SUMMARY
Recent evidence indicates that mouse and human embryonic stem (ES) cells are fixed at different developmental stages, with the former positioned earlier. We show that a narrow concentration of the naturally occurring short chain fatty acid, sodium butyrate, supports the extensive self-renewal of mouse and human ES cells, while promoting their convergence toward an intermediate stem cell state. In response to butyrate human ES cells regress to an earlier developmental stage characterized by a gene expression profile resembling that of mouse ES cells, preventing precocious Xist expression, while retaining the ability to form complex teratomas in vivo. Other histone deacetylase inhibitors (HDACi) also support human ES cell self-renewal. Our results indicate that HDACi can promote ES cell self-renewal across species, and demonstrate that ES cells can toggle between alternative states in response to environmental factors.
Introduction
Embryonic stem (ES) cells might be regarded as a tissue culture artifact (Smith, 2001), plucked from the pre-implantation blastocyst and supported using culture conditions bearing little resemblance to conditions in vivo. Even leukemia inhibitory factor (LIF), the quintessential extracellular regulator of mouse ES cell self-renewal, is not required for development other than for implantation (Stewart et al., 1992; Ware et al., 1995). Nowhere is the lack of a physiological context for ES cells more pressing than in attempts to define their cellular identity, also known as the “stem cell state” (Buszczak and Spradling 2006). One definition of the ES cell state is the epigenetic state that endows ES cells with the unique option to self-renew or to differentiate into any cell type in the body.
It has been proposed that an epigenetic event may be rate limiting in the derivation of new ES cell lines (Smith, 2001; Thomson et al., 1998), and may further operate in selecting for ES cells that adapt to standard culture conditions. An epigenomic bottleneck might similarly explain the inefficiency inherent in generating new ES-like cells - induced pluripotent stem (iPS) cells (Takahashi and Yamanaka, 2006; Nakagawa et al., 2007, Park et al., 2007, Takahashi et al., 2007; Yu et al., 2007) and epiblast stem cells (EpiSC) (Brons et al., 2007; Tesar et al., 2007). Stated differently, our current understanding of the ES cell state may be influenced by the manner in which ES cells are derived and maintained. Consistent with this interpretation was the recent demonstration that the efficiency of iPS cell formation is enhanced upon addition of valproic acid, an inhibitor of histone deacetylases, to the culture medium (Huangfu et al., 2008).
Despite their uncertain in vivo corollary, studies using in vitro cultured ES cells have enhanced our understanding of how stem cells work (Bernstein et al., 2006; Ivanova et al., 2006; Chambers et al., 2007, Ying et al., 2008). To self-renew, ES cells must stably propagate their epigenetic patterns through cell division. Fundamental differences exist in the mechanisms by which mouse ES cells (mESC) and human ESC (hESC) self-renew. mESC respond to LIF-triggered activation of STAT3, whereas hESC do not (Daheron et al., 2004). hESC respond to super-physiological levels of bFGF (Levenstein et al., 2006) and to Activin A (Vallier et al., 2005; Xiao et al., 2006) to maintain pluripotency. The differences in culture conditions for mESC and hESC might be explained by the recent finding that mouse and human ES cells are not developmental equals, with the former representing an earlier developmental stage (Brons et al., 2007; Tesar et al., 2007).
ES cells and cancer cells employ overlapping signaling networks (Dreesen and Brivanlou, 2007, Wong et al., 2008, Ben-Porath et al., 2008), raising the possibility that understanding self-renewal in ES cells might bring new insights to cancer therapy. Recently, histone deacetylase (HDAC) inhibitors (HDACi) have emerged as an important new class of anti-cancer drug (Xu et al., 2007). Here we show, for the first time, that both naturally occurring and synthetic HDACi promote ES cell self-renewal using defined, serum-free culture conditions. HDACi induce a shift in the gene expression profiles of human and mouse ES cells toward a state intermediate between ES cell and EpiSC. Our results define an alternative ES cell state, and point to the existence of an evolutionarily conserved self-renewal program.
Results
A narrow concentration of sodium butyrate maintains hESCs in an undifferentiated state
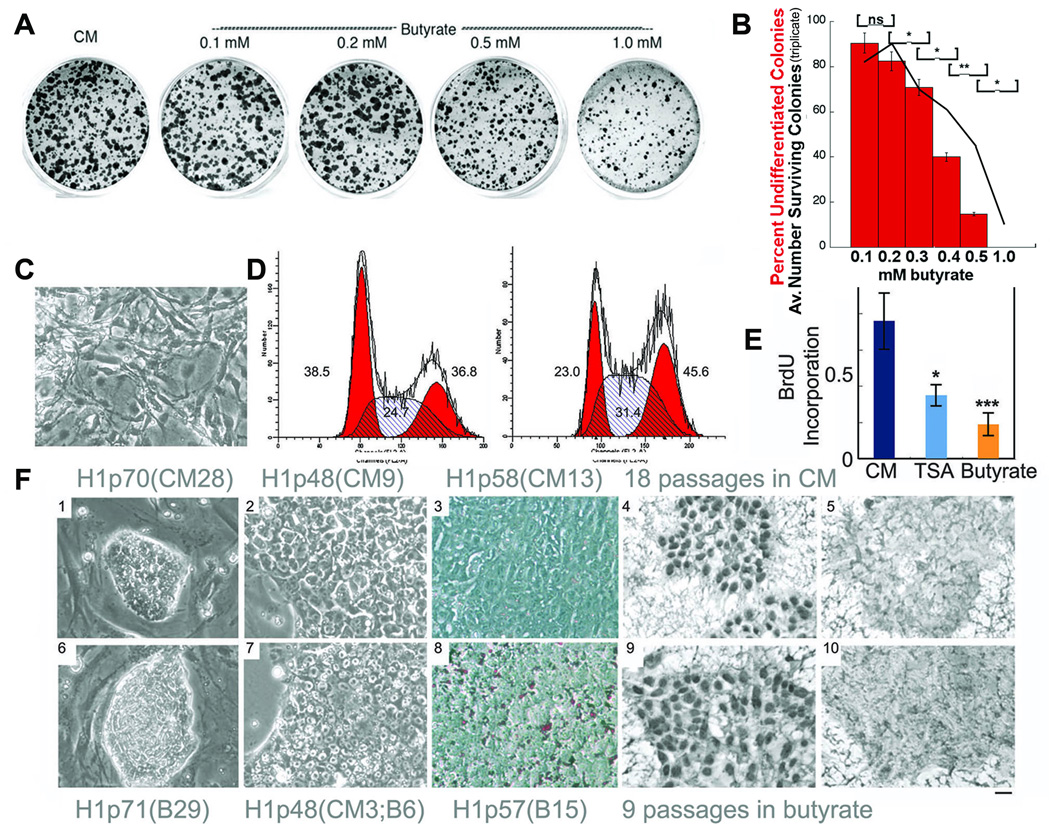
H1 cells cultured on Matrigel coated dishes were switched from standard culture conditions (conditioned medium that includes FGF2; CM) to feeder-free culture in hESC medium (hESM) without feeder conditioning with a range of sodium butyrate concentrations. hESM contains a proprietary serum replacer (KSR-Invitrogen), but lacks FGF2 or other growth factors. We found that the butyrate concentrations used in most publications (>1.0 mM; Boffa et al., 1978) were toxic in hESC cultures, and did not allow for sustained growth (Fig. 1 A,B). However lower butyrate concentrations (0.2 – 0.3 mM) induced a transient wave of differentiation (Fig. 1C), followed by the outgrowth of approximately 50% of the original cell population as undifferentiated hESC that could be maintained in culture indefinitely (at least 33 passages over 7 months). ES cells are thought to lack a G1 checkpoint (Savatier et al., 1994; Becker et al., 2007). H1 cells cultured in hESM with 0.2 mM butyrate accumulated cells in S and G2, consistent with a relative G2/M block (Fig. 1D). A similar accumulation of cells in G2/M was recently reported for γ-aminobutyric acid (Andäng et al., 2008). Butyrate-converted hESC divided at about one-quarter the rate of cells cultured according to standard conditions as measured by BrdU incorporation (Fig. 1E). Notably, 0.2 mM butyrate was not associated with a significant induction of p21 by real time quantitative PCR (qPCR; data not shown). Once acclimated, butyrate treated cells formed tightly clustered colonies that adhered firmly to Matrigel and lacked the small percentage of differentiated cells that normally accompany H1 cells cultured in CM (Fig. 1F). Butyrate also induced the appearance of lipid droplets as determined by Oil Red O staining (Fig. 1F panel 8).
Figure 1.
Butyrate supports the self-renewal of H1 cells. A. H1 cells acclimated to growth in CM (which contains 2ng/ml FGF2-see methods) were cultured on Matrigel-coated 35 mm dishes for 4 days in conditioned medium (CM) or in hESM (which lacks conditioning or FGF2) at the indicated concentrations of butyrate. Undifferentiated colonies were scored based on alkaline phosphatase staining. B. Quantitative results from a repeat of the experiment shown in panel a), performed in triplicate. Black line indicates numbers of colonies per dish. * = p<0.05, ** = p<0.01. Red bars indicate percentages of alkaline phosphatase-positive colonies. Error bars denote standard errors of the mean. C. Appearance of differentiated cells that appear transiently after switching from CM to butyrate for two passages. D. Cell cycle profiles of H1 cells cultured in CM (left) versus butyrate (right). Numbers indicate percentages of cells in G1/S/G2. Note increases in percentages of cells in S and G2 in the butyrate cultures. These findings are representative of 2 independent experiments. E. Bromodeoxyuridine (BrdU) incorporation in H1 cells cultured in CM, trichostatin A (TSA) (10 nM) (*P<0.05; CM vs. TSA) and butyrate (0.2mM) (***P<0.001; CM vs. butyrate) F. Morphology of H1 cells cultured in CM (1–5) or butyrate (6–10). 1,6: on feeders; 2–5 and 7–10: on MatrigelTM; 3, 8: Oil red O stained; 4,9: Pou5F1 (Oct4) stained; 5,10: secondary control antibody staining companions to Pou5f1 staining. Note that in F1, H1p70(CM28) is 70 passages total, the last 28 of which were without feeder in CM, in F6, H1p71(B29) is 71 passages total, the last 29 of which were in butyrate and for F7, H1p48(CM3;B6) is 48 passages total, and within the last 9 passages, the first 3 were without feeder in CM and the final 6 passages were in butyrate. This format is followed for all subsequent figures. The size bar indicates 38 µm in C and F1 & 6 and 15 µm in F2–5 &7–10.
We confirmed the undifferentiated features of butyrate-converted H1 cells by assessing a series of ES cell specific markers and by measuring telomerase activity (Fig. 1F panel 9 and Supplemental Fig. 1). In total, six different hESC lines [H1 (National Institutes of Health designation WA01), H9 (WA09), H13 (WA13), BG02, BG03, hSF6 (UC06)] and a rhesus ES cell line (rh366.4) were converted to butyrate dependent, MEF/CM-independent culture conditions in hESM supplemented with 0.2–0.3 mM sodium butyrate but lacking FGF2 or other growth factors (Fig. 2A, Supplemental Table 1). We observed differences among hESC lines in their responses to butyrate. BG03 cells were readily converted to butyrate, with little evidence of differentiation. H1 cells were more fastidious, requiring dense plating prior to butyrate exposure and 1:1 passaging (for at least 2 passages) after butyrate addition. The concomitant presence of feeder cells eased the conversion of some (H9) but not all (H1, BG02, BG03) hESC lines. The emergence of karyotypic abnormalities, common in hESC cultures (Denning et al., 2006, Ware et al., 2006, Baker et al., 2007), occurred in butyrate cultures at rates similar to CM (Supplemental Table 1 and Supplemental Fig. 2). Although trisomy 17 is a common abnormality in hESC cultures (Baker et al., 2007), we have not observed trisomy 17 in butyrate treated hESCs.
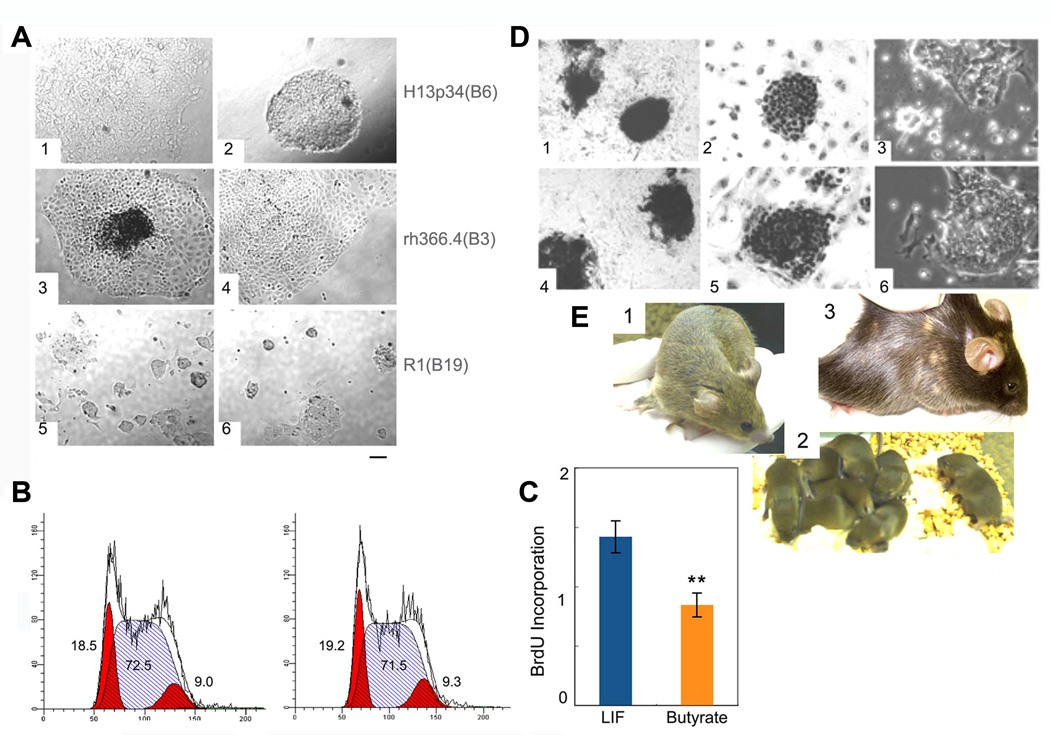
Figure 2.
Butyrate supports ES cell self-renewal across species. A. H13 cells (1, in CM, 2 in butyrate), rh366.4 rhesus ES cells (3 in CM, 4 in butyrate), R1 mouse ES cells (5 in LIF, 6 in butyrate). B. Cell cycle profile of R1 cells cultured in LIF (left) or butyrate (right). C. BrdU uptake in R1 cells cultured in LIF versus butyrate. ** denotes P<0.01. D. Alkaline phosphatase staining (1, 4), Pou5f1 (Oct4) staining (2, 5), and phase contrast microscopy (3, 6) of R1 cells cultured in LIF (1 – 3) or 0.2 mM butyrate (4 −6). E. Chimeric mouse generated from R1 ES cells cultured for 3 passages in butyrate plus LIF on feeders (panel 1), and its progeny (panel 2), indicating 100% germ line transmission (note all are brown). Chimeric mouse from a mEpiSC line #5, a gift of Paul Tesar and Ron McKay (Tesar et al., 2007) cultured for 18 passages on feeders with the addition of butyrate for the last 8 passages (panel 3). The size bar indicates 38 µm for panels A and D.
Notwithstanding the well established differences between the self-renewal programs of mouse and human ES cells, butyrate also promoted the extensive self-renewal of a mESC line (R1) (Fig. 2 A,D). mESCs cultured in butyrate-containing medium without LIF (see Methods) adopted a very similar appearance to mESCs cultured in LIF. In both conditions, scattered differentiated cells at the colony periphery surrounded undifferentiated cells located centrally. In mESC, butyrate slowed the rate of cell division without perturbing the cell cycle (Fig. 2 B,C). Based on the resulting hypothesis that butyrate induced ES cell self-renewal arises from a common, evolutionarily conserved mechanism, we predicted that butyrate induced self-renewal of mESC could occur in the absence of LIF signaling. Confirming this prediction we showed that mESC lacking either the LIF receptor specific subunit or its shared gp130 partner could be converted to butyrate culture conditions (Supplemental Fig. 3).
Differentiation potential of butyrate treated ES cells
Teratomas generated by butyrate-converted hESC were complex, containing prominent carrot-shaped cells with rod-shaped cytoplasmic granules consistent with melanin containing pigmented ectoderm (both in H1 and BG02 cells) while the CM exposed counterparts rarely contained identifiable melanin (Supplemental Fig. 4). To more rigorously evaluate the differentiation potential of ES cells exposed to butyrate we used mESC. We added butyrate to mESC (R1) cultured on feeders supplemented with LIF for three passages, and these cells contributed to multiple tissues in two high level chimeras, including the germline (Fig. 2E panels 1&2). In addition, two different gene-targeted mESC clones that had not previously contributed to coat color chimerism using standard culture conditions were able to generate high-level chimeras when butyrate was added to MEF-containing cultures (Supplemental Table 2). These clones did not contribute to the germline, likely due to preexisting abnormalities. Additionally, we were able to derive a new C57BL/6 mESC line in the presence of butyrate on MEFs that contributed extensively to coat color chimerism (Supplemental Table 2). While mESCs cultured in butyrate in MEF-free conditions did not generate chimeras (data not shown), our results demonstrate that adding butyrate to MEF-containing cultures can maintain and may even augment the ability of mESCs to contribute to various tissues in vivo.
Butyrate-induced transcriptional response in hESCs
We used whole genome arrays to compare the transcriptomes of feeder-free H1 cells cultured according to three conditions (each in triplicate): 1) Standard culture conditions [CM with 2 ng/ml bFGF on Matrigel; H1p48(CM9)] (Group A); 2) Cells from the same pool as Group A converted to butyrate for 6 passages [hESM with 0.2 mM butyrate on Matrigel; H1p48(CM3;B6)] (Group B) and 3) Cells from the same pool as Group B cultured in butyrate for 4 passages, then reverted back to standard culture conditions for 3 passages [H1p49(CM3;B4;CM3)] (Group C). Groups A and B and groups B and C were directly compared on Agilent arrays. A supervised cluster analysis readily distinguished the 3 groups (Supplemental Fig. 5). Butyrate significantly regulated 479 genes; 250 were up-regulated and 229 down-regulated (https://depts.washington.edu/iscrm/GS_data/gsdata.html). The large number of down-regulated transcripts is consistent with a recognized but poorly understood role for butyrate in gene repression (Rada-Iglesias et al., 2007). The top 15 upregulated and downregulated genes are shown in Supplemental Table 3. qPCR confirmed the differential regulation of all 14 representative genes tested (Supplemental Fig. 6A). Butyrate strongly induced several embryonic and germ cell associated transcripts including Dppa5 (Esg1), Piwil2, Bnc1, Lrrc8e, Mbd3, and Ecat8 while downregulating Tcf3 and Gata6 (Supplemental Table 4). Dppa5, Piwil2, Ecat8, Ddx25 and Ddx43 all encode RNA binding proteins while Mbd3 is a part of the Nucleosome Remodeling and Histone Deacetylation (NuRD) co-repressor complex associated with cell fate decisions and pluripotency (Kaji et al., 2007). Dppa5 and Ddx43/HAGE, the first and second most strongly induced genes, are spaced 60 kilobases apart on chromosome 6, and Ecat1 is positioned between them. While Ecat1 was not represented on the microarray we found that it is also induced by butyrate (Supplemental Fig. 6 B,C).
Several members of the canonical Wnt signaling pathway were down-regulated by butyrate, including Wnt3, Tcf3, Frzb, and Sfrp2, as were β-catenin targets Sp5 (Weidinger et al., 2000), Gad1 (Li et al., 2004), Fst (Yao et al., 2004), Lefty1 (Tabibzadeh and Hemmati-Brivanlou, 2006), Pitx2 (Zirn et al., 2006), and Id2 (Willert et al., 2002). An increase in Inhba (INHBA homodimerizes to form ACTIVIN A) and reduced expression of Lefty1 and Fst indicate that the TGFβ pathway may be recruited to maintain self-renewal of butyrate treated hESC (Vallier et al., 2005, Xiao et al., 2006, Eiselleova et al., 2008, Xu et al., 2008). Butyrate treatment also resulted in down-regulation of Tcf3, a repressor of Nanog in mESCs (Pereira et al., 2006; Yi et al., 2008). While butyrate did not induce a significant change in Nanog levels, we found Nanog to be consistently (albeit subtly) elevated in hESCs cultured in butyrate relative to CM (Supplemental Fig. 1B). NANOG stabilizes ES cells in culture, underpinning the epigenetic erasure unique to pluripotent and germ cells (Chambers et al. 2007). A subtle rise in Nanog may reflect the absence of differentiating cells in butyrate-acclimated cultures.
Withdrawal from feeder cells was not necessary to elicit the butyrate transcriptional program, since Dppa5, Ecat1 and Piwil2 were all induced in H13 cells, three passages after adding butyrate to feeder-containing cultures (Supplemental Fig. 6B). Approximately 85% of genes that were differentially expressed in response to butyrate (Group B versus Group A) (Fig. 3A) returned promptly to near baseline levels after reverting back to standard culture conditions (Group C versus Group A) (Fig. 3B). Conversely, some butyrate-responsive genes remained persistently altered following 3 passages in CM (Fig. 3B and indicated in Supplemental Table 4). Persistently induced genes included Dppa5, Ddx43/HAGE and Ecat1 (Supplemental Fig. 6C). The basis for this persistent effect on the expression of some genes long after butyrate withdrawal suggests the presence of distinct mechanisms for regulating these genes. A significant overlap in butyrate-regulated genes occurred between H1 cells and a second hESC line, BG02 (Fig. 3C). The similar transcriptional responses between H1 and BG02 were highly significant (p<0.0001).
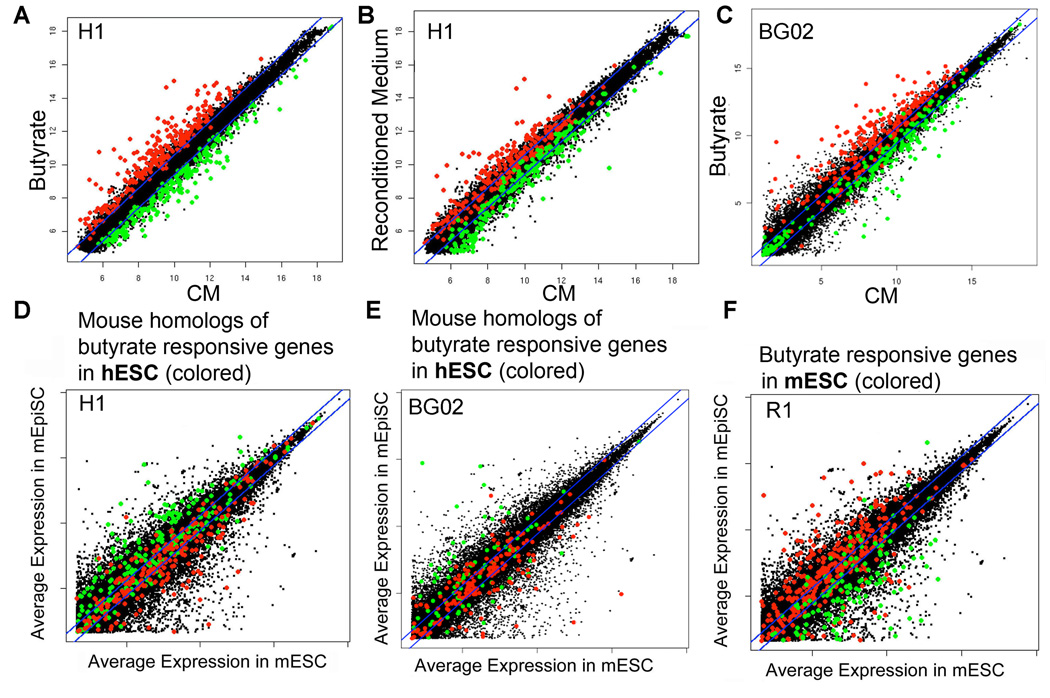
Figure 3.
Transcriptional responses to butyrate in human and mouse ES cells. Colored dots depict genes that are significantly upregulated (red) or downregulated (green) in response to butyrate in the hESC lines H1 (panels A–D) and BG02 (panel E) and the mESC line R1 (panel F). Panel A - Scatter plot depicting the transcriptional response of H1 cells cultured for 6 passages in butyrate versus H1 cells maintained in CM (see text for details). Panel B - reversion toward the original pattern of expression after returning butyrate treated H1 cells back to CM for 3 passages (“Reconditioned Medium”). Panel C – Scatter plot depicting the transcriptional response to butyrate in BG02 cells (black dots), with red and green dots identifying those genes that were butyrate-regulated in H1 cells, to highlight genes that were coordinately regulated in both hESC lines. Panels D–F contain scatter plots (in black) depicting average expression levels in mEpiSCs versus mESC (from Tesar et al., 2007 - identical for all three panels). Panels D and E overlay butyrate responsive homologous genes in H1 and BG02 cells, respectively. Colored dots indicate homologous genes in the hESC lines that were significantly upregulated (red) or downregulated (green) in response to butyrate. Panel F overlays butyrate responsive genes in mESCs. Colored dot overlays indicate genes in mESCs (R1 cells) that were significantly upregulated (red) or downregulated (green) in response to butyrate. Note that butyrate pulls the gene expression profile of hESCs toward mESCs (X axis) and away from mEpiSCs (Y axis), while pushing mESCs toward EpiSCs, thus the relative orientation of red and green dots between panels D and E versus F is reversed. T-tests indicate that these changes are highly significant (Panel D: t=-9.921, panel E: t=-4.88, panel F: t=11.139, all p<0.001, see Supplemental Experimental Procedures).
A distinct transcriptional response to butyrate in mESC
We also compared the transcriptional profiles of mESC cultured according to our standard MEF-free culture conditions (medium identical to hESM except 20% fetal bovine serum substitutes for serum replacer, plus the addition of LIF), versus butyrate (using the same medium without LIF). Supervised cluster analysis distinguished the two groups (Supplemental Fig. 7). Strikingly, there was very little overlap between the lists of butyrate-regulated genes in mESCs and hESCs (https://depts.washington.edu/iscrm/GS_data/gsdata.html). Even genes that were dramatically induced by butyrate in hESC (Dppa5, Ddx43/HAGE, Piwil2) were unchanged or even very modestly down-regulated (in the case of the Piwil2 homologue Mili) in mESC.
Butyrate induces the convergence of hESC and mESC toward a common developmental intermediate
Two recent reports described the derivation of epiblast stem cells (EpiSCs) from post implantation mouse blastocysts, and that EpiSCs more closely resemble hESC than do mESC (Brons et al., 2007; Tesar et al., 2007). We reasoned that the contrasting effects of butyrate on the transcriptional profiles of hESC versus mESC might be reconciled if butyrate brought both hESC and mESC nearer one another toward a developmental stage intermediate between mESC and mouse EpiSC (mEpiSC). To test this hypothesis we correlated butyrate-induced transcriptional responses in hESCs and mESCs with the published transcriptional profiles of mESC versus mEpiSC. Figure 3 panels D–F present identical scattergrams (black dots) comparing the relative levels of gene expression from published mRNA microarrays of mESC (X axis) versus EpiSC (Y axis) (Tesar et al., 2007). In Fig. 3 panels D and E, mouse homologues of genes that are significantly induced or repressed by butyrate in H1 and BG02 cells are overlaid as red or green dots, respectively. Note that butyrate-induced genes in hESC (red) are homologous to genes that tend to be more highly expressed in mESC and localize nearer the X axis, whereas homologues of butyrate-repressed transcripts (green) tend to be more abundant in EpiSC. This correlation is consistent with the interpretation that butyrate shifts the transcriptional program of hESC away from EpiSC, and toward mESC. Strikingly, Fig. 3F shows the opposite pattern in the mESC line R1: genes that are significantly induced by butyrate in mESC (red) tend to be more highly expressed in EpiSCs, whereas butyrate-repressed genes tend to be expressed at higher levels in mESCs. Fig. 4 shows results for ES cell-associated genes, comparing butyrate responses in hESCs with the relative expression levels of their mouse homologues in mESC vs. EpiSC. These patterns were highly significant, and support the initial observation that mEpiSC and hESC equate to a similar embryonic stage with mESC positioned earlier (Brons et al., 2007; Tesar et al., 2007), and that butyrate advances mESC and retracts hESC toward a developmental state intermediate between mESC (inner cell mass, ICM) and hESC (epiblast).
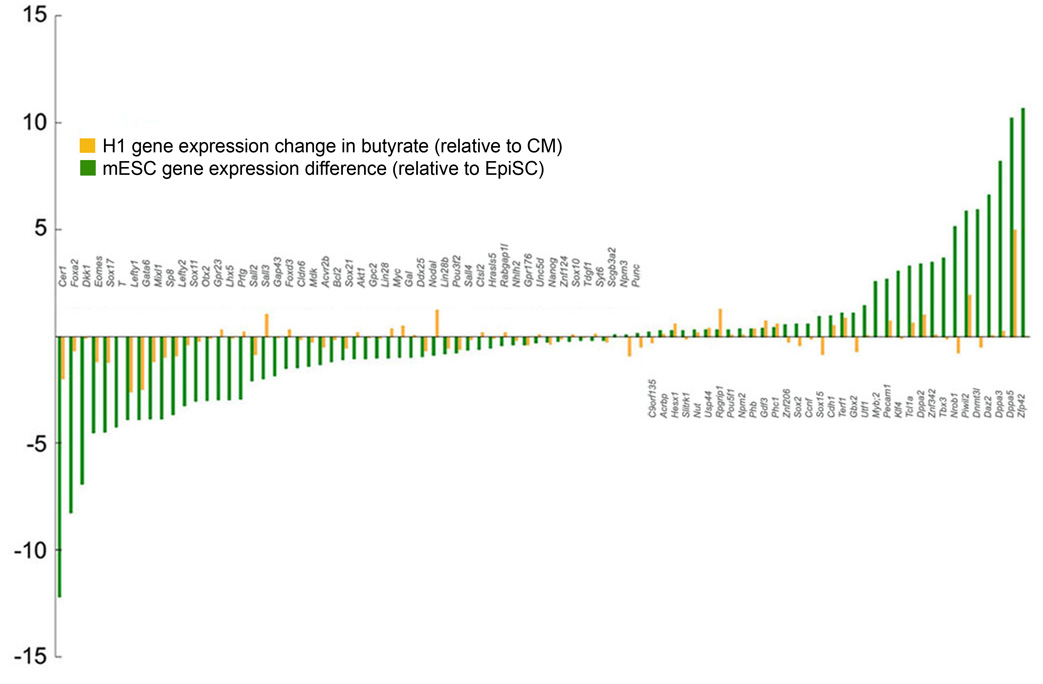
Figure 4.
Differences in expression levels of 87 ES cell-related genes between H1 cells cultured in butyrate [H1p48(CM3;B6)] versus CM [H1p48(CM9)] (butyrate/CM – orange bars) and mESC versus EpiSCs (ES/EpiSC – green bars from Tesar et al 2007). Y axis indicates Log 2 fold change. Spearman's rho correlation coefficient for the 2 data sets is 0.42, P< 10−4.
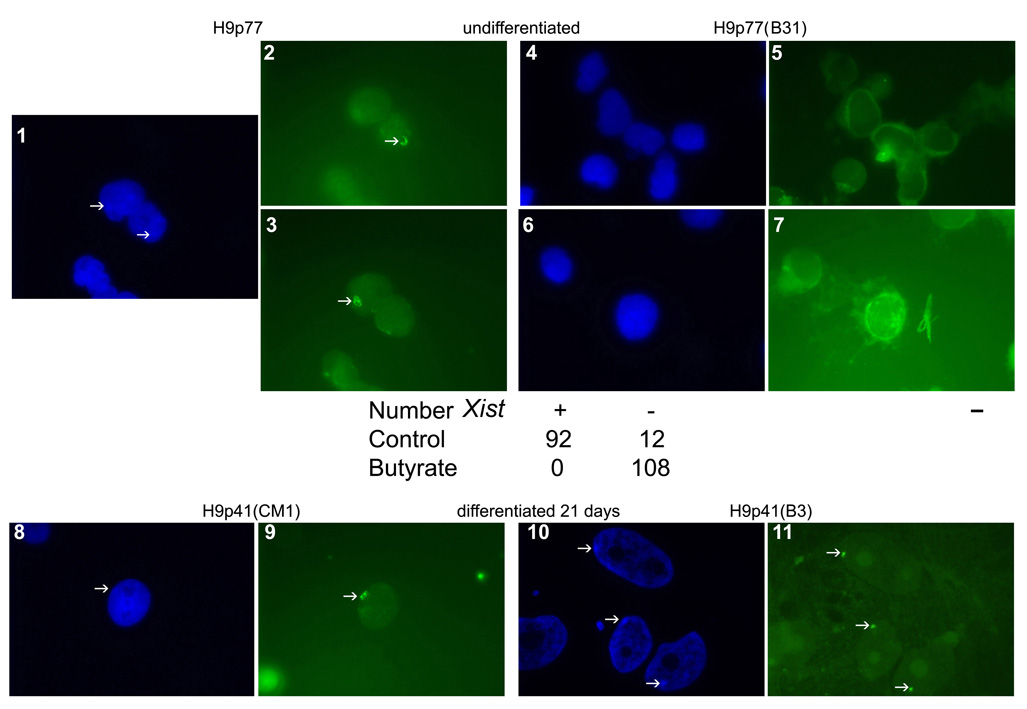
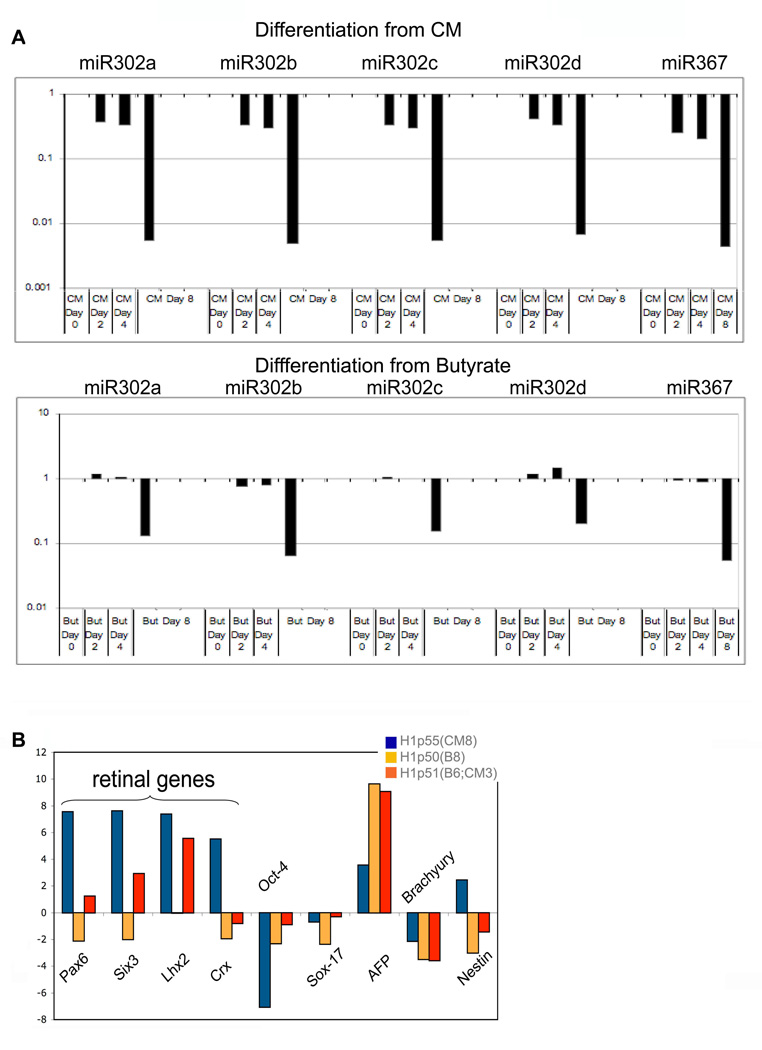
To further examine the hypothesis that butyrate exposure pulls hESC and mEpiSC backward toward an earlier developmental stage, we performed additional studies. Some female hESC lines harbor an inactive X chromosome, reflected in the presence of Xist bodies (International Stem Cell Initiative, 2007: Hall et al., 2008). Xist expression is abundant in later passage H9 cells (p77) cultured on feeders (92 of 104 cells scored [88.5%]), but was undetectable in H9 cells cultured for 31 passages in hESM plus butyrate (0 of 108 cells scored [0%]) (Fig. 5, panels 1–7). However Xist was expressed in butyrate-treated H9 cells after 21 days of differentiation (Fig. 5, panels 8–11). We also found that butyrate treated hESC differentiated more gradually than hESC cultured in CM, as evidenced by a slower decline in 302 family miRNAs (Fig. 6A), and by a delay in directed differentiation toward retinal neurons (Lamba et al., 2006; Fig. 6B). This butyrate induced differentiation delay was reversed in part by returning butyrate-exposed cells to CM for three passages prior to differentiation (Fig. 6B).
Figure 5.
Butyrate cultures are associated with a lack of Xist. Dapi nuclear stain indicating the presumptive presence of the condensed X chromatin (arrows, 1) and accompanying Xist expression (arrows 2 & 3) in later passage H9 cells. The same cells grown for the last 31 passages in butyrate on feeders did not show condensation of the X chromatin (4 and 6) or Xist (5 and 7). Below panels 1–7 are the corresponding counts of Xist positive versus negative cells on the coverslips. Dapi staining of earlier passage H9 cells grown on feeders with one passage in CM shows evidence of X-inactivation upon differentiation for 21 days (8, Dapi and 9, Xist). Cells grown for 3 passages in butyrate followed by differentiation for 21 days in the absence of butyrate showed clear evidence of appropriate Xist body formation induced by differentiation (10, Dapi and 11, Xist). Arrows depict location of condensed chromatin (Dapi) and Xist bodies (green Xist). The size bar indicates 5 µm.
Figure 6.
hESCs cultured in butyrate differentiate more slowly than hESCs cultured in CM. A. Time course of 302 family member miRNA expression in H1 cells during an 8-day course of differentiation. Top panels: H1 cells cultured in CM prior to differentiation [H1p69(CM14)]. Bottom panels: H1 cells cultured in butyrate prior to differentiation [H1p67(B15)]. Note that butyrate treated H1 cells exhibit a slower decline in 302 family miRNAs with differentiation compared to H1 cells cultured in CM. B. qRTPCR of differentiation associated transcripts in H1 cells cultured according to a previously published neuroretinal differentiation protocol (Lamba et al., 2006). Blue bars: H1 cells cultured in CM for 8 passages; orange bars: H1 cells cultured in butyrate for 8 passages; red bars: H1 cells cultured in butyrate for 6 passages and reverted to CM for 3 passages. Pax6, Six3, Lhx2, Crx are associated with retinal differentiation.
We also tested butyrate’s effect on mEpiSC (Tesar et al. 2007; Supplemental Fig. 8). Hallmark functional differences between mESC and mEpiSC include the latter’s non-responsiveness to LIF and markedly reduced ability to generate chimeras (Brons et al., 2007; Tesar et al., 2007). While butyrate did not induce LIF dependency nor feeder independency in mEpiSCs (not shown), its presence did allow an mEpiSC line provided by Tesar and McKay to generate a single coat color chimera in 14 pups (48 blastocysts injected; Fig. 2E panel 3).
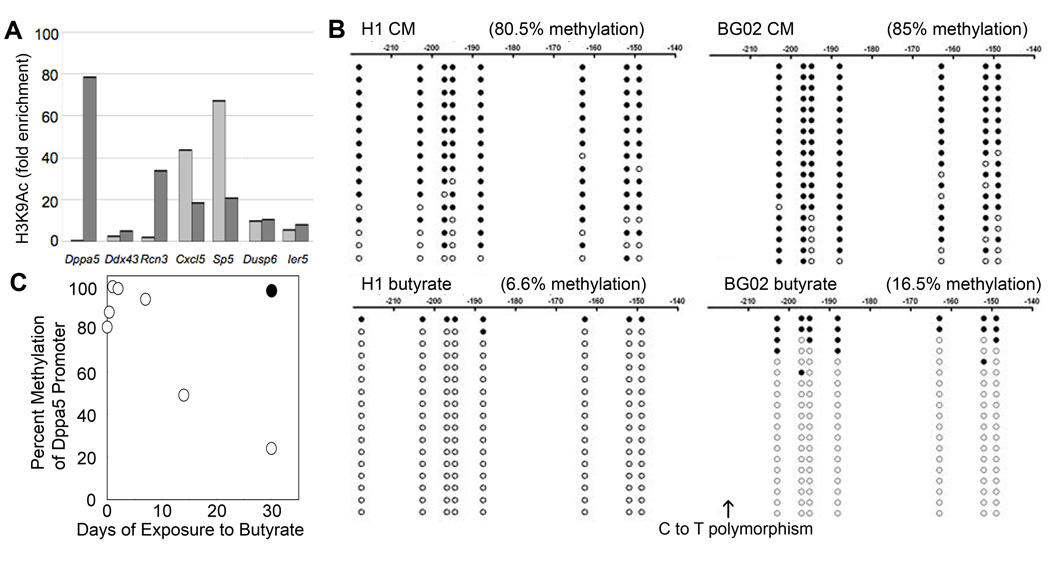
Butyrate induces H3K9 acetylation and CpG demethylation at the Dppa5 promoter
We examined the epigenetic responses of a number of butyrate-regulated genes using chromatin immunoprecipitation (ChIP) assays. The promoters of some (Dppa5, Ddx43, Rcn3) but not all (Cxcl5) butyrate-induced genes displayed a corresponding rise in H3K9 acetylation, whereas repressed genes showed little change (Dusp6, Ier5) or a decline (Sp5) in H3K9 acetylation (Fig 7A). Bisulfite sequencing of the Dppa5 promoter showed a striking decline in DNA methylation in H1 cells treated with butyrate, and a very similar response occurred in BG02 cells (Fig. 7B). For both hESC lines, the residual methylation in butyrate treated cells was concentrated among a few clones, consistent with a butyrate-induced inhibition of Dppa5 methylation following DNA replication. Serial monitoring of the Dppa5 promoter at various timepoints following butyrate exposure revealed no change in DNA methylation by day 7, however significant declines were observed on days 14 and 28 (Fig. 7C). These kinetics are also consistent with a replication dependent decline in DNA methylation.
Figure 7.
Epigenetic responses to butyrate. A. H3K9 acetylation in the promoters of butyrate-responsive genes. Light grey bars indicate BG02 cells cultured in CM [BG02p74(CM35)]; dark grey bars indicate BG02 cells cultured in butyrate [BG02p79(CM29;B24)]. An antibody directed against total histone H3 provided a control. B. Bisulfite sequencing of the Dppa5 promoter in H1 cells (left) and BG02 cells (right). Closed circles indicate the presence of methylation and open circles the absence of methylation. Numbers indicate the distance upstream from the transcription start site. Comparisons were made between H1p77(CM43) vs. H1p84(B42) (left panels) and BG02p49(CM10) vs. BG02p76(CM29;B18) (right panels). C. Changes in Dppa5 promoter methylation at various time points following a switch from CM [H1p63(CM8)] to butyrate. Open circles indicate methylation levels in butyrate treated cells whereas the closed circle depicts Dppa5 promoter methylation in the same cells continuously cultured in CM.
HDACi and ES cell self-renewal
Other HDACi also supported hESC self-renewal. Trichostatin A (TSA) supported H1 cells for more than 30 passages (Supplemental Fig. 9A) whereas valproic acid (0.5 mM) was effective only in maintaining hESC previously converted to butyrate culture conditions. Each HDACi induced a subtly different, but reversible colony morphology, most apparent at the colony edges. Butyrate, butyryl CoA (the retained derivative of butyrate uptake in mitochondria), suberoylanilide hydroxamic acid (vorinostat/SAHA) and TSA exposed cells plated tightly with sharply demarcated edges, whereas some cell spread at the edges was seen in CM. Stretching of cells at the colony edge was also observed in response to valproic acid. hESC cultured in TSA or valproic acid divided more rapidly than in butyrate but not as rapidly as in CM, and were free of lipid-containing vacuoles (data not shown). These findings indicate that as a class, HDACi can promote the self-renewal of ES cells. Suggesting a common mechanism of action, TSA induced a similar but not identical transcriptional response in a panel of 18 butyrate responsive genes (Supplemental Fig. 9B). In contrast to butyrate, TSA did not promote improved chimera formation in mice and did not appear to support the long-term maintenance of mESC without LIF or feeders. We conclude that while not all HDACi share butyrate’s full spectrum of activity, HDAC inhibition is at the core of butyrate-induced ES cell self-renewal.
Discussion
We show that ES cells can extensively self-renew in response to butyrate, without need for feeder conditioning or recombinant growth factors. ES cells are exquisitely sensitive to butyrate and self-renewal occurs only within a narrow concentration range, with higher concentrations (prevalent in the literature) inducing differentiation. Since other HDACi (TSA, valproic acid and vorinostat) also promote ES cell self-renewal our results point to the existence of a core machinery for ES cell self-renewal that is under HDAC control and which can be activated upon HDAC inhibition. Nonetheless we do note differences in the response of ES cells to various HDACi. For example, TSA did not allow mESC to develop appropriately in the embryo to generate chimeras, and valproic acid could only maintain hESCs in culture that had already been converted to butyrate. We cannot discern whether these differences reflect butyrate’s potency or range of HDAC inhibition, or additional activities beyond HDAC inhibition. Within cells butyrate is converted to butyryl CoA, which we show can also support hESC self-renewal. Butyryl CoA is integral to mammalian metabolism both for immediate energy and for energy storage, and can be utilized to build triacylglycerols and phospholipids, the likely contents of the Oil Red O+ droplets present in butyrate-treated cells.
The transcriptional response to butyrate in hESCs was distinctive, as exemplified by Dppa5. Butyrate induced a number of embryonic/cancer/testes- associated genes in hESCs, including Dppa5 and its neighbors Ddx43/HAGE and Ecat1. A CpG island upstream of Dppa5, unmethylated in sperm and testes, is densely methylated in peripheral blood (Shen et al., 2007) and in hESCs cultured in CM (Fig. 7). Butyrate treatment dramatically reduced methylation of this CpG island, providing the first example, to our knowledge, of CpG demethylation in response to an HDACi (Cameron et al., 1999). This demethylation was highly context dependent since other induced genes (Ddx43, Rcn3, and Cxcl5) were only modestly demethylated and Sp5, a repressed gene, was moderately hypermethylated in response to butyrate (preliminary observations). Butyrate also prevented the appearance of Xist expression in later passage H9 cells, suggesting an inhibitory effect on X inactivation. In general, the induction of butyrate responsive genes is rapidly reversed following butyrate withdrawal (Davie, 2003). However, another unusual feature of the butyrate response observed here was the very gradual return of a subset of butyrate-regulated genes back to baseline after reverting hESC back to CM (Fig. 3B, Supplemental Fig. 6C and Supplemental Table 4). This slow return to baseline demonstrates the existence of a mechanism for the prolonged transcriptional memory of butyrate exposure.
The most surprising finding from our study was the contrast between butyrate’s ability to support ES cell self-renewal across species, while eliciting virtually non-overlapping transcriptional responses in hESCs versus mESCs. Our gene expression analysis strongly supports the conclusion that HDACi push mESC forward and pull hESC backward toward a developmental corollary intermediate between ESC and EpiSC. Other reports support the existence of alternative ES cell-like states. Culturing mESCs in medium conditioned by HepG2 cells was found to elicit a gene expression profile similar to early primitive ectoderm, with induction of Fgf5 and repression of Rex1, Stella/Dppa3 and Pecam1 (Rathjen et al., 1999). These early primitive ectoderm-like (EPL) cells could not form chimeric mice, yet reverted back to an ES cell-like state (regaining chimera forming ability) upon return to LIF. Very recently, another group used a GFP reporter under control of a Stella/Dppa3 promoter to identify large numbers of STELLA-negative, PECAM1-negative ES cells in LIF-containing cultures (Hayashi et al., 2008). While chimera-forming ability was not tested, the STELLA-negative population reverted to more of an ES cell-like state when cultured in the presence of feeders or TSA. In contrast to these published reports, the alternative ES cell state induced by butyrate appears to be better able to generate chimeric mice. While butyrate might also enhance the poor chimera forming ability of EpiSC, it does not convert EpiSC to mESC, consistent with the notion that EpiSC fall beyond the range of inter-convertible ES cell states (Hayashi et al., 2008).
Whether the effects described here have a corollary in vivo is not known. A potential physiological underpinning is suggested by the conservation of butyrate responsiveness from mouse to human ES cells and mEpiSC. Our data suggest that butyrate may fine-tune peri-implantation development. That butyrate-treated mouse ES cells retain the ability to contribute to chimeric mice, including transmission to the germline, indicates that butyrate effects are fully reversible and exert no obvious harm on embryonic development. Butyrate exposure improves the ability of previously unsuccessful mESC clones to generate chimeras, thus, butyrate confers an ability to survive and contribute appropriately in the context of the blastocyst. That these mESC clones, in the absence of butyrate, are unable to generate chimeras (although pups are born) suggests that they do not integrate or participate in normal development and can only respond appropriately with the assistance of butyrate.
Our results demonstrate that butyrate and other HDACi can shape the ES cell state. Cross-species convergence toward a common ES cell state raises the possibility of an as yet undiscovered physiological signaling axis that involves butyrate or analogous molecules that can potently regulate development.
Experimental Procedures
hESC culture –
Initial cultures of hESC [H1 (NIH code WA01), H7 (WA07), H9 (WA09), H13 (WA13), hSF6 (UC06), BG02 and BG03] and non-human primate (rh366.4) ES cells were grown on a feeder layer of γ-irradiated (3000 rads) primary mouse embryonic fibroblasts (MEF) (Abbondanzo et al., 1993). For cultures without feeders, cells were plated on Matrigel (BD Biosciences) diluted according to manufacturer’s instructions. Human ESC culture medium (hESM) consisted of DMEM/F12 containing GlutaMax™ supplemented with 20% serum replacer (SR), 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 50 U/ml penicillin, 50 mg/ml streptomycin, (all from Invitrogen, Carlsbad, CA, USA), 0.1 mM β-mercaptoethanol (Sigma, St. Louis, MO, USA). Conditioned medium (CM) was made by incubating hESM in the presence of MEF as described in Supplemental Methods. CM also included 2 ng/ml basic FGF (FGF2, Peprotech, Rocky Hill, NJ, USA). hESC cultured in the presence of 0.2 – 0.3 mM sodium butyrate (Sigma) were cultured in hESM in the absence of MEF conditioning or FGF2. Concentrations of other HDACi were: 10 nM TSA, 0.5 mM valproic acid, 10 µM butyryl CoA (all from Sigma) and 400 nM vorinostat (Cayman Chemical, Ann Arbor, MI, USA). Nomenclature for culture conditions follows the convention: cell line, total passage number (# passages off of feeders where CM indicates culture on Matrigel in conditioned medium and B indicates culture in butyrate – generally on Matrigel with no feeder or added FGF; followed by # passages under a different growth medium; etc). Thus, H1p62(CM4;B6;CM3) would be H1 grown for 62 passages overall. Of the last 13 passages, all were on Matrigel without feeders and passage 49–52 were in CM, passages 53–59 were in butyrate and passages 60–62 were in CM. Occasionally, hESC were cultured in butyrate on MEFs. When this was done it is listed in the text.
mESC culture
mESC were cultured in the same medium described for hESC, except the serum replacer in human medium was substituted with 20% FBS (ES qualified, Invitrogen). MEF – free cultures were performed on gelatin coated dishes using medium supplemented with 1000 units/ml mouse LIF (ESGRO, Chemicon) or butyrate (0.2mM). On occasion, butyrate was added to mESC cultured on MEF. Cultures of LIF receptor null mESC (a gift of Austin Smith), gp130 null mESC (a gift of Ian Chambers) and derivation of mESC were performed as described in Supplemental Experimental Procedures.
mEpiSC Culture
Mouse EpiSC#5 (a gift of Paul Tesar and Ron McKay) was cultured as described (Tesar et al., 2007). In addition, new mEpiSC lines were derived as previously described (Tesar et al., 2007).
Immunohistochemistry and Flow Cytometry
Maintenance of an undifferentiated phenotype was established by immunohistochemistry using antibodies for Oct-4 (R&D Systems; 1:200 dilution) and SSEA-4 (Chemicon; 1:50 dilution) and by staining for alkaline phosphatase (AP) activity using a Black Alkaline Phosphatase Substrate Kit II (Vector Laboratories). Flow cytometry was performed using a FACScan flow cytometer and Cell Quest software (BD Biosciences) and the following antibodies: anti-SSEA4, anti-TRA-1-60 and TRA-1-81 (Chemicon), and anti-SSEA-3 (R&D Systems). For cell cycle analysis cells were harvested, washed with phosphate-buffered saline (PBS), and fixed in 70% ethanol. Fixed cells were stained with propidium iodide (PI) and DNA content was measured by the intensity of the fluorescence produced by PI. Data were analyzed with the Modfit 3.0 software (Verity House Software).
Quantitative PCR
Total RNA was purified using the RNeasy Micro Kit (Qiagen) following the manufacturer-specified protocol. Reverse transcription of total RNA was performed using random hexamers with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative PCR was performed in triplicate using TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems) or SYBR Green PCR Master Mix (Applied Biosystems) in 25 µl reactions in an Applied Biosystems 7900HT Fast Real-Time PCR System. Settings and primer sequences are described in Supplemental Experimental Procedures.
Microarray Analysis
Agilent whole human genome arrays were hybridized with total RNA from H1 cells cultured in each of the following conditions (2 arrays per condition). A) Cultured on Matrigel on CM; B) cultured for 6 passages in butyrate; C) cultured for 3 passages in butyrate, followed by 3 passages in CM. Group A was compared to B in independent triplicate and B was compared to C in independent triplicate. Thus, by experimental design and Agilent platform, Groups A and C served as controls relative to B. Agilent whole mouse genome microarrays were hybridized with total RNA from R1 cells cultured on gelatin without feeder support in LIF or R1 cells cultured in 0.2 mM butyrate without LIF or feeder support. These were run in independent quadruplicate.
Genes were defined as differentially expressed if they showed both a change in expression at a false discovery rate of 0.2 as analyzed by an empirical paired t-test and a 1.5 fold change in expression level. Genes were matched across the mouse and human data sets using Homologene. See the Supplemental Experimental Procedures for more details.
Bisulfite-sequencing analysis of promoter methylation and Chromatin immunopreciptation (ChIP) analysis of promoter acetylation
Bisulfite-sequencing to assess Dppa5 promoter methylation was performed as reported previously (Shen et al., 2007). ChIP analysis to assess promoter acetylation was performed as described (Nelson et al., 2006) and utilized an anti-acetyl-histone H3 antibody which recognizes lysine residues 9 and 14 (Millipore).
Supplementary Material
Supplemental Figure 1. Butyrate treated H1 cells grown in hESM on Matrigel maintain an undifferentiated state relative to cells in CM. A. Flow cytometry profile of H1 cells cultured in butyrate (top) or CM (below). Profiles shaded in gray indicate control staining with secondary antibodies alone. B. Pou5f1 (Oct 4) and Nanog mRNA levels in H1 cells cultured in butyrate [orange bars, H1p48(CM3;B6)] or CM [blue bars, H1p48(CM9)]. C. Telomerase activity assay of H1 cells cultured in CM or butyrate in the presence (+) or absence (−) of heat inactivation. HFF: human foreskin fibroblasts. Cells were exposed to ≥ 3 passages in butyrate in all assays to compare to cells grown in CM.
Supplemental Figure 2. Schematic depiction of hESC karyotypes during culture in HDACi versus standard conditions. A. H1 cells cultured on Matrigel starting on passage 39. B. H7 cells cultured continuously on feeders. C. H13 cells cultured continuously on feeders. D. BG02 cells cultured on Matrigel beginning at passage 31. E. BG02 cells cultured on Matrigel plus butyrate starting at passage 41. F. BG02 cells cultured on Matrigel beginning at passage 39. Open circles indicate normal karyotype. ? indicates unknown karyotype. Closed circles indicate abnormal karyotypes. Hatched circles indicate inferred abnormal karyotype. Numbers of circles represent numbers of karyotypes examined.
Supplemental Figure 3. mESC do not require gp130 mediated signaling to affect self-renewal in response to HDACi. Phase contrast appearance (top panels) and alkaline phosphatase staining (lower panels) of a. R1 cells, b. Lifr−/− mESC, or c. gp130 null mESC. In panel c, the lower panels are alkaline phosphatase labeled (left) and Pou5f1 labeled (right) under the respective unstained image. Control cultures for R1 and Lifr−/− cells used IL6/sIL6 receptor (to bypass LIFR and signal through gp130) whereas gp130 null mESC used parietal endoderm conditioned medium (that can sustain mESC without functional gp130 signaling) to maintain self-renewal. All cultures were performed in the absence of feeders. Control cultures [using IL6 + sIL6R (A, B) or parietal endoderm conditioned medium (C)] were compared to cells cultured for 4 passages in butyrate (without IL6/sIL6R or parietal endoderm conditioned medium). Note that differentiating cells could be detected in all culture conditions. Image areas were selected to reflect the inherent heterogeneity irrespective of culture conditions. The size bar indicates 38 µM.
Supplemental Figure 4. hESC cultured in butyrate retain full differentiation capacity. A. Teratomas excised at day 49 – left panel generated from BG02p66(CM35) and the right panel generated from BG02p80(CM29;B20). The arrows placed in the butyrate tumor indicate the presence of melanin. B. Top panels: Trimmed paraffin blocks of teratomas shown in panel A. Bottom panels: Trimmed paraffin blocks of teratomas generated 10 weeks after injection of BG02 cells cultured in CM (left) or 8 weeks for cells cultured in butyrate without CM and FGF2 (right). Arrows indicate avascular areas. Triangles point toward melanin containing spots. C. Histological sections of teratomas from H1 cells cultured in CM (left) or butyrate (right). The top 3 images in each column are the same view at increasing magnification. The number of days between implantation and harvest is indicated at the top of the columns. Representatives of the 3 germ lineages are indicated EN: Endoderm. EC: ectoderm. M: mesoderm. The size bar indicates 250 µm in the upper panels, 100 µm in the second and fourth panels, and 50 µm in the third panels. Note the pigment in the ectoderm of the butyrate tumors. D. A section from BG02p80(CM29;B20) 7 week tumor shown in panel B highlighting melanin expression (upper arrow). Lower arrow indicates cells labeled with an antibody directed against synaptophysin, a glycoprotein present in the membrane of neuronal presynaptic vesicles and an early neuroectodermal marker. The size bar indicates 12.5 mm in panel D.
Supplemental Figure 5. A relatively small number of genes are regulated by butyrate. Unsupervised analysis (panels A & C) showing all genes and supervised analysis (panels B & D) showing only significantly altered expression from Agilent mRNA arrays probed with mRNA from H1 cells cultured in CM [hES-CM; H1p48(CM9)], H1 cells from the same pool cultured in butyrate for 6 passages [hES-But; H1p48(CM3;B6)] or H1 cells from the same pool converted to butyrate for 4 passages followed by CM for 3 passages [hES-rCM; H1p49(CM3;B4;CM3)]. Note that genome-wide expression patterns are only modestly affected by butyrate (C).
Supplemental Figure 6. Characterizing the transcriptional response to butyrate. A. qRTPCR confirmation of 14 transcripts identified by microarray as being differentially regulated by butyrate [H1p48(CM;B6) relative to H1p48(CM9)] B. Addition of butyrate to H13 cells cultured on feeders (without added bFGF when butyrate was present and with 2 ng/ml added bFGF when grown in CM) induces Dppa5, Piwil2 and Ecat1. Butyrate was added for the last 3 passages. C. qRTPCR demonstrates the persistence of Ddx43/HAGE, Dppa5 and Ecat1 expression 3 passages after returning H1 cells cultured in butyrate for 4 passages back to CM for 3 passages [H1p49(CM3;B4;CM3)].
Supplemental Figure 7. Unsupervised (left) and supervised (right) profiles from whole mouse genome Agilent arrays using mRNA from R1 mESC cultured in LIF versus butyrate for four passages. Arrays were run independently, in quadruplicate.
Supplemental Figure 8. Morphology of EpiSCs cultured in standard hESC-like conditions for 12 passages (left), or cultured under the same conditions except for the addition of butyrate for 3 passages (right). Size bar indicates 38 5m.
Supplemental Figure 9. hESC respond to a variety of HDACi. A. Phase contrast appearance of hESC cultured in CM (BG02), sodium butyrate (BG02), butyryl CoA (BG03), trichostatin A (BG02), valproic acid (H1) and vorinostat/SAHA (BG02) at the indicated concentrations and passage numbers. Size bar indicates 38 µm. B. Transcriptional response of hESC (BG02) cultured in 0.2 mM butyrate (without feeders) for 38 passages or 10 nM trichostatin A for 34 passages relative to CM.
Acknowledgments
We thank Daniel G. Miller, William Noble, Richard Young, Matt Guenther, Paul Tesar, Ron McKay, Piper Treuting, Edith Wang and Julian Simon for useful discussions. We thank Austin Smith for LIFR -/- mESC, and Ian Chambers for gp130 -/- mESC, Paul Tesar and Ron McKay for mEpiSC and Jeff Okada, Jennifer Potter, Chris Cavanaugh Jennifer Hesson for technical support. We also thank Robert Hall at the University of Washington Center for Array Technologies and William Howald at the Mass Spectrometry Center for their expertise and assistance.
Supported by NIH grant 1P01GM081619 and through the Institute for Stem Cell and Regenerative Medicine (ISCRM) of the University of Washington. PP is supported by a Michael Smith Foundation for Health Research Career Investigator Award and NIH grant GM076990.
Footnotes
Data information
Data deposition The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE15112 (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE15112).
Additional information from our microarray analysis is available at https://depts.washington.edu/iscrm/GS_data/gsdata.html.
Competing interests The authors have no competing financial interests.
REFERENCES
- Abbondanzo SJ, Gadi I, Stewart CL. Derivation of embryonic stem cell lines. Meth Enzymol. 1993;225:803–823. doi: 10.1016/0076-6879(93)25052-4. [DOI] [PubMed] [Google Scholar]
- Andäng M, Hierling-Leffler J, Moliner A, Lundgren TK, Castelo-Branco G, Nanou E, Pozas E, Brvia V, Halliez S, Nishimaru H, Wilbertz J, Arenas E, Koltzenburg M, Charnay P, El Manira A, Ibañez CF, Ernfors P. Histone H2AX-dependent GABA(A) receptor regulation of stem cell proliferation. Nature. 2008;451:460–464. doi: 10.1038/nature06488. [DOI] [PubMed] [Google Scholar]
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, LIan JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J. Cell Physiol. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boffa LC, Vidali G, Mann RS, Allfrey VG. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J. Biol. Chem. 1978;253:3364–3366. [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Spradling AC. Searching chromatin for stem cell identity. Cell. 2006;125:233–236. doi: 10.1016/j.cell.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Dahéron L, Opitz SL, Zaehres H, Lensch MS, Andrews PW, Itskovitz-Eldor J, Daley GQ. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 2004;22:770–778. doi: 10.1634/stemcells.22-5-770. [DOI] [PubMed] [Google Scholar]
- Davie JR. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- Denning C, Allegrucci C, Priddle H, Barbadillo-Muñoz MD, Anderson D, Self T, Smith NM, Parkin CT, Young LE. Common culture conditions for maintenance and cardiomyocyte differentiation of the human embryonic stem cell lines, BG01 and HUES-7. Int. J. Dev. Biol. 2006;50:27–37. doi: 10.1387/ijdb.052107cd. [DOI] [PubMed] [Google Scholar]
- Dreesen O, Brivanlou AH. Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev. 2007;3:7–17. doi: 10.1007/s12015-007-0004-8. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiselleova L, Peterkiva I, Neradil J, Slaninova I, Hampl A, Dvorak P. Comparative study of mouse and human feeder cells for human embryonic stem cells. Int.J. Dev. Biol. 2008;52:353–363. doi: 10.1387/ijdb.082590le. [DOI] [PubMed] [Google Scholar]
- Hall LL, Byron M, Butler J, Becker KA, Nelson A, Amit M, Itskovitz-Eldor J, Stein J, Stein G, Ware C, Lawrence JB. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J. Cell. Physiol. 2008;6:445–452. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa LopesSM, Tang F, Surani MA. Dynammic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008 doi: 10.1038/nbt.1502. (Oct. 12 ePub) [DOI] [PubMed] [Google Scholar]
- International Stem Cell Initiative. Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Blum B, Brooking J, Chen KG, Choo AB, Churchill GA, Corbel M, Damjanov I, Draper JS, Dvorak P, Emanuelsson K, Fleck RA, Ford A, Gertow K, Gertsenstein M, Gokhale PJ, Hamilton RS, Hampl A, Healy LE, Hovatta O, Hyllner J, Imreh MP, Itskovitz-Eldor J, Jackson J, Johnson JL, Jones M, Kee K, King BL, Knowles BB, Lako M, Lebrin F, Mallon BS, Manning D, Mayshar Y, McKay RD, Michalska AE, Mikkola M, Mileikovsky M, Minger SL, Moore HD, Mummery CL, Nagy A, Nakatsuji N, O’Brien CM, Oh SK, Olsson C, Otonkoski T, Park KY, Passier R, Patel H, Patel M, Pedersen R, Pera MF, Piekarczyk MS, Pera RA, Reubinoff BE, Robins AJ, Rossant J, Rugg-Gunn P, Schulz TC, Semb H, Sherrer ES, Siemen H, Stacey GN, Stojkovic M, Suemori H, Szatkiewicz J, Turetsky T, Tuuri T, van den Brink S, Vintersten K, Vuoristo S, Ward D, Weaver TA, Young LA, Zhang W. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Lamba DA, Karl MO, Ware CB, Reh TA. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2006;103:12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenstein ME, Ludwig TE, Xu RH, Llanas RA, VanDenHeuvel-Kramer K, Manning D, Thomson JA. Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells. 2006;24:568–574. doi: 10.1634/stemcells.2005-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CM, Kim CE, Margolin AA, Guo M, Zhu J, Mason JM, Hensle TW, Murty VV, Grundy PE, Fearon ER, D’Agati V, Licht JD, Tycko B. CTNNB1 mutations and overexpression of Wnt/beta-catenin target genes in WT1-mutant Wilms' tumors. Am. J. Pathol. 2004;165:1943–1953. doi: 10.1016/s0002-9440(10)63246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts.Nat. Biotechnol. 2007;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2007;451:135–136. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pereira L, Yi F, Merrill BJ. Repression of Nanog Gene Transcription by Tcf3 Limits Embryonic Stem Cell Self-Renewal. Mol. Cell Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Enroth S, Ameur A, Koch CM, Clelland GK, Respuela-Alonso P, Wilcox S, Dovey OM, Ellis PD, Langford CF, Dunham I, Komorowski J, Wadelius C. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007;17:708–719. doi: 10.1101/gr.5540007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen J, Lake JA, Bettess MD, Washington JM, Chapman G, Rathjen PD. Formation of a primitive ectoderm like cell population, EPL cells, from ES cells in response to biologically derived factors. J. Cell Sci. 1999;112:601–612. doi: 10.1242/jcs.112.5.601. [DOI] [PubMed] [Google Scholar]
- Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9:809–818. [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmend S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG. Embryo-derived stem cells: of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Köntgen F, Abbondanzo SJ. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- Tabibzadeh S, Hemmati-Brivanlou A. Lefty at the crossroads of "stemness" and differentiative events. Stem Cells. 2006;24:1998–2006. doi: 10.1634/stemcells.2006-0075. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar SA, Gliniak BC, McKenna HJ, Papayannopoulou T, Thoma B, Cheng L, Donovan PJ, Peschon JJ, Bartlett PF, Willis CR, Wright BD, Carpenter MK, Davison BL, Gearing DP. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development. 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Blau CA. A comparison of NIH-approved human ESC lines. Stem Cells. 2006;24:2677–2684. doi: 10.1634/stemcells.2005-0452. [DOI] [PubMed] [Google Scholar]
- Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. The Sp1-related transcription factors Sp5 and Sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Curr. Biol. 2000;15:489–500. doi: 10.1016/j.cub.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Willert J, Epping M, Pollack JR, Brown PO, Nusse RA. transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev. Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Yuan X, Sharkis SJ. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells. 2006;24:1476–1486. doi: 10.1634/stemcells.2005-0299. [DOI] [PubMed] [Google Scholar]
- Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- Yao HH, Matzuk MM, Jorgez CJ, Menke DB, Page DC, Swain A, Capel B. Follistatin operates downstream of Wnt4 in mammalian ovary organogenesis. Dev. Dyn. 2004;230:210–215. doi: 10.1002/dvdy.20042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Pereira L, Merrill BJ. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells. 2008;26:1951–1960. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zirn B, Samans B, Wittmann S, Pietsch T, Leuschner I, Graf N, Gessler M. Target genes of the WNT/beta-catenin pathway in Wilms tumors. Genes Chromosomes Cancer. 2006;45:565–574. doi: 10.1002/gcc.20319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Butyrate treated H1 cells grown in hESM on Matrigel maintain an undifferentiated state relative to cells in CM. A. Flow cytometry profile of H1 cells cultured in butyrate (top) or CM (below). Profiles shaded in gray indicate control staining with secondary antibodies alone. B. Pou5f1 (Oct 4) and Nanog mRNA levels in H1 cells cultured in butyrate [orange bars, H1p48(CM3;B6)] or CM [blue bars, H1p48(CM9)]. C. Telomerase activity assay of H1 cells cultured in CM or butyrate in the presence (+) or absence (−) of heat inactivation. HFF: human foreskin fibroblasts. Cells were exposed to ≥ 3 passages in butyrate in all assays to compare to cells grown in CM.
Supplemental Figure 2. Schematic depiction of hESC karyotypes during culture in HDACi versus standard conditions. A. H1 cells cultured on Matrigel starting on passage 39. B. H7 cells cultured continuously on feeders. C. H13 cells cultured continuously on feeders. D. BG02 cells cultured on Matrigel beginning at passage 31. E. BG02 cells cultured on Matrigel plus butyrate starting at passage 41. F. BG02 cells cultured on Matrigel beginning at passage 39. Open circles indicate normal karyotype. ? indicates unknown karyotype. Closed circles indicate abnormal karyotypes. Hatched circles indicate inferred abnormal karyotype. Numbers of circles represent numbers of karyotypes examined.
Supplemental Figure 3. mESC do not require gp130 mediated signaling to affect self-renewal in response to HDACi. Phase contrast appearance (top panels) and alkaline phosphatase staining (lower panels) of a. R1 cells, b. Lifr−/− mESC, or c. gp130 null mESC. In panel c, the lower panels are alkaline phosphatase labeled (left) and Pou5f1 labeled (right) under the respective unstained image. Control cultures for R1 and Lifr−/− cells used IL6/sIL6 receptor (to bypass LIFR and signal through gp130) whereas gp130 null mESC used parietal endoderm conditioned medium (that can sustain mESC without functional gp130 signaling) to maintain self-renewal. All cultures were performed in the absence of feeders. Control cultures [using IL6 + sIL6R (A, B) or parietal endoderm conditioned medium (C)] were compared to cells cultured for 4 passages in butyrate (without IL6/sIL6R or parietal endoderm conditioned medium). Note that differentiating cells could be detected in all culture conditions. Image areas were selected to reflect the inherent heterogeneity irrespective of culture conditions. The size bar indicates 38 µM.
Supplemental Figure 4. hESC cultured in butyrate retain full differentiation capacity. A. Teratomas excised at day 49 – left panel generated from BG02p66(CM35) and the right panel generated from BG02p80(CM29;B20). The arrows placed in the butyrate tumor indicate the presence of melanin. B. Top panels: Trimmed paraffin blocks of teratomas shown in panel A. Bottom panels: Trimmed paraffin blocks of teratomas generated 10 weeks after injection of BG02 cells cultured in CM (left) or 8 weeks for cells cultured in butyrate without CM and FGF2 (right). Arrows indicate avascular areas. Triangles point toward melanin containing spots. C. Histological sections of teratomas from H1 cells cultured in CM (left) or butyrate (right). The top 3 images in each column are the same view at increasing magnification. The number of days between implantation and harvest is indicated at the top of the columns. Representatives of the 3 germ lineages are indicated EN: Endoderm. EC: ectoderm. M: mesoderm. The size bar indicates 250 µm in the upper panels, 100 µm in the second and fourth panels, and 50 µm in the third panels. Note the pigment in the ectoderm of the butyrate tumors. D. A section from BG02p80(CM29;B20) 7 week tumor shown in panel B highlighting melanin expression (upper arrow). Lower arrow indicates cells labeled with an antibody directed against synaptophysin, a glycoprotein present in the membrane of neuronal presynaptic vesicles and an early neuroectodermal marker. The size bar indicates 12.5 mm in panel D.
Supplemental Figure 5. A relatively small number of genes are regulated by butyrate. Unsupervised analysis (panels A & C) showing all genes and supervised analysis (panels B & D) showing only significantly altered expression from Agilent mRNA arrays probed with mRNA from H1 cells cultured in CM [hES-CM; H1p48(CM9)], H1 cells from the same pool cultured in butyrate for 6 passages [hES-But; H1p48(CM3;B6)] or H1 cells from the same pool converted to butyrate for 4 passages followed by CM for 3 passages [hES-rCM; H1p49(CM3;B4;CM3)]. Note that genome-wide expression patterns are only modestly affected by butyrate (C).
Supplemental Figure 6. Characterizing the transcriptional response to butyrate. A. qRTPCR confirmation of 14 transcripts identified by microarray as being differentially regulated by butyrate [H1p48(CM;B6) relative to H1p48(CM9)] B. Addition of butyrate to H13 cells cultured on feeders (without added bFGF when butyrate was present and with 2 ng/ml added bFGF when grown in CM) induces Dppa5, Piwil2 and Ecat1. Butyrate was added for the last 3 passages. C. qRTPCR demonstrates the persistence of Ddx43/HAGE, Dppa5 and Ecat1 expression 3 passages after returning H1 cells cultured in butyrate for 4 passages back to CM for 3 passages [H1p49(CM3;B4;CM3)].
Supplemental Figure 7. Unsupervised (left) and supervised (right) profiles from whole mouse genome Agilent arrays using mRNA from R1 mESC cultured in LIF versus butyrate for four passages. Arrays were run independently, in quadruplicate.
Supplemental Figure 8. Morphology of EpiSCs cultured in standard hESC-like conditions for 12 passages (left), or cultured under the same conditions except for the addition of butyrate for 3 passages (right). Size bar indicates 38 5m.
Supplemental Figure 9. hESC respond to a variety of HDACi. A. Phase contrast appearance of hESC cultured in CM (BG02), sodium butyrate (BG02), butyryl CoA (BG03), trichostatin A (BG02), valproic acid (H1) and vorinostat/SAHA (BG02) at the indicated concentrations and passage numbers. Size bar indicates 38 µm. B. Transcriptional response of hESC (BG02) cultured in 0.2 mM butyrate (without feeders) for 38 passages or 10 nM trichostatin A for 34 passages relative to CM.