Abstract
The retinoic acid–inducible gene I (RIG-I) and melanoma differentiation–associated antigen 5 (MDA-5) helicases sense viral RNA in infected cells and initiate antiviral responses such as the production of type I IFNs. Here we have shown that RIG-I and MDA-5 also initiate a proapoptotic signaling pathway that is independent of type I IFNs. In human melanoma cells, this signaling pathway required the mitochondrial adapter Cardif (also known as IPS-1) and induced the proapoptotic BH3-only proteins Puma and Noxa. RIG-I– and MDA-5–initiated apoptosis required Noxa but was independent of the tumor suppressor p53. Triggering this pathway led to efficient activation of mitochondrial apoptosis, requiring caspase-9 and Apaf-1. Surprisingly, this proapoptotic signaling pathway was also active in nonmalignant cells, but these cells were much less sensitive to apoptosis than melanoma cells. Endogenous Bcl-xL rescued nonmalignant, but not melanoma, cells from RIG-I– and MDA-5–mediated apoptosis. In addition, we confirmed the results of the in vitro studies, demonstrating that RIG-I and MDA-5 ligands both reduced human tumor lung metastasis in immunodeficient NOD/SCID mice. These results identify an IFN-independent antiviral signaling pathway initiated by RIG-I and MDA-5 that activates proapoptotic signaling and, unless blocked by Bcl-xL, results in apoptosis. Due to their immunostimulatory and proapoptotic activity, RIG-I and MDA-5 ligands have therapeutic potential due to their ability to overcome the characteristic resistance of melanoma cells to apoptosis.
Introduction
The elimination of dangerous host cells is essential to avoid disease. Cancer cells and virus-infected cells are examples for potentially dangerous host cells and they share certain properties: both express specific antigens, and both need to evade immune and nonimmune control mechanisms in order to persist (1, 2). To eliminate virus-infected cells, the host combines two defense strategies: (a) in infected host cells, receptor-mediated sensing of virus triggers endogenous immune responses or apoptosis; (b) in immune cells, sensing of virus activates innate and adaptive immune responses, protecting neighboring cells and stimulating cytotoxic immune cells such as NK cells and CD8+ T cells, which kill infected cells via externally induced cell lysis and apoptosis. The same type of responses are sought for tumor control. Therefore, an effective approach to tumor therapy may be achievable using components that trigger these antiviral responses.
Melanoma, the most aggressive form of skin cancer, is particularly amenable to immunotherapy. Melanoma-associated antigens are well defined; treatment with immunomodulatory substances such as IFN-α and IL-2 is part of established protocols; and new immunotherapeutic strategies are being evaluated, but so far with limited success (3–5). Melanoma cells are remarkably resistant to apoptosis induction by standard cytotoxic therapies, and there is an urgent need for new therapeutics that effectively activate apoptosis in melanoma cells.
Antiviral responses are initiated through the detection of viral nucleic acids by innate pattern recognition receptors. TLRs and RIG-I–like helicases (RLHs), comprising retinoic acid–inducible gene I (RIG-I), melanoma differentiation–associated antigen 5 (MDA-5), and LGP2 (laboratory of genetics and physiology 2), are the two major families of receptors for detecting RNA viruses. RIG-I is localized in the cytosol and recognizes 5′-triphosphate RNA (pppRNA) generated by viral RNA polymerases in the cytosol of cells. Of note, pppRNA is also generated physiologically in the nucleus, but due to processing (splicing, adding a 5′ cap, additional modifications), it loses its RIG-I ligand activity before it is released to the cytosol (6, 7). Active pppRNA can be generated by in vitro transcription. While long double-stranded RNA (300–2,000 bp) even in the absence of a 5′-triphosphate moiety seems to activate RIG-I, MDA-5 preferentially detects very long double-stranded RNA as present during replication of certain viruses (8). Polyinosinic-polycytidylic acid [poly(I:C)], a synthetic and artificial mimic of long double-stranded RNA, is a strong activator of MDA-5 (9). Upon recognition of RNA ligands, RIG-I and MDA-5 bind to the adapter protein IFN-β promoter stimulator 1 (IPS-1) (also known as CARDIF, MAVS, or VISA) located in the outer mitochondrial membrane (10–13). The interactions of RIG-I or MDA-5 with IPS-1 initiate signaling pathways that elicit the activation of transcription factors including IFN regulatory factor 3 (IRF-3) and NF-κB, resulting in IFN production, activation of NF-κB target genes, and the secondary induction of IFN-stimulated genes (14).
We hypothesized that triggering RIG-I and MDA-5 with their cognate RNA ligands will not only stimulate type I IFN and NF-κB–dependent inflammatory cytokines, but at the same time may trigger endogenous apoptosis as part of an antiviral host response. Tumor cells could be more susceptible to this kind of death signal than nonmalignant cells, since many alterations required for tumor formation can also result in increased vulnerability to certain apoptotic stimuli (a phenomenon termed “synthetic lethality” or “oncogene addiction”) (15, 16). Increased immunostimulation and sensitivity toward apoptosis cooperate, and the feasibility of a combined chemo- and immunotherapy is currently being investigated in clinical applications (17). The results presented here support this hypothesis. We identify an IFN-independent proapoptotic signaling pathway that is activated by RIG-I and MDA-5 in normal cells as well as in tumor cells. We demonstrate that this pathway leads to apoptosis in melanoma cells, whereas primary cells are less susceptible due to an intact Bcl-xL counterregulatory pathway, resulting in preferential tumor cell death in vitro and in immunodeficient mice in vivo.
Results
pppRNA and poly(I:C) induce apoptosis in melanoma cells.
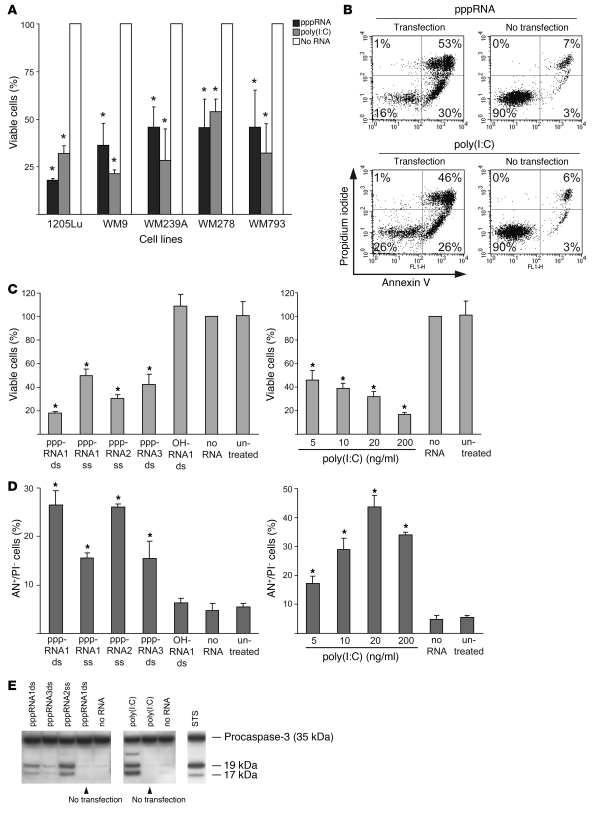
We tested the ability of RIG-I and MDA-5 ligands to induce cell death in human melanoma cell lines. Five cell lines derived from advanced melanomas (vertical growth phase or metastatic origin) were analyzed. Activation of RIG-I and MDA-5 by pppRNA1 and poly(I:C) strongly reduced viability from 100% in controls to 20%–50% within 24 hours (Figure 1A). Viability was reduced due to induction of apoptosis as determined by staining with annexin V. Apoptosis strictly required intracellular delivery, as neither pppRNAs nor poly(I:C) without transfection were active (Figure 1B). Different pppRNAs were tested, and all reduced cell viability (Figure 1C). The 5′-triphosphate moiety was required, since synthetic RNAs carrying a free OH group at the 5′ end (e.g., OH-RNA1) had no effect (Figure 1C, left panel). Strong dose-dependent reduction of viability was observed for poly(I:C) (Figure 1C, right panel). Reduced viability was reflected in an increased number of cells undergoing apoptosis (Figure 1D). Confirming the onset of apoptosis, caspase-3 was activated in cells transfected with pppRNAs or poly(I:C) but not in cells exposed to pppRNA or poly(I:C) in the absence of transfection reagent (Figure 1E). Together, these results show high sensitivity of human melanoma cell lines toward apoptosis induction by pppRNAs or poly(I:C) when delivered to the cytosol.
Figure 1. pppRNA and poly(I:C) induce apoptosis in melanoma cells.
(A) The viability of different melanoma cell lines was determined after transfection of in vitro–transcribed RNA (pppRNA) or poly(I:C) (20 ng/ml). Cells were analyzed 17 (pppRNA) or 24 hours [poly(I:C)] after transfection. Viability of cells treated with transfection reagent alone (mock-transfected cells; No RNA) was set to 100%. *P ≤ 0.05 compared with the same cell line treated with transfection reagent alone. (B) Fluorescence-activated cell sorting (FACS) analysis of apoptotic 1205Lu cells treated with pppRNA or poly(I:C) (200 ng/ml) for 24 hours either complexed with a liposomal transfection reagent (Transfection) or not (No transfection). Percentages indicate the portion of cells in each quadrant that defines annexin V– or propidium iodide–positive or –negative cells. Results are representative of 3 independent experiments. (C) Left: Viability of the metastatic melanoma cell line 1205Lu was measured 24 hours after transfection of different short in vitro–transcribed RNAs (pppRNA1–3) in double-stranded (ds) or single-stranded (ss) form or synthetic unconjugated RNAs (OH) with the same sequence. Sequences of pppRNAs are shown in Methods. Right: Viability of 1205Lu cells transfected with different doses of poly(I:C) (20–200 ng/ml) for 24 hours. Viability of mock-transfected cells was set to 100%. (D) FACS analysis of apoptotic 1205Lu cells treated with pppRNA or poly(I:C) as described in C. Annexin V–positive and propidium iodide–negative cells (AN+/PI–) are depicted. In A, C, and D, the mean ± SD of 3 independent experiments is shown. *P ≤ 0.05 compared with mock-transfected cells in C and D. (E) pppRNAs (pppRNA1–3) or poly(I:C) (5 ng/ml) were used with or without transfection reagent, and 1205Lu cells were analyzed 12 (pppRNA) or 24 hours [poly(I:C)] after treatment. Activation of caspase-3 was assessed by immunoblotting with an antibody specific for procaspase-3 (35 kDa) and its active cleaved subunits (17, 19 kDa). WM9 melanoma cells treated with 1 μM staurosporine (STS) for 5 hours served as a positive control for caspase cleavage. One representative of 3 independent experiments is shown.
Proapoptotic signaling of pppRNA and poly(I:C) is mediated by the cytosolic RNA receptors RIG-I and MDA-5.
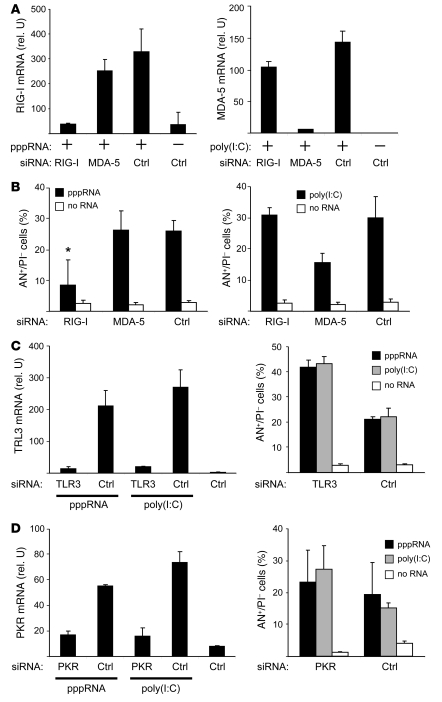
siRNAs effectively targeting RIG-I and MDA-5 were designed (sequences are shown in Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI37155DS1), and inhibition of the target was confirmed (Figure 2A; mRNA reduction in RIG-I and MDA-5 levels, >90%). In pppRNA-treated melanoma cells, inhibition of RIG-I led to a decrease in apoptotic cells almost to baseline levels, whereas inhibition of MDA-5 had no major effect on pppRNA-induced apoptosis (Figure 2B, left panel). In contrast, apoptosis induction by poly(I:C) was reduced by inhibition of MDA-5 but not of RIG-I (Figure 2B, right panel). Next, the involvement of TLR3 and RNA-activated protein kinase (PKR) was tested. TLR3 and PKR can be activated by poly(I:C) and PKR by pppRNA (18, 19). The siRNAs effectively silenced TLR3 (Figure 2C, left panel) and PKR (Figure 2D, left panel), but inhibition of neither TLR3 nor PKR rescued cells from apoptosis induction by pppRNA or poly(I:C). In fact, apoptosis was even enhanced by inhibition of TLR3 or PKR (Figure 2, C and D, right panels). RIG-I, MDA-5, TLR3, and PKR mRNA was expressed at low levels in melanoma cells and was strongly upregulated by pppRNA and poly(I:C) (Figure 2, A, C, and D). Taken together, these results suggest that proapoptotic signaling induced by pppRNA and poly(I:C) is triggered by RIG-I and MDA-5, respectively, while TLR3 and PKR, despite being upregulated, do not contribute to apoptosis induction.
Figure 2. Apoptosis induction in melanoma cells requires RIG-I and MDA-5.
(A) 1205Lu cells were treated with siRNAs specific for RIG-I or MDA-5 or with control siRNA (Ctrl) for 48 hours. Then cells were treated (+) with pppRNA or poly(I:C) (3 ng/ml) or with transfection reagent alone (–). Expression of RIG-I and MDA-5 mRNA was analyzed by quantitative RT-PCR. Mean ± SD of 3 independent experiments is shown. rel. U, relative units. (B) 1205Lu cells were treated with siRNAs and pppRNA or poly(I:C) (5 ng/ml) as described for A and analyzed for apoptosis by FACS. Annexin V–positive and propidium iodide–negative cells are represented. Mean ± SD of 3 independent experiments is shown. *P ≤ 0.05 compared with control siRNA–transfected cells treated with pppRNA. (C) 1205Lu cells were treated with siRNA specific for TLR3 or control siRNA and pppRNA, poly(I:C), or transfection reagent alone as described for A. Left: Quantification of TLR3 mRNA by quantitative RT-PCR. Right: Analysis of apoptotic cells. Mean ± SD of 3 independent experiments is shown. (D) 1205Lu cells were treated with siRNA specific for PKR or control siRNA and pppRNA, poly(I:C), or transfection reagent alone as described for A. Left: Quantification of PKR mRNA by quantitative RT-PCR. Right: Analysis of apoptotic cells. Mean ± SD of 3 independent experiments is shown.
Apoptosis induction by pppRNA and poly(I:C) involves IPS-1 but is independent of IFN signaling.
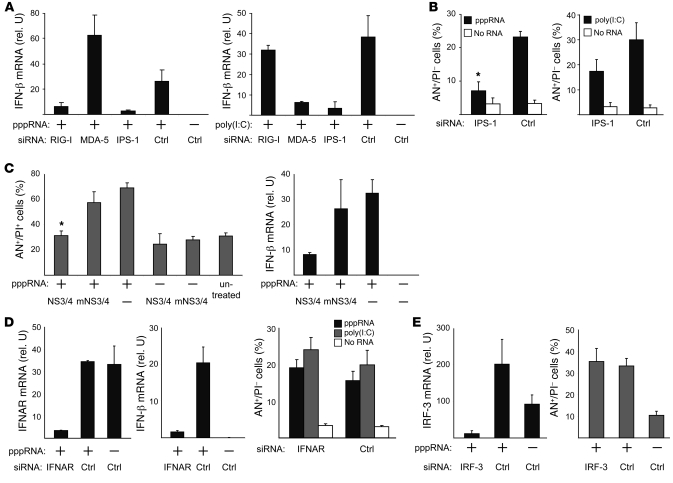
The RNA ligands pppRNA and poly(I:C) both induced IFN-β expression in melanoma cells (Figure 3A). Silencing of RIG-I and MDA-5 confirmed that induction of IFN-β by pppRNA and poly(I:C) required RIG-I and MDA-5, respectively, and that both required IPS-1 (Figure 3A). Similar to IFN-β induction, silencing of IPS-1 also decreased pppRNA- and poly(I:C)-induced apoptosis (Figure 3B); residual apoptosis induction was higher for poly(I:C). IPS-1–dependent apoptosis induction was confirmed by transfection of cells with NS3-4A, a multifunctional serine protease derived from hepatitis C virus that inactivates IPS-1 by cleavage (11). Expression of NS3-4A but not the inactive mutant (mNS3-4A) in melanoma cells reduced pppRNA-induced cell death to control levels (Figure 3C). Analysis of IFN-β expression confirmed the inhibitory function of NS3-4A in this system (Figure 3C).
Figure 3. Apoptosis as well as IFN-β induction by pppRNA and poly(I:C) are mediated via IPS-1 in melanoma, but apoptosis is independent of IFN-β secretion.
(A) 1205Lu cells were treated with pppRNA or poly(I:C) (3 ng/ml) 48 hours after transfection of siRNAs specific for RIG-I, MDA-5, or IPS-1 or control siRNA. IFN-β expression was analyzed by quantitative RT-PCR. (B) 1205Lu cells were treated with pppRNA or poly(I:C) (5 ng/ml) 48 hours after transfection of siRNAs specific for IPS-1 or control siRNA, and rates of apoptosis were determined by FACS. Annexin V–positive and propidium iodide–negative cells are represented. *P ≤ 0.05 compared with control siRNA–transfected cells treated with pppRNA1ds. (C) 1205Lu cells were transfected with vectors expressing wild-type NS3-4A (NS3/4) or an inactive mutant form (mNS3/4) (51). Twenty-four hours after vector transfection, cells were treated with pppRNA. Left: Apoptotic and dead cells (AN+/PI+) were measured. *P ≤ 0.05 compared with mNS3/4- or mock-transfected cells treated with pppRNA. Right: Analysis of IFN-β expression by quantitative RT-PCR. (D) 1205Lu cells were treated with siRNA specific for the type I IFN receptor (IFNAR) or control siRNA and pppRNA, poly(I:C), or transfection reagent alone as described for A. Left: quantification of IFNAR mRNA. Middle: Quantification of IFN-β expression. Right: FACS analysis of apoptotic cells (AN+/PI–) treated with pppRNA, poly(I:C), or transfection reagent alone. (E) 1205Lu cells were treated with an IRF-3–specific siRNA or control siRNA and pppRNA as described for A. Left: Analysis of IRF-3 mRNA expression. Right: Analysis of apoptotic cells (AN+/PI–). For all panels, mean ± SD of 3 independent experiments is shown.
To analyze the role of IFN signaling in apoptosis induction, we used siRNA to silence the IFN-α/β receptor (IFNAR), aiming at interrupting the downstream signaling of type I IFNs. An effective IFNAR-targeting siRNA was established (mRNA reduction, >90%; Figure 3D, left panel). Inhibition of IFNAR was functional, since it resulted in a strong decrease in IFN-β (amplified through autocrine feedback loop via IFNAR) (Figure 3D, middle panel). In the absence of IFNAR and IFN-β, pppRNA and poly(I:C) induced apoptosis to the same extent (Figure 3D). Similarly siRNA-mediated silencing of the transcription factor IRF-3 (essential for the RIG-I– and MDA-5–mediated induction of type I IFN) had no effect on pppRNA- and poly(I:C)-induced apoptosis (Figure 3E). Together, these data suggest that RIG-I– and MDA-5–mediated apoptosis induction requires IPS-1 but is independent of type I IFN signaling and IRF-3 expression.
RIG-I and MDA-5 activate the mitochondrial apoptosis pathway.
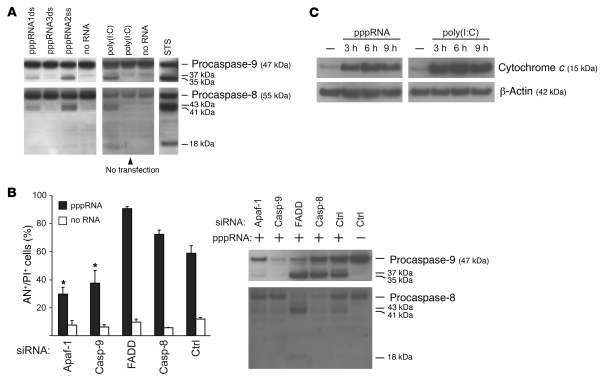
To further characterize the apoptotic signaling triggered by RIG-I and MDA-5, we analyzed the contributions of caspase-9, an essential mediator in the mitochondrial (intrinsic) apoptotic pathway, and of caspase-8, which is required for the receptor-mediated (extrinsic) apoptotic pathway (20). Immunoblot analysis revealed active subunits of caspase-9 and caspase-8 in melanoma cells treated with pppRNA or poly(I:C) (Figure 4A). Cleavage of caspase-8 or -9, although they are initiator caspases, can be caused by active effector caspases (e.g., caspase-3) at later stages of apoptosis. To clarify which apoptotic pathway was initially activated, we silenced critical molecules involved in these pathways by RNAi: caspase-9 and its adapter protein Apaf-1 as components of the mitochondrial pathway; and caspase-8 and the adapter protein FADD as essential mediators of the receptor-mediated pathway.
Figure 4. RIG-I and MDA-5 activate the mitochondrial apoptosis pathway.
(A) Transfection of pppRNA or poly(I:C) activates the initiator caspase-9 and -8. pppRNAs (pppRNA1–3) or poly(I:C) (5 ng/ml) were used with or without transfection reagent, and 1205Lu cells were analyzed 12 (pppRNA) or 24 hours [poly(I:C)] after RNA treatment. Caspase activation was assessed by immunoblotting with antibodies that are specific for both procaspases and their respective active subunits. WM9 melanoma cells treated with 1 μM staurosporine (STS) for 5 hours served as a positive control for caspase cleavage. Blots are representative of 3 independent experiments. The last lane of the upper-left blot was run on the same gel but was not contiguous, as indicated by the white line. (B) 1205Lu cells were treated with pppRNA or transfection reagent alone 48 hours after transfection of siRNAs specific for Apaf-1, caspase-9, FADD, caspase-8, or control siRNA. Left: Apoptotic and dead cells (AN+/PI+) were analyzed. Mean ± SD of 3 independent experiments is shown. *P ≤ 0.05 compared with control siRNA–transfected cells treated with pppRNA. Right: Analysis of caspase-8 and -9 activation 12 hours after pppRNA transfection by immunoblotting as described for A. Blots are representative of 2 independent experiments. (C) 1205Lu cells were treated with pppRNA or poly(I:C) (5 ng/ml) or left untreated. Cytosolic protein fractions were prepared at the indicated time points (3, 6, or 9 hours after transfection) and analyzed for the presence of cytochrome c in the cytosol by immunoblotting. β-Actin served as loading control. Blots are representative of 2 independent experiments.
Functional assays showed that RIG-I– and MDA-5–triggered apoptosis is dependent on caspase-9 and Apaf-1, while inhibition of caspase-8 or FADD did not reduce cell death (Figure 4B, left panel). Immunoblots of caspase-9 and caspase-8 were carried out to confirm that the siRNAs had the desired effect (Figure 4B, right panel). Apaf-1 is required for activation of caspase-9, and, accordingly, in melanoma cells treated with Apaf-1–specific siRNA, activation of caspase-9 was strongly decreased (procaspase-9 was present, but active caspase-9 subunits were reduced). Melanoma cells treated with caspase-9–specific siRNA showed the expected reduction of both procaspase-9 and its active subunits. Cell death was reduced in cells in which Apaf-1 or caspase-9 was knocked down (Figure 4B, left panel). This correlated with a reduced activation of caspase-8 in these samples (Figure 4B, right panel, first two lanes), suggesting that caspase-8 was cleaved subsequent to caspase-9 activation. To confirm the involvement of the mitochondrial pathway, we measured cytochrome c release from mitochondria, a critical event in mitochondrial apoptosis, in cytosolic extracts. Cytosolic cytochrome c was detectable 3 hours after transfection, with peak levels at 6 hours (Figure 4C). These results suggest that RIG-I and MDA-5 activate the mitochondrial pathway requiring Apaf-1 and caspase-9 and that activation of caspase-8 is secondary to Apaf-1 and caspase-9 activation.
Induction of Noxa contributes to RIG-I– and MDA-5–mediated apoptosis.
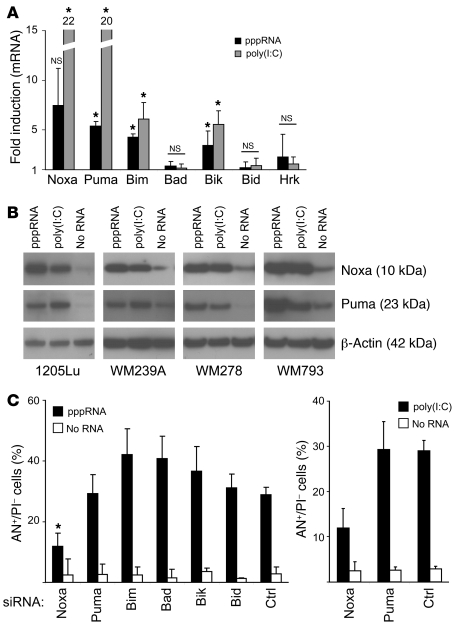
Mitochondrial apoptosis is regulated by proteins of the Bcl-2 family, and important initiators of mitochondrial apoptosis are the BH3-only proteins of the Bcl-2 family. Analysis of BH3-only proteins during RIG-I– and MDA-5–mediated apoptosis showed strong transcriptional induction of Noxa and Puma with RNA levels increasing 5- to 20-fold. Smaller but significant increases were observed for Bim and Bik, whereas Bad, Bid, and Hrk mRNA levels were not changed (Figure 5A). To confirm these observations on the protein level, we performed immunoblotting in several melanoma cell lines. Induction of Noxa and Puma was observed in cells exposed to either pppRNA or poly(I:C) (Figure 5B). RNAi-mediated silencing of Noxa strongly reduced the amount of apoptotic cells in pppRNA-treated melanoma cells, whereas the percentage of apoptotic cells in Puma-, Bim-, Bad-, Bik-, or Bid-inhibited cells was not reduced compared with the control (Figure 5C, left panel). Similar observations were made with poly(I:C) (Figure 5C, right panel). These data suggest that expression of Noxa, Puma, Bik, and Bim is increased during RIG-I– or MDA-5–mediated apoptosis. The presence of Noxa was critically required for apoptosis, whereas other BH3-only proteins were not individually required.
Figure 5. RIG-I and MDA-5 induce expression of the proapoptotic BH3-only members of the Bcl-2 family Noxa, Puma, Bim, and Bik.
(A) 1205Lu cells were treated with pppRNA or poly(I:C) (3 ng/ml) for 24 hours, and levels of Noxa, Puma, Bim, Bad, Bik, Bid, and Hrk mRNA were measured by quantitative RT-PCR. Relative mRNA levels compared with mock-transfected cells are depicted. Mean ± SD of 3 independent experiments is shown. *P ≤ 0.05 and NS compared with mock-transfected cells. (B) The melanoma cell lines 1205Lu, WM239A, WM278, and WM793 were treated with pppRNA or poly(I:C) (5 ng/ml), and Noxa and Puma proteins were quantified by immunoblotting. β-Actin served as loading control. Blots are representative of 2 independent experiments. (C) Left: Expression of the BH3-only proteins Noxa, Puma, Bim, Bad, Bik, and Bid was inhibited by treatment with respective siRNAs for 48 hours. Thereafter, cells were treated with pppRNA (left), poly(I:C) (5 ng/ml; right), or transfection reagent alone and analyzed for apoptotic cells by FACS. Annexin V–positive and propidium iodide–negative cells are represented. Mean ± SD of 3 independent experiments is shown. *P ≤ 0.05 compared with control siRNA–transfected cells treated with pppRNA.
Role of the tumor suppressor p53 in RIG-I– and MDA-5–mediated apoptosis.
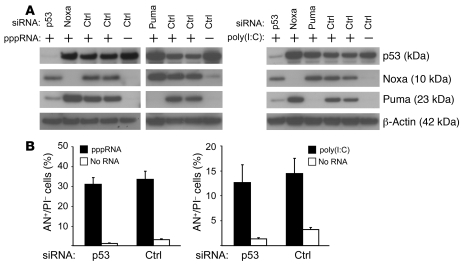
Noxa and Puma, the two BH3-only proteins most strongly induced by pppRNA, were originally isolated as target genes upregulated by the tumor suppressor p53. The tumor suppressor p53 is rarely mutated in melanoma, in contrast to many other malignancies, and serves as an important regulator of apoptosis (21, 22). However, expression studies revealed no increase in p53 on the mRNA (data not shown) and protein levels (Figure 6A) upon treatment with pppRNA or poly(I:C). Induction of Noxa was not affected when p53 was silenced, whereas Puma induction required p53 (Figure 6A). Inhibition of p53 by RNAi did not rescue cells from RIG-I– and MDA-5–triggered apoptosis (Figure 6B). Together, the data suggest that apoptosis induction by RIG-I and MDA-5 is not mediated by p53 and induction of Noxa occurs independent of p53, whereas Puma requires p53 for its induction.
Figure 6. Role of p53 in apoptosis induced by RIG-I and MDA-5.
(A) 1205Lu cells were treated with pppRNA or poly(I:C) 48 hours after transfection of siRNAs specific for p53, Noxa, Puma, or control siRNA and analyzed by immunoblotting. Left: Treatment with pppRNA. Blots are representative of 3 independent experiments. Right: Treatment with poly(I:C) (20 ng/ml). Blots are representative of 2 independent experiments. β-Actin served as loading control. (B) 1205Lu cells were transfected with a p53-specific siRNA or control siRNA for 48 hours, treated with pppRNA (left) or poly(I:C) (5 ng/ml; right) or transfection reagent alone, and analyzed for apoptotic cells (AN+/PI–). Mean ± SD of 3 independent experiments is shown.
Melanoma cells are more sensitive to RIG-I– and MDA-5–induced apoptosis than primary cells.
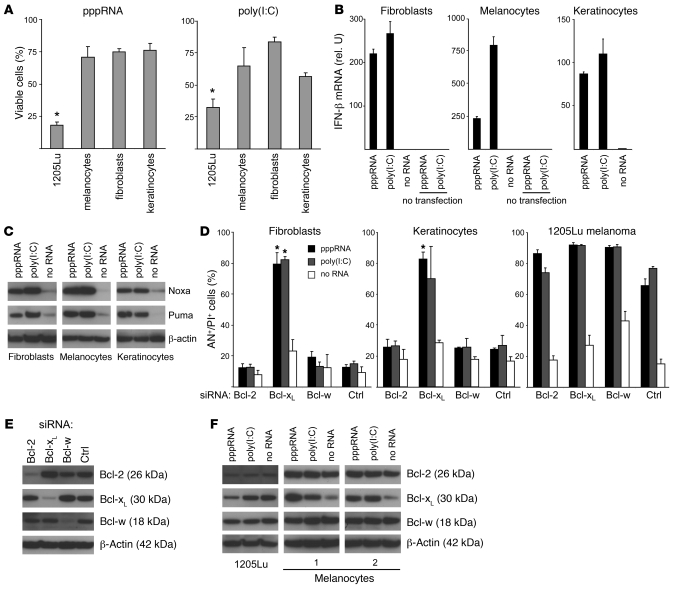
We next compared healthy primary cells of the skin with melanoma cells to evaluate tumor specificity of apoptosis induction by RIG-I and MDA-5. Primary human melanocytes, primary fibroblasts, and primary keratinocytes were significantly less sensitive to pppRNA and poly(I:C) compared with melanoma cells (Figure 7A). pppRNA and poly(I:C) also induced expression of IFN-β in primary cells, and transfection into the cytosol was required for IFN induction (Figure 7B). Interestingly, despite resistance of these cells to apoptosis, pppRNA and poly(I:C) also induced expression of Noxa and Puma in primary cells (Figure 7C). Therefore, we considered the possibility that mechanisms within the Bcl-2 family or downstream may rescue primary cells from apoptosis.
Figure 7. Increased apoptotic sensitivity of melanoma cells to RNA ligands of RIG-I and MDA-5.
(A) Cell viability of 1205Lu melanoma cells was compared with that of human melanocytes, primary human fibroblasts, or primary human keratinocytes. Cells were transfected with pppRNA or poly(I:C) (20 ng/ml) for 24 hours. The viability of mock-transfected cells was set to 100% for each cell type. Mean of 3 transfections of 1205Lu is indicated; the mean ± SEM of 2 or 3 donors measured in triplicate for primary cells is shown. *P ≤ 0.05 compared with melanocytes, fibroblasts, or keratinocytes. (B) Primary cells were treated with pppRNA, poly(I:C) (3 ng/ml) with or without transfection reagent, or with transfection reagent alone. IFN-β expression was analyzed by quantitative RT-PCR 17 hours after transfection. Mean ± SD of triplicate measurements of RNAs pooled from 3 (melanocytes) or 2 donors (fibroblasts and keratinocytes) is shown. (C) Primary cells were transfected with pppRNA for 17 hours or poly(I:C) (10 ng/ml) for 24 hours, and expression of Puma and Noxa protein was quantified by immunoblotting. Blots are representative of 3 independent experiments. (D) Primary fibroblasts, primary keratinocytes, or 1205Lu melanoma cells were treated with pppRNA, poly(I:C) (3 ng/ml), or transfection reagent alone 48 hours after transfection of siRNAs specific for antiapoptotic Bcl-2, Bcl-xL, Bcl-w, or control siRNA. Cell death was determined by FACS 17 hours after treatment with pppRNA or poly(I:C). Annexin V– and propidium iodide–positive cells are represented. Mean ± SD of 3 experiments with different donors for primary cells or different passages of 1205Lu cells is shown. *P ≤ 0.05, primary cells compared with cells transfected with control siRNA and the respective stimulus, pppRNA or poly(I:C). (E) Primary fibroblasts were treated with the indicated siRNAs and analyzed 48 hours after transfection by immunoblotting. Blots are representative of 3 independent experiments. (F) Expression of Bcl-2, Bcl-xL, and Bcl-w upon transfection with RIG-I and MDA-5 ligands. Melanocytes of 2 donors or 1205Lu melanoma cells were treated with pppRNA for 17 hours or poly(I:C) (10 ng/ml) for 24 hours. Expression was quantified by immunoblotting. Blots are representative of 3 independent experiments for 1205Lu melanoma cells. In C, E, and F, β-actin served as loading control.
A small molecule BH3-only mimetic that inhibits antiapoptotic Bcl-2, Bcl-xL, and Bcl-w led to induction of apoptosis of human fibroblasts when used together with pppRNA (data not shown). Therefore, siRNAs were used to specifically silence Bcl-2, Bcl-xL, and Bcl-w. Knockdown of Bcl-xL, but not of Bcl-2 or Bcl-w, strongly increased sensitivity of fibroblasts and keratinocytes to apoptosis, resulting in approximately 80% cell death after treatment with pppRNA or poly(I:C) (Figure 7D). Knockdown of Bcl-xL, Bcl-2, or Bcl-w in the absence of pppRNA or poly(I:C) did not lead to substantial induction of apoptosis. In contrast to primary cells, knockdown of Bcl-xL, Bcl-2, or Bcl-w in tumor cells did not substantially increase the sensitivity toward pppRNA or poly(I:C) (Figure 7D, right panel); furthermore, whereas in primary cells only Bcl-xL influenced apoptosis, no specific difference between Bcl-xL, Bcl-2, and Bcl-w was observed in tumor cells (Figure 7D). All siRNAs effectively and specifically silenced Bcl-2, Bcl-xL, and Bcl-w (Figure 7E). siRNA-mediated silencing in primary melanocytes could not be achieved for technical reasons. However, expression analysis of antiapoptotic Bcl-2 proteins in melanocytes and in melanoma cells revealed that melanocytes upregulated Bcl-xL but not Bcl-2 and Bcl-w upon pppRNA or poly(I:C) transfection, whereas melanoma cells were unable to upregulate Bcl-xL, further supporting an essential role of Bcl-xL in tumor selectivity of pppRNA- or poly(I:C)-induced apoptosis (Figure 7F). Together, these results show that RIG-I or MDA-5 activate proapoptotic signaling not only in tumor cells but also in primary cells, but primary cells are rescued from apoptosis by upregulation or activation specifically of Bcl-xL, which counteracts proapoptotic effects mediated by Noxa and Puma, a safeguard mechanism that seems to be defective in melanoma cells.
Tumor-specific induction of apoptosis leads to tumor growth inhibition in severely immunodeficient mice.
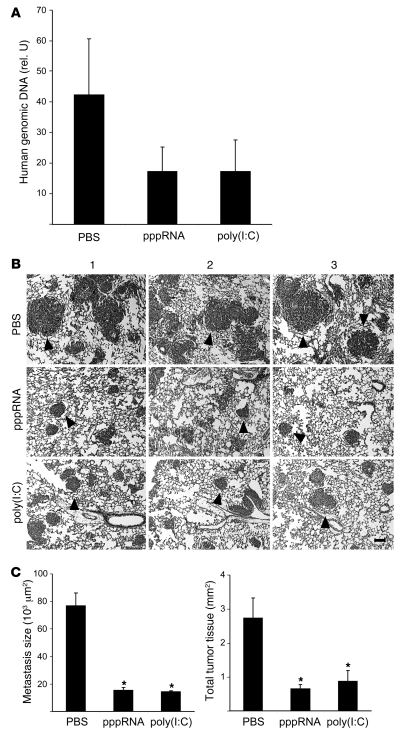
Next, we tested whether the higher sensitivity of tumor cells for apoptosis induction can be translated in tumor treatment in vivo. The tumor-specific proapoptotic activity of RIG-I and MDA-5 RNA ligands was analyzed in NOD/SCID immunodeficient mice, which have a strongly impaired cellular immune system including NK cells (23). Lung tumor metastases were induced with cells of the 1205Lu melanoma line injected intravenously. pppRNA and poly(I:C) were complexed with the polyethylenimine (PEI) derivative jetPEI as described in Methods and treatment was carried out on days 3, 6, and 9 after tumor inoculation. On day 10, mice were sacrificed, and the extent of lung metastasis formation was quantified by determining the amount of human genomic DNA. The level of human DNA was 50% lower in mice treated with pppRNA or poly(I:C) (Figure 8A). At a later time point (day 24), histological analysis of lung metastasis was carried out. Treatment with pppRNA or poly(I:C) reduced the size of metastases and the total amount of tumor tissue (Figure 8, B and C). Together, the data show that efficacy and specificity of RIG-I– and MDA-5–driven apoptosis is sufficient to reduce tumor lung metastasis formation in vivo in an immunodeficient background.
Figure 8. Therapeutic efficacy of RIG-I and MDA-5 ligands in immunodeficient mice.
(A) Groups of 3 NOD/SCID mice were challenged with 4 × 105 1205Lu melanoma cells and treated intravenously on days 3, 6, and 9 with pppRNA, poly(I:C), or PBS complexed with jetPEI as described in Methods. Human genomic DNA, representative for lung metastases, was measured in triplicate by quantitative PCR in lungs isolated at day 10. The relative amount of genomic DNA was expressed as a ratio of the amount of murine genomic DNA determined in the same lung sample. Mean ± SEM of each group is depicted. (B) Groups of 3 NOD/SCID mice were challenged with 4 × 105 1205Lu melanoma cells and treated intravenously on days 3, 6, 9, and 20 with pppRNA, poly(I:C), or PBS as described in Methods. Analysis was done on day 24. Representative lung sections after H&E staining of individual mice of each group are shown. 1205Lu metastases are indicated by black arrowheads. Scale bar: 100 μm. (C) Left: Metastasis size was calculated from the diameter in histological sections as described in Methods. Mean metastasis size was determined for each mouse, and the mean of each group ± SEM is shown. Right: Total tumor burden was calculated as the sum of all metastasis areas in each mouse as described in Methods. Mean ± SD of each group is shown. *P ≤ 0.05 compared with PBS-treated mice.
Discussion
Infection of cells with RNA viruses leads to the induction of type I IFN and apoptosis if the virus does not make use of escape strategies. RIG-I and MDA-5 are the two cytosolic receptors responsible for the detection of viral RNA and for the induction of type I IFN. However, the contribution of RIG-I and MDA-5 to virus-induced apoptosis has not been analyzed. Here we demonstrate that RIG-I and MDA-5 trigger two distinct and functionally independent signaling pathways, one leading to type I IFN induction and the other to induction of proapoptotic BH3-only proteins. We show that both pathways rely on the RIG-I/MDA-5 adapter protein Cardif (IPS-1) in the outer mitochondrial membrane, but then diverge. Both pathways were active in primary and in tumor cells. Both primary cells and tumor cells produced type I IFNs; however, in melanoma cells, triggering of RIG-I and MDA-5 resulted in massive cell death, whereas nonmalignant cells of the skin remained intact. The antiapoptotic Bcl-2 protein Bcl-xL was identified as being responsible for the prevention of cell death in primary cells, a safeguard mechanism found to be lost in melanoma cells.
Mitochondrial apoptosis is regulated at the outer mitochondrial membrane by the pro- and antiapoptotic members of the Bcl-2 family. The functional activity of the proapoptotic BH3-only subgroup is regulated on the transcriptional level and by posttranslational modification and inhibits the function of antiapoptotic Bcl-2 proteins by protein-protein interactions (24–26), and thus BH3-only proteins are critical initiators of mitochondrial apoptosis (27). Our results show that activation of RIG-I and MDA-5 dramatically increases the expression of the proapoptotic BH3-only proteins Noxa and Puma, and to a lesser extent the expression of Bim and Bik. Silencing the expression of Noxa versus Puma revealed an essential role for Noxa but not for Puma in the initiation of RIG-I– and MDA-5–mediated mitochondrial apoptosis. Since other BH3-only proteins may compensate for the function of individually silenced BH3-only proteins, our data do not rule out that elevated levels of Puma, Bim, and Bik or posttranslational modifications contribute to proapoptotic activity, and such synergistic activity has been reported (28).
Our data are in agreement with studies by others who described that transcriptional induction of Noxa is involved in virus-mediated apoptosis (29, 30). Noxa and Puma were originally identified as target genes regulated by the tumor suppressor p53 (31, 32). There is good evidence that p53, induced by type I IFN, contributes to apoptosis in virally infected cells (33). However, our results now provide evidence that RIG-I and MDA-5 trigger a p53-independent alternative pathway for the induction of Noxa. This is in agreement with p53-independent induction of Noxa upon viral infection postulated by others (29). Unlike Noxa, we found that induction of Puma by RIG-I and MDA-5 depended on p53; consistent with the results of silencing Puma, silencing of p53 did not affect RIG-I– and MDA-5–induced apoptosis.
The lack of involvement of the type I IFN system in the induction of apoptosis by RIG-I and MDA-5 is surprising. The transcription factor IRF-3, which has a major role in the type I IFN pathway, was reported to be involved in apoptosis induction by certain viruses including Sendai virus, encephalomyocarditis virus (EMCV), and reovirus, which are all sensed by RIG-I or MDA-5 (30, 34, 35). However, induction of Noxa by Sendai virus is independent of IRF-3 (36), which is in line with our results indicating IRF-3– and IFN receptor–independent induction of apoptosis by RIG-I and MDA-5. The use of intact viruses may be responsible for seemingly inconsistent results in the literature (contribution or no contribution of type I IFN to apoptosis induction by virus). Type I IFN influences virus replication and infectivity, and thus the number of RNA ligand molecules available for RIG-I or MDA-5 stimulation is reduced in the presence of type I IFN. Furthermore, virus-specific escape strategies, e.g., based on viral proteins that modulate apoptosis or IFN pathways, complicate the identification of antiviral signaling pathways. Thus, viral RNA mimetics are advantageous for analyzing the basic principles of antiviral responses as revealed in our study. It is important to note that RIG-I and MDA-5 are capable to induce apoptosis independently of two key antiviral proapoptotic pathways, the type I IFN system and p53 (37, 38). As a consequence, apoptosis induction by RIG-I and MDA-5 remains functional even in situations where the virus applies strategies to downregulate type I IFN (11, 39) or p53 (38).
Surprisingly, activation of RIG-I or MDA-5 also induced Noxa and Puma in primary cells, although these cells did not undergo apoptosis. Noxa, relevant for apoptosis in melanoma, specifically inactivates antiapoptotic Mcl-1 (and A1) (40). Screening of antiapoptotic Bcl-2 molecules revealed that Bcl-xL, but not Bcl-2 or Bcl-w, compensated for Mcl-1 inactivation by Noxa in primary cells. This is in agreement with a study that described apoptosis induction in murine fibroblasts only when two antiapoptotic Bcl-2 proteins were inactivated simultaneously, i.e., inactivation of Mcl-1 by Noxa and inactivation of Bcl-xL by Bad (28). Thus, the Bcl-xL–based survival mechanism may represent a common way on which normal cells rely to avoid immediate cell death when infected with viruses, e.g., in order to produce IFN. As a consequence, tumor cells are specifically susceptible to RIG-I– or MDA-5–mediated apoptosis, since they have lost the ability to counteract apoptosis via Bcl-xL. Tumor cells develop in the absence of viral infection and thus seem to be unprepared for proapoptotic signaling upon recognition of viral infection. Apoptosis induction by RIG-I or MDA-5 activation can specifically hit such a tumor cell, since rescue by Bcl-xL is not functional, as it is in healthy cells.
Since RIG-I and MDA-5 can be activated in most cell types, protection of cells during virus infection is highly relevant to protect the host from excessive apoptosis induction. An important function of Bcl-xL appears to be to intercept the proapoptotic signal triggered through these virus receptors. In this context, it is important to note that small molecules (known as BH3-only mimetics) targeting antiapoptotic Bcl-2 proteins including Bcl-xL are currently being developed for cancer therapy (41, 42). Inhibition of Bcl-xL by those molecules may abrogate the protection against exaggerated apoptosis induction in the case of ongoing viral infections activating RIG-I or MDA-5, e.g., influenza or hepatitis.
With short 5ι-triphosphate RNAs, ligands for RIG-I are available that can be generated synthetically. They share certain structural features with siRNAs, and the requirements for cytosolic delivery are similar. Several siRNA molecules are in clinical studies, and short pppRNAs may benefit from formulations developed for the clinical application of siRNAs. In a preclinical mouse model, PEI alone did not affect the counts of leukocytes, platelets, and erythrocytes. PEI-complexed pppRNAs reduced the counts of leukocytes and platelets but not of erythrocytes, presumably due to systemic IFN induction, but no other obvious toxicities were observed (43). While pppRNA is a relatively new compound, poly(I:C) has been known for decades and was tested in multiple clinical trials for cancer therapy (44). However, only in the last few years has it become evident that there are at least 3 distinct receptors involved in the recognition of poly(I:C): PKR, TLR3, and MDA-5. Our results demonstrate that PKR and TLR3 can be dispensable for apoptosis induction by poly(I:C), and of these 3 only MDA-5 induces apoptosis. Of note, in all clinical trials performed to date, poly(I:C) was used without a formulation that allows cytosolic delivery; however, this type of delivery is required for MDA-5 activation and thus apoptosis induction. Thus, although poly(I:C) has been around for long time, the identification and characterization of the corresponding receptors may now lead to an improved formulation of poly(I:C) for tumor therapy.
In fact, here we found that both poly(I:C) and pppRNA when delivered by PEI have strong antitumor activity in immunodeficient mice, demonstrating an antitumor effect that does not require the immune system. This observation is consistent with the higher sensitivity of malignant versus nonmalignant cells seen in our study in vitro, providing the basis for the applicability of these ligands for tumor therapy. The signaling steps leading to the upregulation of proapoptotic Noxa do not rely on p53. In a more general context, it is interesting to note that inactivation of p53 is present in more than 50% of all cancer types and contributes to resistance to chemo- and radiotherapy (45, 46) but importantly will not protect tumor cells from RIG-I– and MDA-5–induced apoptosis. Our study is in agreement with previous results from another group that saw preferential induction of apoptosis by poly(I:C) in various tumor cell lines, including Adriamycin-resistant tumor cells, compared with normal fibroblasts and primary hepatocytes, but only when liposomal delivery was used (47).
Although this work focuses on the direct induction of tumor cell apoptosis, indirect attack of the tumor by immune cells is expected to contribute to the overall antitumor activity of RIG-I and MDA-5 ligands in vivo. In fact, in another study we found that pppRNA delivered by PEI in vivo strongly activated NK cells and that activated NK cells represent an additional arm of antitumor activity of pppRNA in immunocompetent mice (43). This effect was absent in the immunodeficient mice used in the work presented here.
In conclusion, we identified a signaling pathway of apoptosis that physiologically contributes to the elimination of virus-infected cells and for which all melanoma cells tested so far are highly vulnerable. With well-defined, potent synthetic RNA ligands for virus-sensing receptors, tumor cells could be attacked where they are not prepared to be hit and where they are more vulnerable than healthy cells.
Methods
Reagents and antibodies.
Anti–caspase-3, anti–caspase-8 (1C12), anti–caspase-9, anti–Bcl-xL, anti–Bcl-w, and HRP-conjugated secondary antibodies were obtained from New England Biolabs. Anti–cytochrome c (clone 7H8.2C12) was from BD Biosciences. Anti-Noxa (N-15) antibody was from Santa Cruz Biotechnology Inc. Anti–Bcl-2 (Ab-1) and anti-p53 (Ab-6) antibodies were from Merck Biosciences. Anti–IPS-1 antibody was obtained from Bethyl Laboratories Inc. Anti–β-actin (AC-15) and anti-Puma (bbc3) antibodies were purchased from Sigma-Aldrich. PCR primers and siRNAs were purchased from MWG Biotech.
Immunostimulatory and siRNAs.
Poly(I:C) was purchased from Amersham Biosciences. 5′-Triphosphate–conjugated RNAs (pppRNAs) were transcribed in vitro from DNA templates as described in ref. 6. They contained a T7 RNA Polymerase consensus promoter sequence followed by the sequence of interest to be transcribed (MEGAshortscript Kit; Ambion). Reactions were treated with DNAse I (Ambion) to digest template DNA, and RNA was purified by phenol/chloroform extraction, alcohol precipitation, and, subsequently, spinning through a mini Quick Spin Oligo Column (Roche). Sequences of single-stranded (ss) pppRNAs were: pppRNA1, 5′-GGCAUGCGACCUCUGUUUGA-3′; pppRNA2, 5′-GCUACAUCCUGUCCAUUCAA-3′; pppRNA3, 5′-GGCGGGGCGCGGCGGGGCGC-3′. For double-stranded (ds) pppRNAs, the corresponding complementary strand was transcribed separately and annealed after purification. Because of high sequence homology to human Bcl-2 mRNA, double-stranded pppRNA1 was tested for RNAi-mediated Bcl-2–silencing capacity, but Bcl-2 protein levels (Figure 7F) and RNA levels (data not shown) were not altered.
siRNAs were designed according to published guidelines (48, 49). 3′ Overhangs were carried out as two deoxythymidine residues (dTdT). Sequences of specific siRNAs are listed in Supplemental Table 1. Nonsilencing control siRNAs were designed to contain random sequences that do not match within the human genome. The sequences of the 19-nt sense strand of control siRNAs were 5′-GCGCAUUCCAGCUUACGUA-3′ and 5′-GCGCUAUCCAGCUUACGUA-3′.
Cell culture.
Human melanoma cell lines were a gift of M. Herlyn (Wistar Institute, Philadelphia, Pennsylvania, USA). All were isolated from clinically and histologically defined lesions. WM793 and WM278 were derived from primary melanomas (stage 1 and stage 2, respectively); metastatic cell lines WM239A and WM9 were derived from lymph nodes; and 1205Lu was isolated from a lung metastasis. They were maintained in a culture medium consisting of MCDB153 (Sigma-Aldrich) with 20% Leibovitz’s L-15 (PAA Laboratories), 2% FBS (PAA Laboratories), 1.68 mM CaCl2 (Sigma-Aldrich), and 5 μg/ml insulin (Sigma-Aldrich). For analysis, cells were detached with 0.2% EDTA in PBS. Keratinocytes and fibroblasts were isolated from neonatal human foreskins and cultivated as described previously (50).
Transfection procedures.
pppRNAs were transfected at a concentration of 1 μg/ml. Unless otherwise indicated, pppRNA1ds was used. The concentrations of poly(I:C) are indicated in the figure legends. Typically, pppRNAs or poly(I:C) were transfected in 3.5-cm dishes using 2.5 μl Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. Unless otherwise indicated, analysis was done 17 hours after transfection. siRNAs were transfected at 20 nM with 1.25 μl Lipofectamine RNAiMAX. Plasmids were transfected at 1 μg/ml with 3 μl FuGENE 6 (Roche) according to the manufacturer’s protocol. In some experiments, siRNA-treated cells were transfected with pppRNA or poly(I:C). In these experiments, medium was changed 48 hours after siRNA treatment. Thereafter, cells were retransfected with pppRNA or poly(I:C) mixed with 20 nM of siRNAs and complexed with 2.5 μl Lipofectamine RNAiMAX.
Quantification of viable cells.
Viable cells were quantified in 12-well dishes by using a fluorometric assay (CellTiter-Blue Cell Viability Assay; Promega). Viable cells with intact metabolism were identified by their ability to reduce cell-permeable resazurin to fluorescent resorufin. Medium was replaced with 375 μl of culture medium and 75 μl of CellTiter-Blue reagent. After 1 hour incubation at 37°C, fluorescence was measured.
Quantification of apoptotic cells and cell death.
Adherent and supernatant cells were analyzed by staining with FITC-labeled Annexin V (Roche) and propidium iodide (Sigma-Aldrich). Annexin V staining was performed according to the manufacturer’s instructions. Propidium iodide was added to a final concentration of 0.5 μg/ml. Cells were analyzed by fluorescence-activated cell sorting (FACS) analysis using CellQuest software (BD).
RNA extraction and quantification.
Total RNA was extracted from cells using TRIzol (Invitrogen) as described by the manufacturer and analyzed by quantitative RT-PCR. RNA (1 μg) was reverse transcribed using Expand Reverse Transcriptase (Roche) and poly(dT) oligonucleotide (Roche) according to the manufacturer’s protocol. Quantitative PCR was performed using the LightCycler TaqMan Master Kit (Roche) together with the Universal Probe Library system (Roche). Relative gene expression was expressed as a ratio of the expression level of the gene of interest to that of hypoxanthine phosphoribosyltransferase (HPRT) RNA determined in the same sample.
Protein preparation and immunoblot analysis.
Adherent and supernatant cells were lysed in buffer containing 50 mM Tris, pH 7.4, 0.25 M NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.1 mM EGTA, 5 mM Na3VO4, 50 mM NaF, and protease inhibitors (Complete, Mini, EDTA-free; Roche). Gel electrophoresis and blotting were carried out with 5–10 μg denatured protein lysate by using the Xcell SureLock Mini-Cell apparatus with 4–12% gels in MES SDS buffer and PVDF membranes according to the manufacturer’s protocol (Invitrogen). After blocking (Western Blocking Reagent in PBS; Roche), blots were incubated with primary antibodies for 1 hour at room temperature or overnight at 4°C. Following washes with 0.1% Tween 20 (Sigma-Aldrich) in PBS and 1 hour of incubation with HRP-conjugated secondary antibodies, blots were washed and visualized (ECL Plus Western Blotting Detection System; Amersham, GE Healthcare). Protein levels of β-actin were analyzed as a control for constant loading and transfer.
Preparation of cytosolic extracts.
Cells were detached, washed, and incubated for 15 minutes in a digitonin-containing buffer (200 μg/ml in PBS) on ice to lyse cell membranes, keeping mitochondria intact. Lysates were centrifuged (1,000 g) for 5 minutes at 4°C to pellet membranes and mitochondria. Ten micrograms of the supernatant (cytosolic fraction) was subjected to immunoblotting.
Tumor engraftment and RNA treatment in vivo.
JAX NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice were purchased from Charles River Laboratories. Mice were 9 weeks of age at the onset of experiments. Animal studies were approved by the local regulatory agency (Regierung von Oberbayern, Munich, Germany). Experiments were done in groups of 3 mice. For the induction of lung metastases, 4 × 105 1205Lu melanoma cells were injected into the tail vein. Mice were treated with RNAs on days 3, 6, 9, and 20 after tumor cell inoculation. RNAs were complexed with jetPEI (Biomol) according to the manufacturer’s protocol. Briefly, 10 μl of in vivo jetPEI was mixed with 50 μg of pppRNA or poly(I:C) at an N/P ratio of 10:1 in a volume of 200 μl 5% glucose solution and incubated for 15 minutes. Subsequently, the complexes were administered by retro-orbital injection. Tumor load at day 10 was measured by quantifying human genomic DNA or by histologic analysis at day 24.
Analysis of lung metastasis.
For metastasis analysis at day 10, we isolated genomic DNA from lungs. Mouse lungs were reduced to small pieces and digested overnight at 56°C in a buffer containing 10 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 1% SDS, 0.5 mg/ml Pronase E (Sigma-Aldrich), and 150 μg/ml Protease K (Sigma-Aldrich). Genomic DNA was purified by phenol/chloroform extraction. The amount of human and murine DNA was determined by quantitative PCR using the LightCycler TaqMan Master Kit (Roche) together with the Universal Probe Library system (Roche). A 72-bp portion in the second intron of the human β-actin gene (forward primer, 5′-CGCCCTTTCTCACTGGTTC-3′; reverse primer, 5′-TCCAAAGGAGACTCAGGTCAG-3′; probe 29) and a 69-bp portion in the first intron of murine β-actin gene (forward primer, 5′-CAGCCAACTTTACGCCTAGC-3′; reverse primer, 5′-GGGCCCACGAGTGTCTAC-3′; probe 05) was amplified. The amount of human DNA was normalized to the amount of murine DNA determined in the same sample. Species specificity was controlled by genomic DNA prepared from B16 (murine) or 1205Lu (human) cell lines, and no cross-species reactivity of PCR reactions was observed. Metastasis analysis at day 24 was carried out by histological analysis. Melanoma metastases were visualized in histological sections by H&E staining. Eight visual fields per mouse lung were analyzed for number and diameter of metastases. The size of metastases was defined by the area and calculated from the diameters assuming round metastases (size = πr2). Total tumor burden per mouse was calculated as the sum of all metastasis areas in 8 visual fields of one mouse.
Statistics.
For statistical analysis, 2-tailed Student’s t test was used to assess the significance of mean differences. Differences were considered significant at a P value of 0.05 or less.
Supplementary Material
Acknowledgments
We thank Claudia Kammerbauer and Ursula Nägele for excellent technical assistance. This work was supported by German Cancer Aid (Deutsche Krebshilfe), Mildred Scheel Foundation grant 107805 to R. Besch and C. Berking; DFG grant GK 1202 to T. Hohenauer, D. Senft, S. Rothenfusser, R. Besch, C. Berking, and S. Endres; Hiege-Stiftung gegen Hautkrebs to R. Besch and T. Hohenauer; DFG grant Ro 2525/3-1 to S. Rothenfusser; DFG grant Be 2189/2-3 to C. Berking; DFG grants SFB-TR 36 and 198 En 169/7-2 to S. Endres; and the Wilhelm-Sander-Stiftung to G. Häcker. G. Hartmann received support through grants BMBF BioFuture 0311896, SFB 704, SFB 670, KFO115, and KFO177.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: IPS-1, IFN-β promoter stimulator 1; IRF-3, IFN regulatory factor 3; MDA-5, melanoma differentiation–associated antigen 5; PEI, polyethylenimine; PKR, RNA-activated protein kinase; poly(I:C), polyinosinic-polycytidylic acid; pppRNA, 5′-triphosphate RNA; RIG-I, retinoic acid–inducible gene I; RLH, RIG-I–like helicase.
Citation for this article: J. Clin. Invest. 119:2399–2411 (2009). doi:10.1172/JCI37155
Hendrik Poeck’s present address is: III Medizinische Klinik, Klinikum rechts der Isar, Technische Universität München, Munich, Germany.
Georg Häcker’s present address is: Institute for Medical Microbiology and Hygiene, Universitätsklinikum Freiburg, Freiburg, Germany.
See the related Commentary beginning on page 2127.
References
- 1.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Tschopp J., Thome M., Hofmann K., Meinl E. The fight of viruses against apoptosis. Curr. Opin. Genet. Dev. 1998;8:82–87. doi: 10.1016/S0959-437X(98)80066-X. [DOI] [PubMed] [Google Scholar]
- 3.Boon T., Coulie P.G., Van den Eynde B.J., van der Bruggen P. Human T cell responses against melanoma. Annu. Rev. Immunol. 2006;24:175–208. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- 4.Kirkwood J.M., et al. High-dose interferon alfa-2b significantly prolongs relapse-free and overall survival compared with the GM2-KLH/QS-21 vaccine in patients with resected stage IIB-III melanoma: results of intergroup trial E1694/S9512/C509801. J. Clin. Oncol. 2001;19:2370–2380. doi: 10.1200/JCO.2001.19.9.2370. [DOI] [PubMed] [Google Scholar]
- 5.Parmiani G., et al. Immunotherapy of melanoma. Semin. Cancer Biol. 2003;13:391–400. doi: 10.1016/j.semcancer.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Hornung V., et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 7.Pichlmair A., et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′ phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 8.Kato H., et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gitlin L., et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai T., et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 11.Meylan E., et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 12.Xu L.G., et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Sun Q., et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Johnson C.L., Gale M., Jr. CARD games between virus and host get a new player. Trends Immunol. 2006;27:1–4. doi: 10.1016/j.it.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Evan G.I. Can’t kick that oncogene habit. Cancer Cell. 2006;10:345–347. doi: 10.1016/j.ccr.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Hartwell L.H., Szankasi P., Roberts C.J., Murray A.W., Friend S.H. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 17.Zitvogel L., Apetoh L., Ghiringhelli F., Kroemer G. Immunological aspects of cancer chemotherapy. Nat. Rev. Immunol. 2008;8:59–73. doi: 10.1038/nri2216. [DOI] [PubMed] [Google Scholar]
- 18.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 19.Nallagatla S.R., et al. 5′-Triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 20.Green D.R. Apoptotic pathways: paper wraps stone blunts scissors. Cell. 2000;102:1–4. doi: 10.1016/S0092-8674(00)00003-9. [DOI] [PubMed] [Google Scholar]
- 21.Besch R., Berking C., Kammerbauer C., Degitz K. Inhibition of urokinase-type plasminogen activator receptor induces apoptosis in melanoma cells by activation of p53. Cell Death Differ. 2007;14:818–829. doi: 10.1038/sj.cdd.4402065. [DOI] [PubMed] [Google Scholar]
- 22.Smalley K.S., et al. An organometallic protein kinase inhibitor pharmacologically activates p53 and induces apoptosis in human melanoma cells. Cancer Res. 2007;67:209–217. doi: 10.1158/0008-5472.CAN-06-1538. [DOI] [PubMed] [Google Scholar]
- 23.Shultz L.D., et al. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J. Immunol. 1995;154:180–191. [PubMed] [Google Scholar]
- 24.Sattler M., et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 25.Certo M., et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 26.Chen L., et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 27.Huang D.C., Strasser A. BH3-only proteins-essential initiators of apoptotic cell death. Cell. 2000;103:839–842. doi: 10.1016/S0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 28.Willis S.N., et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y., Leaman D.W. Involvement of Noxa in cellular apoptotic responses to interferon, double-stranded RNA, and virus infection. J. Biol. Chem. 2005;280:15561–15568. doi: 10.1074/jbc.M412630200. [DOI] [PubMed] [Google Scholar]
- 30.Lallemand C., et al. Single-stranded RNA viruses inactivate the transcriptional activity of p53 but induce NOXA-dependent apoptosis via post-translational modifications of IRF-1, IRF-3 and CREB. Oncogene. 2007;26:328–338. doi: 10.1038/sj.onc.1209795. [DOI] [PubMed] [Google Scholar]
- 31.Oda E., et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 32.Nakano K., Vousden K.H. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell. 2001;7:683–694. doi: 10.1016/S1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 33.Takaoka A., et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature. 2003;424:516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 34.Peters K., Chattopadhyay S., Sen G.C. IRF-3 activation by sendai virus infection is required for cellular apoptosis and avoidance of persistence. J. Virol. 2008;82:3500–3508. doi: 10.1128/JVI.02536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holm G.H., et al. Retinoic acid-inducible gene-I and interferon-beta promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J. Biol. Chem. 2007;282:21953–21961. doi: 10.1074/jbc.M702112200. [DOI] [PubMed] [Google Scholar]
- 36.Elco C.P., Guenther J.M., Williams B.R., Sen G.C. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3. J. Virol. 2005;79:3920–3929. doi: 10.1128/JVI.79.7.3920-3929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz-Fontela C., et al. Resistance to viral infection of super p53 mice. Oncogene. 2005;24:3059–3062. doi: 10.1038/sj.onc.1208477. [DOI] [PubMed] [Google Scholar]
- 38.Groskreutz D.J., et al. Respiratory syncytial virus decreases p53 protein to prolong survival of airway epithelial cells. J. Immunol. 2007;179:2741–2747. doi: 10.4049/jimmunol.179.5.2741. [DOI] [PubMed] [Google Scholar]
- 39.Brzozka K., Finke S., Conzelmann K.K. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J. Virol. 2006;80:2675–2683. doi: 10.1128/JVI.80.6.2675-2683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willis S.N., Adams J.M. Life in the balance: how BH3-only proteins induce apoptosis. Curr. Opin. Cell Biol. 2005;17:617–625. doi: 10.1016/j.ceb.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oltersdorf T., et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 42.Vogler M., Dinsdale D., Dyer M.J., Cohen G.M. Bcl-2 inhibitors: small molecules with a big impact on cancer therapy. Cell Death Differ. 2009;16:360–367. doi: 10.1038/cdd.2008.137. [DOI] [PubMed] [Google Scholar]
- 43.Poeck H., et al. 5′-triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat. Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- 44.Thompson K.A., et al. Results of a double-blind placebo-controlled study of the double-stranded RNA drug polyI:polyC12U in the treatment of HIV infection. Eur. J. Clin. Microbiol. Infect. Dis. 1996;15:580–587. doi: 10.1007/BF01709367. [DOI] [PubMed] [Google Scholar]
- 45.Igney F.H., Krammer P.H. Death and anti-death: tumour resistance to apoptosis. Nat. Rev. Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 46.Hollstein M., et al. Somatic point mutations in the p53 gene of human tumors and cell lines: updated compilation. Nucleic Acids Res. 1996;24:141–146. doi: 10.1093/nar/24.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirabayashi K., et al. Inhibition of cancer cell growth by polyinosinic-polycytidylic acid/cationic liposome complex: a new biological activity. Cancer Res. 1999;59:4325–4333. [PubMed] [Google Scholar]
- 48.Reynolds A., et al. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 49.Ui-Tei K., et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berking C., et al. Transforming growth factor-beta1 increases survival of human melanoma through stroma remodeling. Cancer Res. 2001;61:8306–8316. [PubMed] [Google Scholar]
- 51.Li X.D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.