Abstract
Since their discovery in the early 2000s, microRNAs (miRNAs) and their penchant for RNA interference have taken the scientific community by storm, working their way into virtually every corner of biological inquiry. The very nature of their action, the ability to simultaneously extinguish the expression of a multitude of genes and negate their functions, immediately suggested therapeutic promise. In this issue of the JCI, a step toward the realization of this promise is described. Taulli et al. demonstrate that the miRNAs miR-1/miR-206, which are routinely lost in advanced, poorly differentiated rhabdomyosarcoma (RMS) but characteristically expressed in the mature skeletal muscle from which these tumors arise, restore the myogenic differentiation program and block the tumorigenic phenotype (see the related article beginning on page 2366). Their data support the notion that these small RNAs, effectively functioning as “micro-sheriffs” by restoring myogenic law and order, hold substantial clinical potential as differentiation therapy for RMS and perhaps other solid tumors. miRNA reexpression therapy constitutes a novel approach to handcuff oncogenes and arrest tumor development.
Sometimes you can tell a large story with a tiny subject.
— Eliot Porter
One of the largest, most intriguing scientific stories concerns the ability of developing organisms to orchestrate the expression of a vast multitude of genes to achieve the formation of the various tissues and organs required for life, despite the fact that every cell has the same complement of genetic material. One obvious explanation that has emerged is that every tissue has its own set of transcription factors that regulate tissue-specific gene expression and development. With the discovery of epigenetics, we now know that promoters of many genes, including transcription factors, can be turned on and off early during development, through covalent chromatin modifications. Most recently, the discovery of “tiny” tissue-specific microRNAs (miRNAs) has provided further complexity and coordination to the regulation of developmental gene sets, framing a broader picture in which epigenetic programming of specific transcription factors and miRNAs during development define tissue specificity.
miRNAs are small, noncoding RNAs (22–23 nucleotides long), shown to regulate gene expression at translational and posttranslational levels (1). Processed from longer transcripts by Drosha and Dicer, miRNAs mostly bind to the 3′ untranslated regions of target genes and inhibit gene expression translationally and/or by destabilizing the target mRNA (2). miRNAs, encoded in intergenic/intronic noncoding regions once thought to be “junk” DNA, contribute to differentiation when expressed in a tissue-specific manner (2) and even possess the power to shift global mRNA expression patterns with respect to differentiation signatures (3). Such miRNAs are responsible for tissue integrity and homeostasis, behaving functionally as “micro-sheriffs” (Table 1). Like a sheriff, miRNAs maintain law and order in specific tissues, by stringently regulating the expression of their target genes, especially oncogenes. Accumulating evidence now suggests that such miRNAs are assigned their working “zip codes” early during development by epigenetic modifications of miRNA promoters (4).
Table 1 .
miRNAs are expressed in a tissue-specific manner and play an important role in maintaining tissue-specific functions and cellular differentiation
With respect to cancer, miRNAs are often located in genomic unstable regions and therefore are typically downregulated in tumors (reviewed in ref. 5); moreover, inhibiting miRNA biogenesis tends to enhance tumorigenesis (6). Downregulation may be achieved through mutation or by epigenetic silencing of the miRNA, resulting in loss of tissue-specific miRNA synthesis and overexpression of pro-proliferation genes (i.e., oncogenes); these miRNAs normally function as tumor suppressors (7). Of course, miRNAs can also act as oncogenes (8, 9). Many investigators have posited that reexpression of specific miRNAs may have therapeutic anticancer value (10, 11). In a report published in the current issue of the JCI, Taulli et al. demonstrate that expression of specific miRNAs regulating skeletal muscle development, miR-1/miR-206 (also known as myomiRs), is reduced in rhabdomyosarcoma (RMS) (12). RMS tumors, the most common soft tissue sarcomas in pediatric patients and young adults, are thought to arise from the skeletal muscle lineage, coexpressing markers of proliferation and myogenic differentiation (13). Reexpression of these myomiRs to physiological levels suppressed many aspects of the transformed phenotype and induced myogenic differentiation (12), raising the possibility that miRNA reexpression may represent effective differentiation therapy for RMS and perhaps other cancer types.
miRNA reexpression as differentiation therapy in cancer
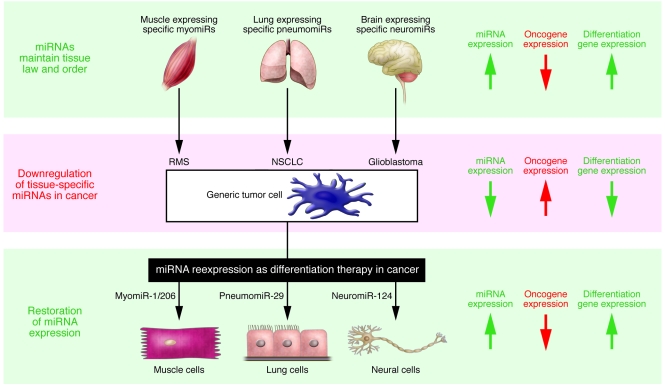
The discovery of miRNAs has added an entirely new dimension to antitumor therapeutic approaches; the potential of noncoding RNAs as drugs for cancer patients is both intriguing and compelling. As miRNA expression seems to be altered in many human diseases, including cancer, the miRNA revolution has already set the stage for “miRNA reexpression therapy” (Figure 1). Reexpressing lost miRNA in a cell can deliver a dramatic effect, because miRNAs regulate a vast number of genes and pathways. Among the many genes that miRNAs can regulate are oncogenes and tumor suppressors, targets of drugs currently used in the clinic. Although a few miRNAs are overexpressed in cancer and seem to function as oncogenes themselves (miR-17-92, miR-155), a greater number of miRNAs have been shown to be downregulated in cancer and have the potential to act as tumor suppressors (5, 9) (i.e., Let-7, miR-15/miR-16, miR-1/miR-206, miR-29, miR-124, miR-143/miR-145; see Table 1 and Figure 1). miRNA reexpression and downregulation have both been shown to have antitumor effects (9, 14). Silencing an oncogenic miRNA could allow reexpression of tumor suppressor genes, while reexpressing a tumor suppressor miRNA could downregulate multiple oncogenes (4).
Figure 1. A model describing miRNA reexpression as differentiation therapy in cancer.
Tissue-specific miRNAs play an important role in cell differentiation and are often deregulated in many human diseases, including cancer. Reexpressing such “micro-sheriff” miRNAs in a cell can deliver a dramatic impact, because miRNAs regulate a vast number of genes and pathways, including oncogenes and tumor suppressor genes. Tissue-specific tumor-suppressing miRNAs have the potential to promote the redifferentiation of tumor cells to their normal counterparts and solid malignancies to their original tissue types. The lung-specific miRNA pneumomiR-29, downregulated in non–small cell lung cancer (NSCLC), suppresses tumorigenicity by normalizing atypical patterns of methylation in non–small cell lung cancer cells (15). Taulli et al. now report in this issue of JCI that reexpression of myomiR-1/206 in RMS cells reduces oncogenic phenotypes and induces myogenic differentiation by downregulating cancer-associated genes and upregulating muscle-specific genes (12). Remarkably, introduction of striated muscle-specific myomiR-1 (downregulated in RMS) or brain-specific neuromiR-124 (downregulated in glioblastoma) into HeLa cells, a very well-characterized cervical carcinoma line, can shift the mRNA expression profile toward that of the tissue in which these miRNAs were originally enriched (3); this finding suggests that particularly powerful tissue-specific miRNAs may actually be able to drive virtually any cancer cell (shown here as a “generic tumor cell”) toward a specific differentiated state. Arrows on the right indicate the status of tissue-specific miRNA expression and the corresponding expression patterns of oncogenic genes and differentiation genes, which are opposing.
Reexpression, to physiological levels, of tissue-specific miRNAs that are lost in cancer can induce the dedifferentiation of cancer cells (12, 14). Strikingly, transfection of the myomiR miR-1 or the neural-specific miR-124 in HeLa cells downregulated hundreds of genes and shifted the mRNA expression profile toward that of the tissue in which they were originally enriched (3). Transfection of miR-124 induced neuronal phenotypic changes, by affecting the expression of neuronal-specific markers in stem cells derived from both mouse neural/brain tumor and human glioblastoma multiforme (11). It was also recently demonstrated that miR-29, a ubiquitous miRNA previously shown to suppress tumorigenicity by normalizing atypical patterns of methylation in non–small cell lung cancer cells (15), acts as an enhancer of myogenic differentiation and a suppressor of RMS (16) and is aberrantly expressed in RMS (17). Collectively, these data suggest that tissue-specific, tumor suppressor miRNAs have the potential to promote the redifferentiation of tumor cells to their normal counterparts and solid malignancies to their original tissue types.
MyomiR-1/206 reexpression effectively targets RMS
The two major histological types of RMS tumors are alveolar RMS, the more aggressive subtype defined by specific translocations involving the genes paired box 3 (PAX3) or paired box 7 (PAX7) and forkhead box O1 (FKHR also known as FOXO1a), and embryonal RMS, which is less genetically well defined (18). These tumors differ in occurrence, location, patient age, and, most importantly, prognosis. It is widely recognized that RMS development represents a failure of myoblast differentiation (19). A subset of miRNAs, the myomiRs, are specifically expressed in striated muscle and have been shown to help regulate myogenesis, muscle growth, cardiac function, and hypertrophy (20). MyomiRs consist of three bicistronic pairs of miRNAs (miR-1-1/miR-133a-2, miR-1-2/miR-133a-1, and miR-206/miR-133b) (12). All myomiRs have very similar sequences, can target the same mRNAs, and function prominently in the development of both skeletal muscle and heart. miR-206 is a skeletal muscle–specific miRNA expressed during development and postnatally, although expression levels in mature muscle are low compared with those for miR-1 (reviewed in ref. 21). Notably, miR-1/miR-206 expression can promote differentiation of muscles, whereas myomiR-133 expression appears to promote myoblast proliferation (21). MyomiR-1/206 are also known to target the hepatocyte growth factor/scatter factor receptor MET (22, 23). miR-206 is upregulated during perinatal skeletal muscle development in mice and increases in myotubes over the course of differentiation. In chicken embryos, miR-206 is expressed in the somatic myotome (21). In zebrafish, miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization (24, 25). Interestingly, changes in miR-1 abundance have also been implicated in cardiac muscle disease, including arrhythmia and heart failure (21).
In their new study, Taulli et al. demonstrate that myomiR-1/206 expression/function is very low or undetectable in both primary embryonal and alveolar RMS (12) (Figure 1). Moreover, upon growth factor deprivation, which normally promotes a switch from proliferation to differentiation, miR-1/miR-206 could not be induced in RMS cell lines. Strikingly, Taulli and colleagues went on to show that forced expression of pre–miR-1 and –miR-206 reduced the ability of the RMS to grow in soft agar and induced myogenic differentiation (12) (Figure 1). An inducible lentivirus vector was employed to confirm that miR-206 could promote in vitro molecular and morphological differentiation, cell cycle exit and apoptosis, and impede invasiveness. The role of miR-206 as a tumor suppressor was confirmed by transplantation of the transduced RMS cells into immunocompromised mice, in which expression of miR-206 was shown to promote differentiation and inhibit tumor growth under a variety of conditions. The authors also used microarray analysis to show that reexpression of miR-206 can differentiate RD18 RMS cells by switching the global mRNA expression profiles back to their original myogenic phenotype. Upregulated genes included many muscle-specific genes, such as those encoding titin, muscle creatine kinase, myosin light chain, troponin C, myomesin 2, and tropomyosin 2. Many of the detected downregulated genes have been associated with cancer, including those involved with the cell cycle, metabolism, and DNA repair. Thus, miR-206 was sufficient to force neoplastic cells into resuming and completing the full myogenic program.
miR-1/miR-206 are known to target MET (22, 23), which is overexpressed in many cancers including RMS. Notably, deregulated MET signaling in genetically engineered mice induces embryonal RMS (26). MET is known to be rapidly downregulated at the onset of myogenic differentiation (27). Taulli and colleagues show that MET downregulation correlates with miR-206 upregulation in normal myogenic cells and, in fact, directly suppresses MET expression in RMS cells (12). This was confirmed through reexpression of constitutively active oncogenic Tpr-Met, which lacks the MET 3′ untranslated region, in miR-206–expressing RMS cells, abrogating myomiR-mediated differentiation. The authors correctly concluded that MET is a key target for the anticancer effect of miR-1/miR-206 in RMS, and their data suggest that reexpression of miR-1/miR-206 to its physiological levels may be useful as a therapeutic differentiation approach for RMS by restoring a normal muscle phenotype (12).
miR-1/miR-206 has been reported to possess activities that strongly influence other solid tumors. miR-206 was shown to target estrogen-responsive genes, including those encoding estrogen receptor, steroid receptor coactivators (SRC1, SRC3) and GATA binding protein 3 (GATA3), and has been suggested as a biomarker for basal-like breast cancers (28). Introduction of miR-206 into estrogen-dependent MCF-7 breast cancer cells inhibited cell growth in a dose- and time-dependent manner. miR-1 expression was also markedly reduced in primary human hepatocellular carcinomas (HCCs) compared with normal liver tissues. miR-1 reexpression in HCC cells downregulated its direct/indirect targets, including forkhead box P1 (FOXP1), histone deacetylase 4 (HDAC4), and MET, and inhibited cell growth and proliferation (23). Recently miR-1 has been implicated in lung cancer therapy (22). miR-1 is expressed in the normal bronchial epithelium but downregulated in human primary lung cancer tissues and cell lines, along with its activator C/EBPα (CCAAT/enhancer–binding protein). Notably, HDAC inhibitor treatment of lung cancer cells induced the expression of repressed miR-1, downregulating oncogenes such as MET, PIM1, FOXP1, and HDAC4, and reversing the tumorigenicity of lung cancer cells. Interestingly, miR-1 reexpression made a lung cancer cell line more sensitive to the chemotherapeutic doxorubicin by inducing apoptosis (22).
microRNA reexpression therapy: the pros, the cons, and the promise
Like siRNAs, miRNAs are easy to synthesize and can potentially target any gene, including otherwise non-druggable targets. However, miRNA therapy has many advantages over the originally envisioned RNA interference–based therapeutics (siRNA therapy). The major advantage of miRNA therapy is that miRNA reexpression can influence the expression of hundreds of genes involved in many cellular pathways. While siRNA therapy is more gene-specific, miRNA therapy can target an array of different gene products, more closely resembling the action of the so-called “dirty drugs” used in the clinic today; in fact, both the sense and antisense strands of miRNAs might target different mRNAs. Moreover, dirty drugs typically target only a handful of gene products, whereas miRNAs can target hundreds of genes, casting miRNAs as “super” dirty drugs. Of course, as with any such drug, the advantage of hitting a broad spectrum of targets is also a potential disadvantage. However, miRNAs are evolutionarily conserved, and targeting the upregulation or downregulation of a tissue-specific tumor suppressor miRNA or oncogenic miRNA, respectively, to its “physiological level” may incite fewer of the nonspecific, off-target effects often associated with artificial siRNAs or currently available dirty drugs.
miRNA therapy shares many of the disadvantages of siRNA-therapy, including delivery limitations, instability, and off-target effects. A major obstacle to effective miRNA-based therapy is the requirement for successful delivery. Unlike many other drugs, miRNAs do not freely diffuse into cells; therefore, miRNAs may require special delivery approaches to achieve the desired effect. Moreover, small RNAs tend to be unstable and might be degraded upon entering a cell; new methods may be required to stabilize these small sequences. Another factor is that double-stranded RNA and unmethylated CpG sequences are potentially immunogenic; their presence might increase IFN production and induce an immune response in patients. Moreover, in the case of miRNA reexpression therapy of cancer, preventing miRNA expression from exceeding physiological levels also represents a therapeutic challenge. Ultimately, miRNA-based gene therapy (i.e., delivering miRNAs encoded in a vector) has many of the same disadvantages as gene therapy (29–31). However, developing smarter/safer ways to deliver stable miRNAs/miRNA inhibitors into cells should go a long way toward overcoming the majority of these limitations, and preclinical models will be extremely helpful in this regard.
Although successful use of small RNAs as therapeutic agents represents a substantial challenge, miRNA therapeutics also carry great expectations based on the ability of a single miRNA to specifically target multiple oncogenes, many of which are being individually targeted by drugs already in the clinic or under preclinical development. But cancer remains a complex disease and patients with the same diagnosis will harbor gene mutations and polymorphic variants in a myriad of permutations; therefore, incorporation of personalized medicine approaches will continue to be advisable (reviewed in ref. 32). Cancer differentiation therapy has seen success in hematological cancers (e.g., acute promyelocytic leukemia) (5, 33) but not yet for solid tumors. The recent work of Taulli et al. and others (10, 11) on solid tumors demonstrates that coaxing the micro-sheriff out of retirement and back into town represents a promising and novel approach for arresting tumor growth and reestablishing cellular law and order.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: HDAC, histone deacetylase; miRNA, microRNA; RMS, rhabdomyosarcoma.
Citation for this article: J. Clin. Invest. 119:2119–2123 (2009). doi:10.1172/JCI40107
See the related article beginning on page 2366.
References
- 1.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 2.Mishra P.J., Bertino J.R. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim L.P., et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 4.Chuang J.C., Jones P.A. Epigenetics and microRNAs. Pediatr. Res. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 5.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 7.Garzon R., Fabbri M., Cimmino A., Calin G.A., Croce C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B., Pan X., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 9.Hammond S.M. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Negrini M., Ferracin M., Sabbioni S., Croce C.M. MicroRNAs in human cancer: from research to therapy. J. Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 11.Silber J., et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taulli R., et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J. Clin. Invest. . 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wachtel M., et al. Subtype and prognostic classification of rhabdomyosarcoma by immunohistochemistry. J. Clin. Oncol. 2006;24:816–822. doi: 10.1200/JCO.2005.03.4934. [DOI] [PubMed] [Google Scholar]
- 14.Garofalo M., Condorelli G., Croce C.M. MicroRNAs in diseases and drug response. Curr. Opin. Pharmacol. 2008;8:661–667. doi: 10.1016/j.coph.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Fabbri M., et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H., et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciarapica R., et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8:172–175. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 18.Merlino G., Khanna C. Fishing for the origins of cancer. Genes Dev. 2007;21:1275–1279. doi: 10.1101/gad.1563707. [DOI] [PubMed] [Google Scholar]
- 19.Merlino G., Helman L.J. Rhabdomyosarcoma--working out the pathways. Oncogene. 1999;18:5340–5348. doi: 10.1038/sj.onc.1203038. [DOI] [PubMed] [Google Scholar]
- 20.Chen J.F., et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy J.J. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasser M.W., et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J. Biol. Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Datta J., et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Mishima Y., et al. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharp R., et al. Synergism between INK4a/ARF inactivation and aberrant HGF/SF signaling in rhabdomyosarcomagenesis. Nat. Med. 2002;8:1276–1280. doi: 10.1038/nm787. [DOI] [PubMed] [Google Scholar]
- 27.Anastasi S., et al. A natural hepatocyte growth factor/scatter factor autocrine loop in myoblast cells and the effect of the constitutive Met kinase activation on myogenic differentiation. . J. Cell Biol. 1997;137:1057–1068. doi: 10.1083/jcb.137.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams B.D., Cowee D.M., White B.A. Mol. Endocrinol. 2009. The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-alpha (ER{alpha}) signaling and a luminal phenotype in MCF-7 breast cancer cells. Online publication ahead of print. doi:10.1210/me.2009-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wurdinger T., Costa F.F. Molecular therapy in the microRNA era. Pharmacogenomics J. 2007;7:297–304. doi: 10.1038/sj.tpj.6500429. [DOI] [PubMed] [Google Scholar]
- 30.Bumcrot D., Manoharan M., Koteliansky V., Sah D.W. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang V., Wu W. MicroRNA-based therapeutics for cancer. BioDrugs. 2009;23:15–23. doi: 10.2165/00063030-200923010-00002. [DOI] [PubMed] [Google Scholar]
- 32.Mishra P.J. MicroRNA polymorphisms: a giant leap towards personalized medicine. Personalized Medicine. 2009;6:119–125. doi: 10.2217/17410541.6.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohno R., et al. Multi-institutional study of all-trans-retinoic acid as a differentiation therapy of refractory acute promyelocytic leukemia. Leukaemia Study Group of the Ministry of Health and Welfare. Leukemia. 1993;7:1722–1727. [PubMed] [Google Scholar]
- 34. Labourier, E., et al. 2009. microRNAs as potential diagnostic and prognostic markers of disease. http://www.ambion.com/techlib/posters/miRNA_0405.html . [Google Scholar]