Abstract
Germ cells are the only cells that transmit genetic information to the next generation, and they therefore must be prevented from differentiating inappropriately into somatic cells1. A common mechanism by which germline progenitors are protected from differentiation-inducing signals is a transient and global repression of RNA polymerase II (RNAPII)-dependent transcription1. In both Drosophila and Caenorhabditis elegans embryos, the repression of messenger RNA transcription during germ cell specification correlates with an absence of phosphorylation of Ser 2 residues in the carboxy-terminal domain of RNAPII (hereafter called CTD)2, a critical modification for transcriptional elongation3. Here we show that, in Drosophila embryos, a small protein encoded by polar granule component (pgc) is essential for repressing CTD Ser 2 phosphorylation in newly formed pole cells, the germline progenitors. Ectopic Pgc expression in somatic cells is sufficient to repress CTD Ser 2 phosphorylation. Furthermore, Pgc interacts, physically and genetically, with positive transcription elongation factor b (P-TEFb), the CTD Ser 2 kinase complex, and prevents its recruitment to transcription sites. These results indicate that Pgc is a cell-type-specific P-TEFb inhibitor that has a fundamental role in Drosophila germ cell specification. In C. elegans embryos, PIE-1 protein segregates to germline blastomeres, and is thought to repress mRNA transcription through interaction with P-TEFb4–7. Thus, inhibition of P-TEFb is probably a common mechanism during germ cell specification in the disparate organisms C. elegans and Drosophila.
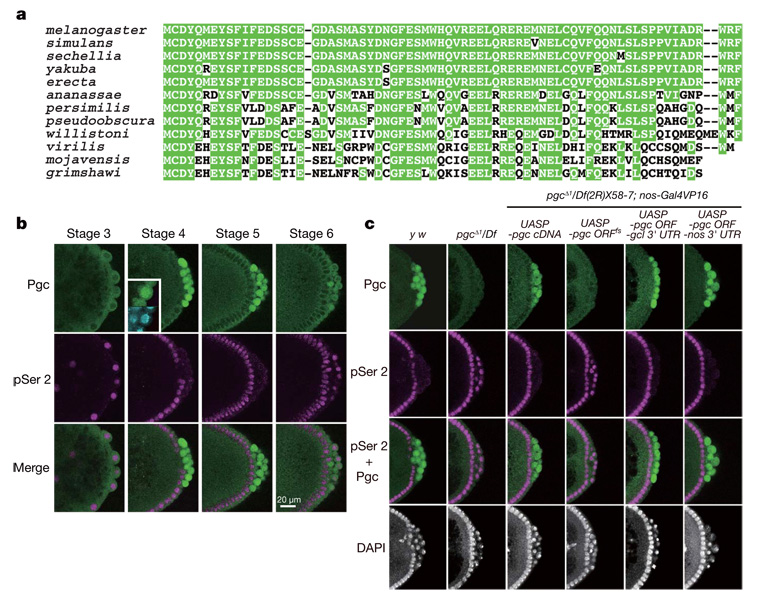
pgc RNA, a component of the Drosophila germ plasm8, has been implicated in the repression of CTD Ser 2 phosphorylation in early pole cells9,10. However, the mechanism underlying pgc-mediated transcriptional repression has been unknown. Initial characterization of its nucleotide sequence suggested that pgc RNA acts as a non-coding RNA8. Nevertheless, we have noticed that an AUG triplet, beginning at nucleotide 117 of the 0.7-kilobase (kb) transcript, is in a favourable context to serve as a translation initiation site8. Completion of the Drosophila genomic sequence revealed an error in our original manual sequencing in a region where a strong stem structure can form (an additional C exists between nucleotides 178 and 179). In the revised pgc sequence, the open reading frame (ORF) beginning at nucleotide 117 is capable of encoding a 71-amino-acid polypeptide (Fig. 1a). We found that this polypeptide sequence is conserved in 12 Drosophila species for which genomes have been sequenced (Fig. 1a), although no homologous sequences have been identified in other animal groups, even in dipteran insects.
Figure 1. Pgc expression in pole cells is essential for the repression of CTD Ser 2 phosphorylation.
a, Pgc sequences of 12 Drosophila species. Conserved amino acid residues are highlighted by green shading. Note that no apparent Pgc orthologues can be found in other animal groups. b, Pgc expression in pole cells is complementary to pSer 2. Panels show the posterior pole of wild-type embryos immunostained for Pgc (green) and pSer 2 (magenta). Nuclei were counter-stained with 4,6-diamidino-2-phenylindole (DAPI; cyan). Pgc is concentrated in the nucleus (inset). c, Posterior poles of stage-4 embryos immunostained for pSer 2 (magenta) and Pgc (green). Nuclei were counter-stained with DAPI. Maternal genotypes are indicated. Note that pgc ORFfs RNA accumulates normally in the germ plasm (Supplementary Fig. 2).
Antibodies raised against the 71-amino-acid polypeptide showed immunoreactivity in wild-type pole cells, but not in pole cells lacking pgc (Fig. 1b, c). The signals were first detected in pole cells at stage 4, when pole cells are formed. Although pgc RNA is detectable in pole cells until mid-embryogenesis8, the Pgc immunoreactivity in pole cells dropped markedly at stage 6. We did not detect the Pgc signal in somatic cells. These dynamic patterns of Pgc expression suggest that pgc RNA translation and Pgc protein stability are regulated. Notably, double staining of wild-type embryos for Pgc and CTD phosphorylated at Ser 2 (pSer 2) revealed that the Pgc expression in pole cells was complementary to pSer 2 (Fig. 1b).
To investigate the function of pgc in repressing CTD Ser 2 phosphorylation, we generated pgcΔ1, a chromosomal null for pgc (Supplementary Fig. 1). Homozygous or hemizygous pgcΔ1 females were fertile, and embryos from pgcΔ1 mothers (hereafter termed pgc− embryos) formed normal numbers of pole cells (Fig. 1c, Supplementary Fig. 2 and data not shown). However, pgc− pole cells failed to repress CTD Ser 2 phosphorylation during stages 4–5 (Fig. 1c). These pole cells degenerated from stage 10 onwards, and few or no pole cells coalesced into the gonads (Supplementary Fig. 2). Consequently, about 80% of the pgc− embryos developed into sterile adults. These defects were rescued by the expression of intact pgc RNA, but not a frame-shift version of pgc RNA (pgc ORFfs), during oogenesis (Fig. 1c and Supplementary Fig 2 and Supplementary Fig 3). We next made hybrid transgenes in which the pgc ORF was fused with the nanos (nos) or germ cell-less (gcl) 3′ UTR, which contains RNA localization signals that mediate accumulation in the germ plasm. Expression of these fusion genes during oogenesis in pgcΔ1 hemizygotes promoted Pgc expression in early pole cells and repressed CTD Ser 2 phosphorylation (Fig. 1c). Pole cell death was also rescued in these embryos (Supplementary Fig. 2 and data not shown), which developed into fertile adults. These results indicate that Pgc protein is essential for the repression of CTD Ser 2 phosphorylation in pole cells
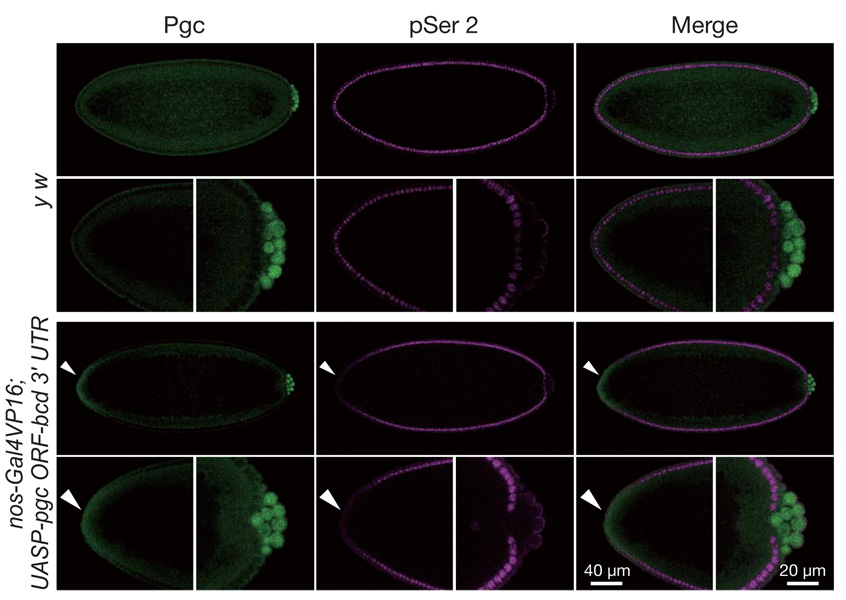
When Pgc was misexpressed at the anterior of the blastoderm embryos, CTD Ser 2 phosphorylation and zygotic expression of a hunchback (hb)-lacZ reporter was repressed in somatic cells where ectopic Pgc was detected (Fig. 2 and Supplementary Fig. 4). Furthermore, Pgc expression in Drosophila S2 cells repressed CTD Ser 2 phosphorylation (Supplementary Fig. 5). These results demonstrate that Pgc can prevent CTD Ser 2 phosphorylation even in somatic cells, and further suggest that the target of Pgc action is a general component of the RNAPII-dependent transcription machinery.
Figure 2. Pgc is sufficient to repress CTD Ser 2 phosphorylation in somatic cells.
Embryos were immunostained for Pgc (green) and pSer 2 (magenta). High magnifications of anterior and posterior parts of the embryos are also shown. To misexpress Pgc at the anterior somatic cell region, a hybrid gene, in which the pgc ORF was fused with the bicoid (bcd) anterior localization signal, was expressed in oogenesis. In embryos expressing pgc-bcd 3′ UTR mRNA, CTD Ser 2 phosphorylation was repressed in regions expressing ectopic Pgc (arrowheads). Many embryos (50–80%) expressing the pgc ORF-bcd 3′ UTR transgene died with variable defects, but we never observed a bicaudal phenotype, suggesting that anterior misexpression of Pgc is incapable of recruiting germ plasm components required for posterior development.
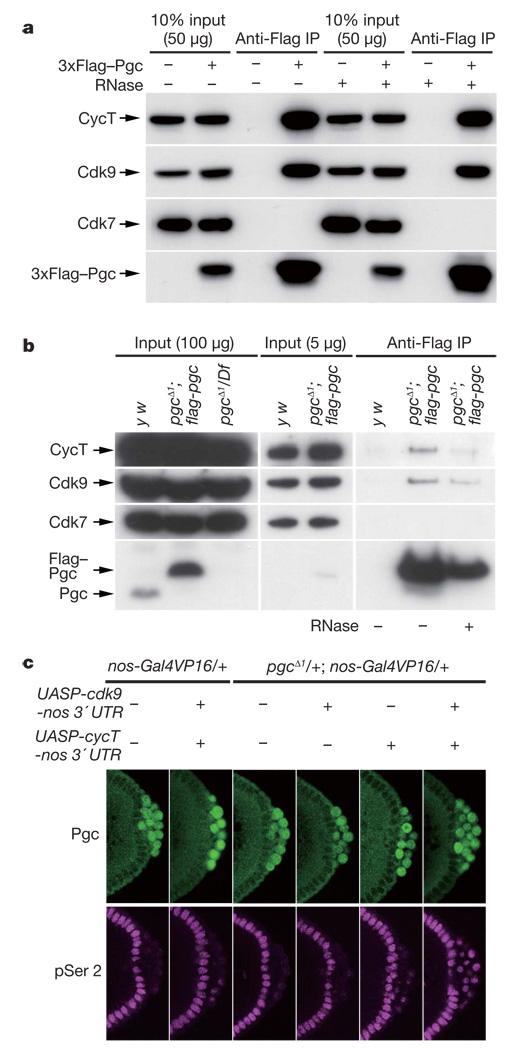
Positive transcription elongation factor-b (P-TEFb) is responsible for CTD Ser 2 phosphorylation in vivo3,11–13, making it a candidate Pgc target. Drosophila P-TEFb consists of a kinase subunit, Cdk9, and a regulatory subunit, Cyclin T (CycT)13–15. Supporting a link between P-TEFb and Pgc, both Cdk9 and CycT were co-immunoprecipitated with 3×Flag–Pgc from S2 cell lysates, but another CTD kinase, Cdk7 (a component of TFIIH that preferentially phosphorylates CTD Ser 5; ref. 3), was not (Fig. 3a). A reciprocal immunoprecipitation also confirmed a specific interaction between Pgc and P-TEFb (Supplementary Fig. 6). To examine their interaction in pole cells, we used a Flag-tagged pgc transgene that fully rescued the pgcΔ1 mutant. Lysates from pools of embryos enriched for stages 4–5 were immunoprecipitated with anti-Flag antibody. Cdk9 and CycT were specifically co-immunoprecipitated with Flag–Pgc from the lysates of the transgenic embryos but not from control lysates (Fig. 3b). RNase treatment of lysates did not affect the Pgc–P-TEFb interaction (Fig. 3a, b). These results demonstrate that Pgc forms a complex with both CycT and Cdk9.
Figure 3. Pgc interacts, physically and genetically, with P-TEFb.
a, Cdk9 and CycT, but not Cdk7, were co-immunoprecipitated with Pgc from the lysates of S2 cells expressing 3×Flag–Pgc. b, Lysates of y w or pgc− embryos expressing the flag–pgc transgene were immunoprecipitated with anti-Flag antibody, and bound proteins were analysed by western blotting. c, P-TEFb was overexpressed in pole cells by expressing cycT-nos 3′ UTR and cdk9-nos 3′ UTR transgenes in oogenesis. Overexpression of P-TEFb in pole cells caused precocious CTD pSer 2 phosphorylation (magenta), even in the presence of Pgc expression (green). This precocious CTD pSer 2 phosphorylation was strongly induced when P-TEFb was overexpressed in the pgcΔ1 heterozygous background.
Pull-down assay showed that maltose-binding protein (MBP)–Pgc fusion protein, but not control MBP, interacted with the P-TEFb complex in vitro (Supplementary Fig. 7a). To examine which subunit of P-TEFb interacts with Pgc, in-vitro-synthesized Cdk9 or CycT was individually tested for pull-down assay. Cdk9, but not CycT, was specifically pulled down by MBP–Pgc (Supplementary Fig. 7b, c), indicating that Cdk9 alone is sufficient for the direct interaction with Pgc in vitro. Taken together, these results suggest that Pgc can form a ternary complex with Cdk9 and CycT.
To investigate the significance of the interaction between Pgc and P-TEFb in vivo, we overexpressed P-TEFb in pole cells. Although overexpression of P-TEFb failed to induce CTD Ser 2 phosphorylation in pole cells at stage 4, when Pgc expression is highest (Fig. 1b), it caused precocious CTD Ser 2 phosphorylation in a few pole cells at stage 5 (Fig. 3c). Furthermore, precocious CTD Ser 2 phosphorylation was strongly induced when both transgenes were expressed in the pgcΔ1 heterozygous background (Fig. 3c). Many pole cells in these embryos degenerated during embryogenesis, and the gonads often included few or no pole cells (Supplementary Table 1). The over-expression of P-TEFb in pole cells promoted the ectopic expression of somatic genes that are misexpressed in pgc− pole cells (Supplementary Fig. 8)9,10. By contrast, no ectopic expression of even skipped (eve) or fushi tarazu (ftz), which are not misexpressed in pgc− pole cells9, was detected in P-TEFb-overexpressing pole cells (data not shown). Thus, overexpression of P-TEFb in pole cells mimics pgc mutant phenotypes in a pgc dosage-dependent manner. These data demonstrate that Pgc represses CTD Ser 2 phosphorylation in pole cells by interfering with P-TEFb action.
Pgc alone was unlikely to inhibit the kinase activity of P-TEFb, as MBP–Pgc, which interacted with P-TEFb in vitro (Supplementary Fig. 7), did not affect CTD phosphorylation by P-TEFb in an in vitro kinase assay (Supplementary Fig. 9). The recruitment of P-TEFb to paused promoter regions is crucial for the productive elongation of most nascent transcripts13. We therefore examined the effects of Pgc expression in salivary glands on the distribution of P-TEFb. In control polytene chromosomes, Cdk9 and CycT co-localized at numerous sites, which were also positive for pSer 5, a marker for active transcription3 (Fig. 4a), indicating that P-TEFb is recruited to active promoter regions as reported16. By contrast, when Pgc was expressed in salivary glands, the P-TEFb signals on the polytene chromosomes were severely decreased; instead, P-TEFb was mostly detected on the cell debris (arrowheads in Fig. 4a). Western blot analysis showed that Pgc expression caused an ~40% reduction in the level of pSer 2, whereas the levels of Cdk9 and CycT were virtually unchanged (Fig. 4b). Pgc expression in salivary glands also caused a reduction in the levels of pSer 5 (to ~65%), similar to the effects of RNA-interference-mediated cdk9 knockdown in Drosophila larvae17. These results suggest that Pgc sequesters P-TEFb and prevents its recruitment to active promoters.
Figure 4. Pgc prevents P-TEFb recruitment.
a, Salivary gland polytene chromosome squashes prepared from third-instar larvae carrying either the sgs3-Gal4 driver alone or sgs3-Gal4 plus two copies of UAST-pgc ORF transgenes immunostained for pSer 5, CycT and Cdk9, and counter-stained with DAPI. Arrowheads indicate P-TEFb signals on cell debris in the Pgcexpressing salivary gland squash. b, Western blot analysis of salivary gland lysates from sgs3-Gal4 or sgs3-Gal4; 2×UAST-Pgc third-instar larvae. c, Real-time PCR analyses of ChIP experiments on the Hsp70 and Hsp27 gene regions in heatshocked (HS) or non-heat-shocked (NHS) S2 cell transfectants. The proportion of Pgc-expressing S2 cells after CuSO4 induction was about 70%. Each result shows an average of at least three independent experiments with the standard error of the mean.
We next examined the effects of Pgc expression on the recruitment of P-TEFb to transcription sites by chromatin immunoprecipitation (ChIP) assay. Consistent with previous reports12,18, heat-shock treatment promoted the rapid recruitment of CycT and Cdk9 to the Hsp70 and Hsp27 gene regions in control S2 cells (Fig. 4c). By contrast, Pgc expression caused a significant reduction in the levels of both CycT and Cdk9 on these genes after heat shock (Fig. 4c; P<0.01). RNAPII distribution on these gene regions was not significantly affected by Pgc expression (Fig. 4c; P>0.4), similar to the effects of P-TEFb inactivation in Drosophila cells12. These results support the idea that Pgc represses CTD Ser 2 phosphorylation by inhibiting the recruitment of P-TEFb to transcription sites.
P-TEFb is a key regulator of RNAPII-dependent transcription of most cellular genes3,13–15 as well as HIV-1 transcription and replication 13,19,20. In mammals, P-TEFb is inhibited by the 7SK RNA–HEXIM1 complex, which suppresses the Cdk9 kinase activity and prevents P-TEFb from binding to transcription templates12,21–24. Pgc acts through an analogous—but not identical—mechanism, as Pgc alone failed to inhibit the kinase activity of P-TEFb in vitro (Supplementary Fig. 9). Furthermore, 7SK RNA is unique to vertebrates13, and the Pgc–P-TEFb interaction is RNase insensitive (Fig. 3a, b). We speculate that additional Pgc-like P-TEFb inhibitors may exist in somatic cells. In C. elegans germline blastomeres, PIE-1 is proposed to prevent CTD Ser 2 phosphorylation through interaction with CycT6,7. Thus, P-TEFb seems to be a common regulatory target for the repression of mRNA transcription during germ cell specification in both flies and nematodes. Despite their analogous functions, Pgc and PIE-1 are unrelated in sequence and therefore must have arisen independently in evolution. This is consistent with the hypothesis that germ cell specification by the maternally inherited germ plasm evolved independently among diverse animal groups25. The inhibition of somatic transcriptional programmes is also crucial for the establishment of mouse germ cells, which are specified through an epigenetic mechanism1,26–28. Whether P-TEFb is targeted in mammalian germ cell progenitors as well will be an interesting question in the future.
METHODS SUMMARY
Fly strains and transgenic constructs
The fly strains used in this study are described in FlyBase, unless otherwise noted. rF139 is a homozygous-viable P[ry+PZ] insertion that is located ~13 kb distal to the 5′ side of the pgc locus and was obtained from the Berkeley Drosophila Genome Project (BDGP). Imprecise excision of the rF139 transposon generated a line (pgc#1) in which an~15-kb genomic region containing the entire pgc locus was deleted. The pgc#1 chromosome also has the deletion of an essential gene, gp150, which was subsequently rescued by introducing an ~17.5-kb genomic fragment containing the gp150 locus, thus generating pgcΔ1, a chromosomal null for pgc and T3dh (Supplementary Fig. 1). pgc cDNA was amplified by PCR and cloned into pUASP to generate UASP-pgc cDNA. UASP-pgc ORFfs contains a 2-base deletion at the fifth codon. The full-length nos and gcl 3′ UTRs and the EcoRV/StuI fragment of the bcd 3′ UTR region29 were used to create chimaeric gene constructs.
Immunostaining and in situ hybridization
Immunostaining and in situ hybridization of embryos were performed using standard procedures. To detect Pgc and pSer 2 signals, the vitelline membranes of fixed embryos were hand peeled, as the reactivity of these antibodies was found to be sensitive to methanol, which is generally used for devitellinization. Salivary glands from climbing third-instar larvae cultured at 27 °C were fixed in 2% paraformaldehyde (PFA) in PBS containing 0.1% Triton-X100 for 30 s followed by 45% acetic acid/1.85% PFA for 5 min before being squashed onto coated microscope slides. Polytene chromosomes were pre-treated with PBS containing 0.1% SDS for 10 min before antibody staining. The specificities of newly raised anti-Cdk9 and anti-CycT antibodies were examined by western blot analyses of lysates from S2 cells depleted of Cdk9 or CycT by RNA interference (Supplementary Fig. 10).
METHODS
Transgenic constructs
The Flag–Pgc genomic rescue plasmid was constructed by inserting the Flag sequence immediately downstream of the initiation codon in a 3.6-kb HincII fragment containing the entire pgc gene region. All the constructs were verified by sequencing the PCR-amplified DNA regions. P-element-mediated germline transformation was performed using a standard method with y w flies as recipients30. nos-Gal4VP16 and sgs3-Gal4 were used as drivers to express UAS constructs during oogenesis and in salivary glands, respectively.
Cell culture
S2 cells were grown at 25 °C in Schneider medium (Invitrogen) supplemented with 2 mM glutamine and 10% fetal calf serum. Cell transfection was performed using Effectene (Qiagen) or siLentFect (BioRad) reagents. To induce protein expression from pMT-based constructs, CuSO4 was added to the medium at a final concentration of 0.5 mM. To establish stable lines, the cells were co-transfected with pCoBlast (Invitrogen) and cultured in the presence of blasticidin.
Antibodies
The following antibodies were used: rabbit and rat anti-Pgc (this study), rabbit and rat anti-Vas (laboratory stocks), rabbit anti-β-galactosidase (Cappel), mouse anti-RNAPII H5, H14 (Covance) and CTD4H8 (Upstate), rabbit anti-CTD pSer 2 (Abcam), mouse anti-α-tubulin (Sigma), mouse anti- Flag M2 (Sigma), mouse anti-Cdk7 (ref. 31), rat anti-Cdk9 (ref. 12), rabbit anti-Cdk9 (this study), and rat and rabbit anti-CycT (this study). Signals were detected using Alexa Fluor-conjugated secondary antibodies (Invitrogen), or biotinylated anti-rabbit IgGs and the ABC kit (Vector Lab). Images were taken with a laser confocal microscope (Leica TCS SP2 AOBS) or a compound microscope with Nomarski optics (Leica DMR), and processed using Adobe Photoshop. For western analyses, signals were detected by the ECL (Amersham) or Super Signal West Dura (Pierce) system. The quantification of western signals (Fig. 4b) was performed using a luminescent image analyser, LAS3000-UVmini (Fujifilm). Quantified data were normalized to the α-tubulin levels.
Antibody generation
The Pgc-coding region was cloned into pMAL (NEB) and pGEX (Amersham) to produce MBP and glutathione S-transferase (GST) fusion proteins, respectively. MBP–Pgc was used as the antigen to raise antibodies, and GST–Pgc protein was used for affinity purification. The full-length Drosophila Cdk9-coding region and a region of CycT (amino acids 550–1097) were expressed as His-tagged proteins, and used as antigens to raise antibodies. A region of Cdk9 (amino acids 1–50) was expressed as a GST fusion protein, and used for affinity purification.
Immunoprecipitation and pull-down assay
Cells and embryos were homogenized in TNG150 (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 1% Triton-X100) containing Complete EDTA-free (Roche). The supernatants were pre-cleared with Sepharose CL-6B (Sigma) and Protein G Sepharose beads (GE Healthcare). The cleared supernatants were mixed with anti-Flag beads and incubated at 4 °C overnight in the presence or absence of RNases (10 µg RNase A (Sigma) and 100U RNase T1 (Roche) per mg of protein in the lysates). The beads were washed with TNG150. Bound proteins were eluted with 1 mg ml−1 3×Flag peptide (Sigma) in TNG150, and analysed by western blotting.
For the pull-down experiments, 1 mg of MBP–Pgc or control MBP protein was crosslinked to ~100 µl of NHS Sepharose beads (GE Healthcare). The P-TEFb complex was purified from S2 cells expressing Flag–CycT. Flag–Cdk9 was synthesized using the TNT T7 Quick Kit (Promega). Flag–CycT was synthesized using the PROTEIOS system (Toyobo). Samples were mixed with ~20 µl of bait-coupled beads in TNG150 at 4 °C overnight. The beads were washed with TNG150 and bound proteins were analysed by western blotting with the anti-Flag antibody.
CTD kinase assay
The CTD-coding region of Drosophila RNAPII was cloned into pGEX to generate the GST–CTD construct. For the experiment shown in Supplementary Fig. 9a, cleared lysates from S2 cells expressing Flag–CycT were immunoprecipitated with anti-Flag beads. The beads were washed and equilibrated with the kinase buffer (20 mM HEPES-KOH (pH 7.6), 55 mM KCl and 5 mM MgCl2). The P-TEFb-bound beads were then divided into 13 tubes and incubated with or without MBP–Pgc or MBP in 10-µl volumes for 15 min at 25 °C. Subsequently, the kinase reaction was performed by adding 10 µl kinase buffer containing 1 µg GST–CTD, 20 nM ATP and 10 µCi [γ-32P]ATP (3,000 Ci mmol−1); in some tubes 50 nM DRB was also included. After a 20-min incubation at 25 °C, the supernatants were filtrated and analysed by SDS–PAGE followed by autoradiography.
For the experiment shown in Supplementary Fig. 9b, different amounts of lysates from mock-transfected or Flag–CycT-expressing S2 cells were immunoprecipitated with the same amount of anti-Flag beads. The beads were incubated with or without 1 µg of MBP–Pgc fusion proteins for 20 min at 25 °C before the kinase assay was performed.
ChIP and real-time PCR
A control or pMT-Pgc plasmid-transfected cells were cultured in 10-cm culture dishes at a density of 1×107 cells ml−1 in the presence of 0.5 mM CuSO4 for 4 h. Cells (1×108) were subsequently subjected to heat shock at 37 °C for 5 min. Heat shocked (HS) and non-heat shocked (NHS) cells were fixed with 1% paraformaldehyde for 10 min at room temperature. After quenching crosslinking by 200 mM glycine, the cells were washed in ice-cold PBS containing 0.5 mM EDTA, incubated in NE buffer32 on ice for 30 min, and resuspended in RIPA150 (ref. 33) containing protease inhibitors.
The fixed chromatin was sheared into lengths of <500 bp by sonication. The chromatin lysates were pre-cleaned with Sepharose CL-6B and Protein G beads. Cleared lysates from the equivalent of 1 × 107 cells were mixed with antibodies and incubated at 4 °C overnight. Subsequently, the lysates were incubated with ~10 µl Protein G beads at 4 °C for 5 h. The beads were washed and reverse crosslinked as described33. The samples were then treated with RNaseA followed by proteinase K. The supernatants were extracted and ethanol precipitated in the presence of glycogen.
The relative amount of immunoprecipitated DNA was quantified using realtime PCR (ABI Prism 7000) with the qPCR Master Mix Plus for SYBR Green I kit (Eurogentec). Primer sequences used were as described18. Quantifications were performed on samples from at least three independent immunoprecipitations from three separate chromatin preparations. Probability was calculated by the Student’s t-test.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank K. Zinn for the lambda phage genomic clone containing the gp150 region, P. Rørth and E. R. Gavis for plasmids, J. T. Lis, D. H. Price and S. Larochelle for antibodies, the Berkeley Drosophila Genome Project and the Bloomington Drosophila stock center for fly stocks, and J. Nakayama, M. Ukai-Tadenuma and H. R. Ueda for technical advice on ChIP analysis. This work was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, and Japan Society of the Promotion of Science, Japan and the RIKEN President Discretionary Fund (to A.N.), and by grants from CIHR and NICHD (to P.L.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- 3.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nature Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 4.Seydoux G, et al. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- 5.Mello CC, et al. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- 6.Batchelder C, et al. Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1. Genes Dev. 1999;13:202–212. doi: 10.1101/gad.13.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F, Barboric M, Blackwell TK, Peterlin BM. A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev. 2003;17:748–758. doi: 10.1101/gad.1068203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- 9.Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr. Biol. 2004;14:159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 10.Deshpande G, Calhoun G, Schedl P. Overlapping mechanisms function to establish transcriptional quiescence in the embryonic Drosophila germline. Development. 2004;131:1247–1257. doi: 10.1242/dev.01004. [DOI] [PubMed] [Google Scholar]
- 11.Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16:2135–2146. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni Z, Schwartz BE, Werner J, Suarez J-R, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 13.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, et al. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng J, Marshall NF, Price DH. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 1998;273:13855–13860. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- 16.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 17.Eissenberg JC, Shilatifard A, Dorokhov N, Michener DE. Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment. Mol. Genet. Genomics. 2007;277:101–114. doi: 10.1007/s00438-006-0164-2. [DOI] [PubMed] [Google Scholar]
- 18.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9- associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 20.Barboric M, Peterlin BM. A new paradigm in eukaryotic biology: HIV Tat and the control of transcriptional elongation. PLoS Biol. 2005;3:e76. doi: 10.1371/journal.pbio.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 23.Michels AA, et al. MAQ1 and 7SK RNA interact with CDK9/Cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yik JHN, et al. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinate action of HEXIM1 and 7SK RNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 25.Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- 26.Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 27.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 28.Seki Y, et al. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- 29.Macdonald PM, Struhl G. Cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- 30.Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 31.Larochelle S, Pandur J, Fisher RP, Salz HK, Suter B. Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 1998;12:370–381. doi: 10.1101/gad.12.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agata Y, et al. Histone acetylation determines the developmentally regulated accessibility for T cell receptor γ gene recombination. J. Exp. Med. 2001;193:873–880. doi: 10.1084/jem.193.7.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.