SUMMARY
In several growth factor receptors, the intracellular juxtamembrane (JM) region participates in autoinhibitory interactions that must be disrupted for tyrosine kinase activation. Using alanine scanning mutagenesis and crystallographic approaches, we define a domain within the JM region of the epidermal growth factor receptor (EGFR) that instead plays an activating – rather than autoinhibitory – role. Mutations in the C-terminal 19 residues of the EGFR JM region abolish EGFR activation. In a crystal structure of an asymmetric dimer of the tyrosine kinase domain, the JM region of an acceptor monomer makes extensive contacts with the C-lobe of a donor monomer, thus stabilizing the dimer. We describe how an uncharacterized lung cancer mutation in this JM activation domain (V665M) constitutively activates EGFR by augmenting its capacity to act as an acceptor in the asymmetric dimer. This JM mutant promotes cellular transformation by EGFR in vitro and is tumorigenic in a xenograft assay.
INTRODUCTION
The EGF receptor (EGFR) has served as an important model for elucidating the activation mechanisms of receptor tyrosine kinases (RTKs), and is also the target for several cancer therapeutics in current clinical use (Ciardiello and Tortora, 2008; Sergina and Moasser, 2007). Like other RTKs (Linggi and Carpenter, 2006), EGFR comprises an extracellular ligand-binding region, a single transmembrane helix, and a cytoplasmic region that contains the tyrosine kinase domain (TKD). Activation of the TKD is essential for cellular responses to growth factors, and mutations that enhance EGFR tyrosine kinase activity are frequently oncogenic in human tumors (Lee et al., 2006; Sergina and Moasser, 2007; Sharma et al., 2007). For example, deletions or substitutions in the extracellular region activate EGFR in glioblastoma, and small deletions or point mutations within the TKD activate EGFR in non-small cell lung cancer (NSCLC).
Structural analyses of extracellular regions have shown how ligand binding facilitates EGFR dimerization (Lemmon, 2009), a critical step for ligand-dependent RTK activation (Schlessinger, 2000). Crystallographic and mutational studies suggest that this leads to allosteric activation of the TKD through the formation of an asymmetric dimer (Zhang et al., 2006), in which the C-lobe of one TKD abuts the N-lobe of its dimerization partner and promotes conformational changes that stabilize an activated state (Zhang et al., 2006).
The TKD accounts for only ~50% of the EGFR intracellular region. The remainder comprises the 225 amino acid carboxy terminal (CT) region (residues 961-1186) and the 38 amino acid cytoplasmic juxtamembrane (JM) region (residues R645-I682). The CT region harbors most of the autophosphorylation sites, and contains a putative autoinhibitory domain (Walton et al., 1990) that has not been characterized. The JM region of EGFR contains two threonines (T654 and T669) that are known to be phosphorylated with inhibitory consequences (Heisermann et al., 1990; Welsh et al., 1991). In addition, the JM region includes receptor trafficking signals (Bao et al., 2000; He et al., 2002; Hsu and Hung, 2007; Morrison et al., 1996), a putative calmodulin-binding site (Martin-Nieto and Villalobo, 1998), and a polybasic region (Aifa et al., 2005; McLaughlin et al., 2005). In previous studies using large deletion mutations, we found that a portion of the JM region is required for kinase activation (Thiel and Carpenter, 2007). Our observations suggest that the JM region of EGFR plays an activating role, contrasting starkly with the autoinhibitory function described for JM regions of other RTKs (Hubbard, 2004). Here, we define the minimal JM activation domain (JMAD) using alanine scanning mutagenesis, and evaluate reported lung cancer mutations in this region for their influence on EGFR activity. We also describe a crystal structure of the EGFR TKD that contains the entire JM region. This structure reveals key interactions responsible for function of the JMAD in TKD activation, and suggests structural mechanisms for EGFR activation by JM mutations in NSCLC.
RESULTS and DISCUSSION
Scanning Alanine Mutagenesis of the EGF Receptor Juxtamembrane Region
We previously showed that deletions in the JM region abolish both EGF-induced activation of intact EGFR and autophosphorylation of an intracellular domain (ICD) construct (Figure 1A) overexpressed in Cos-7 cells (Thiel and Carpenter, 2007). Deleting amino acids 645-662 or 645-676 from the JM region reduced ICD autophosphorylation in cells by ~95%. These JM residues are absent from published crystal structures of the EGFR TKD, which utilized constructs beginning at residue 672 (Stamos et al., 2002; Wood et al., 2004; Zhang et al., 2006). To determine which amino acids within the JM region contribute to TKD activation, we used scanning alanine mutagenesis in the context of an epitope-tagged ICD construct (Figure 1A). The influence of mutations on ICD autophosphorylation should reflect only alterations of TKD activation. Indirect effects arising from mutation of JM trafficking motifs will not complicate interpretation of these studies as they might with intact EGFR (He et al., 2002; Kil et al., 1999; Lin et al., 2001; Song Jae Kil, 2000). We showed previously that mutating K721 (thus abrogating ATP-binding) abolishes in vivo tyrosine autophosphorylation of EGFR ICD constructs (Thiel and Carpenter, 2007), illustrating that tyrosine phosphorylation of the ICD reflects intrinsic kinase activity levels.
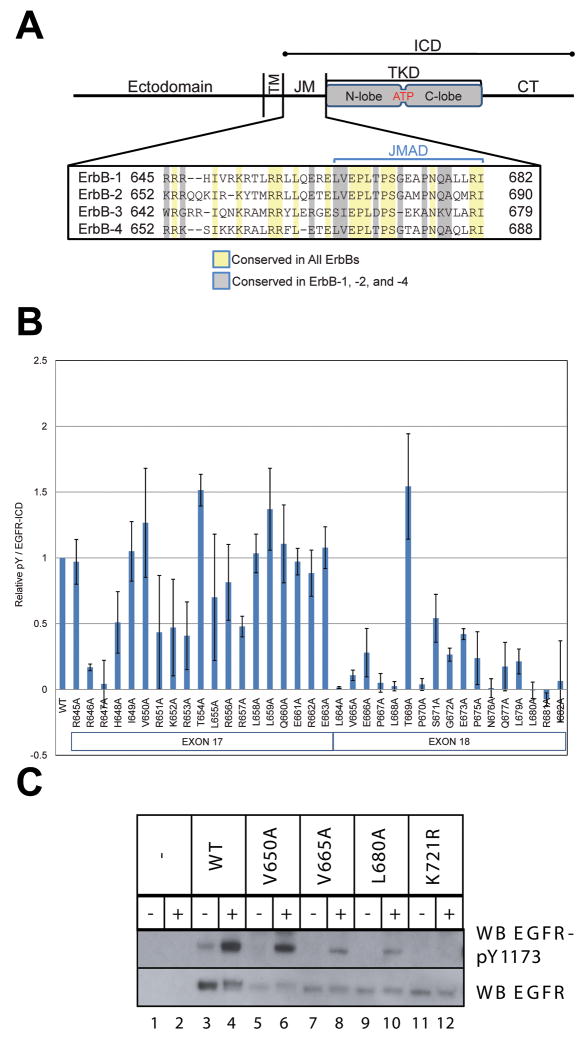
Figure 1. Effect of Scanning Alanine Mutagenesis of the JM Region on Tyrosine Phosphorylation.
A. Schematic of full-length EGFR, with the transmembrane (TM), tyrosine kinase domain (TKD), and carboxy-terminal (CT) regions marked. The intracellular domain (ICD) construct includes the JM, TKD, and CT regions. An alignment of JM region sequences across the human ErbB receptor family is shown, with the putative JM activation domain (JMAD) indicated. B. Scanning alanine mutagenesis of the JM region using ICD-flag constructs. Cos-7 cells transiently expressing wild-type or mutated ICD-flag constructs were analyzed by western blotting and densitometry for both phosphotyrosine content and flag-epitope expression. The graph demonstrates the ratio of phosphotyrosine to flag expression for each construct relative to WT. Results represent the means of 2 experiments ± standard deviation. C. Effect of JM alanine substitutions on tyrosine autophosphorylation of full-length EGFR. NIH3T3 cells transiently expressing full-length EGFR with the indicated mutations were serum-starved and then treated (+) or not (−) with EGF for 10 min. Cell lysates were subjected to SDS-PAGE and immunoblotting, detecting phospho-EGFR with anti-EGFR-pY1173 (upper panel) and receptor levels with anti-EGFR (lower panel).
Of the 36 alanine substitutions made in the JM region, 19 led to a significant reduction (≥50%) of ICD tyrosine phosphorylation. Fifteen of these ‘sensitive’ positions are located in the C-terminal section of the JM (Figure 1B, Supplementary Figure 1), between residues 664 and 682 – in a region encoded by exon 18 (which also encodes a portion of the TKD N-lobe). Within this sequence, T669 (a known MAP kinase phosphorylation site: see below) is the only site at which alanine substitution does not impair kinase activity. Interestingly, four of the loss-of-function mutations are either similar (L664A, V665A) or identical (P675A, L680A) to changes present in the JM region of ErbB-3 (Figure 1A), a member of the EGFR family thought to have an inactive TKD (Guy et al., 1994). As shown in Figure 1A, the boundary between exon 17 and exon 18 is conserved in all ErbB receptors, and the four receptors share more identity in the part of the JM region encoded by exon 18 than by exon 17.
Analysis of JM Mutations in Full-Length EGFR
We also analyzed three JM mutations in full-length EGFR: V650A, V665A, and L680A (Figure 1C), alongside catalytically inactive K721R EGFR (to establish a baseline for loss-of-function). Each EGFR variant was transiently expressed in NIH3T3 cells, and tyrosine phosphorylation at Y1173, a known autophosphorylation site, was measured by western blotting in the absence and presence of EGF. The V650A mutant resembles wild-type EGFR in its EGF-dependent autophosphorylation (Figure 1C), consistent with the failure of this mutation to reduce ICD autophosphorylation (Figure 1B). By contrast, activation of the V665A and L680A EGFR mutants is significantly compromised (as seen in the ICD context), and autophosphorylation of K721R EGFR is undetectable. To control for the possibility that reduced activation of V665A and L680A EGFR arises from impaired trafficking of the receptor to the cell surface, we bound Alexafluor-conjugated EGF to 293 cells expressing each mutated receptor (or wild-type EGFR) and assessed cell-surface fluorescence using flow cytometry. The full-length receptor mutants were expressed on the cell surface at levels within 10% of the wild-type receptor (Supplementary Figure 2A).
The JM region contributes directly to EGFR dimerization
In an effort to understand how the JM region exerts its positive influence on EGFR activation, we crystallized a form of the EGFR TKD that extends from residue 645 to 998 (EGFR645-998), and includes the entire JM region. Previous crystallographic studies of EGFR TKD utilized protein that lacks residues 645-671 (Stamos et al., 2002; Wood et al., 2004; Yun et al., 2007; Zhang et al., 2006). The best crystals were obtained using a form of EGFR645-998 harboring an inactivating K721M mutation. The structure of EGFR645-998(K721M) was solved to 2.8Å resolution using molecular replacement as described in Experimental Procedures (Table 1).
Table 1.
Data collection and refinement statistics.
| PDB code | 3GOP |
|---|---|
| Data collection | |
| Space group | P43212 |
|
| |
| Cell dimensions | |
| a, b (Å) | 62.1 |
| c (Å) | 177.2 |
| α, β, γ, (°) | 90 |
| Resolution (Å) | 50–2.8 |
| Rsyma | 0.103 (0.306) |
| I/σ | 20.8 (7.86) |
| Completeness (%) | 98.8 (100) |
| Redundancy | 7.6 (7.8) |
|
| |
| Refinement | |
| Resolution (Å) | 50–2.8 |
| No. Reflections | 9,127 |
| Rwork/Rfree | 20.7–25.9 |
| No. atoms | 2,402 |
| Protein | 2,396 |
| Water | 6 |
|
| |
| Bfactors | |
| Protein | 27.39 |
|
| |
| Rms deviations | |
| Bond length (Å) | 0.016 |
| Bond angles (°) | 1.52 |
| Ramachandran statistics (%)c | 100 |
highest resolution shell data is shown in parentheses.
Rsym = Σ|Ih − <Ih>|/ΣIh, where <Ih> = average intensity over symmetry equivalent measurements.
R factor = Σ|Fo − Fc|/ΣFo, where summation is over data used in the refinement; Rfree includes 10% of the data excluded from the refinement.
Ramachandran plots were calculated using RAMPAGE (Lovell et al., 2003). Percentage denotes residues in allowed regions.
As shown in Figure 2A, EGFR645-998(K721M) forms the same asymmetric dimer in crystals as that seen repeatedly (Figure 2B) for active conformations of the EGFR TKD (Stamos et al., 2002; Yun et al., 2007; Zhang et al., 2006), and the ErbB4 TKD in both active (Qiu et al., 2008) and inactive (inhibitor-bound) states (Wood et al., 2008). In this asymmetric dimer, the C-lobe of a ‘donor’ TKD (yellow) abuts the N-lobe of an ‘acceptor’ TKD (green), contacting a surface of the acceptor made up from elements of the αC helix, the β4/β5 loop, and an N-terminal extension of the N-lobe (marked in Figure 2B) that includes part of the JM region. Remodeling of these structural elements upon leads to allosteric activation of the acceptor (Zhang et al., 2006). Well-defined electron density was seen for EGFR645-998(K721M) beginning at R653, so this structure lacks only the sequence 645RRRHIVRK652 that immediately follows the transmembrane domain. The extended JM region of the acceptor TKD appears to ‘cradle’ the C-lobe of the donor in the asymmetric dimer of EGFR645-998 (Figure 2A), and the details of this cradling interaction provide satisfying explanations for the alanine scanning mutagenesis results presented in Figure 1B, as described below.
Figure 2. The Acceptor JM Region ‘Cradles’ the C-lobe of the Donor in an Asymmetric TKD Dimer.
A. Cartoon representation of the crystal structure of EGFR645-998 harboring an inactivating K721M mutation. The TKD forms a crystallographic dimer that closely resembles the asymmetric TKD dimer seen for protein that lacks the complete JM region (Zhang et al., 2006), shown in B. The ‘Acceptor’ TKD is colored green, and ‘Donor’ TKD yellow. The N-lobe and C-lobe of donor and acceptor molecules are labeled, as are key helices in the donor C-lobe. Helix αC in the acceptor N-lobe is also marked. The N-terminal part of the JM region of the acceptor ‘cradles’ the donor C-lobe. The short N-terminal α-helix of the acceptor (residues 654-663) projects away from the donor, and makes no direct contacts. B. Structure of the active asymmetric dimer of the wild-type EGFR TKD lacking residues 645-671 from the JM region (PDB ID 2GS6), from Zhang et al. (2006). Features labeled in A are also labeled for this structure. C. Detailed view of side-chains in the acceptor JM region (green) that make contact with the C-lobe of the donor TKD. The orientation for this figure is shown in the cartoon representation in the inset, and the region illustrated is boxed. All side-chains present in the crystal structure from T654-R681 are shown. Those side-chains colored light grey could be replaced by alanine with no effect on ICD activity in Figure 1B (including T669). Side-chains colored green could not be replaced by alanine without significant loss of activity. Residue labels boxed in green correspond to the N-terminal half of the JM activation domain, where alanine substitutions have the greatest effect. Note that the N-terminal helix of the acceptor structure makes no direct contact with the donor.
Although nearly all of the donor/acceptor interactions seen in previous active structures (Zhang et al., 2006) are precisely maintained in the EGFR645-998(K721M) dimer (Figure 2A), it is important to note that the K721M TKD itself adopts an inactive-like conformation (Wood et al., 2004; Zhang et al., 2006). This is not unexpected, since K721 is a catalytically crucial residue in EGFR that must form an ion pair with a glutamate (E738) in the αC helix (and contact ATP) in the normal active kinase (Huse and Kuriyan, 2002). Loss of the K721/E738 ion pair in EGFR645-998(K721M) is likely to disrupt the communication of interactions required for normal allosteric activation upon formation of the asymmetric TKD dimer. Indeed, there are poorly defined regions of electron density within the P-loop (residues 696-699), within the loop that connects strand β3 and the αC helix (residues 724-729) and within the activation loop (residues 833-851), suggesting significant disorder in these regions as a result of reduced restraints on the αC helix in the K721M mutant. Thus, the K721M-mutated acceptor TKD in Figure 2A maintains an inactive-like conformation despite retaining nearly all interactions with the donor that normally promote allosteric activation. This is evident from the slightly different relationships between the N- and C-lobes of each monomer in the structures shown in Figures 2A and 2B. The fact that EGFR645-998(K721M) forms the asymmetric dimer shown in Figure 2 despite adopting an inactive conformation suggests that the JM region plays a crucial role in stabilizing this normally-activating mode of TKD association. A similar situation can be inferred for ErbB-4, where an inhibitor-bound inactive conformation also crystallized as an asymmetric dimer when part of the JM region was included in the protein (Wood et al., 2008), but not when the JM region was absent (Qiu et al., 2008).
The acceptor JM region ‘cradles’ the donor C-lobe to stabilize the asymmetric dimer
As shown in Figure 2C, most of residues 664-671 (T669 is an exception) in the JM region of the acceptor (green) make intimate contact with the C-lobe of the donor (yellow). Consistent with this, alanine substitution at these positions had a strong inhibitory effect on EGFR ICD activation (Figure 1B). Residues 664-671 of the acceptor JM region occlude 488Å2 on the donor C-lobe surface, with a reasonably high shape complementarity (Sc) of 0.61 (Lawrence and Colman, 1993). Alanine substitutions at L664, V665, L668 or S671 will reduce van der Waal’s interactions between the JM region and donor C-lobe (or hydrogen bonding interactions in the case of S671). Replacing E666 with alanine will break electrostatic interactions with R949 in helix I of the donor C-lobe, and may also alter local structure by breaking an E666/R662 ion pair. Similarly, alanine substitution at P667, P670 or G672 could alter local structure (and/or disrupt van der Waal’s interactions) and thus impair JM interactions with the donor C-lobe. The data in Figure 1B show that each of these changes impairs EGFR ICD autophosphorylation as the structure would predict. Of equal importance, where Figure 1B indicates no (or little) inhibitory effect of alanine substitution (T654-E663, and T669), Figure 2C shows that the side-chains in the JM region are not engaged in contacts with the donor C-lobe. These ‘insensitive’ side-chains are colored light gray in Figure 2C.
The α-helix at the N-terminus of the acceptor JM region (residues 653-663) makes no direct contact with the donor C-lobe. Its axis is almost perpendicular to the C-lobe surface in Figures 2A and C, and it makes contacts with symmetry-related molecules in the crystal. This helix is part of a reported binding site for calmodulin that encompasses residues 645-660 (Martin-Nieto and Villalobo, 1998). The independence of this N-terminal helix in the structure presented here, and the disorder of the polybasic region that precedes it (residues 645-653), argue that the reported interactions of the EGFR JM region with calmodulin (Martin-Nieto and Villalobo, 1998) and with negatively-charged membrane surfaces (McLaughlin et al., 2005) could occur coincident with the cradling of the donor C-lobe seen in Figure 2A. Thus, proposed modes of JM-mediated EGFR regulation that involve these interactions (McLaughlin et al., 2005) may occur in parallel with the JM contributions to donor:acceptor interactions seen here.
Effects of Threonine Phosphorylation within the JM Region
The JM region contains two threonines that are known to be phosphorylated in presumed regulatory feedback loops. T654 is phosphorylated by protein kinase C (Hunter et al., 1984), leading to impaired EGF-induced EGFR phosphorylation. We showed previously (Thiel and Carpenter, 2007) that a phosphomimetic mutation at this position (T654D) reduces ICD autophosphorylation, whereas a T654A mutation elevates ICD phosphorylation (Figure 1B). T669 phosphorylation by MAP kinase also negatively regulates EGFR kinase activity (Li et al., 2008; Northwood et al., 1991; Takishima et al., 1991). As shown in Figure 3A, a phosphomimetic T669D mutation again prevents ICD tyrosine phosphorylation, whereas a T669A mutation leads to elevated tyrosine phosphorylation compared to the wild-type ICD (Figures 3A and 1B). Thus, phosphorylation of T669 is likely to exert the same influence on EGFR ICD activity as T654 phosphorylation, supporting a mechanism for EGFR regulation by post-translational modification of T669 within the putative JM activation domain.
Figure 3. Effects of mutations at JM phosphorylation sites.
A. Effect of a phosphomimetic mutation at T669 on ICD tyrosine Phosphorylation. The indicated flag-tagged ICD constructs were transiently expressed in Cos-7 cells. Lysates were subjected to SDS-PAGE and immunoblotting as in Figure 1C, but using anti-Flag to normalize for ICD expression levels.
B. Regulation of constitutively active EGFR-L834R by a JM loss-of-function mutant. 293 cells transiently expressing indicated ICD constructs were lysed and subjected to SDS-PAGE and immunoblotted with anti-EGFR-pY-1173 and anti-Flag.
Since the side-chains of both T654 and T669 are solvent-exposed in Figure 2C, and make no contacts with the donor C-lobe, the inhibitory effect of their phosphorylation does not reflect a simple (direct) disruption of JM-mediated interactions between receptors. In the case of T669, phosphorylation could have an indirect effect, by altering JM structure or stability. For T654 phosphorylation, the structure presented here does not suggest a clear possibility. Interestingly, alignment of EGFR (ErbB-1) and ErbB-3 JM sequences (Figure 1A) shows that the EGFR T669, which is conserved in both ErbB-2 and -4, is an aspartate in ErbB-3, consistent with reports that ErbB-3 has little or no kinase activity (Guy et al., 1994).
Regulation of L834R Tyrosine Phosphorylation by JM Alanine Mutation
Clinical studies of patients with non-small cell lung cancer (NSCLC) have identified a number of mutations in the EGFR TKD that are associated with increased sensitivity to EGFR-targeted tyrosine kinase inhibitors (Sharma et al., 2007). Many of these mutations cause constitutive (ligand-independent) activation of the EGF receptor when studied in reconstituted cellular systems (Choi et al., 2007; Jiang et al., 2005), and appear to do so by disrupting autoinhibitory interactions within the TKD itself (Zhang et al., 2006). The L834R mutation, located in the kinase activation loop, is one of the most clinically frequent and well characterized examples. To determine whether EGFR activation by such mutations requires the JM-mediated interactions shown in Figure 2C, we combined the L834R mutation with a loss-of-function JM region mutation (L680A). A doubly mutated ICD construct containing L834R plus the JM L680A mutation was expressed in Cos-7 cells. Figure 3B shows that kinase activation due to the L834R mutation is blocked by the L680A mutation, indicating that the oncogenic activated (L834R) kinase remains dependent on JM function and, presumably, on formation of the asymmetric TKD dimer.
Activating EGFR JM Mutations in Non-Small Cell Lung Cancer
In addition to the TKD mutations, several publications have described relatively rare EGFR exon 18 JM point mutations in NSCLC patients (Chou et al., 2005; Pallis et al., 2007; Tsao et al., 2005). However, the impact of these mutations on receptor activity was not assessed, and in some cases normal patient tissue was not examined for presence of the mutation. Thus, their relationship to NSCLC is currently not clear. To determine how these patient-derived JM mutations affect receptor kinase activity, equivalent changes were made in ICD constructs and expressed in Cos-7 cells (Figure 4A). The L834R mutation was analyzed in parallel as a positive control. As shown in Figure 4A, most of the patient-derived JM mutations (L664P, P667S, L668P, P670L/S, and N676D) reduced ICD autophosphorylation, suggesting that they may not be relevant in NSCLC. However, two mutations (V665M and L679F) significantly increased ICD tyrosine phosphorylation, to levels similar to those seen with the L834R mutation.
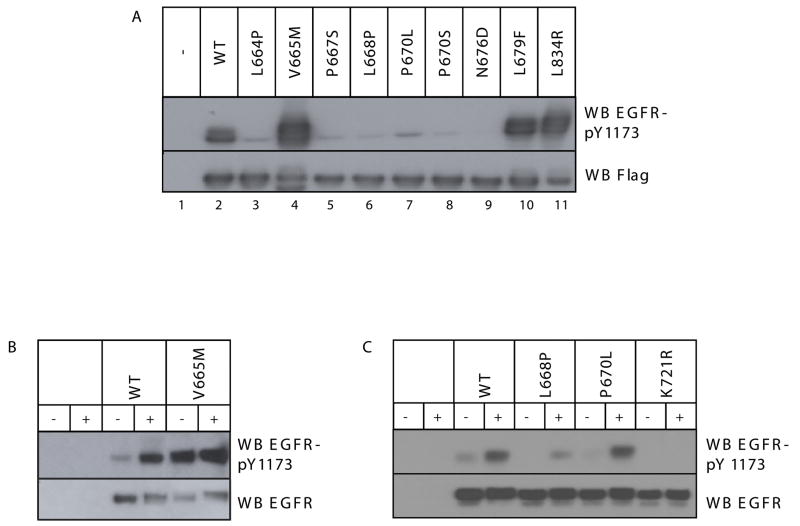
Figure 4. Effects of Clinically-Observed JM Mutations on EGFR Activity.
A. The noted clinically-observed JM mutations were introduced into the Flag-tagged EGFR ICD. The resulting constructs were transiently expressed in Cos-7 cells, and ICD autophosphorylation was assessed as described in Figure 1. B. and C. The V665M, L668P, and P670L mutations were also introduced into full-length EGFR, and the influence on EGF-dependent (+) and EGF-independent (−) EGFR autophosphorylation was assessed as described for Figure 1C. The K721R inactivating mutation was included as a negative control (C).
We also introduced the V665M, L668P and P670L mutations into full-length EGFR, and measured tyrosine phosphorylation of transiently expressed receptor in the absence and presence of EGF (Figure 4B and 4C). Consistent with the ICD studies, the V665M mutation promotes ligand-independent EGFR autophosphorylation so that, even without ligand, V665M EGFR is as heavily phosphorylated as the fully activated wild-type receptor. This effect cannot be explained by differences in cell-surface expression of the mutant compared to wild-type receptor (Supplementary Figure 2A). Thus, the V665M mutation resembles other activating NSCLC mutations in its effects on full-length EGFR (Choi et al., 2007; Jiang et al., 2005), suggesting that it could represent an oncogenic JM mutation with relevance in NSCLC. By contrast, the L668P mutation reduced EGF-induced receptor autophosphorylation, consistent with its effect in the ICD context. The P670L mutation had a less marked negative effect on full-length EGFR than on ICD autophosphorylation, suggesting a possible effect of this proline substitution on ICD stability.
V665M-mutated EGFR Promotes Cellular Transformation and Tumorigenesis
To evaluate the proliferative and oncogenic potential of the V665M mutation, we tested NIH3T3 cells stably expressing wild-type, V665M, L834R, or D813A (catalytically-inactive) EGFR for colony formation in soft agar in the absence and presence of EGF. As shown in Figure 5A, in the presence of EGF the V665M EGFR mutant supports anchorage-independent growth more effectively than the wild-type receptor. Moreover, cells expressing V665M EGFR formed more colonies following EGF addition than cells expressing the well-described NSCLC mutant L834R.
Figure 5. EGFR-V665M is transforming in NIH3T3 cells.
A. NIH3T3 cells stably expressing the indicated wild-type or mutated forms of EGFR (or pBabe-puro vector only) were seeded at a density of 8×103 cells per well in a standard colony forming assay (see Experimental Procedures). Cells were incubated for 3 weeks in the absence or presence of 50ng/ml EGF, and colonies were then counted. The results are presented as the means of 3 experiments ± standard deviation. To control for possible differences in expression level of the mutants, NIH3T3 stably expressing each mutant were lysed and analyzed by SDS-PAGE and immunoblotting with anti-EGFR. B. Upper panel: Example of tumors isolated from nude mice 3 months after s.c. injection with NIH3T3 cells stably expressing wild-type or V665M EGFR or the pBabe-puro vector. Lower panel: Volumes of tumors derived from the cells indicated above were evaluated 3 months after s.c. injection. Open circles represent single mice with a total of 4 mice per cell type analyzed.
To determine whether the observed increase in anchorage-independent growth by the V665M mutation reflects oncogenic transformation, we injected NIH3T3 cells stably expressing equivalent levels of wild-type or V665M EGFR subcutaneously into nude mice. The results, shown in Figure 5B, demonstrate that 3 months after injection, cells expressing V665M EGFR formed more (and larger) tumors than cells expressing wild-type EGFR, regardless of the number of cells injected. These results indicate that the JM activation domain is a significant factor in the regulation of EGF receptor kinase activity at the biological, as well as biochemical, level. The appearance of the V665M mutation in NSCLC patients also indicates that mutations in the JM activation domain are clinically important.
Mechanism of EGFR Activation by the V665M Mutation
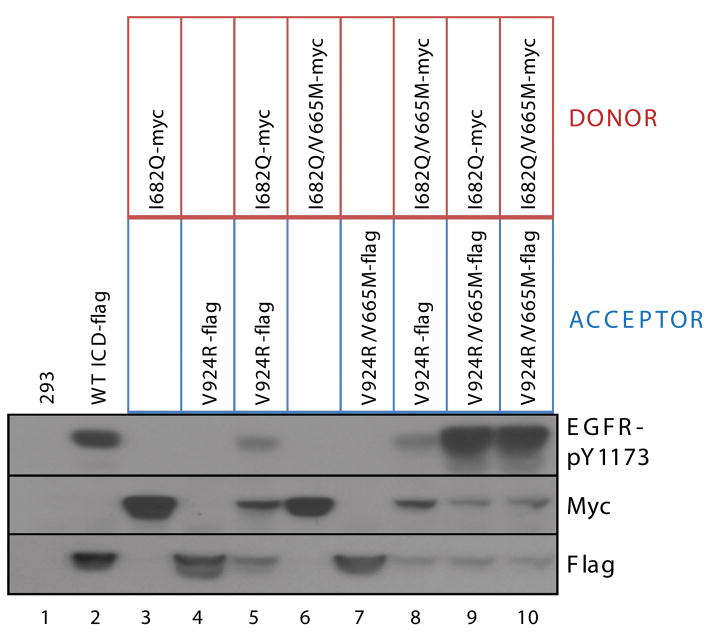
In mechanism for EGFR TKD activation proposed by Zhang et al. (2006), the C-lobe of the donor contacts the N-lobe of the acceptor in the asymmetric dimer (Figure 2B), and promotes conformational changes in the acceptor that lead to its allosteric activation. A crucial question is whether the V665M mutation promotes constitutive EGFR TKD activation by augmenting the function of the donor or acceptor. We took advantage of mutations (Zhang et al., 2006) that force receptor molecules to act solely as either donor monomers (I682Q) or acceptor monomers (V924R) to ask whether the V665M-mutated EGFR ICD functions more effectively than wild-type as donor, acceptor, or both. In the experiment shown in Figure 6, the donor ICD is myc-tagged, and the acceptor ICD is FLAG-tagged. Alone, neither becomes autophosphorylated (lanes 3 and 4), but substantial activity is seen when the two are co-expressed (lane 5). Lanes 8–10 (Figure 6) demonstrate that maximal ICD autophosphorylation (greater than wild-type levels) is seen when the V665M mutation is present in the acceptor monomer (V924R/V665M), regardless of whether it is co-expressed with the I682Q donor or the doubly-mutated (I682Q/V665M) donor. If the V924R/V665M acceptor is expressed without a donor monomer (Figure 6: lane 7), there is no activation, indicating that the large increase in activation observed in lanes 9 and 10 requires donor:acceptor interaction. In contrast, coexpression of an I682Q/V665M donor with a V924R acceptor (lane 8) does not increase tyrosine phosphorylation compared to the control (lane 5). These data establish that the V665M JM mutation enhances the capacity of the ICD to act as an acceptor monomer, but does not greatly influence its ability to function as a donor. This is consistent with our previous finding that an intact JM region is required for EGFR ICD to function as an acceptor in the asymmetric dimer (Thiel and Carpenter, 2007). The V665M mutation seen in NSCLC appears to promote JM function in this context.
Figure 6. The V665M Mutation must be present in the Acceptor Molecule to Activate EGFR.
ICD constructs containing mutations that limit them to functioning only as a donor (I682Q) or acceptor (V924R) in the asymmetric TKD dimer were used to assess the mechanism by which the V665M mutation activates EGFR. The noted combinations of ICD constructs were coexpressed in 293 cells as indicated, and lysates were subjected to SDS-PAGE and immunoblotting for phospho-EGFR (with anti-EGFR-pY1173) and for protein levels with anti-Myc, and anti-Flag as appropriate. Autophosphorylation levels are greater than wild-type (lane 2) only when the V665M mutation is present in the acceptor TKD (lanes 9 and 10).
Structural explanation for EGFR activation by V665M and L679F mutations
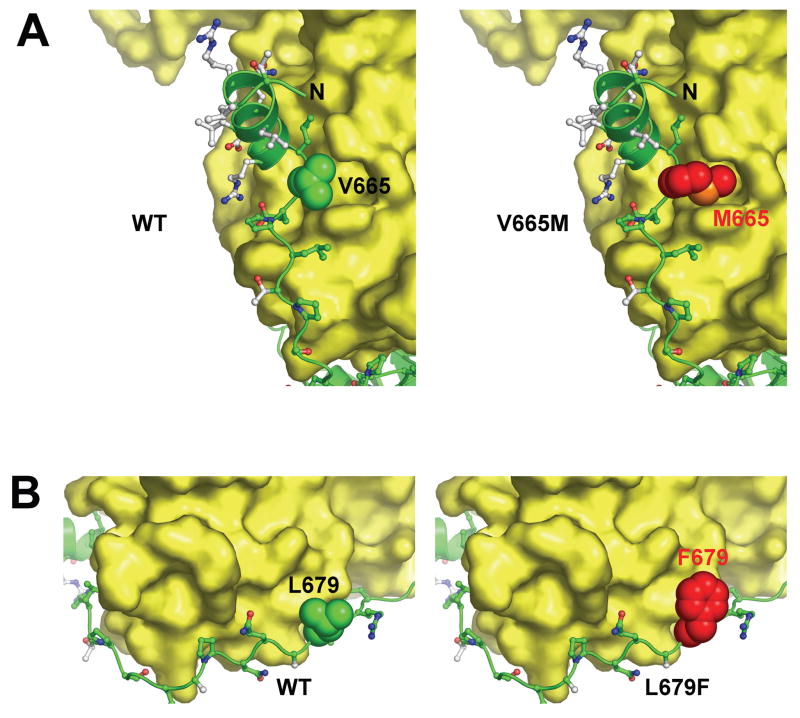
Details of the interface between the JM region of the acceptor and C-lobe of the donor (Figure 2C) suggest a possible structural basis for the EGFR activation by the V665M and L679F mutations found in NSCLC patients. The side-chain of V665 projects into a cavity on the surface of the C-lobe that is lined by aliphatic portions of Q788 and Y789 from helix E and Q825 from the β7/β8 loop (between the catalytic loop and activation loop of the TKD). The V665 side-chain does not fill this cavity (Figure 7A). However, substituting V665 with a methionine – as modeled in the right-hand panel of Figure 7A – would fill the cavity completely, and is likely to stabilize the association of the acceptor’s JM region with the donor C-lobe at this location. We also considered the possibility that projection of a methionine side-chain into the cavity shown in Figure 7A might alter the positions of its lining residues (Q788, Y789 and Q825). In particular, since Q825 lies between two key regulatory elements in the kinase, such alterations could allosterically regulate kinase activity. However, in overlays of all known active and inactive EGFR TKD structures (including the structure described here), the positions of these three residues do not change. A direct allosteric effect of the V665M mutation therefore seems unlikely.
Figure 7. Mechanism of Activating JM Mutations.
A. Contacts between the acceptor JM domain and donor C-lobe are shown with the V665 side-chain represented as space-filling spheres. The V665 side-chain fails to fill a cavity on the donor C-lobe formed by the side-chains of Q788, Y789 and Q825. However, as shown in the right-hand panel, a methionine (red) at position 665 – modeled for this figure - would completely fill the cavity, increasing van der Waal’s contacts between acceptor and donor. B. Similarly, the L679F mutation is likely to promote interactions between the JM region (green) and the donor C-lobe, as indicated by modeling a phenylalanine at this position.
A second activating mutation in the JM region among those found in NSCLC patients is seen at L679, in a region that has been visualized in all active EGFR TKD structures (Stamos et al., 2002; Yun et al., 2007; Zhang et al., 2006), and is unaltered in the structure presented here. Mutation of L679 to phenylalanine could improve packing of this region of the JM region with a PQPP sequence between helices αG and αH in the C-lobe of the donor (Figure 7B). Again, this region is identically placed in the active and inactive configurations of the kinase.
CONCLUSIONS
Crystallographic studies together with mutagenesis and biochemical data have shown that for several RTKs (c-Kit, EphB2, and Flt3) the intracellular JM region plays an autoinhibitory role, sterically hindering substrate access to the nucleotide binding pocket (Chan et al., 2003; Griffith et al., 2004; Wybenga-Groot et al., 2001). In these RTKs, phosphorylation of key tyrosines within the JM region results in a conformational shift that reverses the autoinhibition - leading to kinase activation (Binns et al., 2000; Mol et al., 2003).
We show here that the JM region of the EGF receptor instead plays an activating (rather than autoinhibitory) role. Alanine substitutions at most positions in the C-terminal half of the JM region (encoded by exon 18) lead to a loss of kinase activity. We term this part of the JM region the JM activation domain (JMAD). By solving the crystal structure of an EGFR TKD with its JM region intact, we showed that the entire 19-amino acid JMAD (residues 664-682) of an acceptor in the asymmetric TKD dimer makes intimate contact with the C-lobe of the donor. The most straightforward explanation for how the JMAD contributes to EGFR activation is that it enhances formation of the asymmetric dimer, thus promoting allosteric activation of the acceptor TKD. Indeed, although wild-type EGFR672-998 is monomeric at concentrations up to 50–100μM [data not shown and (Zhang et al., 2006)], adding the JM region causes EGFR645-998 to aggregate in AUC studies (not shown). This might reflect JMAD-stabilized formation of head-to-tail TKD polymers similar to those seen in crystals. Phosphorylation of T669 within the JMAD is likely to disrupt its local structure, thus inhibiting EGFR activity by reducing the strength of acceptor/donor interactions.
Identification of an important regulatory region within the ICD but outside the TKD itself is of particular significance, because regions outside the kinase domain are rarely examined for mutations in clinical screens (Sharma et al., 2007). Where broader screens have been performed, a limited number of apparently rare JM mutations in clinical tumor samples have been reported, but not characterized. We found that two of these previously-reported clinical EGFR JM mutations (V665M and L679F) activate the receptor. In both cases, the degree of EGFR activation is similar to that seen for well-characterized oncogenic mutations (such as L834R) in the TKD itself (Sharma et al., 2007). Biochemical studies and structural arguments suggest that the V665M mutation stabilizes acceptor/donor interactions. The existence of such a clinically-observed mutation within the JMAD that we also show to be activating and transforming highlights the biological importance of this domain in regulating the EGF receptor kinase activity and cellular responsiveness. This activation domain is encoded by the same exon as a significant portion of the N-lobe of the TKD, and could be considered as a part of the TKD proper.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, Cell lines, and Plasmids
NIH3T3, Cos-7, and 293 cells were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals) at 37°C at 5% CO2. EGF was purchased from R&D Systems.
Monoclonal anti-Flag was purchased from Sigma. Polyclonal anti-EGFR pY1173 was purchased from Santa Cruz Biotechnologies. Polyclonal anti-EGFR was purchased from Millipore. Anti-EGFR Ab-3 was purchased from LabVision. The respective HRP-conjugated secondary antibodies were purchased from Zymed. Alexa-488 goat-anti mouse was purchased from Molecular Probes. For immunoblot detection, ECL reagent from Perkin Elmer Life Sciences was used. Stripping and reprobing of blots was performed according to manufacturer’s recommendations.
The plasmid encoding EGFR-ICD-flag was cloned as previously described (Thiel and Carpenter, 2007) and its sequence verified. Site-directed mutagenesis was employed to make all mutants with the use of PfuUltra polymerase (Stratagene) (Table S1 for primer sequences). Flag-EGFR plasmid was a gift from Tony Burgess (Ludwig Institute for Cancer Research) and was fully sequenced. The pBABE retroviral vectors used in EGF receptor expression for soft agar assays were purchased from Addgene. Site-directed mutagenesis of pBABE-EGFR was employed to make pBABE-EGFR-V665M. All mutations were confirmed by fully sequencing each construct.
Cell Culture and Transfections
Transient transfection for expression in mammalian cells was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. 2μg of expression plasmid was used per 60mm dish. Transient expression of full-length EGF receptor in NIH3T3 cells was carried out using a DNA:Fugene6 (Roche) ratio of 1.5μg:4.5μl, according to the manufacturer’s instructions. To assess ligand-dependent EGFR activation, transfected cells were serum starved overnight and treated with 50ng/ml EGF.
Cell lysis and Immunoblotting
Cells at 80–90% confluence were either untreated or treated with EGF as indicated, and then washed with phosphate buffered saline. Cells were scraped and lysed in ice-cold lysis buffer containing 1% Triton, 10% glycerol, 50mM Hepes, pH 7.2, and 100mM NaCl, 1mM EDTA, 1mM sodium orthovanadate, 1mM phenylmethylsulfonylfluoride, 10ug/mL aprotinin, and 25ug/mL leupeptin. Lysates were cleared by centrifugation at 13,000 rpm for 10 minutes at 4°C. SDS sample buffer was added to lysates and samples were boiled for 5 minutes. Samples were then subjected to SDS-PAGE on 8% polyacrylamide gels, transferred to PVDF and immunoblotted.
Retrovirus-mediated gene transduction and colony formation assay
The pBABE-puro retroviral vector containing the coding sequences for wild-type EGFR, V665M, L834R, or D813A were used to produce virus-containing supernatants from transfected Phoenix cells. Viral supernatant was used to transduce NIH3T3 cells in the presence of 5μg/ml polybrene. Equal amounts of retrovirus containing wild-type EGFR, V665M, L834R, D813A, or vector alone (m.o.i.=0.1) were used for infection. Cells expressing equal concentrations of receptor were sorted via FACS by staining with a non-ligand interfering ectodomain antibody (anti-EGFR Ab-3, LabVision) and then seeded at 8×103 cells per well in the 0.4% top layer of a 12-well plate. Cells were cultured in 10% FBS with or without 50ng/ml EGF. Medium was changed every two days and colonies were counted at 3 weeks using an Oxford Optronix Gelcount.
In vivo tumorigenicity
To determine the capacity of NIH3T3 cells to form tumors in vivo when stably expressing pBabe-puro vector, wild-type, or V665M EGFR, nude mice (4 mice per cell type) received four dorsal s.c. injections, one in each quadrant, with either 1.25×105, 2.5×105, 5×105, or 106 tumor cells in 200 μl PBS. Tumor growth was then analyzed over time, as plotted in Figure 5. Three months post tumor cell injection, mice were sacrificed and the number and size of tumors were evaluated. Tumor volumes were calculated using the following formula: tumor volume (mm3) = (length × width2)/2 (Chen et al., 2005). All animal protocols were approved by Vanderbilt University Medical Center Institutional Animal Care and Use Committee.
Expression and purification of EGFR645-998
A PCR-amplified cDNA fragment encoding amino acids 645-998 of EGFR was subcloned into pFastBac1, modifying the amino terminus to include a hexa-histidine tag in the N-terminal sequence MHHHHHHGR… (where the final R is R645 of EGFR). The K721M mutation was incorporated using the Quikchange kit (Stratagene). Baculovirus generation and protein expression in Sf9 cells employed the Bac-to-Bac expression system (Invitrogen) as recommended by the manufacturer. Baculovirus-infected Sf9 cells were harvested 3 days after infection, were lysed by sonicating in 50mM Tris, pH 8.0, 150mM NaCl, 10% glycerol, containing 5mM β-mercaptoethanol and 1x protease inhibitor cocktail (Roche). Cell debris was removed by centrifugation at 40,000 × g for 30 minutes. The supernatant was incubated with Ni-NTA agarose (Qiagen) for 1 hour at 4°C and bound protein was eluted using stepwise imidazole washes. Eluted protein was bound to a cation exchange column (S2: Bio-Rad) in 20mM Tris pH 8.0, 200mM NaCl and 2mM DTT and eluted with a linear gradient to 1M NaCl in the same buffer. Fractions containing EGFR TKD were diluted in 10mM phosphate buffer (pH 8), loaded on a hydroxyapatite column (CHT2-I: Bio-Rad), and eluted with a linear gradient to 500mM Na/K phosphate (pH 8.0). Fractions containing protein were concentrated and gel filtered on a Superdex 200 column (GE Healthcare) in 20mM Tris pH 8.0, 250mM NaCl and 2mM DTT.
Crystallization and structure determination
EGFR645-998(K721M) was concentrated to 5mg/ml and crystallized using the hanging drop method. Crystals (~10μm ×10μm ×40μm) grew in 1 day in 100mM Tris pH8.5, 100mM KCl and 10% PEG3350 at 21°C. Crystals were cryo-protected in mother liquor containing 15% PEG and 20% glycerol and frozen in liquid nitrogen. Diffraction data were collected at the CHESS F1 beamline, where crystals diffracted to 2.8Å (Table 1). Data were processed using HKL2000 (Otwinowski and Minor, 1997), and the structure was solved by molecular replacement (MR) using Phaser implemented in the CCP4 package (Storoni et al., 2004). The N- and C-lobes of the inactive EGFR TKD (PDB code 2GS7) were found in simultaneous but independent searches. Density for the JM region of EGFR645-998(K721M) was immediately visible, and the structure of the protein was built and rebuilt using the program COOT (Emsley and Cowtan, 2004). Manual model building was alternated with successive rounds of refinement using the programs DM and Refmac in CCP4 (CCP4 (Collaborative Computational Project Number 4), 1994), and inspection of composite omit maps generated using CNS (Brunger et al., 1998). TLS refinement was incorporated in the final stages (Winn et al., 2001). Figures for publication were made using MacPymol (DeLano, 2002).
Supplementary Material
Supplementary Figure 1. Scanning alanine mutagenesis of the JM region using ICD-flag constructs. Cos-7 cells transiently expressing wild-type or mutant ICD-flag constructs were analyzed by SDS-PAGE and immunoblotting with anti-phosphotyrosine and anti-Flag (as described in Experimental Procedures). The blots were then analyzed densitometrically for both phosphotyrosine and flag-epitope expression.
Supplementary Figure 2. Cell Surface Expression of Full-length EGF Receptor Mutants. A. 293 cells transiently expressing equivalent levels of N-terminal Flag-tagged WT, V665A, L680A, or V665M EGF receptors were treated with Alexa-EGF and analyzed using flow cytometry. Geometric mean fluorescence values were collected and Alexa-EGF binding was quantified. All mutants were normalized to WT. B. NIH3T3 cells transiently expressing equivalent levels of the constructs indicated above were stained with anti-EGFR followed by Alexa488-labeled anti-mouse. Cells were fixed with 4%PFA and mounted with glass coverslips. Images were acquired using a Zeiss fluorescence microscope (see Supplemental Experimental Procedures).
Acknowledgments
M.R.-B. was supported by U.S. Army Medical Research and Materiel Command fellowship W81XWH-06-1-0758. A.P. was supported by NIH grant DK065138, and by a Merit award from the Department of Veterans Affairs. S.H.C. and K.M. were supported by predoctoral fellowships from the Great Rivers Affiliate of the American Heart Association (Awards 0715334U and 0715300U). D.A. was supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Fund and by NIH Training Program in Developmental Biology (T32HD007516). Work in the Lemmon laboratory was supported by NIH grant 5R01-CA079992. This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the NSF and the NIGMS under NSF award DMR-0225180, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award RR-01646 from the NIH. Thanks to Dr. Doug Sheffler and Dr. Allison Limpert for image preparation assistance. The atomic coordinates for EGFR645-998(K721M) have been deposited in the RCSB Protein Data Bank under ID code 3GOP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aifa S, Aydin J, Nordvall G, Lundstrom I, Svensson SPS, Hermanson O. A basic peptide within the juxtamembrane region is required for EGF receptor dimerization. Exp Cell Res. 2005;302:108–114. doi: 10.1016/j.yexcr.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Bao J, Alroy I, Waterman H, Schejter ED, Brodie C, Gruenberg J, Yarden Y. Threonine phosphorylation diverts internalized epidermal growth factor receptors from a degradative pathway to the recycling endosome. J Biol Chem. 2000;275:26178–26186. doi: 10.1074/jbc.M002367200. [DOI] [PubMed] [Google Scholar]
- Binns KL, Taylor PP, Sicheri F, Pawson T, Holland SJ. Phosphorylation of Tyrosine Residues in the Kinase Domain and Juxtamembrane Region Regulates the Biological and Catalytic Activities of Eph Receptors. Mol Cell Biol. 2000;20:4791–4805. doi: 10.1128/mcb.20.13.4791-4805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chan PM, Ilangumaran S, La Rose J, Chakrabartty A, Rottapel R. Autoinhibition of the Kit receptor tyrosine kinase by the cytosolic juxtamembrane region. Mol Cell Biol. 2003;23:3067–3078. doi: 10.1128/MCB.23.9.3067-3078.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Su Y, Fingleton B, Acuff H, Matrisian LM, Zent R, Pozzi A. Increased plasma MMP9 in integrin alpha1-null mice enhances lung metastasis of colon carcinoma cells. Int J Cancer. 2005;116:52–61. doi: 10.1002/ijc.20997. [DOI] [PubMed] [Google Scholar]
- Choi SH, Mendrola JM, Lemmon MA. EGF-independent activation of cell-surface EGF receptors harboring mutations found in gefitinib-sensitive lung cancer. Oncogene. 2007;26:1567–1576. doi: 10.1038/sj.onc.1209957. [DOI] [PubMed] [Google Scholar]
- Chou TY, Chiu CH, Li LH, Hsiao CY, Tzen CY, Chang KT, Chen YM, Perng RP, Tsai SF, Tsai CM. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11:3750–3757. doi: 10.1158/1078-0432.CCR-04-1981. [DOI] [PubMed] [Google Scholar]
- Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Griffith J, Black J, Faerman C, Swenson L, Wynn M, Lu F, Lippke J, Saxena K. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol Cell. 2004;13:169–178. doi: 10.1016/s1097-2765(03)00505-7. [DOI] [PubMed] [Google Scholar]
- Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway KL., 3rd Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci U S A. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Hobert M, Friend L, Carlin C. The epidermal growth factor receptor juxtamembrane domain has multiple basolateral plasma membrane localization determinants, including a dominant signal with a polyproline core. J Biol Chem. 2002:277. doi: 10.1074/jbc.M104646200. [DOI] [PubMed] [Google Scholar]
- Heisermann GJ, Wiley HS, Walsh BJ, Ingraham HA, Fiol CJ, Gill GN. Mutational removal of the Thr669 and Ser671 phosphorylation sites alters substrate specificity and ligand-induced internalization of the epidermal growth factor receptor. J Biol Chem. 1990;265:12820–12827. [PubMed] [Google Scholar]
- Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- Hunter T, Ling N, Cooper JA. Protein kinase C phosphorylation of the EGF receptor at a threonine residue close to the cytoplasmic face of the plasma membrane. Nature. 1984;311:480–483. doi: 10.1038/311480a0. [DOI] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Jiang J, Greulich H, Jänne PA, Sellers WR, Meyerson M, Griffin JD. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65:8968–8974. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- Kil SJ, Hobert M, Carlin C. A leucine-based determinant in the epidermal growth factor receptor juxtamembrane domain is required for the efficient transport of ligand-receptor complexes to lysosomes. J Biol Chem. 1999;274:3141–3150. doi: 10.1074/jbc.274.5.3141. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, Yoshimoto K, King JC, Nghiemphu P, Yuza Y, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009 Oct 31; doi: 10.1016/j.yexcr.2008.10.024. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Huang Y, Jiang J, Frank SJ. ERK-dependent threonine phosphorylation of EGF receptor modulates receptor downregulation and signaling. Cell Signal. 2008;20:2145–2155. doi: 10.1016/j.cellsig.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Martin-Nieto J, Villalobo A. The human epidermal growth factor receptor contains a juxtamembrane calmodulin-binding site. Biochemistry. 1998;37:227–236. doi: 10.1021/bi971765v. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Smith SO, Hayman MJ, Murray D. An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J Gen Physiol. 2005;126:41–53. doi: 10.1085/jgp.200509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Lim KB, Sridhar V, Zou H, Chien EY, Sang BC, Nowakowski J, Kassel DB, Cronin CN, McRee DE. Structure of a c-kit product complex reveals the basis for kinase transactivation. J Biol Chem. 2003;278:31461–31464. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]
- Morrison P, Chung KC, Rosner MR. Mutation of Di-leucine residues in the juxtamembrane region alters EGF receptor expression. Biochemistry. 1996;35:14618–14624. doi: 10.1021/bi961630+. [DOI] [PubMed] [Google Scholar]
- Northwood IC, Gonzalez FA, Wartmann M, Raden DL, Davis RJ. Isolation and characterization of two growth factor-stimulated protein kinases that phosphorylate the epidermal growth factor receptor at threonine 669. J Biol Chem. 1991;266:15266–15276. [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pallis AG, Voutsina A, Kalikaki A, Souglakos J, Briasoulis E, Murray S, Koutsopoulos A, Tripaki M, Stathopoulos E, Mavroudis D, et al. ‘Classical’ but not ‘other’ mutations of EGFR kinase domain are associated with clinical outcome in gefitinib-treated patients with non-small cell lung cancer. Br J Cancer. 2007;97:1560–1566. doi: 10.1038/sj.bjc.6604068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Tarrant MK, Choi SH, Sathyamurthy A, Bose R, Banjade S, Pal A, Bornmann WG, Lemmon MA, Cole PA, et al. Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Sergina NV, Moasser MM. The HER family and cancer: emerging molecular mechanisms and therapeutic targets. Trends Mol Med. 2007;13:527–534. doi: 10.1016/j.molmed.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Song Jae Kil CC. EGF receptor residues Leu(679), Leu(680) mediate selective sorting of ligand-receptor complexes in early endosomal compartments. J Cell Physiol. 2000:185. doi: 10.1002/1097-4652(200010)185:1<47::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Takishima K, Griswold-Prenner I, Ingebritsen T, Rosner MR. Epidermal growth factor (EGF) receptor T669 peptide kinase from 3T3-L1 cells is an EGF-stimulated “MAP” kinase. Proc Natl Acad Sci U S A. 1991;88:2520–2524. doi: 10.1073/pnas.88.6.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc Natl Acad Sci U S A. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- Walton GM, Chen WS, Rosenfeld MG, Gill GN. Analysis of deletions of the carboxyl terminus of the epidermal growth factor receptor reveals self-phosphorylation at tyrosine 992 and enhanced in vivo tyrosine phosphorylation of cell substrates. J Biol Chem. 1990;265:1750–1754. [PubMed] [Google Scholar]
- Welsh JB, Gill GN, Rosenfeld MG, Wells A. A negative feedback loop attenuates EGF-induced morphological changes. J Cell Biol. 1991;114:533–543. doi: 10.1083/jcb.114.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Wood ER, Shewchuk LM, Ellis B, Brignolac P, Brasheard RL, Caferroe TR, Dickerson SH, Dickson HD, Donaldson KH, Gaul M, et al. 6-ethynylthieno[3,2-d]- and 6-ethynylthieno[2,3-d]pyrimidin-4-anilines as tunable covalent modifiers of ErbB kinases. Proc Natl Acad Sci U S A. 2008;105:2773–2778. doi: 10.1073/pnas.0708281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- Wybenga-Groot LE, Baskin B, Ong SH, Tong J, Pawson T, Sicheri F. Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell. 2001;106:745–757. doi: 10.1016/s0092-8674(01)00496-2. [DOI] [PubMed] [Google Scholar]
- Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Scanning alanine mutagenesis of the JM region using ICD-flag constructs. Cos-7 cells transiently expressing wild-type or mutant ICD-flag constructs were analyzed by SDS-PAGE and immunoblotting with anti-phosphotyrosine and anti-Flag (as described in Experimental Procedures). The blots were then analyzed densitometrically for both phosphotyrosine and flag-epitope expression.
Supplementary Figure 2. Cell Surface Expression of Full-length EGF Receptor Mutants. A. 293 cells transiently expressing equivalent levels of N-terminal Flag-tagged WT, V665A, L680A, or V665M EGF receptors were treated with Alexa-EGF and analyzed using flow cytometry. Geometric mean fluorescence values were collected and Alexa-EGF binding was quantified. All mutants were normalized to WT. B. NIH3T3 cells transiently expressing equivalent levels of the constructs indicated above were stained with anti-EGFR followed by Alexa488-labeled anti-mouse. Cells were fixed with 4%PFA and mounted with glass coverslips. Images were acquired using a Zeiss fluorescence microscope (see Supplemental Experimental Procedures).