct open, partially closed bullet, and closed conformations for 3D reconstruction with 8-fold symmetry (Figure 3). The reconstructions had resolutions of approximately 10 Å for the open, 9.5 Å for the bullet and 11 Å for the closed form.
Figure 3. Surface views of the three reconstructed Mm-ADP•AlF3 complexes with domain fitting.
3D reconstructions, displayed at a threshold of one sigma, of the open (A and D), bullet (B and E) and closed (C and F) Mm-ADP•AlF3 complexes, with their fitted domains, viewed from the top and the side. The structures used for fitting were the domains of the thermosome crystal structure (PDB accession number 1A6D). Dotted line shows apical rotation, helices 10 and 11 are indicated.
The open state of Mm-ADP•AlF3 resembles the apo-Mm map but contains almost complete density for the apical domains, suggesting that nucleotide binding stabilises about one third of the complexes in a symmetric, open conformation. The α-helical protrusions in the apical domains, which form the lid in the closed structure, were too far apart to form intra-ring contacts and were instead pointing upwards towards the bulk solution.
The bullet map was asymmetric, with one ring in an open state (bottom ring in Figure 3E) and the other in a partially closed conformation in which the apical domains have rotated clockwise when viewed from outside the complex. Because of this clockwise rotation and a closing of the ring, the apical domains in the partially closed state form an additional inter-subunit contact (Figure 3B, compared to 3A). The partially closed ring has a much larger volume and central opening than the fully closed ring (Figure 3C). We estimate the enclosed volume as 250,000-290,000 Å3 in the partially closed ring versus 130,000 Å3 in the fully closed ring. In the closed map, both rings are tightly closed with intra-ring contacts made between all domains.
The open, bullet and closed conformations observed in this study resemble those previously observed at much lower resolution in the α-only thermosome and TF55 structures (Schoehn et al., 2000a & 2000b). At the resolution of the present maps, it was possible to examine the domain rotations between open and closed rings of the Mm chaperonin by fitting the thermosome atomic structure into the cryo-EM maps of the open, bullet and closed conformations. Because of the large differences between the closed conformation, observed by X-ray crystallography, and the open and bullet conformations, all domains had to be fitted as separate rigid bodies. Initially the three domains of one subunit in each of the two chaperonin rings were manually fitted into the open and bullet maps using PYMOL (www.pymol.org). The positions of the 6 individual domains in both maps were then refined using the automated fitting program URO (Navaza et al., 2002) (Figure 3A, B, D and E). The hand of all the maps was based on the crystal structure of the thermosome. The connectivity between the three domains was used to assess the accuracy of the fits. However, due to the large movements of the apical domains, the distances between the intermediate-apical connections in the open rings were too large, suggesting that there is some refolding in that part of the structure. All the other connections in the fitted structures had backbone distances within 6 Å of each other. The long loop region preceding the descending α-helical limb (helix 10) of the protrusion and the tip of helix 10 protruded from the density in the open and partially closed rings, but this part of the apical domain has been shown by NMR to be flexible (Heller et al., 2004).
For the closed Mm-ADP•AlF3 map it was possible to fit the whole thermosome hexadecamer crystal structure as a single rigid body (Figure 3C and F), showing that the structures are identical and supporting the idea that this conformation exists in solution during the nucleotide cycle. This closed conformation has previously been seen in CCT in the presence of nucleotide (Llorca et al., 2001).
Analysis of the domain rotations between the open, partially closed and fully closed rings of Mm chaperonin
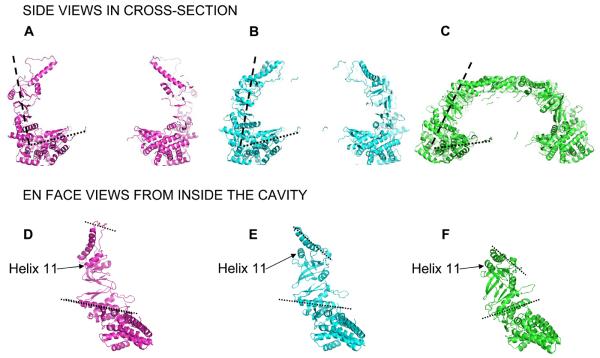
The open ring of the bullet map (lower ring in Figure 3E) was very similar to the rings in the open structure (Figure 3D). Therefore one of the rings from the open map was used for further analysis. The angle between the equatorial domains and the intermediate and apical domains is approximately 90° in the open ring (Figure 4A). This produces a jaw-like opening of the ring relative to the fully closed form (Figure 4C), causing an expansion in diameter in the open ring at the level of both intermediate and apical domains, to approximately 156 and 170 Å respectively (Figure 4A and 4C). Due to the expansion and rotation of the intermediate domains, the nucleotide binding sites in the open rings are open to the bulk solution. The conformation of the apical domains indicates that the surface facing the folding chamber is now lined by helix 11, which is thought to contain a substrate-binding site (Spiess et al., 2006) (Figure 4D). This is consistent with apo-GroEL, in which the folding cavity is lined by helices H and I, where helix I is the equivalent of helix 11 and has been shown to be a preferred site of binding for the stringent substrate mitochondrial malate dehydrogenase (Elad et al., 2007).
Figure 4. Analysis of domain rotations in Mm-ADP•AlF3 rings.
Fitted coordinates of the open (A and D), partially closed (B and E) and closed rings (C and F). A, B and C show cross-sectioned side views and individual subunits are viewed from inside the cavity in D, E and F. The dashed lines in A, B and C indicate the angle between the equatorial domain and the intermediate/apical domains. The dashed lines in D, E and F show the change in angle between intermediate and apical domains. The dashed lines in the intermediate domains (lower dashed line) run along helix 14 which contains an aspartate residue required for ATP hydrolysis.
In the partially closed ring (Figure 3B and 4B) the jaws have closed slightly with a 25° smaller angle between the equatorial domains and the intermediate and apical domains than in the open ring (Figure 4B versus 4A). The intermediate domains have a slightly different orientation to the open ring, bringing helix 14 bearing an aspartate residue required for ATP hydrolysis slightly closer to the nucleotide-binding pocket. Relative to the open ring, the apical domains move radially inward and rotate clockwise by 30° when viewed from outside the ring (compare Figures 3B to 3A and 4B to 4A). As a result, the substrate binding region on the face of helix 11 and the tip of helix 10 are rotated into the interface between the subunits and now take part in inter-subunit contacts (Figure 3B, E and 4E). This is similar to the apical domain rotations in GroEL when it converts from its open, substrate-binding conformation to its closed, GroES-bound, substrate-folding conformation, suggesting that the highly populated, partially closed ring is a folding active conformation. Moreover, if the partially closed ring serves as the folding chamber, it would mean that there is more space available to fold proteins than previously thought, because the chamber is twice the size of the one in the closed ring. The larger volume of the folding chamber in the partially closed ring helps to account for reports of CCT substrates up to 100 kDa (Spiess et al., 2004). However, even in the partially closed ring the volume would accommodate a maximum substrate of around 80 kDa, suggesting that there may be an alternative mechanism for larger substrates. The folding chamber in the partially closed ring is 40-50% larger than the one formed by GroEL-GroES.
In the closed ring, the entire subunit rotates inward by 20° (Figure 4C), bringing the apical domains together to form the central β-barrel (Figure 3C). This reduces the size of the folding chamber and closes it off entirely from the bulk solution. In addition, the intermediate domains close down over the nucleotide binding sites in the equatorial domains (Figure 4C). The large downward tilt of the equatorial domains in the closed form moves the inter-ring salt bridge, formed by Arg429 and Asp455, radially outwards by 6 Å relative to the open and partially closed forms. Such large rotations of the equatorial domains have not been observed in any of the states of GroEL (Ranson et al., 2001 & 2006) in which the out of register interface between the two rings (1:2 versus 1:1 in the group 2 chaperonins) likely constrains equatorial domain rotation.
Implications for folding states of the Mm complex
In the open ring, the potential substrate binding region on helix 11 is very accessible and lines the folding chamber of the open ring (Figure 4D), analogous to the orientation of substrate binding sites in apo-GroEL. In the partially closed ring the substrate binding region on helix 11 is rotated to form an inter-subunit contact (Figure 4B and E). This partially closed state is associated with a conformational change that leads to the formation of a very large chamber for protein folding, analogous to the GroEL-GroES folding chamber. The large folding chamber formed in the partially closed ring would allow the group 2 chaperonins to bind and productively fold large or elongated substrate proteins. After initial folding of the substrate in this chamber, to a more compact intermediate, when ATP is hydrolysed, the partially closed ring might compact further to form the fully closed ring, forcing further compaction of the substrate. Such a fully closed state may not be achievable when large substrates are involved. This problem of limited space in the fully closed chamber is highlighted when the crystal structure of actin (43 kDa) is docked into the closed thermosome crystal structure (Supplementary Figure 2). A native actin monomer fills the available space in the fully closed chamber with a few clashes, making it difficult to envisage how expanded folding intermediates of actin could be encapsulated in the fully closed chamber. However, actin could be readily encapsulated and folded in the much larger partially closed folding chamber described here.
Experimental Procedures
Protein expression and purification
M. maripaludis cpn60 was expressed and purified as described by Kusmierczyk and Martin (2003b) except that the hydroxyapatite column was replaced by a heparin sulphate column eluted with buffers described for anion exchange chromatography.
Sample preparation
For negative stain EM, the Mm complex was diluted to a final concentration of 0.2 mg/ml in 50 mM Tris-HCl (pH 7.4), 150 mM KCl or 50 mM KCl, 10 mM MgCl and 1mM DTT and incubated at 24°C. For cryo-EM of apo-Mm, the complex was diluted to final concentration of 3 mg/ml in 50 mM Tris-HCl (pH 7.4), 150 mM KCl, 10 mM MgCl and 1 mM DTT and incubated at 37°C. For the Mm-ADP•AlF3 complex, KF was added to the sample above followed by KAl(SO4)2 and ADP, giving final concentrations of 5 mM, 0.5 mM and 1 mM respectively. This sample was incubated for 10 minutes at 37°C and then centrifuged for 5 minutes prior to making cryo-EM grids.
Electron microscopy
For negative stain EM, 3.5 μl of 0.2 mg/ml Mm were applied to glow-discharged carbon coated grids and stained with 5 μl of 2% uranyl acetate or 2% sodium vanadate (Nano-Van, Molecular Probes, UK). The stained Mm complexes were imaged under low dose conditions at nominal magnifications of 40,000 or 34,000 and images were both recorded on Kodak SO-163 film .
For cryo-EM, 3 μl of the Mm complex alone or with ADP•AlF3 were applied to C-flat grids (r2/2, Protochips Inc., USA). The grids were rendered hydrophilic using a Fischione plasma cleaner and then used for sample preparation in a vitrobot (FEI, Netherlands) at 37°C and 100% humidity. Low dose images were recorded using the automatic data acquisition system LEGINON (Suloway et al., 2005) on a Tecnai F20 FEG electron microscope operated at 120 kV and equipped with a Gatan cold stage (Gatan, USA), and recorded on a Gatan 4K ultrascan CCD camera (Gatan, USA) at final magnifications of 148,500 for the apo (760 frames) and 92,000 for the ADP•AlF3 (560 frames) and between 0.7 and 3.5 μm underfocus.
Image processing
Negative stain micrographs were digitised in a Zeiss SCAI scanner giving a specimen sampling of 3.5 Å/pixel for uranyl acetate and 4.1 Å/pixel for the sodium vanadate stained complexes. Particles were selected using XIMDISP (Crowther et al., 1996) and extracted into 128 by 128 boxes using LABEL (Crowther et al., 1996). 2552 particles were selected for the uranyl acetate and 1552 for the sodium vanadate stained samples. The boxed particles were band-pass filtered between 160 and 7 Å for the uranyl acetate and 180 and 8 Å for the sodium vanadate stained samples, and then normalised in SPIDER (Frank et al., 1996).
The negative stain images were aligned and classified after initial centering by multivariate statistical analysis (MSA) in IMAGIC (van Heel et al., 1996). Selected class averages of the side views were used for multi-reference alignment (MRA) and further classification.
The CCD cryo-EM images were reduced by a factor of two, giving 2.02 Å/pixel for apo-Mm and 3.26 Å/pixel for Mm-ADP•AlF3. The contrast transfer function (CTF) for each CCD image was determined using CTFFIND3 (Mindell and Grigorieff, 2003). Initially a small subset of particles were selected and extracted into 256 by 256 boxes for apo-Mm and 196 by 196 boxes for the Mm-ADP•AlF3. CTF correction was done in SPIDER, and the boxes were then cropped to 196 by 196 for apo-Mm and 150 by 150 for Mm-ADP•AlF3. The boxed images were band-pass filtered between 200 and 6 Å and normalised. Images were centered using negative stain class averages as references and then classified by MSA. Class averages representing 4 different side views of the Mm complex were used as templates for the automatic particle picking using FINDEM (Roseman, 2004), concentrating on selecting mainly side views, which constituted ~10% of the particles.
For the apo-Mm complex, 22,687 particles were selected and extracted into 256 by 256 boxes, CTF- corrected in SPIDER, band-pass filtered between 200 and 4 Å and normalised. The images were then aligned to selected class averages from the hand picked data set and classified using MSA. Due to both the structural flexibility of the apo-Mm complexes and the selection of a significant minority of end views, a careful selection of only the best side view images had to be made, which left 3,946 images. The side view images were then aligned by projection matching to an angular reconstitution model generated in IMAGIC. The resolution of the final map (10 Å) was estimated with the 0.5 FSC criterion.
For the Mm-ADP•AlF3 complexes, 18,925 particles were selected and extracted into 150 by 150 boxes and pre-processed as above. The particles were separated into the closed conformation in one set (~1100 images) and the open and bullet in the other (~7000 images). The images of the closed form were then aligned via projection matching to a 3D model generated from a side view class average using 8-fold symmetry in IMAGIC. For the open/bullet images, a model was similarly generated for each conformation, and competitive projection matching was used to refine the separation and the maps. The resolutions of the final open, bullet and closed maps (10 Å, 9.5 Å and 11 Å respectively) were estimated as before.
Atomic structure fitting
The open, bullet and closed maps were filtered between 20 Å and 9 Å to correct for the over-representation of low frequencies, reducing the low frequencies to 10% of their original amplitude and eliminating the high frequency components. For the closed structure the thermosome (PDB accession number 1A6D) oligomer was manually fitted as a single rigid body into the cryo-EM map using PYMOL (www.pymol.org). For the open and bullet structures, one subunit in each ring of the thermosome structure was separated into equatorial (17-144 and 403-519), intermediate (145-214 and 365-402) and apical (215-364) domains. These 6 domains were manually fitted into the cryo-EM maps using PYMOL and the rigid body fits were refined with URO (Navaza et al., 2002), taking into account the 8-fold symmetry.
The open, bullet and fully closed ADP•AlF3 maps have been deposited in the EMDB with accession numbers EMD-1396, EMD-1397 and EMD-1398.
Supplementary Material
Acknowledgments
We thank Richard Westlake, Dave Houldershaw and Luchun Wang for computing and EM support and Wayne Fenton for comments on the manuscript. We also thank Bridget Carragher, Clint Potter and the Leginon team for their help and access to the NRAMM facility at the Scripps Research Institute.
This work was carried out at the National Resource for Automated Molecular Microscopy, supported by the National Institutes of Health though the National Center for Research Resources' P41 program (RR17573) and at the School of Crystallography, Birkbeck College, supported by a Wellcome Trust programme grant, EU 3D EM Network of Excellence and 3D Repertoire grants.
References
- Bigotti MG, Clarke AR. Cooperativity in the thermosome. J. Mol. Biol. 2005;348:13–26. doi: 10.1016/j.jmb.2005.01.066. [DOI] [PubMed] [Google Scholar]
- Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature. 1994;371:578–86. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–66. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Cliff MJ, Limpkin C, Cameron A, Burston SG, Clarke AR. Elucidation of steps in the capture of a protein substrate for efficient encapsulation by GroE. J. Biol. Chem. 2006;261:21266–21275. doi: 10.1074/jbc.M601605200. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM. MRC image processing programs. J. Struct. Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93:125–38. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- Elad N, Farr GW, Clare DK, Orlova EV, Horwich AL, Saibil HR. Topologies of a substrate protein bound to the chaperonin GroEL. Mol. Cell. 2007;26:415–26. doi: 10.1016/j.molcel.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton WA, Kashi Y, Furtak K, Horwich AL. Residues in chaperonin GroEL required for polypeptide binding and release. Nature. 1994;371:614–619. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and Visualization of Images in 3D Electron Microscopy and Related Fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 2001;70:603–47. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Gutsche I, Essen LO, Baumeister W. Group II chaperonins: new TRiC(k)s and turns of a protein folding machine. J Mol. Biol. 1999;293:295–312. doi: 10.1006/jmbi.1999.3008. [DOI] [PubMed] [Google Scholar]
- Gutsche I, Holzinger J, Rauh N, Baumeister W, May RP. ATP-induced structural change of the thermosome is temperature-dependent. J. Struct. Biol. 2001;135:139–146. doi: 10.1006/jsbi.2001.4373. [DOI] [PubMed] [Google Scholar]
- Heller M, John M, Coles M, Bosch G, Baumiester W, Kessler H. studies on the substrate-binding domains of the thermosome: structural plasticity in the protrusion region. J. Mol. Biol. 2004;336:717–29. doi: 10.1016/j.jmb.2003.12.035. [DOI] [PubMed] [Google Scholar]
- Horovitz A, Willison KR. Allosteric regulation of chaperonins. Curr. Opin. Struct. Biol. 2005;15:646–651. doi: 10.1016/j.sbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Horwich A, Fenton WA, Chapman E, Farr GW. Two Families of Chaperonin: Physiology and Mechanism. Annu. Rev. Cell. Dev. Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- Kafri G, Willison KR, Horovitz A. Nested allosteric interactions in the cytoplasmic chaperonin containing TCP-1. Protein Sci. 2001;10:445–449. doi: 10.1110/ps.44401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp M, Baumeister W, Essen LO. Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin. Cell. 1997;91:263–70. doi: 10.1016/s0092-8674(00)80408-0. [DOI] [PubMed] [Google Scholar]
- Kusmierczyk AR, Martin J. Nested cooperativity and salt dependence of the ATPase activity of the archaeal chaperonin Mm-cpn. FEBS Lett. 2003a;547:201–4. doi: 10.1016/s0014-5793(03)00722-1. [DOI] [PubMed] [Google Scholar]
- Kusmierczyk AR, Martin J. Nucleotide-dependent protein folding in the type II chaperonin from the mesophilic archaeon Methanococcus maripaludis. Biochem. J. 2003b;371:669–73. doi: 10.1042/BJ20030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Smyth MG, Marco S, Carrascosa JL, Willison KR, Valpuesta JM. ATP binding induces large conformational changes in the apical and equatorial domains of the eukaryotic chaperonin containing TCP-1 complex. J. Biol. Chem. 1998;273:10091–4. doi: 10.1074/jbc.273.17.10091. [DOI] [PubMed] [Google Scholar]
- Llorca O, Smyth MG, Carrascosa JL, Willison KR, Radermacher M, Steinbacher S, Valpuesta JM. 3D reconstruction of the ATP-bound form of CCT reveals the asymmetric folding conformation of a type II chaperonin. Nat. Struct. Biol. 1999a;6:639–42. doi: 10.1038/10689. [DOI] [PubMed] [Google Scholar]
- Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature. 1999b;402:693–6. doi: 10.1038/45294. [DOI] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Grantham J, Ritco-Vonsovici M, Willison KR, Carrascosa JL, Valpuesta JM. The ‘sequential allosteric ring’ mechanism in the eukaryotic chaperonin-assisted folding of actin and tubulin. EMBO J. 2001;20:4065–4075. doi: 10.1093/emboj/20.15.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell. 2003;113:369–381. doi: 10.1016/s0092-8674(03)00307-6. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Navaza J, Lepault J, Rey FA, Alvarez-Rua C, Borge J. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr. D. 2002;58:1820–1825. doi: 10.1107/s0907444902013707. [DOI] [PubMed] [Google Scholar]
- Ranson NA, Farr GW, Roseman AM, Gowen B, Fenton WA, Horwich AL, Saibil HR. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell. 2001;107:869–79. doi: 10.1016/s0092-8674(01)00617-1. [DOI] [PubMed] [Google Scholar]
- Ranson NA, Clare DK, Farr GW, Houldershaw D, Horwich AL, Saibil HR. Allosteric signalling of ATP hydrolysis in GroEL-GroES complexes. Nat. Struct. Mol. Biol. 2006;13:147–52. doi: 10.1038/nsmb1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenzon-Segal D, Wolf SG, Shimon L, Willison KR, Horovitz A. Sequential ATP-induced allosteric transition of the cytoplasmic chaperonin containing TCP-1 revealed by EM analysis. Nat. Struct. Mol. Biol. 2005;12:233–237. doi: 10.1038/nsmb901. [DOI] [PubMed] [Google Scholar]
- Roseman AM. FindEM--a fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 2004;145:91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Rutkat K, Rachel R, Pfeifer G, Jaenicke R, Viitanen P, Lorimer G, Buchner J. Symmetric complexes of GroE chaperonins as part of the functional cycle. Science. 1994;265:656–659. doi: 10.1126/science.7913554. [DOI] [PubMed] [Google Scholar]
- Schoehn G, Hayes M, Cliff M, Clarke AR, Saibil HR. Domain rotations between open, closed and bullet-shaped forms of the thermosome, an archaeal chaperonin. J. Mol. Biol. 2000a;301:323–32. doi: 10.1006/jmbi.2000.3952. [DOI] [PubMed] [Google Scholar]
- Schoehn G, Quaite-Randall E, Jimenez JL, Joachimiak A, Saibil HR. Three conformations of an archaeal chaperonin, TF55 from Sulfolobus shibatae. J. Mol. Biol. 2000b;296:813–9. doi: 10.1006/jmbi.2000.3505. [DOI] [PubMed] [Google Scholar]
- Sigler PB, Xu Z, Rye HS, Burston SG, Fenton WA, Horwich AL. Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 1998;67:581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Miller EJ, McClellan AJ, Frydman J. Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol. Cell. 2006;24:25–37. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A New Generation of the IMAGIC Image Processing System. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- Yifrach O, Horovitz A. Allosteric control by ATP of non-folded protein binding to GroEL. J. Mol. Biol. 1996;255:356–361. doi: 10.1006/jmbi.1996.0028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.